Novel Dual-Coenzyme Specificity and Thermostability of Malate Dehydrogenase Identified in the Cyanobacterium Microcystis aeruginosa PCC7806
Abstract
1. Introduction
2. Results
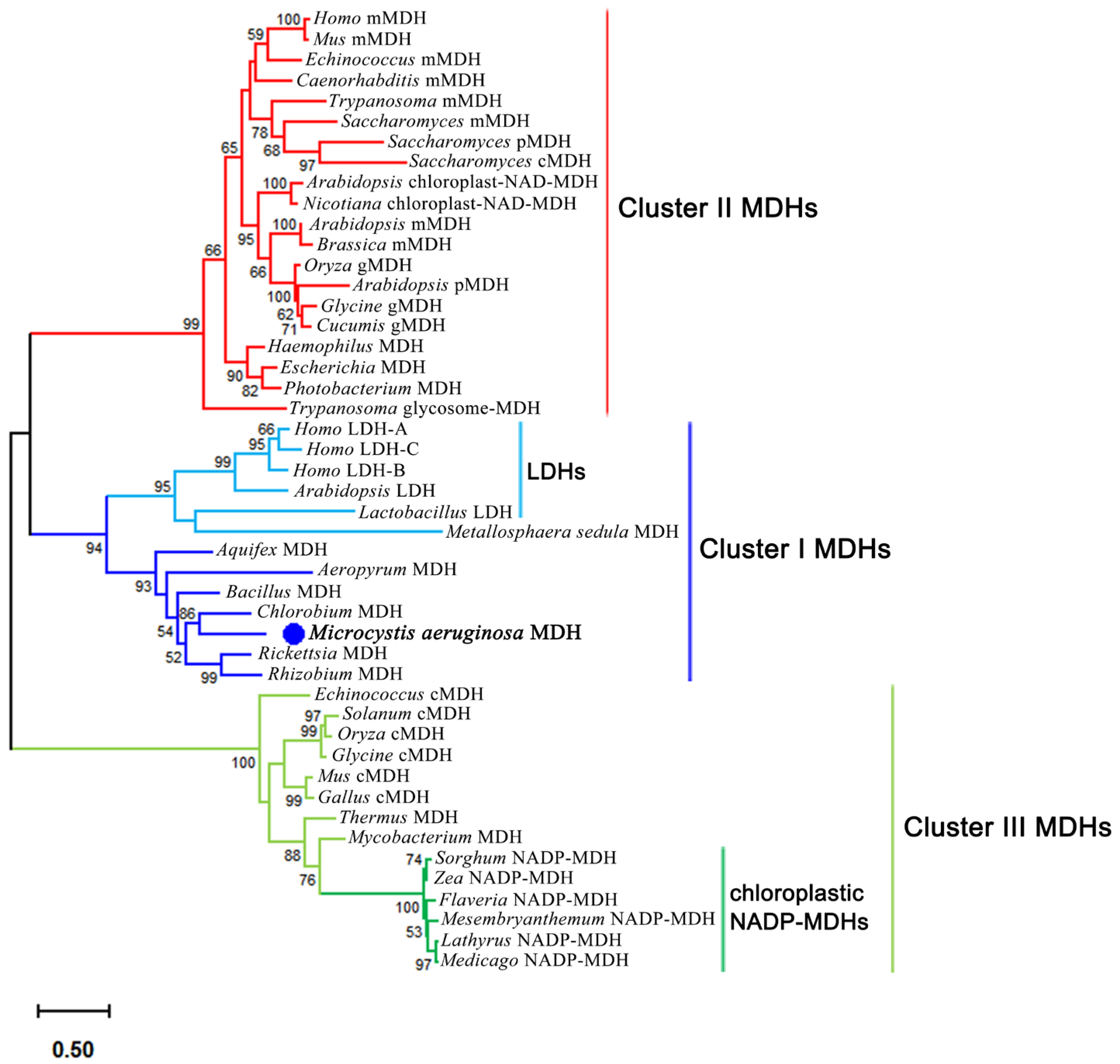
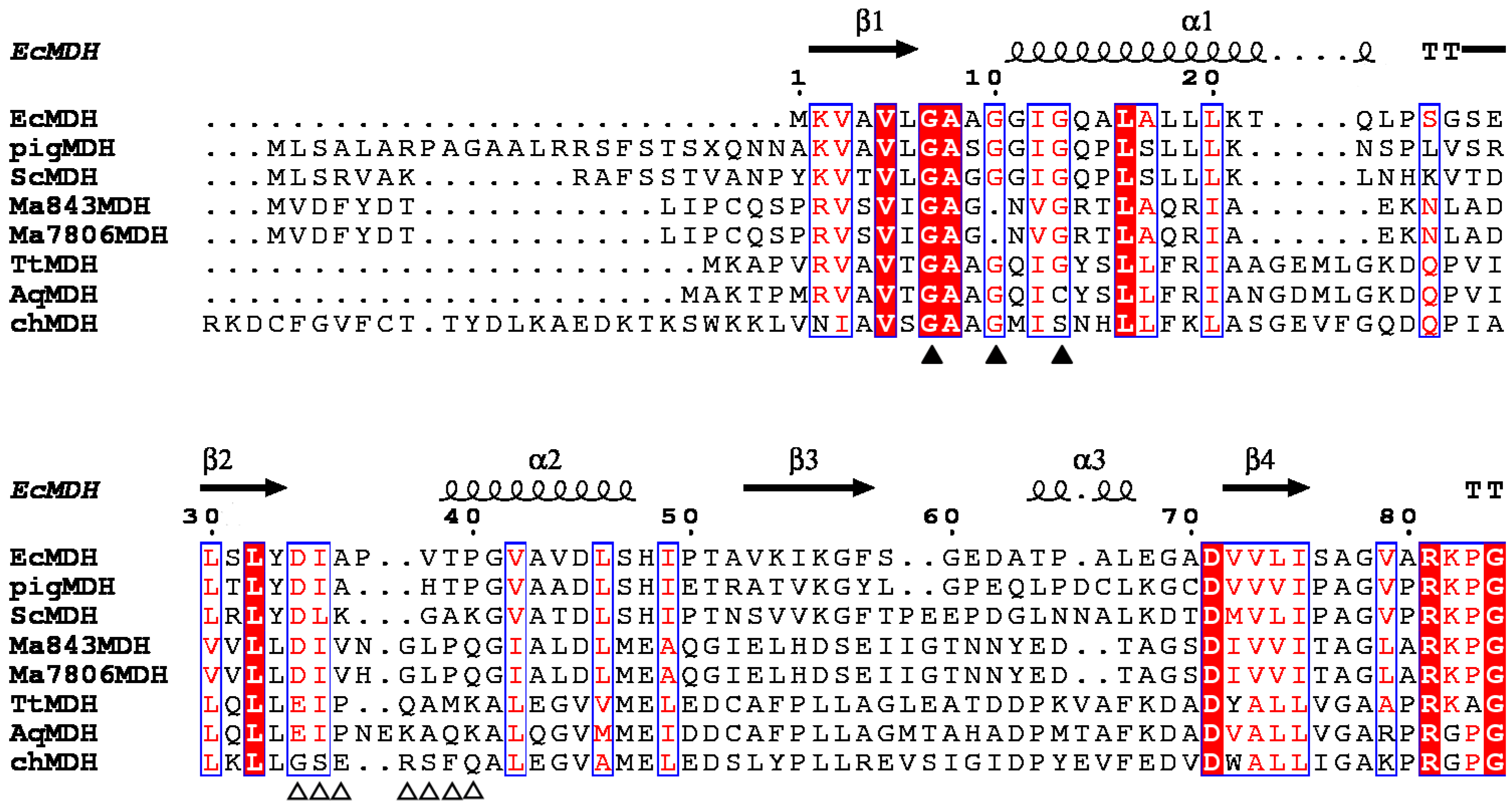
2.1. Bioinformatics Analysis
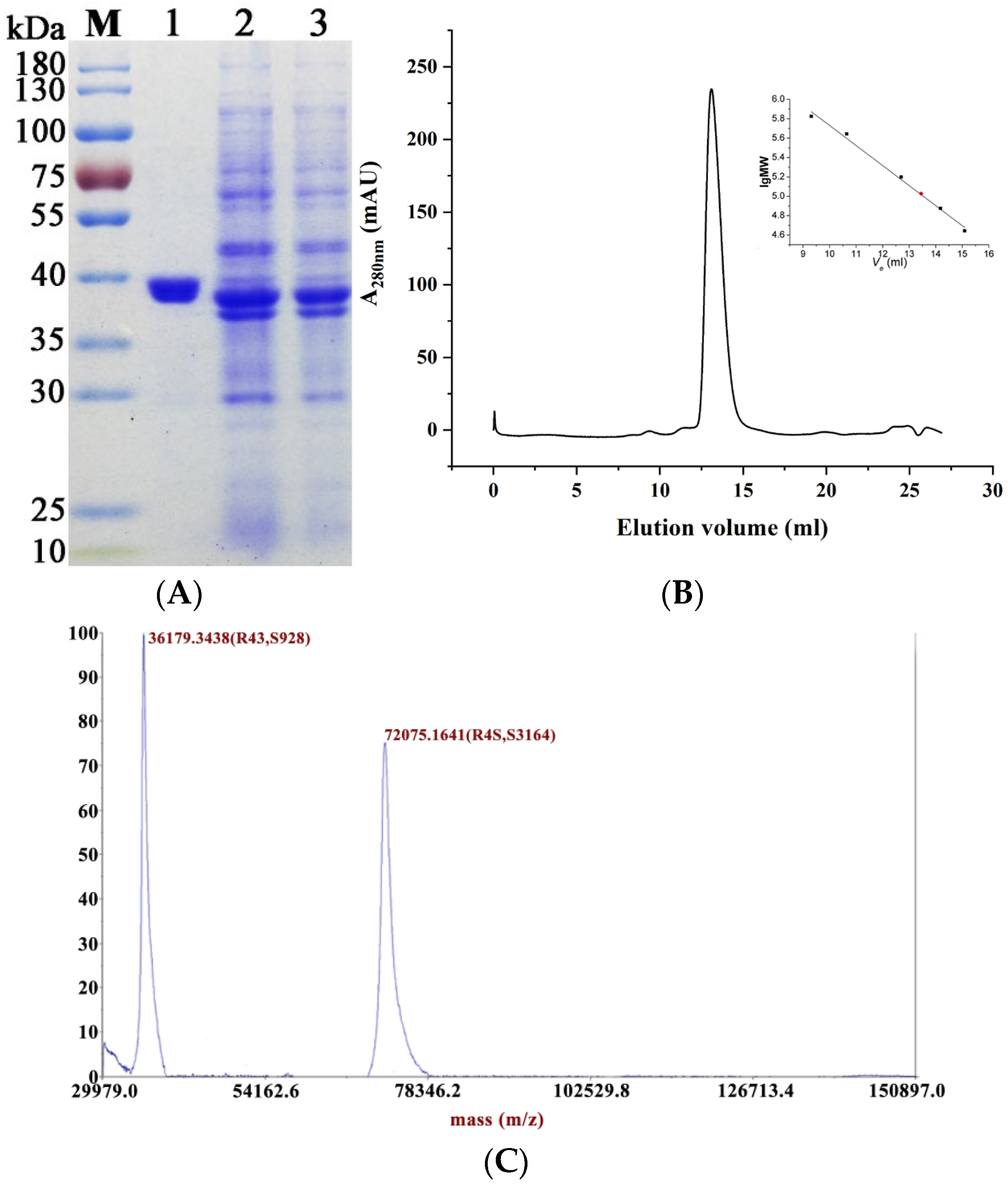
2.2. Expression and Purification of Recombinant MaMDH
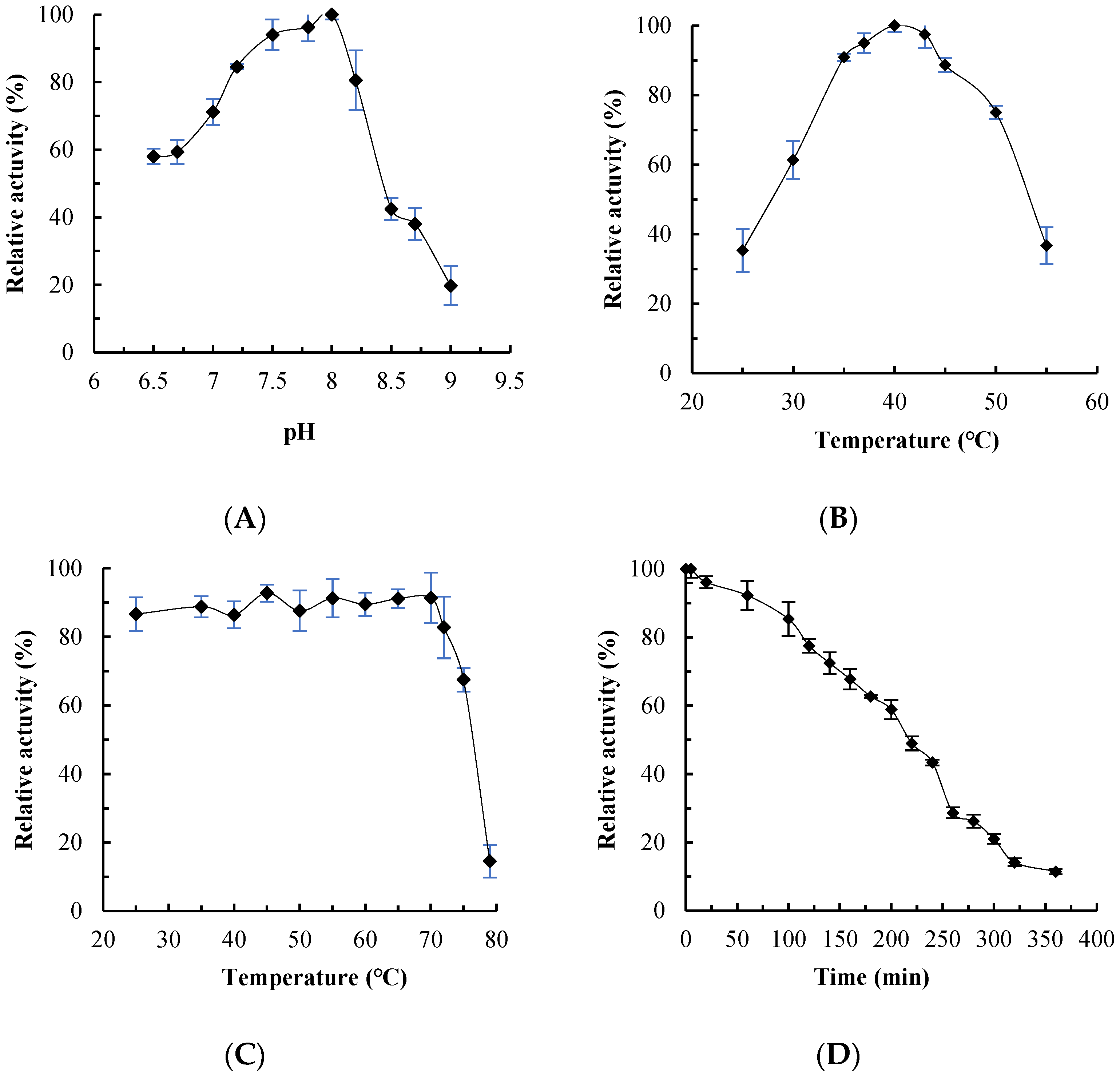
2.3. Effects of pH, Temperature and Metal Ions on MaMDH Activity
2.4. Kinetics Analysis
3. Discussion
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. Molecular Mass Determination
4.3. Enzyme Kinetic Assay
4.4. Enzyme Characterization
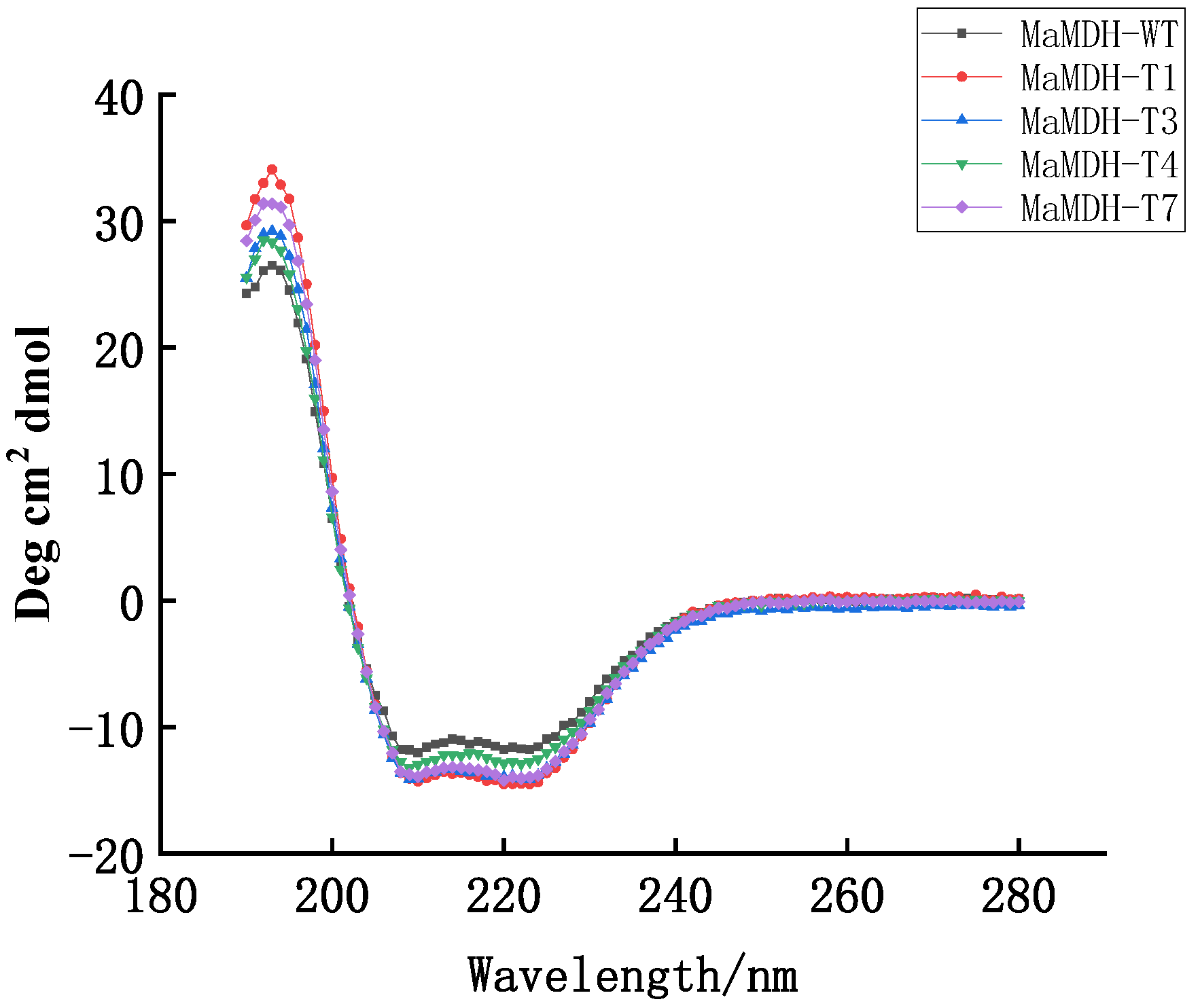
4.5. Site-Directed Mutagenesis and Circular Dichroism Spectroscopy
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baird, L.M.; Berndsen, C.E.; Monroe, J.D. Malate dehydrogenase in plants: Evolution, structure, and a myriad of functions. Essays Biochem. 2024, 68, 221–233. [Google Scholar] [CrossRef]
- Hara, S.; Motohashi, K.; Arisaka, F.; Romano, P.G.N.; Hosoya-Matsuda, N.; Kikuchi, N.; Fusada, N.; Hisabori, T. Thioredoxin-h1 reduces and reactivates the oxidized cytosolic malate dehydrogenase dimer in higher plants. J. Biol. Chem. 2006, 281, 32065–32071. [Google Scholar] [CrossRef] [PubMed]
- Madern, D.; Cai, X.M.; Abrahamsen, M.S.; Zhu, G. Evolution of Cryptosporidium parvum lactate dehydrogenase from malate dehydrogenase by a very recent event of gene duplication. Mol. Biol. Evol. 2004, 21, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Kim, J.H.; Cho, E.J.; Youn, H.D. A nucleocytoplasmic malate dehydrogenase regulates p53 transcriptional activity in response to metabolic stress. Cell Death Differ. 2009, 16, 738–748. [Google Scholar] [CrossRef]
- Gietl, C. Malate dehydrogenase isoenzymes: Cellular locations and role in the flow of metabolites between the cytoplasm and cell organelles. Biochim. Biophys. Acta BBA-Bioenerg. 1992, 1100, 217–234. [Google Scholar] [CrossRef]
- Lee, D.; Hong, J.; Kim, K.J. Crystal structure and biochemical characterization of malate dehydrogenase from Metallosphaera sedula. Biochem. Biophys. Res. Commun. 2019, 509, 833–838. [Google Scholar] [CrossRef]
- Schnarrenberger, C.; Martin, W. Evolution of the enzymes of the citric acid cycle and the glyoxylate cycle of higher plants. Eur. J. Biochem. 2002, 269, 868–883. [Google Scholar] [CrossRef]
- Goward, C.R.; Nicholls, D.J. Malate dehydrogenase: A model for structure, evolution, and catalysis. Protein Sci. 1994, 3, 1883–1888. [Google Scholar] [CrossRef]
- Musrati, R.A.; Kollárová, M.; Mernik, N.; Mikulásová, D. Malate Dehydrogenase: Distribution, Function and Properties. Gen. Physiol. Biophys. 1998, 17, 193–210. [Google Scholar] [CrossRef]
- Edwards, G.E.; Nakamoto, H.; Burnell, J.N.; Hatch, M.D. Pi dikinase and NADP-malate dehydrogenase in C4 photosynthesis: Properties and mechanism of light/dark. Annu. Rev. Plant Physiol. 1985, 36, 236–250. [Google Scholar] [CrossRef]
- Scheibe, R. Malate valves to balance cellular energy supply. Physiol. Plant 2004, 120, 21–26. [Google Scholar] [CrossRef]
- Inga, H.; Jennifer, S.; Corinna, W.; Tatjana, G.; Ingo, V.; Paula, M.; Saijaliisa, K.; Eva-Mari, A.; Marie-Luise, O.; Karl-Josef, D. Multiple strategies to prevent oxidative stress in Arabidopsis plants lacking the malate valve enzyme NADP-malate dehydrogenase. J. Exp. Bot. 2012, 63, 1445–1459. [Google Scholar] [CrossRef]
- Jennifer, S.; Renate, S. Malate valves: Old shuttles with new perspectives. Plant Biol. 2019, 21, 21–30. [Google Scholar] [CrossRef]
- Leverett, B.; Austin, S.; Tan-Arroyo, J. Malate dehydrogenase (MDH) in cancer: A promiscuous enzyme, a redox regulator, and a metabolic co-conspirator. Essays Biochem. 2024, 68, 135–146. [Google Scholar] [CrossRef]
- Shi, C.X.; Wang, Y.K.; Guo, J.; Zhang, D.M.; Zhang, Y.Q.; Zhang, X.Y.; Gong, Z.J. Role of malate dehydrogenase 1 and isocitrate dehydrogenase 1 and their posttranslational modifications in diseases. Biochem. Bioph. Res. Commun. 2025, 754, 151535. [Google Scholar] [CrossRef] [PubMed]
- Wolyniak, M.J.; Frazier, R.H.; Gemborys, P.K.; Loehr, H.E. Malate dehydrogenase: A story of diverse evolutionary radiation. Essays Biochem. 2024, 68, 213–220. [Google Scholar] [CrossRef]
- Cazenave, J.; Wunderlin, D.A.; Bistoni, M.A.; Amé, M.V.; Krause, E.; Pflugmacher, S.; Wiegand, C. Uptake, tissue distribution and accumulation of microcystin-RR in Corydoras paleatus, Jenynsia multidentata and Odontesthes bonariensis: A field and laboratory study. Aquat. Toxicol. 2005, 75, 178–190, Erratum in Aquat. Toxicol. 2006, 77, 439. [Google Scholar] [PubMed]
- Cox, B.; Chit, M.M.; Weaver, T.; Gietl, C.; Bailey, J.; Bell, E.; Banaszak, L. Organelle and translocatable forms of glyoxysomal malate dehydrogenase: The effect of the N-terminal presequence. FEBS J. 2005, 272, 643–654. [Google Scholar] [CrossRef]
- Wali, A.S.; Mattoo, A.K. Malate dehydrogenase from thermophilic Humicola lanuginosa and Mucor pusillus: Purification and comparative properties of the enzymes with differing thermostabilities. Biochem. Cell Biol. 1984, 62, 559–565. [Google Scholar] [CrossRef]
- Nishiyama, M.; Birktoft, J.J.; Beppu, T. Alteration of coenzyme specificity of malate dehydrogenase from Thermus flavus by sitedirected mutagenesis. J. Biol. Chem. 1993, 268, 4656–4660. [Google Scholar] [CrossRef]
- Tomita, T.; Fushinobu, S.; Kuzuyama, T.; Nishiyama, M. Structural basis for the alteration of coenzyme specificity in a malate dehydrogenase mutant. Biochem. Biophys. Res. Commun. 2006, 347, 502–508. [Google Scholar] [CrossRef]
- Takeya, M.; Ito, S.; Sukigara, H.; Osanai, T. Purification and characterisation of malate dehydrogenase from Synechocystis sp. PCC 6803: Biochemical barrier of the oxidative tricarboxylic acid cycle. Front. Plant Sci. 2018, 9, 947, Erratum in Front. Plant Sci. 2021, 12, 699371. https://doi.org/10.3389/fpls.2021.699371. [Google Scholar] [CrossRef]
- Ge, Y.D.; Guo, Y.T.; Jiang, L.L.; Wang, H.H.; Hou, S.L.; Su, F.Z. Enzymatic Characterization and Coenzyme Specificity Conversion of a Novel Dimeric Malate Dehydrogenase from Bacillus subtilis. Protein J. 2023, 42, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Mikulášová, D.; Kollárová, M.; Miginiac-Maslow, M.; Decottignies, P.; Jacquot, J.P.; Kutějová, E.; Mernik, N.; Egyudová, I.; Musrati, R.A.; Horecká, T. Purification and characterization of the malate dehydrogenase from Streptomyces aureofaciens. FEMS Microbiol. Lett. 1998, 159, 299–305. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, Z.D.; Wang, B.J.; Ge, Y.D.; Pan, W.; Wang, J.; Xu, L.; Liu, A.M.; Zhu, G.P. Expression and identification of a thermostable malate dehydrogenase from multicellular prokaryote Streptomyces avermitilis MA-4680. Mol. Biol. Rep. 2011, 38, 1629–1636. [Google Scholar] [CrossRef]
- Maloney, A.P.; Callan, S.M.; Murray, P.G.; Tuohy, M.G. Mitochondrial malate dehydrogenase from the thermophilic, filamentous fungus Talaromyces emersonii. Eur. J. Biochem. 2004, 271, 3115–3126. [Google Scholar] [CrossRef]
- Trípodi, K.E.J.; Podestá, F.E. Purification and characterization of an NAD-dependent malate dehydrogenase from leaves of the crassulacean acid metabolism plant Aptenia cordifolia. Plant Physiol. Biochem. 2003, 41, 97–105. [Google Scholar] [CrossRef]
- Cuevas, I.C.; Podestá, F.E. Purification and physical and kinetic characterization of an NAD+-dependent malate dehydrogenase from leaves of pineapple (Ananas comosus). Physiol. Plant. 2000, 108, 240–248. [Google Scholar] [CrossRef]
- Oikawa, T.; Yamamoto, N.; Shimoke, K.; Uesato, S.; Ikeuchi, T.; Fukioka, T. Purification, characterization, and overexpression of psychrophilic and thermolabile malate dehydrogenase of a novel antarctic psychrotolerant, Flavobacterium frigidimarinis KUC-1. Biosci. Biotechnol. Biochem. 2005, 69, 2146–2154. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.D.; Cao, Z.Y.; Wang, Z.D.; Chen, L.L.; Zhu, Y.M.; Zhu, G.P. Identification and biochemical characterization of a thermostable malate dehydrogenase from the mesophile Streptomyces coelicolor A3(2). Biosci. Biotechnol. Biochem. 2010, 74, 2194–2201. [Google Scholar] [CrossRef]
- Madern, D.; Zaccai, G. Molecular adaptation: The malate dehydrogenase from the extreme halophilic bacterium Salinibacter ruber behaves like a non-halophilic protein. Biochimie 2004, 86, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Bjørk, A.; Dalhus, B.; Mantzilas, D.; Eijsink, V.G.; Sirevåg, R. Stabilization of a tetrameric malate dehydrogenase by introduction of a disulfide bridge at the dimer-dimer interface. J. Mol. Biol. 2003, 334, 811–821. [Google Scholar] [CrossRef] [PubMed]
- Jaindl, M.; Popp, M. Cyclitols protect glutamine synthetase and malate dehydrogenase against heat induced deactivation and thermal denaturation. Biochem. Biophys. Res. Commun. 2006, 345, 761–765. [Google Scholar] [CrossRef] [PubMed]
- Deutch, C.E. L-Malate dehydrogenase activity in the reductive arm of the incomplete citric acid cycle of Nitrosomonas europaea. Antonie Van Leeuwenhoek 2013, 104, 645–655. [Google Scholar] [CrossRef]
- Maheshwari, R.; Bharadwaj, G.; Bhat, M.K. Thermophilic fungi: Their physiology and enzymes. Microbiol. Mol. Biol. Rev. 2000, 64, 461–488. [Google Scholar] [CrossRef]
- Mahmoud, Y.A.G.; Abuelsouod, S.M.; Niehaus, W.G. Purification and characterization of malate dehydrogenase from Cryptococcus neoformans. Arch. Biochem. Biophys. 1995, 322, 69–75. [Google Scholar] [CrossRef]
- Wang, S.Y.; Xu, Z.B.; Ye, X.Y.; Rao, P.F. Purification and characterization of a malate dehydrogenase from Phaseolus mungo. J. Food Biochem. 2005, 29, 117–131. [Google Scholar] [CrossRef]
- Kleiger, G.; Eisenberg, D. GXXXG and GXXXA motifs stabilize FAD and NAD(P)-binding Rossmann folds through Ca-H⋯O hydrogen bonds and van der waals interactions. J. Mol. Biol. 2002, 323, 69–76. [Google Scholar] [CrossRef]
- Scrutton, N.S.; Berry, A.; Perham, R.N. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature 1990, 343, 38–43. [Google Scholar] [CrossRef]
- Ge, Y.D.; Jiang, L.L.; Hou, S.L.; Su, F.Z.; Wang, P.; Zhang, G. Heteroexpression and biochemical characterization of thermostable citrate synthase from the cyanobacteria Anabaena sp. PCC7120. Protein Expr. Purif. 2020, 168, 105565. [Google Scholar] [CrossRef]
| Metal Ions | Relative Activity (%) | Metal Ions and Compounds | Relative Activity (%) |
|---|---|---|---|
| None | 100.00 ± 0.00 | Co2+ | 8.73 ± 0.36 |
| Na+ | 106.23 ± 1.95 | Cu2+ | 79.07 ± 1.62 |
| K+ | 99.52 ± 0.46 | Ca2+ | 130.77 ± 1.99 |
| Li+ | 106.95 ± 1.99 | DMSO (2%) | 97.13 ± 0.54 |
| Rb+ | 115.26 ± 3.22 | DMSO (4%) | 85.75 ± 0.82 |
| Zn2+ | 0.00 ± 0.00 | DMSO (8%) | 69.53 ± 1.93 |
| Mg2+ | 135.84 ± 0.35 | Triton X-100 (2%) | 85.00 ± 0.75 |
| Mn2+ | 128.14 ± 5.12 | Triton X-100 (4%) | 69.31 ± 1.97 |
| Ni2+ | 14.94 ± 0.49 | Triton X-100 (8%) | 50.06 ± 1.19 |
| Enzyme | NADH | NADPH | Specificity | Degree of Alteration | |||||
|---|---|---|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km (A) | Km | kcat | kcat/Km (B) | A/B | B/A | ||
| (μM) | (s−1) | (μM−1·s−1) | (μM) | (s−1) | (μM−1·s−1) | ||||
| MaMDH-WT | 33.140 ± 0.95 | 31.688 ± 1.43 | 0.956 | 113.200 ± 3.46 | 8.710 ± 1.45 | 0.077 | 12.427 | 0.080 | 1.00 |
| MaMDH-T1 D44G | 26.160 ± 2.17 | 22.361 ± 1.75 | 0.855 | 147.700 ± 12.04 | 15.642 ± 1.73 | 0.106 | 8.071 | 0.124 | 1.54 |
| MaMDH-T3 D44G/I45S/G48S | 179.600 ± 11.60 | 22.498 ± 2.37 | 0.125 | 38.230 ± 1.58 | 24.283 ± 1.20 | 0.635 | 0.197 | 5.071 | 63.01 |
| MaMDH-T4 D44G/I45S/H47R/G48S | 177.000 ± 4.83 | 27.131 ± 1.16 | 0.153 | 25.880 ± 1.13 | 26.542 ± 1.04 | 1.026 | 0.149 | 6.691 | 83.15 |
| MaMDH-T7 D44G/I45S/V46E/H47R/G48S/L49F/P50Q | 136.400 ± 6.31 | 46.095 ± 3.06 | 0.338 | 43.110 ± 3.22 | 54.110 ± 2.31 | 1.255 | 0.269 | 3.714 | 46.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ge, Y.; Wu, Y.; Zhou, B.; Wu, X.; Ren, Y.; Zhu, J.; Ge, Y. Novel Dual-Coenzyme Specificity and Thermostability of Malate Dehydrogenase Identified in the Cyanobacterium Microcystis aeruginosa PCC7806. Int. J. Mol. Sci. 2025, 26, 10313. https://doi.org/10.3390/ijms262110313
Ge Y, Wu Y, Zhou B, Wu X, Ren Y, Zhu J, Ge Y. Novel Dual-Coenzyme Specificity and Thermostability of Malate Dehydrogenase Identified in the Cyanobacterium Microcystis aeruginosa PCC7806. International Journal of Molecular Sciences. 2025; 26(21):10313. https://doi.org/10.3390/ijms262110313
Chicago/Turabian StyleGe, Yadong, Yifan Wu, Bo Zhou, Xiaojie Wu, Yu Ren, Jialin Zhu, and Yali Ge. 2025. "Novel Dual-Coenzyme Specificity and Thermostability of Malate Dehydrogenase Identified in the Cyanobacterium Microcystis aeruginosa PCC7806" International Journal of Molecular Sciences 26, no. 21: 10313. https://doi.org/10.3390/ijms262110313
APA StyleGe, Y., Wu, Y., Zhou, B., Wu, X., Ren, Y., Zhu, J., & Ge, Y. (2025). Novel Dual-Coenzyme Specificity and Thermostability of Malate Dehydrogenase Identified in the Cyanobacterium Microcystis aeruginosa PCC7806. International Journal of Molecular Sciences, 26(21), 10313. https://doi.org/10.3390/ijms262110313




