Circulating Molecular Biomarkers for the Diagnosis and Monitoring of NSCLC—A Review
Abstract
1. Introduction
- Pre-Analytical Phase
- Blood Collection:
- Specialized blood collection tubes are used to stabilize nucleic acids and cells, for example
- -
- CTCs: CellSave Preservative Tubes (specific to the CellSearch system)
- -
- RNA: PAXgene Blood RNA Tubes (for RNA stabilization)
- Sample Handling:
- Samples are maintained at room temperature (for Streck tubes) or on ice (for EDTA)
- Centrifugation Protocol:
- Step 1: Whole blood is centrifuged at 1600× g for 10 min at 4 °C to separate plasma
- Step 2: Plasma is transferred carefully without disturbing buffy coat
- Step 3: Second centrifugation at 16,000× g for 10 min at 4 °C to remove residual cells/debris
- Storage: Store plasma at −80 °C until extraction
- Analytical Platforms
- Circulating Tumor Cells (CTCs): Platform: CellSearch System (FDA-approved)
- Cell-Free DNA (cfDNA):
- Detection/Quantification:
- -
- ddPCR (Droplet Digital PCR): Ultra-sensitive mutation detection (e.g., EGFR T790M)
- -
- NGS (Next-Generation Sequencing): Comprehensive mutation profiling
- Cell-Free RNA (cfRNA)/RNA Profiling
- -
- NanoString nCounter System: Multiplexed RNA expression profiling (gene signatures, immune profiling)
- -
- qRT-PCR: Targeted RNA quantification (e.g., fusion transcripts)
- Exosomal Analysis (optional):
- Exosome isolation by ultracentrifugation or commercial kits
- Downstream analysis by ddPCR, NGS, or NanoString
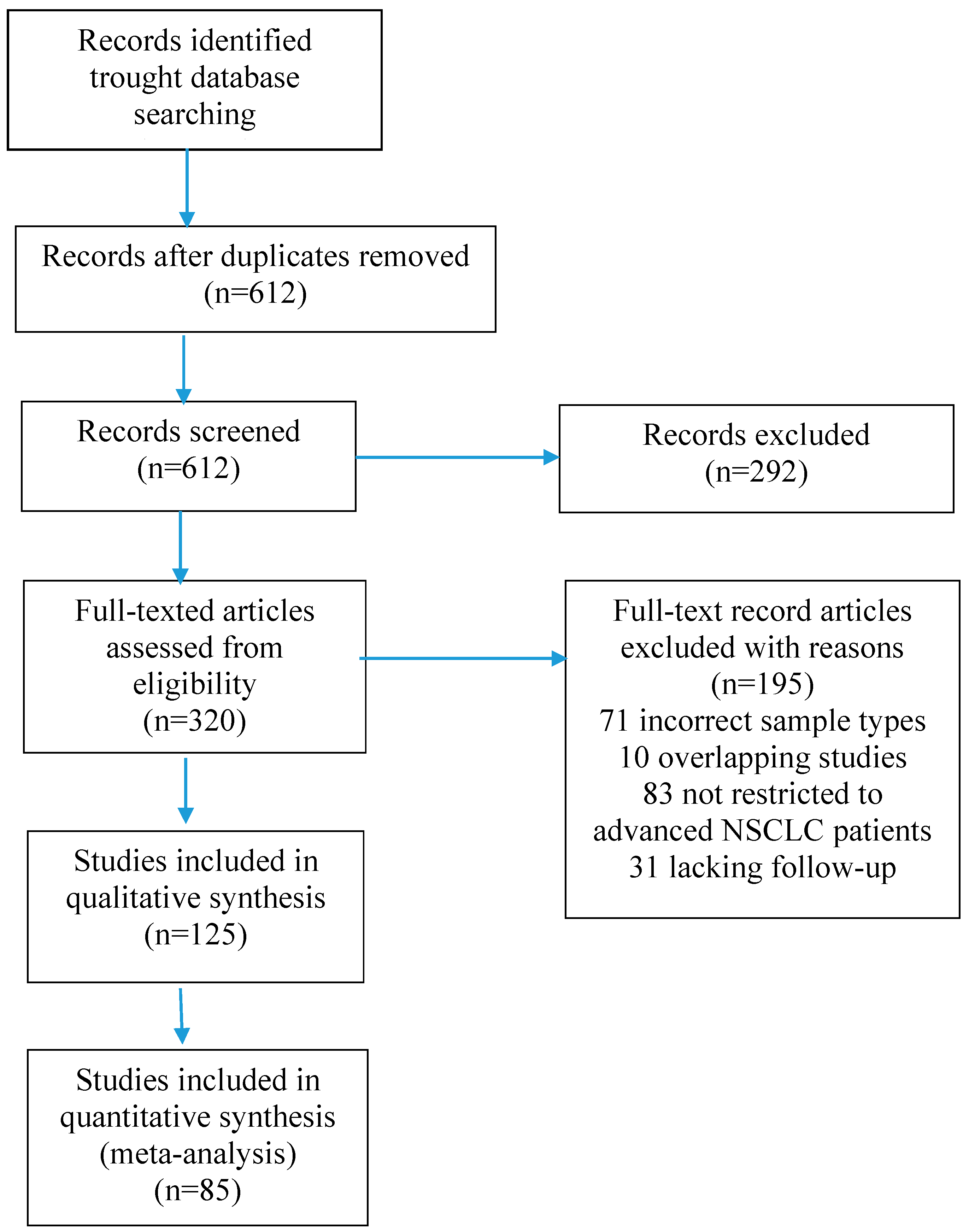
2. Materials and Methods
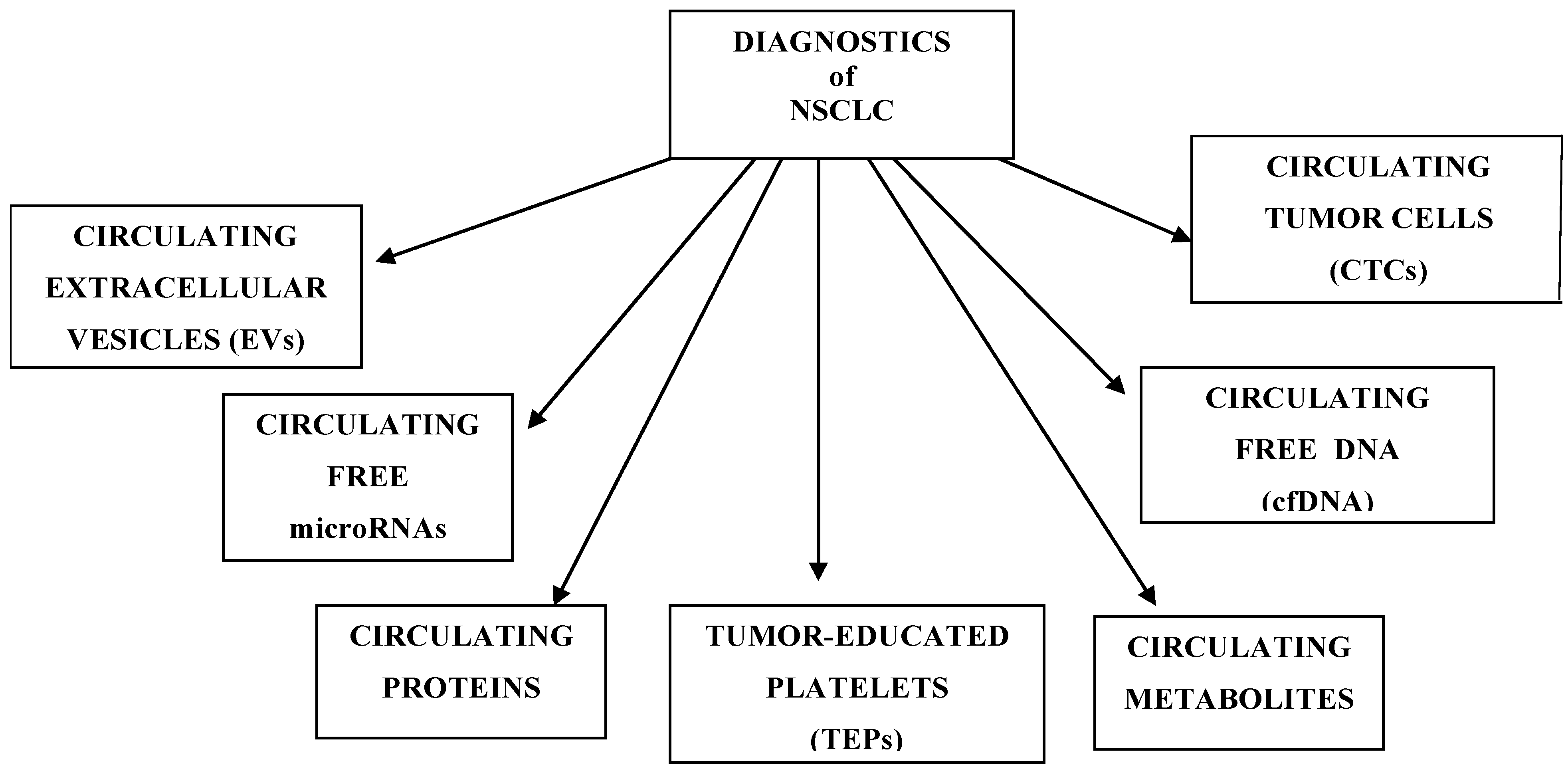
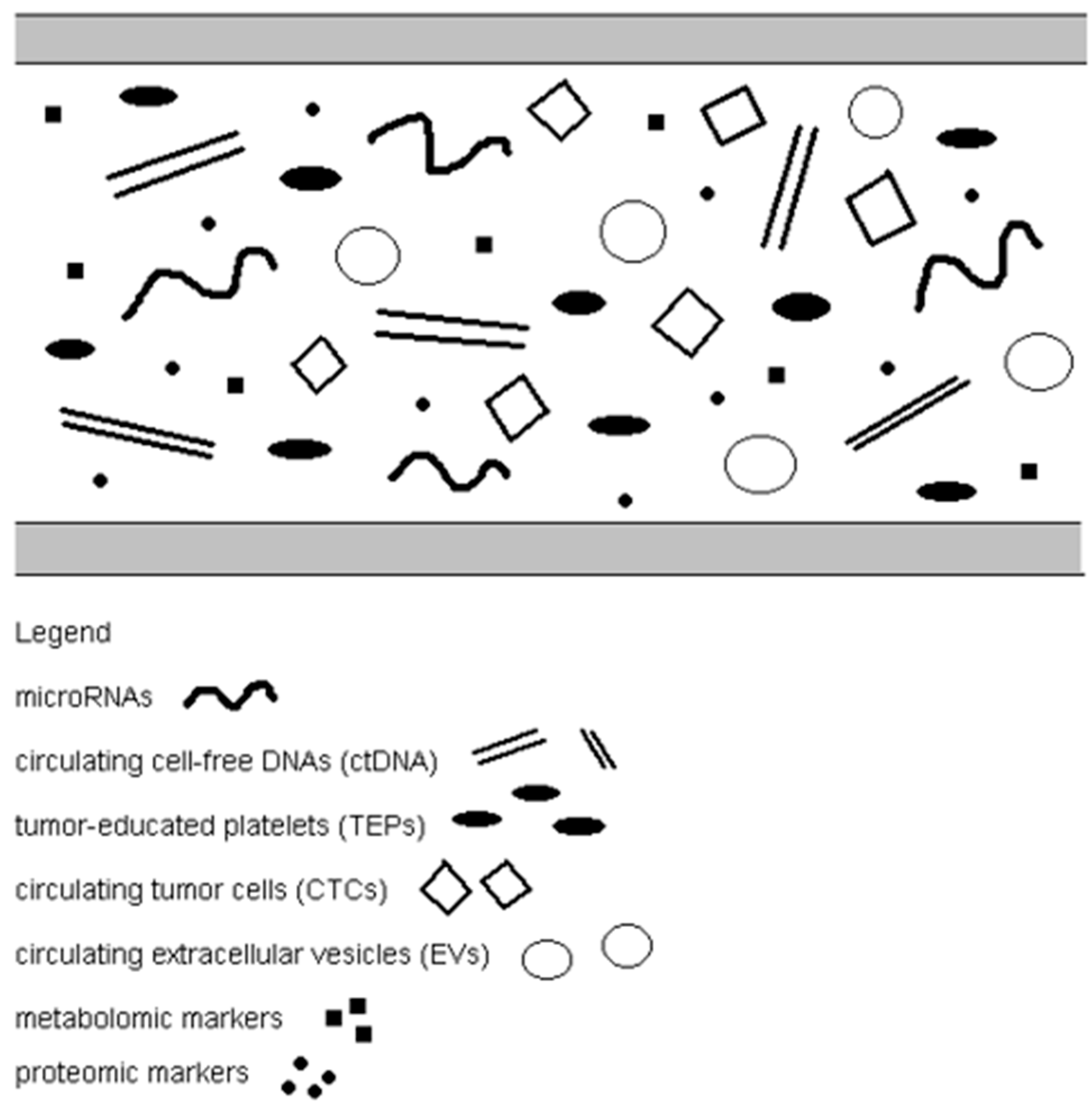
3. Circulating Tumor Cells
4. Circulating Free miRNA
5. Circulating Cell-Free DNA and Circulating Tumor DNA
6. Tumor-Educated Platelets
7. Circulating Extracellular Vesicles
8. Metabolomic and Proteomic Markers
9. Conclusions and Future Perspectives
- -
- ctDNA/cfDNA;
- -
- NGS panels (e.g., EGFR, ALK, KRAS, BRAF);
- -
- Digital PCR (dPCR);
- -
- Methylation-based liquid biopsy;
- -
- Exosome and miRNA-based assays (emerging);
- -
- AI/ML algorithms for data interpretation.
- 1990s—Early Foundations
- 1994: Discovery of circulating tumor DNA (ctDNA) in blood.
- 2000s—Conceptualization and Early Studies
- 2008: First research indicating cfDNA could be used to detect EGFR mutations non-invasively in NSCLC patients.
- 2010–2013—Technological Advances
- 2010: Rise of next-generation sequencing (NGS) enables more precise mutation detection from small DNA fragments.
- 2012: Early clinical studies confirm feasibility of detecting EGFR mutations in plasma of NSCLC patients.
- 2014–2016—Clinical Validation and FDA Recognition
- 2014: First large-scale validation studies of ctDNA for EGFR mutations in NSCLC.
- 2016: FDA approves the cobas® EGFR Mutation Test v2 (Roche), the
- first liquid biopsy test approved for NSCLC.
- 2017–2019—Expanded Panels and MRD
- 2017: Introduction of multi-gene liquid biopsy panels (e.g., Guardant360, FoundationACT) for broader mutation profiling in NSCLC.
- 2019: Liquid biopsy explored for minimal residual disease (MRD) detection and recurrence monitoring.
- 2020: NCCN guidelines include liquid biopsy as an alternative when tissue is insufficient or unavailable.
- 2021–2022: Studies show ctDNA use in early-stage NSCLC for MRD and early relapse prediction.
- 2023–2025—Early Detection and AI Integration
- Growing focus on stage I/II NSCLC detection via ultrasensitive ctDNA and methylation assays.
- 2024–2025 (projected milestones):
- AI/ML-enhanced ctDNA interpretation for early diagnosis and patient stratification.
- 1.
- Cohort Design and Data Collection
- -
- Define clinical questions and endpoints.
- -
- Assemble cohorts: Design multi-center prospective cohorts or leverage existing biobanks with matched clinical data. This includes
- -
- Adequate sample size for statistical power;
- -
- Diverse demographics to ensure generalizability;
- -
- Standardize protocols which harmonize sample collection, processing, and storage across sites to reduce batch effects;
- -
- Multi-omics profiling;
- -
- A data integration framework that sets up secure databases and pipelines for multi-omics data management and preprocessing.
- 2.
- AI-Driven Biomarker Discovery
- -
- Exploratory analysis;
- -
- Feature selection;
- -
- Multi-modal integration;
- -
- Model interpretability;
- -
- Initial validation: Cross-validation within cohorts and retrospective validation on independent datasets.
- 1.
- Prospective Validation and Refinement
- -
- Independent cohort testing;
- -
- Longitudinal validation;
- -
- Analytical validation.
- 2.
- AI Model Refinement and Clinical Contextualization
- -
- Refine predictive models;
- -
- Robustness testing through stress-testing models against confounders and missing data;
- -
- Clinical usability studies.
- 3.
- Pilot Integration into Clinical Workflows
- -
- Integration of biomarker-based AI models into pilot clinical decision support tools with user-friendly interfaces;
- -
- Workflow integration;
- -
- User training;
- -
- Pilot testing by conducting small-scale implementation studies to assess feasibility, clinician acceptance, and preliminary impact on decision-making.
- 1.
- Clinical Validation and Regulatory Submission
- -
- Large-scale clinical trials;
- -
- Health economics analysis through cost–benefit evaluation and reimbursement pathways.
- 2.
- Full-scale Integration into Clinical Decision Support Systems
- -
- System integration;
- -
- Real-time decision support;
- -
- Continuous learning;
- -
- Multi-stakeholder engagement through collaboration with payers, regulators, and patient advocacy groups to ensure broad adoption.
- 3.
- Post-deployment Monitoring and Expansion
- -
- Performance monitoring;
- -
- Scalability and generalizability;
- -
- Knowledge dissemination.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.I.; Kratzer, T.B.; Giaquinto, A.N.; Sung, H.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2025, 75, 10–45. [Google Scholar] [CrossRef]
- Thai, A.A.; Solomon, B.J.; Sequist, L.V.; Gainor, J.F.; Heist, R.S. Lung cancer. Lancet 2021, 398, 535–554. [Google Scholar] [CrossRef]
- Uramoto, H.; Tanaka, F. Recurrence after surgery in patients with NSCLC. Transl. Lung Cancer Res. 2014, 3, 242–249. [Google Scholar] [PubMed]
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speichler, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2019, 20, 71–88. [Google Scholar] [CrossRef]
- Garcia-Pardo, M.; Czarnecka, K.; Law, J.H. Plasma-first: Accelerating lung cancer diagnosis and molecular profiling through liquid biopsy. Ther. Adv. Med. Oncol. 2022, 14, 17588359221126151. [Google Scholar] [CrossRef] [PubMed]
- Szczerba, B.M.; Castro-Giner, F.; Vetter, M.; Krol, I.; Gkountela, S.; Landin, J.; Scheidmann, M.C.; Donato, C.; Scherrer, R.; Singer, J.; et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature 2019, 566, 553–557. [Google Scholar] [CrossRef]
- Au, S.H.; Storey, B.D.; Moore, J.C.; Tang, Q.; Chen, Y.L.; Javaid, S.; Sarioglu, A.F.; Sullivan, R.; Madden, M.W.; O’Keefe, R.; et al. Clusters of circulating tumor cells traverse capillary-sized vessels. Proc. Natl. Acad. Sci. USA 2016, 113, 4947–4952. [Google Scholar] [CrossRef]
- Pantel, K.; Speicher, M.R. The biology of circulating tumor cells. Oncogene 2016, 35, 1216–1224. [Google Scholar] [CrossRef] [PubMed]
- Keup, C.; Mach, P.; Aktas, B.; Tewes, M.; Kolberg, H.C.; Hauch, S.; Sprenger-Haussels, M.; Kimmig, R.; Kasimir-Bauer, S. RNA profiles of circulating tumor cells and extracellular vesicles for therapy stratification of metastatic breast cancer patients. Clin. Chem. 2018, 64, 1054–1062. [Google Scholar] [CrossRef]
- Punnoose, E.A.; Atwal, S.; Liu, W.; Raja, R.; Fine, B.M.; Hughes, B.G.; Hicks, R.J.; Hampton, G.M.; Amler, L.C.; Pirzkall, A.; et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: Association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin. Cancer Res. 2012, 18, 2391–2401. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Warren Mason Mariani, L.; Bromberg, J.E.C.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef]
- Duan, G.C.; Zhang, X.P.; Wang, H.E.; Wang, Z.K.; Zhang, H.; Yu, L.; Xue, W.F.; Xin, Z.F.; Hu, Z.H.; Zhao, Q.T. Circulating tumor cells as a screening and diagnostic marker for early-stage non-small cell lung cancer. Onco Targets Ther. 2020, 13, 1931–1939. [Google Scholar] [CrossRef]
- Marquette, C.H.; Boutros, J.; Benzaquen, J.; Ferreira, M.; Pastre, J.; Pison, C.; Padovani, B.; Bettayeb, F.; Fallet, V.; Guibert, N.; et al. Circulating tumour cells as a potential biomarker for lung cancer screening: A prospective cohort study. Lancet Respir. Med. 2020, 8, 709–716, Erratum in Lancet Respir. Med. 2020, 8, E94. [Google Scholar] [CrossRef]
- Hanssen, A.; Wagner, J.; Gorges, T.M.; Taenzer, A.; Uzunoglu, F.G.; Driemel, C.; Stoecklein, N.H.; Knoefel, W.T.; Angenendt, S.; Hauch, S.; et al. Characterization of different CTC subpopulations in non-small cell lung cancer. Sci. Rep. 2016, 6, 28010. [Google Scholar] [CrossRef] [PubMed]
- Chudasama, D.; Burnside, N.; Beeson, J.; Karteris, E.; Rice, A.; Anikin, V. Perioperative detection of circulating tumour cells in patients with lung cancer. Oncol. Lett. 2017, 14, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Chudasama, D.; Barr, J.; Beeson, J.; Beddow, E.; McGonigle, N.; Rice, A.; Nicholson, A.; Anikin, V. Detection of circulating tumour cells and survival of patients with non-small cell lung cancer. Anticancer Res. 2017, 37, 169–173. [Google Scholar] [CrossRef]
- Qi, Y.; Wang, W. Clinical significance of circulating tumor cells in squamous cell lung cancer patients. Cancer Biomark. 2017, 18, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Zhu, D.; Yang, Y.; Yuan, G.; Xie, H.; Shen, R. The application of nano-enrichment in CTC detection and the clinical significance of CTCs in non-small cell lung cancer (NSCLC) treatment. PLoS ONE 2019, 14, e0219129. [Google Scholar] [CrossRef]
- Castello, A.; Carbone, F.G.; Rossi, S.; Monterisi, S.; Federico, D.; Toschi, L.; Lopci, E. Circulating tumor cells and metabolic parameters in NSCLC patients treated with checkpoint inhibitors. Cancers 2020, 12, 487. [Google Scholar] [CrossRef]
- Shishido, S.N.; Carlsson, A.; Nieva, J.; Bethel, K.; Hicks, J.B.; Bazhenova, L.; Kuhn, P. Circulating tumor cells as a response monitor in stage IV non-small cell lung cancer. J. Transl. Med. 2019, 17, 294. [Google Scholar] [CrossRef]
- Lindsay, C.R.; Blackhall, F.H.; Carmel, A.; Fernandez-Gutierrez, F.; Gazzaniga, P.; Groen, H.J.M.; Hiltermann, T.J.N.; Krebs, M.G.; Loges, S.; López-López, R.; et al. EPAC-lung: Pooled analysis of circulating tumour cells in advanced non-small cell lung cancer. Eur. J. Cancer 2019, 117, 60–68. [Google Scholar] [CrossRef]
- Carlsson, A.; Nair, V.S.; Luttgen, M.S.; Keu, K.V.; Horng, G.; Vasanawala, M.; Kolatkar, A.; Jamali, M.; Iagaru, A.H.; Kuschner, W.; et al. Circulating tumor microemboli diagnostics for patients with non-small-cell lung cancer. J. Thorac. Oncol. 2014, 9, 1111–1119, Erratum in J. Thorac. Oncol. 2020, ahead of print. [Google Scholar] [CrossRef]
- Tay, R.Y.; Fernández-Gutiérrez, F.; Foy, V.; Burns, K.; Pierce, J.; Morris, K.; Priest, L.; Tugwood, J.; Ashcroft, L.; Lindsay, C.R.; et al. Prognostic value of circulating tumour cells in limited-stage small-cell lung cancer: Analysis of the concurrent once-daily versus twice-daily radiotherapy (CONVERT) randomised controlled trial. Ann. Oncol. 2019, 30, 1114–1120. [Google Scholar] [CrossRef]
- Cao, W.; Tang, Q.; Zeng, J.; Jin, X.; Zu, L.; Xu, S. A review of biomarkers and their clinical impact in resected early-stage non-small-cell lung cancer. Cancers 2023, 15, 4561. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.O.; Kim, Y.T.; Jung, K.C.; Yoon, K.J.; Beak-Hui, K.; Chul-Woo, K. TTF-1 mRNA-positive circulating tumor cells in the peripheral blood predict poor prognosis in surgically resected non-small cell lung cancer patients. Lung Cancer 2011, 71, 209–216. [Google Scholar] [CrossRef]
- Bayarri-Lara, C.; Ortega, F.G.; Cueto Ladrón de Guevara, A.; Puche, J.L.; Ruiz Zafra, J.; de Miguel-Pérez, D.; Ramos, A.S.; Giraldo-Ospina, C.F.; Navajas Gómez, J.A.; Delgado-Rodriguez, M.; et al. Circulating tumor cells identify early recurrence in patients with non-small cell lung cancer undergoing radical resection. PLoS ONE 2016, 11, e0148659. [Google Scholar] [CrossRef] [PubMed]
- Frick, M.A.; Feigenberg, S.J.; Jean-Baptiste, S.R.; Aguarin, L.A.; Mendes, A.; Chinniah, C.; Swisher-McClure, S.; Berman, A.; Levin, W.; Cengel, K.A.; et al. Circulating tumor cells are associated with recurrent disease in patients with early-stage non-small cell lung cancer treated with stereotactic body radiotherapy. Clin. Cancer Res. 2020, 26, 2372–2380. [Google Scholar] [CrossRef] [PubMed]
- Shang, R.; Lee, S.; Senavirathne, G.; Lai, E.C. microRNAs in action: Biogenesis, function and regulation. Nat. Rev. Genet. 2023, 24, 816–833. [Google Scholar] [CrossRef]
- Wankhede, D.; Grover, S.; Hofman, P. Circulating Tumor Cells as a Predictive Biomarker in resectable lung cancer: A Systematic Review and Meta-Analysis. Cancers 2022, 14, 6112. [Google Scholar] [CrossRef]
- Fina, E.; Federico, D.; Novellis, P.; Dieci, E.; Monterisi, S.; Cioffi, F.; Margiamelli, G.; Finocchiaro, G.; Alloisio, M.; Veronesi, G. Subpopulations of circulating cells with morphological features of malignancy are preoperatively detected and have differential prognostic significance in non-small cell lung cancer. Cancers 2021, 13, 4488. [Google Scholar] [CrossRef]
- Klein, C.A. Cancer progression and the invisible phase of metastatic colonization. Nat. Rev. Cancer 2020, 20, 681–694. [Google Scholar] [CrossRef]
- Antunes-Ferreira, M.; D’Ambrosi, S.; Arkani, M. Tumor-educated platelet blood tests for non-small cell cancer detection and management. Sci. Rep. 2023, 13, 9359. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.X.; Yang, W.; Wu, J.Z.; Zhou, C.; Liu, S.; Shi, H.B.; Zhou, W.Z. MicroRNA-32-5p inhibits epithelial-mesenchymal transition and metastasis in lung adenocarcinoma by targeting SMAD family 3. J. Cancer 2021, 12, 2258–2267. [Google Scholar] [CrossRef]
- Moretti, F.; D’Antona, P.; Finardi, E.; Barbetta, M.; Dominioni, L.; Poli, A.; Gini, E.; Noonan, D.M.; Imperatori, A.; Rotolo, N.; et al. Systematic review and critique of circulating miRNAs as biomarkers of stage I-II non-small cell lung cancer. Oncotarget 2017, 8, 94980–94996. [Google Scholar] [CrossRef]
- Fehlmann, T.; Kahraman, M.; Ludwig, N.; Backes, C.; Galata, V.; Keller, V.; Geffers, L.; Mercaldo, N.; Hornung, D.; Weis, T.; et al. Evaluating the use of circulating microRNA profiles for lung cancer detection in symptomatic patients. JAMA Oncol. 2020, 6, 714–723. [Google Scholar] [CrossRef]
- Ulivi, P.; Petracci, E.; Marisi, G.; Baglivo, S.; Chiari, R.; Billi, M.; Canale, M.; Pasini, L.; Racanicchi, S.; Vagheggini, A.; et al. Prognostic role of circulating miRNAs in early-stage non-small cell cancer. J. Clin. Med. 2019, 8, 131. [Google Scholar] [CrossRef]
- Sun, L.; Zhou, H.; Yang, Y.; Chen, Y.; Wang, Y.; She, M.; Li, C. Meta-analysis of diagnostic and prognostic value of miR-126 in non-small cell lung cancer. Biosci. Rep. 2020, 40, BSR20200349. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Y.; Yin, Y.; Li, S. Detection of circulating exosomal miR-17-5p serves as a novel non-invasive diagnostic marker for non-small cell lung cancer patients. Pathol. Res. Pract. 2019, 215, 152466. [Google Scholar] [CrossRef] [PubMed]
- Grimolizzi, F.; Monaco, F.; Leoni, F.; Bracci, W.; Staffolani, S.; Bersaglieri, C.; Gaetani, S.; Valentino, M.; Amati, M.; Rubini, C.; et al. Exosomal miR-126 as a circulating biomarker in non-small-cell lung cancer regulating cancer progression. Sci. Rep. 2017, 7, 15277. [Google Scholar] [CrossRef]
- Wu, Q.; Yu, L.; Lin, X.; Zheng, Q.; Zhang, S.; Chen, D.; Pan, X.; Huang, Y. Combination of serum miRNAs with serum exosomal miRNAs in early diagnosis for non-small-cell lung cancer. Cancer Manag. Res. 2020, 12, 485–495. [Google Scholar] [CrossRef]
- Sun, S.; Chen, H.; Xu, C.; Zhang, Y.; Zhang, Q.; Chen, L.; Ding, Q.; Deng, Z. Exosomal miR-106b serves as a novel marker for lung cancer and promotes cancer metastasis via targeting PTEN. Life Sci. 2020, 244, 117297. [Google Scholar] [CrossRef]
- Zhou, Q.; Huang, S.X.; Zhang, F.; Li, S.J.; Liu, C.; Xi, Y.Y.; Wang, L.; Wang, X.; He, Q.C.; Sun, C.C.; et al. MicroRNAs: A novel potential biomarker for diagnosis and therapy in patients with non-small cell lung cancer. Cell Prolif. 2017, 50, e12394. [Google Scholar] [CrossRef]
- Diaz, L.A., Jr.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef]
- Perez-Callejo, D.; Romero, A.; Provencio, M.; Torrente, M. Liquid biopsy based biomarkers in non-smallcell lung cancer for diagnosis and treatment monitoring. Transl. Lung Cancer Res. 2016, 5, 455–465. [Google Scholar] [CrossRef]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef]
- Gale, D.; Heider, K.; Ruiz-Valdepenas, A.; Hackinger, S.; Perry, M.; Marsico, G.; Rundell, V.; Wulff, J.; Sharma, G.; Knock, H.; et al. Residual ctDNA after treatment predicts early relapse in patients with early-stage non-small cell lung cancer. Ann. Oncol. 2022, 33, 500–510. [Google Scholar] [CrossRef]
- Bordi, P.; Del Re, M.; Danesi, R.; Tiseo, M. Circulating DNA in diagnosis and monitoring EGFR gene mutations in advanced non-small cell lung cancer. Transl. Lung Cancer Res. 2015, 4, 584–597. [Google Scholar]
- Moding, E.J.; Nabet, B.Y.; Alizadeh, A.A.; Diehn, M. Detecting liquid remnants of solid tumors: Circulating tumor DNA minimal residual disease. Cancer Discov. 2021, 11, 2968–2986. [Google Scholar] [CrossRef] [PubMed]
- Gottardo, A.; Gristina, V.; Perez, A.; Di Giovanni, E.; Contino, S.; Barraco, N.; Bono, M.; Iannì, G.; Randazzo, U.; Bazan Russo, T.D.; et al. Roles of Tumor-Educated Platelets (TEPs) in the biology of Non-Small Cell Lung Cancer (NSCLC): A systematic review. “Re-discovering the neglected biosources of the liquid biopsy family”. J. Liq. Biopsy 2025, 3, 100136. [Google Scholar] [CrossRef] [PubMed]
- Best, M.G.; Sol, N.; In’t Veld, S.G.J.G.; Vancura, A.; Muller, M.; Niemeijer, A.-L.N.; Fejes, A.V.; Fat, L.-A.T.K.; In’t Veld, A.E.H.; Leurs, C.; et al. Swarm intelligence-enhanced detection of non-small cell lung cancer using Tumor-Educated Platelets. Cancer Cell 2017, 32, 238–252.e9. [Google Scholar] [CrossRef] [PubMed]
- In’t Veld, S.G.J.G.; Arkani, M.; Post, E.; Antunes-Ferreira, M.; D’ambrosi, S.; Vessies, D.C.; Vermunt, L.; Vancura, A.; Muller, M.; Niemeijer, A.-L.N.; et al. Detection and localization of early- and late-stage cancers using platelet RNA. Cancer Cell 2022, 40, 999–1009.e6. [Google Scholar]
- Nilsson, R.J.; Karachaliou, N.; Berenguer, J.; Gimenez-Capitan, A.; Schellen, P.; Teixido, C.; Tannous, J.; Kuiper, J.L.; Drees, E.; Grabowska, M.; et al. Rearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget 2016, 7, 1066–1075. [Google Scholar] [CrossRef]
- Best, M.G.; Sol, N.; Kooi, I.; Tannous, J.; Westerman, B.A.; Rustenburg, F.; Schellen, P.; Verschueren, H.; Post, E.; Koster, J.; et al. RNA-Seq of Tumor-Educated Platelets enables blood-based pan-cancer, Multiclass, and molecular pathway cancer diagnostics. Cancer Cell 2015, 28, 666–676. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Vanni, I.; Alama, A.; Grossi, F.; Dal Bello, M.G.; Coco, S. Exosomes: A new horizon in lung cancer. Drug Discov. Today 2017, 22, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Logozzi, M.; Mizzoni, D.; Di Raimo, R.; Fais, S. Exosomes: A source for new and old biomarkers in cancer. Cancers 2020, 12, 2566. [Google Scholar] [CrossRef] [PubMed]
- Rijavec, E.; Coco, S.; Genova, C.; Rossi, G.; Longo, L.; Grossi, F. Liquid Biopsy in non-Small Cell Lung Cancer: Highlights and Challenges. Cancers 2020, 12, 17. [Google Scholar] [CrossRef]
- Xu, K.; Zhang, C.; Du, T.; Gabriel, A.N.A.; Wang, X.; Li, X.; Sun, L.; Wang, N.; Jiang, X.; Zhang, Y. Progress of exosomes in the diagnosis and treatment of lung cancer. Biomed. Pharmacother. 2021, 134, 111111. [Google Scholar] [CrossRef]
- Wang, N.; Song, X.; Liu, L.; Niu, L.; Wang, X.; Song, X.; Xie, L. Circulating exosomes contain protein biomarkers of metastatic non-small-cell lung cancer. Cancer Sci. 2018, 109, 1701–1709. [Google Scholar] [CrossRef]
- Jakobsen, K.R.; Paulsen, B.S.; Bæk, R.; Varming, K.; Sorensen, B.S.; Jørgensen, M.M. Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J. Extracell. Vesicles 2015, 4, 26659. [Google Scholar] [CrossRef]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-Stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766769. [Google Scholar] [CrossRef]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.W.; Simpson, R.J. Extracellular vesicles in cancer—Implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018, 15, 617–638. [Google Scholar] [CrossRef]
- Schmidt, D.R.; Patel, R.; Kirsch, D.G.; Lewis, C.A.; Vander Heiden, M.G.; Locasale, J.W. Metabolomics in cancer research and emerging applications in clinical oncology. CA Cancer J. Clin. 2021, 71, 333–358. [Google Scholar] [CrossRef]
- Einoch Amor, R.; Levy, J.; Broza, Y.Y.; Vangravs, R.; Rapopor, T.S.; Zhang, M.; Wu, W.; Leja, M.; Behar, J.A.; Haick, H. Liquid biopsy-based volatile organic compounds from blood and urine and their combined data sets for highly accurate detection of cancer. ACS Sens. 2023, 8, 1450–1461. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, X.; Wang, C.; Zhang, H.; Cai, Z. Non-targeted and targeted metabolomics approaches to diagnosing lung cancer and predicting patient prognosis. Oncotarget 2016, 7, 63437–63448. [Google Scholar] [CrossRef] [PubMed]
- Fox, D.B.; Garcia, N.M.G.; McKinney, B.J.; Lupo, R.; Noteware, L.C.; Newcomb, R.; Liu, J.; Locasale, J.W.; Hirschey, M.D.; Alvarez, J.V. NRF2 activation promotes the recurrence of dormant tumour cells through regulation of redox and nucleotide metabolism. Nat. Metab. 2020, 2, 318–334. [Google Scholar] [CrossRef] [PubMed]
- Hensley, C.T.; Faubert, B.; Yuan, Q.; Lev-Cohain, N.; Jin, E.; Kim, J.; Jiang, L.; Ko, B.; Skelton, R.; Loudat, L.; et al. Metabolic heterogeneity in human lung tumors. Cell 2016, 164, 681–694. [Google Scholar] [CrossRef]
- Xu, J.Y.; Zhang, C.; Wang, X.; Zhai, L.; Ma, Y.; Mao, Y.; Qian, K.; Sun, C.; Liu, Z.; Jiang, S.; et al. Integrative proteomic characterization of human lung adenocarcinoma. Cell 2020, 182, 245–261.e17. [Google Scholar] [CrossRef]
- Geyer, P.E.; Voytik, E.; Treit, P.V.; Doll, S.; Kleinhempel, A.; Niu, L.; Müller, J.B.; Buchholtz, M.; Bader, J.M.; Teupser, D.; et al. Plasma proteome profiling to detect and avoid sample-related biases in biomarker studies. EMBO Mol. Med. 2019, 11, e10427. [Google Scholar] [CrossRef]
- Youssef, O.; Sarhadi, V.K.; Armengol, G.; Piirilä, P.; Knuuttila, A.; Knuutila, S. Exhaled breath condensate as a source of biomarkers for lung carcinomas. A focus on genetic and epigenetic markers—A Mini-Review. Genes Chromosomes Cancer 2016, 55, 905–914. [Google Scholar] [CrossRef]
- Zhang, C.; Leng, W.; Sun, C.; Lu, T.; Chen, Z.; Men, X.; Wang, Y.; Wang, G.; Zhen, B.; Qin, J. Urine proteome profiling predicts lung cancer from control cases and other tumors. EBio Med. 2018, 30, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Wang, X.Y.; Han, X.H.; Wang, H.; Qi, J. Diagnostic value of Cyfra21-1, SCC and CEA for differentiation of early-stage NSCLC from benign lung disease. Int. J. Clin. Exp. Med. 2015, 8, 11295–11300. [Google Scholar]
- Jiang, Z.F.; Wang, M.; Xu, J.L. Thymidine kinase 1 combined with CEA, CYFRA21-1 and NSE improved its diagnostic value for lung cancer. Life Sci. 2018, 194, 1–6. [Google Scholar] [CrossRef]
- Doseeva, V.; Colpitts, T.; Gao, G.; Woodcock, J.; Knezevic, V. Performance of a multiplexed dual analyte immunoassay for the early detection of non-small cell lung cancer. J. Transl. Med. 2015, 13, 55. [Google Scholar] [CrossRef]
- Fang, R.; Zhu, Y.; Khadka, V.S.; Zhang, F.; Jiang, B.; Deng, Y. The evaluation of serum biomarkers for non-small cell lung cancer (NSCLC) diagnosis. Front. Physiol. 2018, 9, 1710. [Google Scholar] [CrossRef] [PubMed]
- Zang, R.; Li, Y.; Jin, R.; Wang, X.; Lei, Y.; Che, Y.; Lu, Z.; Mao, S.; Huang, J.; Liu, C.; et al. Enhancement of diagnostic performance in lung cancers by combining CEA and CA125 with autoantibodies detection. Oncoimmunology 2019, 8, e1625689. [Google Scholar] [CrossRef]
- Yang, B.; Li, X.; Ren, T.; Yin, Y. Autoantibodies as diagnostic biomarkers for lung cancer: A systematic review. Cell Death Discov. 2019, 5, 126. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Berry, L.D.; Aisner, D.L.; Sheren, J.; Boyle, T.; Bunn, P.A., Jr.; Johnson, B.E.; Kwiatkowski, D.J.; Drilon, A.; Shol, L.M.; et al. MET IHC is a poor screen for MET amplification or MET Exon 14 mutations in lung adenocarcinomas: Data from a tri-institutional cohort of the lung cancer mutation consortium. J. Thorac. Oncol. 2019, 14, 1666–1671. [Google Scholar] [CrossRef] [PubMed]
- Mino-Kenudson, M.; Schalper, K.; Cooper, W.; Dacic, S.; Hirsch, F.R.; Jain, D.; Lopez-Rios, F.; Tsao, M.S.; Yatabe, Y.; Beasley, M.B.; et al. Predictive biomarkers for immunotherapy in lung cancer: Perspective from the international association for the study of lung cancer pathology committee. J. Thorac. Oncol. 2022, 17, 1335–1354. [Google Scholar] [CrossRef]
- Jahani, M.M.; Mashayekhi, P.; Omrani, M.D.; Meibody, A.A. Efficacy of liquid biopsy for genetic mutations determination in non-small cell lung cancer: A systematic review on literatures. BMC Cancer 2025, 25, 433–446. [Google Scholar] [CrossRef]
- Oxnard, G.R.; Paweletz, C.P.; Kuang, Y.; Mach, S.L.; O’Connell, A.; Messineo, M.M.; Luke, J.; Butaney, M.; Kirschmeier, P.; Jackman, D.M.; et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin. Cancer Res. 2014, 20, 1698–1705. [Google Scholar] [CrossRef]
- Yanagita, M.; Redig, A.J.; Paweletz, C.P.; Dahlberg, S.E.; O’Connell, A.; Feeney, N.; Taibi, M.; Boucher, D.; Oxnard, G.R.; Johnson, B.E. A prospective evaluation of circulating tumor cells and cell-free DNA inEGFR-mutant non–small cell lung cancer patients treated with erlotinib on a phase II trial. Clin. Cancer Res. 2016, 22, 6010–6020. [Google Scholar] [CrossRef]
- Sandfeld-Paulsen, B.; Jakobsen, K.R.; Bæk, R.; Folkersen, B.H.; Rasmussen, T.R.; Meldgaard, P.; Varming, K.; Jorgensen, M.M.; Sorensen, B.S. Exosomal proteins as diagnostic biomarkers in lung cancer. J. Thorac. Oncol. 2016, 11, 1701–1710. [Google Scholar] [CrossRef] [PubMed]
- Lobb, R.J.; van Amerongen, R.; Wiegmans, A.; Ham, S.; Larsen, J.E.; Moller, A. Exosomes derived from mesenchymal non-small cell lung cancer cells promote chemoresistance. Int. J. Cancer 2017, 141, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.; Kim, C.J.; Sunkara, V.; Kim, M.H.; Cho, Y.K. Liquid biopsy in lung cancer: Clinical applications of circulating biomarkers (CTCs and ctDNA). Micromachines 2018, 9, 100. [Google Scholar] [CrossRef] [PubMed]
- Torga, G.; Pienta, K.J. Patient-paired sample congruence between 2 commercial liquid biopsy tests. JAMA Oncol. 2018, 4, 868–870. [Google Scholar] [CrossRef]
| Group/Markers | Significance |
|---|---|
| Circulating tumor cells | Diagnosis, prognostic, monitoring response to treatment |
| Circulating free microRNAs | Early diagnosis, prognostic, metastasis, monitoring, response to treatment |
| Circulating free DNA | Diagnosis and prognostic |
| Tumor-educated platelets | Early diagnosis, monitoring, response to treatment |
| Circulating extracellular vesicles | Diagnosis, metastasis, response to treatment |
| Metabolomic markers | Early diagnosis, prediction, response to treatment |
| Proteomics markers | Early diagnosis, prognostic, monitoring |
| Feature | CellSearch (FDA-Approved) | Microfluidic Platforms (e.g., Celsee, CTC-Chip, Auto-ICell) |
|---|---|---|
| Sensitivity | ~70% (EpCAM + CTCs) | Up to 94–96% |
| Specificity | 93–100% | 100% |
| Cost | €700–€1500/test | Variable; generally higher due to technical complexity |
| CTC phenotype | Primarily EpCAM + (epithelial) CTCs | Epithelial and mesenchymal CTCs |
| Clinical use | Widely used in clinical settings | Emerging; research-focused |
| Turnaround time | 1–3 days | 1–3 days |
| Enrichment strategy | Immunomagnetic (EpCAM-based) | Physical properties, size, deformability |
| Viability assessment | No | Yes |
| Symbol | Expression | Materials | Biomarker |
|---|---|---|---|
| microRNA-23-5p | Up | Plasma | Prediction of survival |
| microRNA-32-5p | Down | Whole blood | Diagnosis, prognosis |
| microRNA-502b-3p | Up | Plasma | Diagnosis, prognosis |
| microRNA-200c | Up | Whole blood | Early diagnosis, prognosis |
| microRNA-150-5p | Up | Whole blood | Early detection, monitoring recurrences |
| microRNA-122-3p | Down | Whole blood | Diagnosis, prognosis |
| microRNA-492a-3p | Down | Plasma | Diagnosis, prognosis |
| microRNA-20a | Down | Whole blood | Prediction of survival |
| microRNA-2223 | Down | Whole blood | Diagnosis, Prognosis |
| microRNA-145 | Up | Plasma | Prediction of survival |
| microRNA-448 | Up | Plasma | Diagnosis, prognosis |
| microRNA-628-3p | Up | Serum | Diagnosis, prognosis |
| microRNA-210 | Up | Plasma | Diagnosis, prognosis |
| microRNA-29c | Down | Whole blood | Early detection |
| microRNA-124 | Up | Plasma | Diagnosis, prognosis |
| microRNA-126 | Down | Serum | Diagnosis, prognosis, response to treatment |
| microRNA-17-5p | Up | Serum | Prediction of survival |
| microRNA-21-5p | Up | Serum | Early detection, prognostic |
| microRNA-141-3p | Up | Serum | Early detection |
| microRNA-222-3p | Up | Serum | Early detection |
| microRNA-486-5p | Down | Serum | Early detection |
| microRNA-146a-5p | Up | Serum | Early detection |
| microRNA-126-3p | Down | Serum | Early detection |
| microRNA-106b | Up | Serum | Monitoring recurrences. metastasis |
| Platform/Module | Typical Mass Accuracy (or Analogous) | Dynamic Range (Linear, Orders of Magnitude) | QA/QC Measures and Challenges |
|---|---|---|---|
| LC-HRMS (Orbitrap, QTOF) | ~1–5 ppm (some <1 ppm for small molecules; ≤10 ppm typical) | ~104 to 105 (4–5 orders) | System suitability tests, internal isotope standards, pooled QC every N samples, blanks, retention time monitoring, drift correction, signal normalization, QC of mass error and retention time deviation; intra-batch CV ≤20–30% |
| GC-MS (high-res and uni-res) | ~1–3 ppm(high-res) or 10s of 100s mDa (low-res) | ~104 (sometimes up to 105) | Retention index calibrants, test mix injections, blanks, drift monitoring, internal standards, replicate injections, detector linearity checks, carry-over monitoring |
| Direct infusion/FIA-MS | ~1–10 ppm (depends on MS) | ~103 to 104 (ion suppression limits range) | Vulnerable to ion suppression/matrix effects; use internal standards, repeated QC injections, drift correction, dilution curves, artifact flagging tools |
| FT-MS/FT-ICR | <<1 pmm (sub-pmm, sometimes 10s of ppb) | ~105 to 106 + (very high) | Requires exceptional stability, regular calibration, lock masses, system suitability, drift monitoring, isotope pattern verification) |
| NMR (1H, 13C) | Chemical shift reproducibility ~0.001–0.005 ppm; spectral resolution ~0.5–1 Hz | ~103 to 104 | Calibration (chemical shift references), shimming, temperaturę stability, instrument checks, replicate measurements, quantitation with standards, QC samples, signal-to-noise monitoring |
| Other/hybrid (ion mobility MS, LC-MS/MS MRM) | Varies; targeted MRM precision high (mDa or better) | Up to 105 or more (targeted) | Calibration curves; low/medium/high-QC samples, reference materials, replicate injections, retention time and transmission monitoring, carry-over checks |
| Biomarker Type | Detection Window | Invasiveness | Cost | Sensitivity | Specificity | Clinical Use Stage |
|---|---|---|---|---|---|---|
| CTCs | Narrow to moderate (detectable in advanced stages) | Low (blood draw) | High | Moderate | High | Limited/research; FDA-approved for prognosis in other cancers (e.g., breast, colon) |
| cfDNA | Moderate (early to late stages) | Low | Moderate to high | High | Moderate to high | Widely used for EGFR mutation testing; approved in NSCLC (liquid biopsy) |
| TEPs | Moderate to broad | Low | Moderate | High | High | Experimental/research |
| EVs | Broad (early detection potential) | Low | Moderate to high | High | High | Experimental/promising for early diagnosis |
| mRNA | Moderate (can vary by stability) | Low | Moderate | Variable | Variable | Research; mRNA panels under evaluation |
| Circulating proteins | Broad (many secreted early) | Low | Low to moderate | Moderate | Moderate | Diagnostic panels in use (e.g., CYFRA 21-1, CEA); not specific alone |
| Circulating metabolites | Moderate (affected by systemic factors) | Low | Low to moderate | Variable | Variable | Research stage; metabolomic signatures under development |
| Year | Key Activities | Milestones |
|---|---|---|
| 1 | Cohort design, multi-omics data generation, AI discovery | Cohort established, biomarker candidates identified |
| 2 | Validation in independent cohorts, model refinement, pilot CDSS | Validated biomarkers, prototype CDSS tested |
| 3 | Clinical trials, regulatory approval, full CDSS integration | Clinical utility proven, CDSS deployed |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jelski, W.; Okrasinska, S.; Rutkowska, W.; Mroczko, B. Circulating Molecular Biomarkers for the Diagnosis and Monitoring of NSCLC—A Review. Int. J. Mol. Sci. 2025, 26, 10278. https://doi.org/10.3390/ijms262110278
Jelski W, Okrasinska S, Rutkowska W, Mroczko B. Circulating Molecular Biomarkers for the Diagnosis and Monitoring of NSCLC—A Review. International Journal of Molecular Sciences. 2025; 26(21):10278. https://doi.org/10.3390/ijms262110278
Chicago/Turabian StyleJelski, Wojciech, Sylwia Okrasinska, Weronika Rutkowska, and Barbara Mroczko. 2025. "Circulating Molecular Biomarkers for the Diagnosis and Monitoring of NSCLC—A Review" International Journal of Molecular Sciences 26, no. 21: 10278. https://doi.org/10.3390/ijms262110278
APA StyleJelski, W., Okrasinska, S., Rutkowska, W., & Mroczko, B. (2025). Circulating Molecular Biomarkers for the Diagnosis and Monitoring of NSCLC—A Review. International Journal of Molecular Sciences, 26(21), 10278. https://doi.org/10.3390/ijms262110278







