Comparative Genomics of Triticum, Secale, and Triticale: Codon Usage Bias in Chloroplast Genomes and Its Implications for Evolution and Genetic Engineering
Abstract
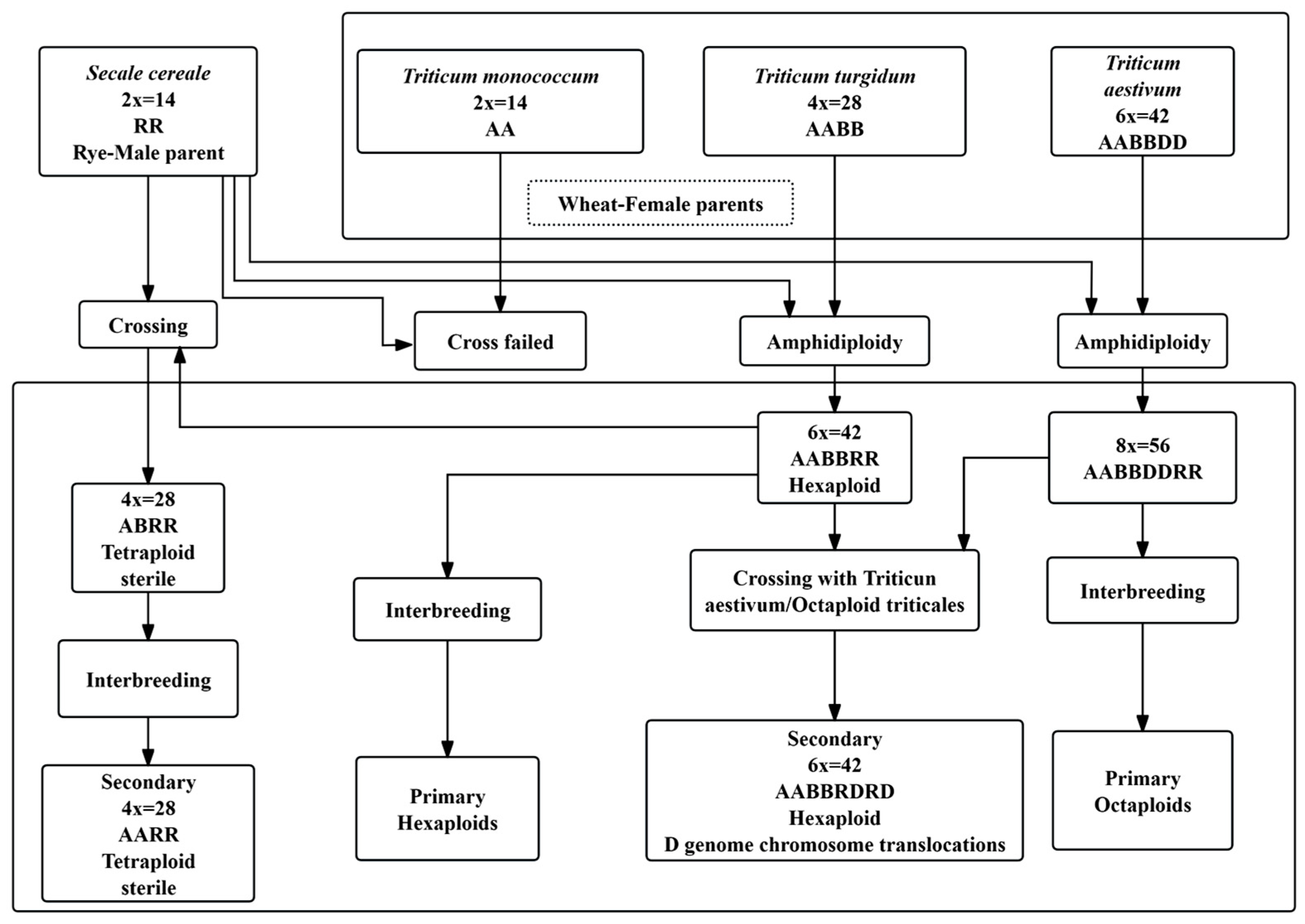
1. Introduction
2. Results
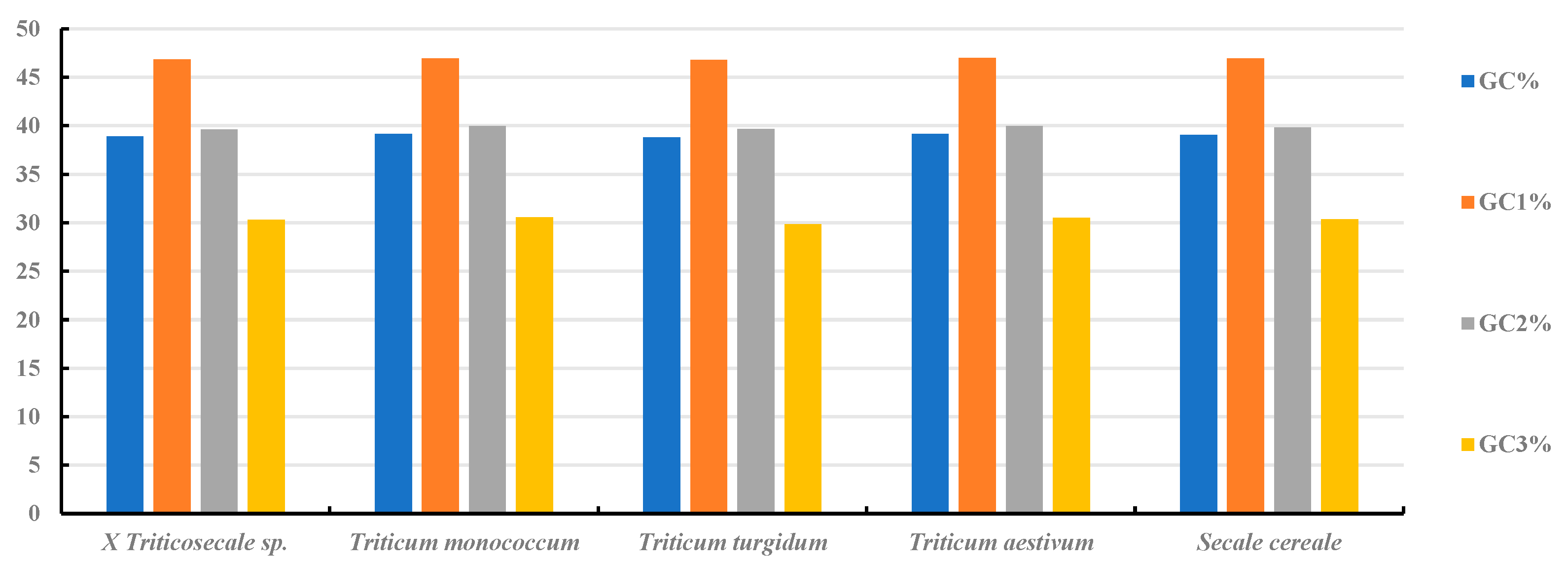
2.1. Nucleotide Composition Analysis of cp Genes in Triticum, Secale, and Triticale
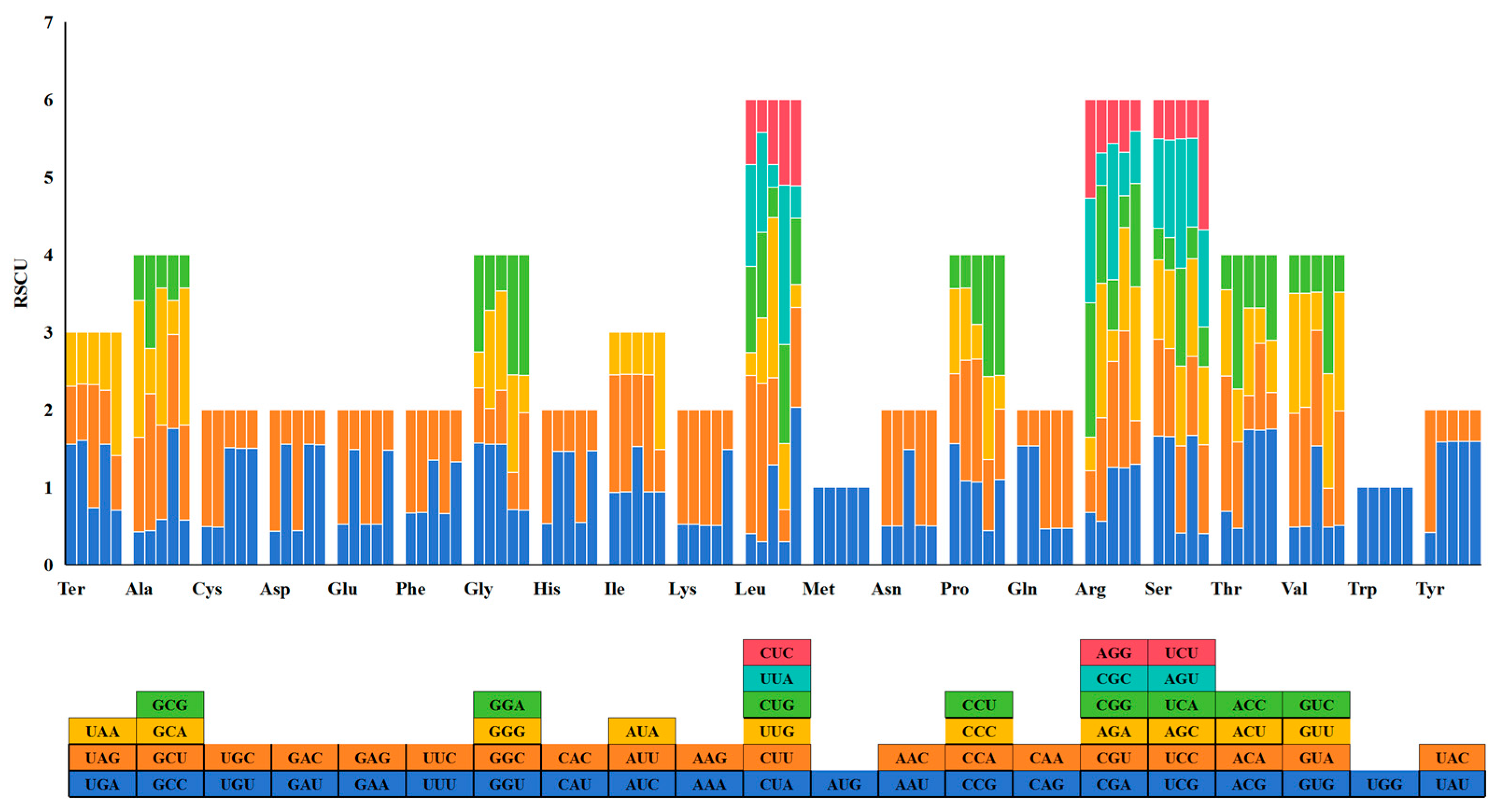
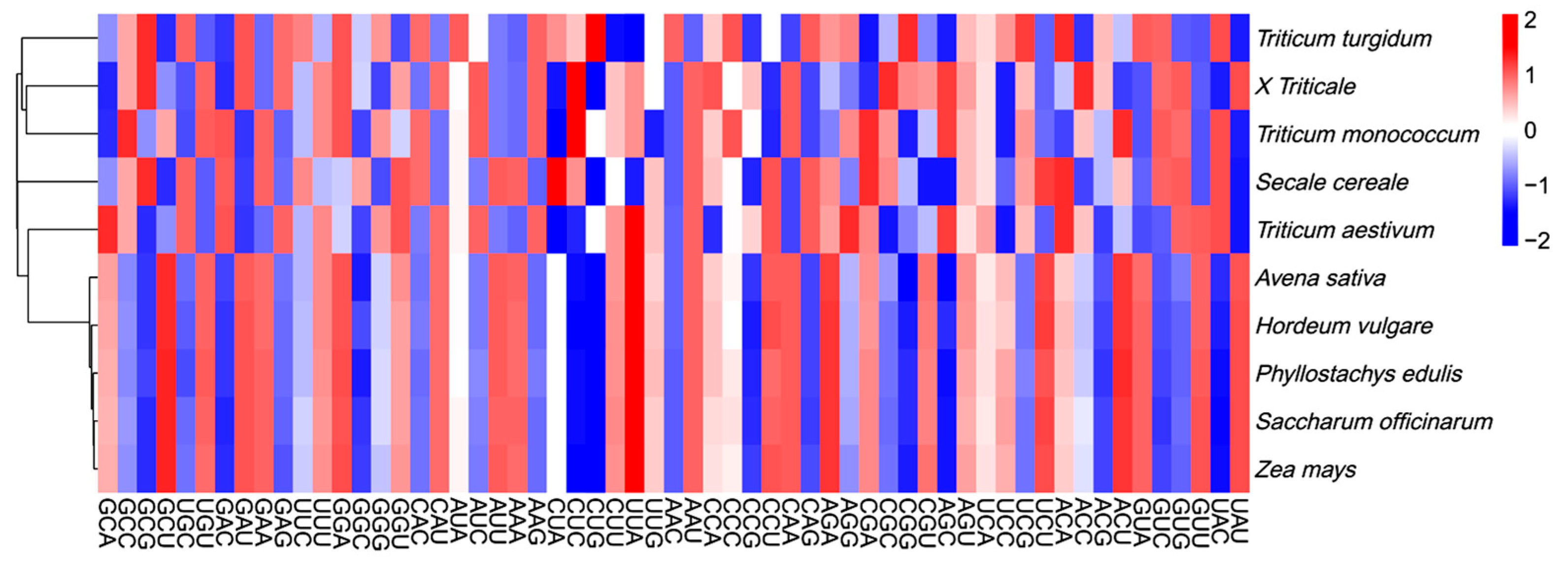
2.2. Relative Synonymous Codon Usage (RSCU) Analysis of Triticum, Secale, and Triticale
2.3. Mutational Pressure Versus Natural Selection in CUB of cp Genes of Triticum, Secale, and Triticale
2.4. Inter-Relationship of CUB and Nucleotide Composition
2.5. Relationship Between Codon Usage Bias and Gene Expression
2.6. ENC-Plot Analysis
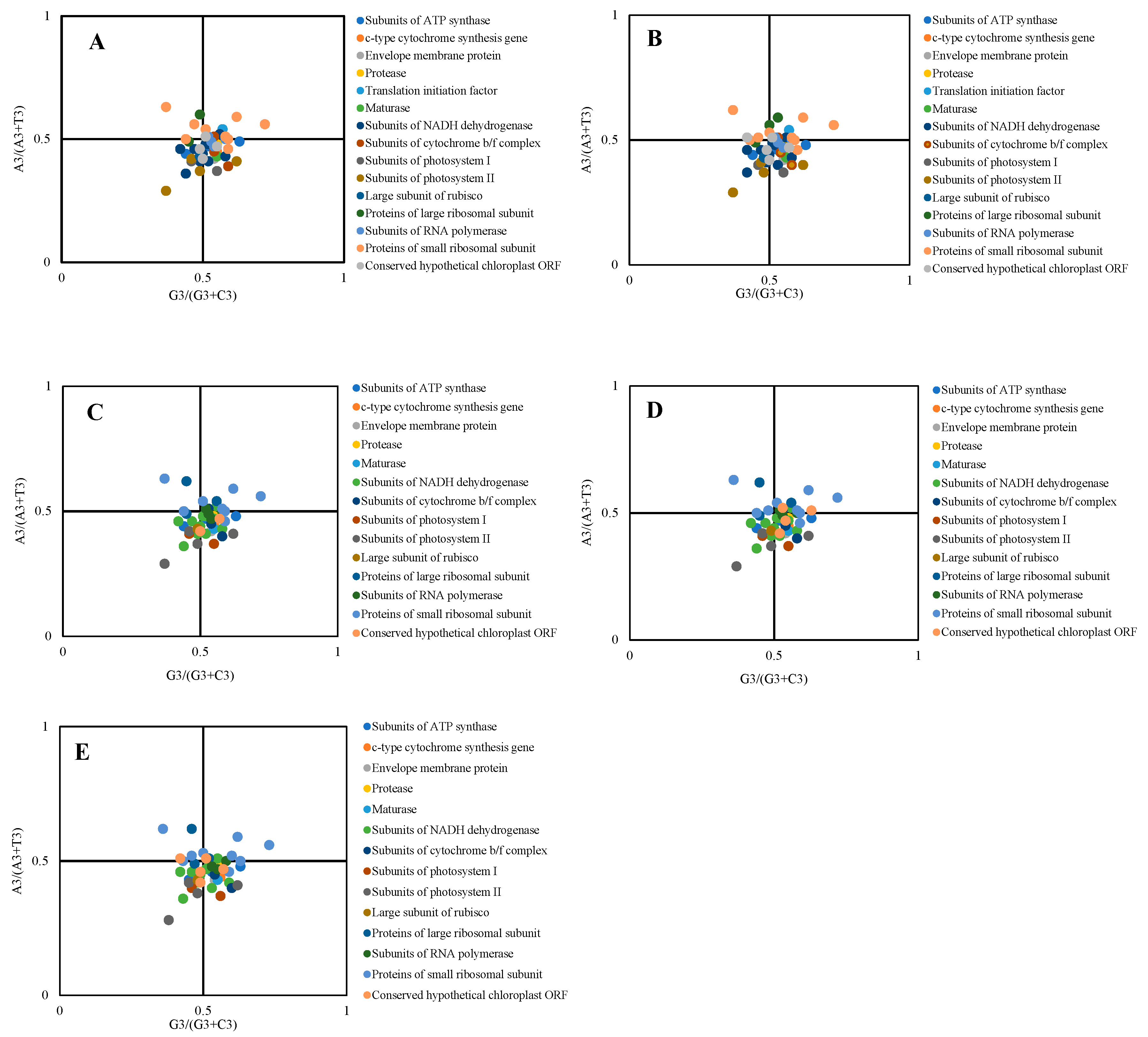
2.7. PR2-Bias-Plot Analysis
2.8. Neutrality Plot
2.9. Optimal Codons
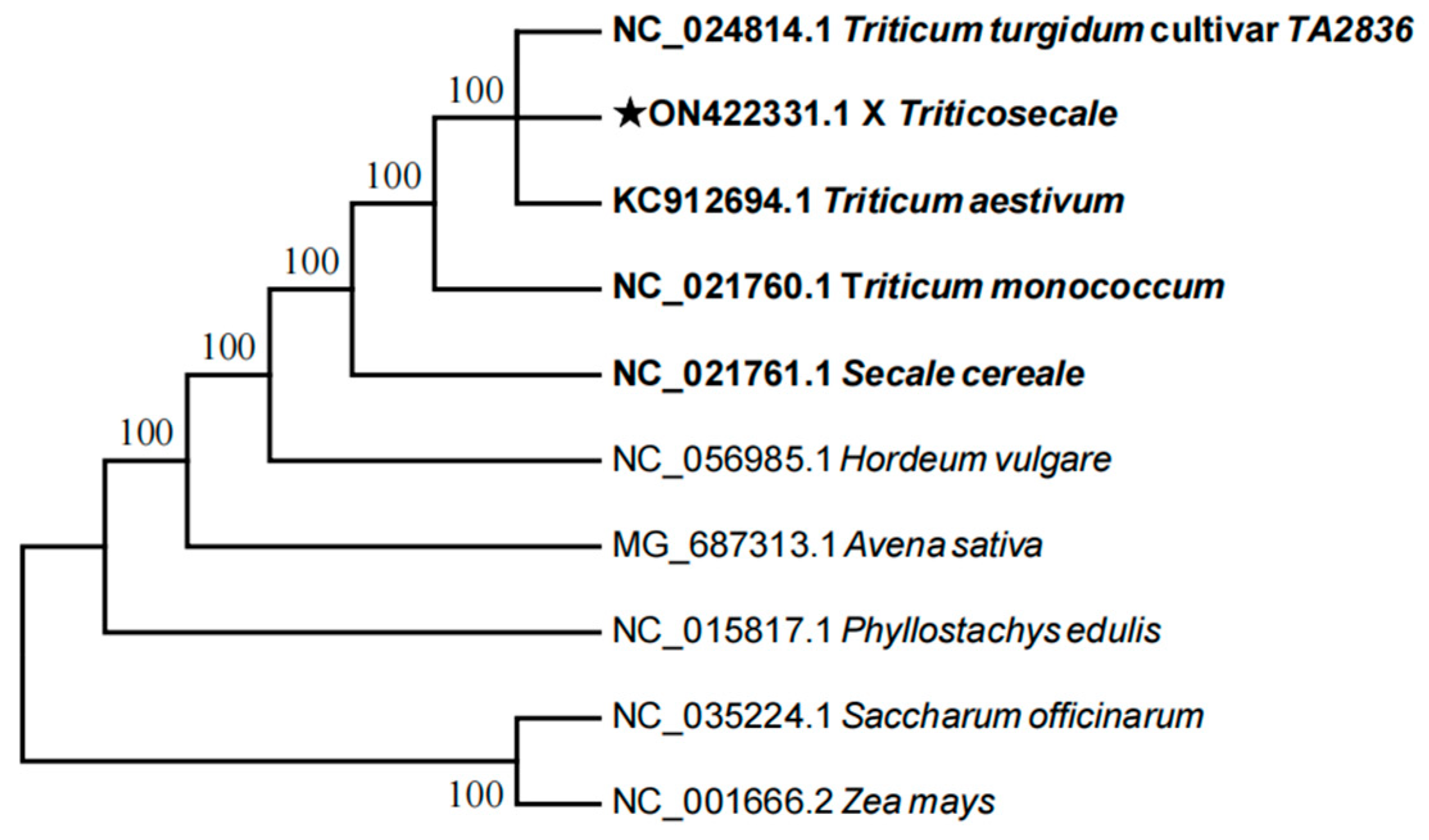
2.10. Phylogenetic Relationships Between Triticale and Its Parental Species Were Inferred from the RSCU Values
3. Discussion
4. Materials and Methods
4.1. Sequence Retrieval
4.2. Analysis of Codon Composition
4.3. Synonymous Codon Usage Order (SCUO) Analysis
4.4. Relative Synonymous Codon Usage (RSCU) Analysis and Measure Independent of Length and Composition (MILC)
4.5. PR-2 Plot
4.6. Neutrality Plot Analysis
4.7. ENC-Plot Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pierre, A.V.; Marion, K.; Sacha, B.; Jotham, R.A.; Gabor, C.; Peter, D.; Felix, K.; Claire, B. Tocopherol cyclase (VTE1) localization and vitamin E accumulation in chloroplast plastoglobule lipoprotein particles. J. Biol. Chem. 2006, 282, 11225–11234. [Google Scholar] [CrossRef] [PubMed]
- Lippold, F.; Dorp, K.V.; Abraham, M.; Hölzl, G.; Wewer, V.; Yilmaz, J.L.; Lager, I.; Montandon, C.; Besagni, C.; Kessler, F.; et al. Fatty Acid Phytyl Ester Synthesis in Chloroplasts of Arabidopsis. Plant Cell 2012, 24, 2001–2014. [Google Scholar] [CrossRef]
- Daniell, H.; Jin, S.; Zhu, X.G.; Matthew, A.G.; Douglas, E.S.; Pamela, S.S. Green giant—A tiny chloroplast genome with mighty power to produce high-value proteins: History and phylogeny. Plant Biotechnol. J. 2022, 219, 430–447. [Google Scholar] [CrossRef]
- Dobrogojski, J.; Adamiec, M.; Luciński, R. The chloroplast genome: A review. Acta Physiol. Plant. 2020, 42, 98. [Google Scholar] [CrossRef]
- Sugiura, M. History of chloroplast genomics. In Advances in Photosynthesis and Respiration; Govindjee, J.T., Gest, H., Allen, J.F., Eds.; Biomedical and Life Sciences; Springer: Dordrecht, The Netherlands, 2003; pp. 1057–1063. [Google Scholar]
- Grevich, J.J.; Daniell, H. Chloroplast Genetic Engineering: Recent Advances and Future Perspectives. Crit. Rev. Plant Sci. 2005, 24, 83–107. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Raubeson, L.A.; Peery, R.; Chumley, T.W.; Dziubek, C.; Fourcade, H.M.; Boore, J.L.; Jansen, R.K. Comparative chloroplast genomics: Analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genom. 2007, 8, 174. [Google Scholar] [CrossRef]
- Kwak, S.-Y.; Lew, T.T.S.; Sweeney, C.J.; Koman, V.B.; Wong, M.H.; Bohmert-Tatarev, K.; Snell, K.D.; Seo, J.S.; Chua, N.-H.; Strano, M.S. Chloroplast-selective gene delivery and expression in planta using chitosan-complexed single-walled carbon nanotube carriers. Nat. Nanotechnol. 2019, 14, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Ruf, S.; Hermann, M.; Berger, I.J.; Carrer, H.; Bock, R. Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat. Biotechnol. 2001, 19, 870–875. [Google Scholar] [CrossRef]
- Murray, E.E.; Lotzer, J.; Eberle, M. Codon usage in plant genes. Nucleic Acids Res. 1989, 17, 477–498. [Google Scholar] [CrossRef]
- Quax, T.E.; Claassens, N.J.; Söll, D.; van der Oost, J. Codon Bias as a Means to Fine-Tune Gene Expression. Mol. Cell 2015, 59, 149–161. [Google Scholar] [CrossRef]
- Parvathy, S.T.; Udayasuriyan, V.; Bhadana, V. Codon usage bias. Mol. Biol. Rep. 2022, 49, 539–565. [Google Scholar] [CrossRef]
- Romero, H.; Zavala, A.; Musto, H. Codon usage in Chlamydia trachomatis is the result of strand-specific mutational biases and a complex pattern of selective forces. Nucleic Acids Res. 2000, 28, 2084–2090. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Cai, X.; Chen, Q.; Zhou, H.; Cai, Y.; Ben, A. Factors Affecting Synonymous Codon Usage Bias in Chloroplast Genome of Oncidium Gower Ramsey. Evol. Bioinform. 2011, 7, 271–278. [Google Scholar] [CrossRef]
- Kurland, C.G. Codon bias and gene expression. FEBS Lett. 1991, 285, 165–169. [Google Scholar] [CrossRef]
- Li, X.Z.; Song, H.; Zhang, Z.H.; Xu, H.F.; Liu, X.; Li, Y.L. Analysis of codon usage bias in the genome of Epichloëgansuensis. Acta Prataculturae Sin. 2020, 29, 67–77. [Google Scholar]
- Gao, S.Y.; Li, Y.Y.; Yang, Z.Q.; Dong, K.H.; Xia, F.S. Codon usage bias analysis of the chloroplast genome of Bothriochloa ischaemum. Acta Prataculturae Sin. 2023, 32, 85–95. [Google Scholar]
- Wang, Y.J.; Xin, L.; Hu, Z.Q.; An, X.N. Current situation of production, consumption and trade of wheat in China. Chin. J. Agric. Resour. Reg. Plan. 2018, 39, 36–45. [Google Scholar]
- Wei, Y.M. Origin, Spread and Evolution of Wheat in China. J. Triticeae Crops 2019, 41, 305–309. [Google Scholar]
- Zhang, X.Y. Effects of Planting Density on Physiology, Stem Characteristics, Yield and Quality of Overwintering Rye. Master’s Thesis, Northeast Agricultural University, Harbin, China, 2019. [Google Scholar]
- Mergoum, M.; Singh, P.K.; Peña, R.J.; Lozano-del, A.J.; Cooper, K.V.; Salmon, D.F.; Macpherson, H.G. Triticale: A “new” crop with old challenges. In Handbook of Plant Breeding; Marisa, L.B., David, H.B., Eds.; Biomedical and Life Sciences; Springer: New York, NY, USA, 2009; pp. 267–287. [Google Scholar]
- Zhu, F. Triticale: Nutritional composition and food uses. Food Chem. 2018, 241, 468–479. [Google Scholar] [CrossRef]
- McGoverin, C.M.; Snyders, F.; Muller, N.; Botes, W.; Fox, G.; Manley, M. A review of triticale uses and the effect of growth environment on grain quality. J. Sci. Food Agric. 2011, 91, 1155–1165. [Google Scholar] [CrossRef]
- Wang, W.Q.; Liu, J.; Tian, X.H.; Du, W.H. Evaluations on the Adaptability of Triticosecale Wittmack ‘Gannong No.4’ in Different Regions of Qinghai Province. Acta Agrestia Sin. 2020, 28, 1626–1634. [Google Scholar]
- Hammouda, D.; Baaziz, N.; Khalfallah, N. Genetic characterization of octoploid (AABBDDRR) and hexaploid (AABBRR) triticales. Eur. Sci. J. 2015, 11, 284–296. [Google Scholar]
- Wang, B.L.; Chen, Q.F.; Men, W.Q.; Tian, X.B.; Miao, J.N.; He, J.Q.; Zhao, Y.; Li, H.H.; Liu, W.X. Analysis of chromosome composition of hexaploid triticale of different plant heights. J. Henan Agric. Univ. 2021, 55, 631–638. [Google Scholar]
- Yang, T.; Chen, X.Z.; Sui, J.S.; Wang, W.; Yang, C.M.; Peng, Z.; Geng, G.D.; Zhang, Q.Q.; Zhang, S.Q. Chromosomic Analysis of Hexaploid Triticaleby Fluorescence in situ Hybridization. J. Southwest Univ. (Nat. Sci. Ed.) 2019, 41, 42–48. [Google Scholar]
- Bin Pek, H.; Klement, M.; Ang, K.S.; Chung, B.K.-S.; Ow, D.S.-W.; Lee, D.-Y. Exploring codon context bias for synthetic gene design of a thermostable invertase in Escherichia coli. Enzym. Microb. Technol. 2015, 75–76, 57–63. [Google Scholar] [CrossRef]
- Butt, A.M.; Nasrullah, I.; Qamar, R.; Tong, Y. Evolution of codon usage in Zika virus genomes is host and vector specific. Emerg. Microbes Infect. 2016, 5, 1–14. [Google Scholar] [CrossRef]
- Zhao, Y.; Zheng, H.; Xu, A.; Yan, D.; Jiang, Z.; Qi, Q.; Sun, J. Analysis of codon usage bias of envelope glycoprotein genes in nuclear polyhedrosis virus (NPV) and its relation to evolution. BMC Genom. 2016, 17, 677. [Google Scholar] [CrossRef] [PubMed]
- Purabi, M.; Othman, B.; Yasmin, R.; Katharina, M.; Ramakrishnan, N.; Jennifer, A.H. Codon usage and codon pair patterns in non-grass monocot genomes. Ann. Bot. 2017, 120, 893–909. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Lee, W.; Hottes, A.K.; Shapiro, L.; McAdams, H.H. Codon usage between genomes is constrained by genome-wide mutational processes. Proc. Natl. Acad. Sci. USA 2004, 101, 3480–3485. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Yengkhom, S.; Uddin, A. Analysis of codon usage bias of chloroplast genes in Oryza species: Codon usage of chloroplast genes in Oryza species. Planta 2020, 252, 67. [Google Scholar] [CrossRef]
- Yuan, X.L.; Li, Y.Q.; Zhang, J.F.; Wang, Y. Analysis of codon usage bias in the chloroplast genome of Dalbergia odorifera. Guihaia 2021, 41, 622–630. [Google Scholar]
- Liu, Q.; Xue, Q. Comparative studies on codon usage pattern of chloroplasts and their host nuclear genes in four plant species. J. Genet. 2005, 84, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Morton, B.R.; Wright, S.I. Selective Constraints on Codon Usage of Nuclear Genes from Arabidopsis thaliana. Mol. Biol. Evol. 2006, 24, 122–129. [Google Scholar] [CrossRef]
- Campbell, W.H.; Gowri, G. Codon Usage in Higher Plants, Green Algae, and Cyanobacteria. Plant Physiol. 1990, 92, 1–11. [Google Scholar] [CrossRef]
- Bhattacharyya, D.; Uddin, A.; Das, S.; Chakraborty, S. Mutation pressure and natural selection on codon usage in chloroplast genes of two species in Pisum L. (Fabaceae: Faboideae). Mitochondrial DNA Part A 2019, 30, 664–673. [Google Scholar] [CrossRef]
- Wang, L.; Roossinck, M.J. Comparative analysis of expressed sequences reveals a conserved pattern of optimal codon usage in plants. Plant Mol. Biol. 2006, 61, 699–710. [Google Scholar] [CrossRef]
- Fages-Lartaud, M.; Hundvin, K.; Hohmann-Marriott, M.F. Mechanisms governing codon usage bias and the implications for protein expression in the chloroplast of Chlamydomonas reinhardtii. Plant J. 2022, 112, 919–945. [Google Scholar] [CrossRef] [PubMed]
- Sueoka, N. Translation-coupled violation of Parity Rule 2 in human genes is not the cause of heterogeneity of the DNA G+C content of third codon position. Gene 1999, 238, 53–58. [Google Scholar] [CrossRef]
- Kawabe, A.; Miyashita, N.T. Patterns of codon usage bias in three dicot and four monocot plant species. Genes Genet. Syst. 2003, 78, 343–352. [Google Scholar] [CrossRef]




| Species Name | A3 | T3 | G3 | C3 | GC3 | |
|---|---|---|---|---|---|---|
| × Triticosecale sp. | A | 0.734 ** | 0.071 | −0.144 | −0.225 | 0.008 |
| T | −0.033 | 0.728 ** | −0.072 | −0.133 | −0.067 | |
| G | 0.164 | −0.008 | 0.373 ** | −0.272 * | −0.311 * | |
| C | −0.102 | 0.016 | −0.333 ** | 0.374 ** | −0.088 | |
| GC | 0.098 | −0.278 * | 0.242 | −0.041 | 0.542 ** | |
| Triticum monococcum | A | 0.737 ** | 0.07 | −0.146 | −0.226 | 0.093 |
| T | −0.031 | 0.727 ** | −0.075 | −0.132 | −0.104 | |
| G | 0.164 | −0.005 | 0.373 ** | −0.272 * | −0.213 | |
| C | −0.104 | 0.009 | −0.335 ** | 0.381 ** | −0.121 | |
| GC | −0.06 | −0.166 | 0.26 | −0.132 | 0.606 ** | |
| Triticum turgidum | A | 0.738 ** | 0.068 | −0.138 | −0.224 | 0.2 |
| T | −0.028 | 0.729 ** | −0.052 | −0.14 | 0.058 | |
| G | 0.164 | −0.001 | 0.381 ** | −0.270 * | −0.177 | |
| C | −0.098 | 0.015 | −0.334 ** | 0.368 ** | −0.13 | |
| GC | 0.029 | −0.184 | 0.28 | −0.073 | 0.554 ** | |
| Triticum aestivum | A | 0.738 ** | 0.066 | −0.146 | −0.225 | 0.109 |
| T | −0.03 | 0.728 ** | −0.073 | −0.137 | −0.181 | |
| G | 0.163 | −0.005 | 0.377 ** | −0.271 * | −0.202 | |
| C | −0.103 | 0.012 | −0.334 ** | 0.378 ** | 0.173 | |
| GC | 0.044 | −0.214 | 0.226 | −0.006 | 0.629 ** | |
| Secale cereale | A | 0.737 ** | 0.071 | −0.149 | −0.226 | 0.094 |
| T | −0.031 | 0.729 ** | −0.073 | −0.13 | −0.081 | |
| G | 0.166 | −0.01 | 0.375 ** | −0.270 * | −0.131 | |
| C | −0.099 | 0.014 | −0.334 ** | 0.377 ** | −0.151 | |
| GC | 0.101 | −0.071 | 0.05 | −0.255 | 0.624 ** |
| Species Name | A | T | G | C | GC | A3 | T3 | G3 | C3 | GC3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| × Triticosecale sp. | SCUO | 0.187 | −0.16 | −0.484 * | 0.229 | −0.022 | −0.144 | 0.178 | −0.441 | 0.051 | −0.221 |
| P | 0.458 | 0.526 | 0.042 | 0.361 | 0.93 | 0.569 | 0.48 | 0.067 | 0.842 | 0.377 | |
| Triticum monococcum | SCUO | 0.208 | −0.175 | −0.494 * | 0.221 | −0.031 | −0.121 | 0.159 | −0.466 | 0.034 | −0.234 |
| P | 0.407 | 0.487 | 0.037 | 0.378 | 0.902 | 0.632 | 0.529 | 0.051 | 0.894 | 0.35 | |
| Triticum turgidum | SCUO | 0.2 | −0.157 | −0.497 * | 0.225 | −0.042 | −0.127 | 0.158 | −0.447 | 0.038 | −0.2 |
| P | 0.426 | 0.534 | 0.036 | 0.368 | 0.87 | 0.614 | 0.532 | 0.063 | 0.88 | 0.426 | |
| Triticum aestivum | SCUO | 0.292 | −0.136 | −0.526 * | 0.07 | −0.16 | −0.136 | 0.187 | −0.543 * | 0.002 | −0.333 |
| P | 0.24 | 0.591 | 0.025 | 0.781 | 0.526 | 0.592 | 0.457 | 0.02 | 0.994 | 0.176 | |
| Secale cereale | SCUO | 0.219 | −0.18 | −0.493 * | 0.244 | −0.037 | −0.125 | 0.158 | −0.457 | 0.042 | −0.211 |
| P | 0.383 | 0.474 | 0.038 | 0.329 | 0.884 | 0.621 | 0.532 | 0.057 | 0.868 | 0.4 |
| × Triticosecale sp. | Triticum monococcum | Triticum turgidum | Triticum aestivum | Secale cereale | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | Codon | ΔRSCU | AA | Codon | ΔRSCU | AA | Codon | ΔRSCU | AA | Codon | ΔRSCU | AA | Codon | ΔRSCU |
| Ala | GCU *** | 0.81 | Ala | GCU *** | 0.79 | Ala | GCA * | 0.13 | Ala | GCU *** | 0.6 | Ala | GCA * | 0.23 |
| Arg | AGA ** | 0.36 | Arg | CGU *** | 1.16 | Ala | GCU *** | 0.66 | Arg | AGA * | 0.21 | Ala | GCU *** | 0.57 |
| Arg | CGU *** | 1.43 | Asp | GAU * | 0.24 | Arg | AGA ** | 0.33 | Arg | CGU *** | 1.1 | Arg | AGA * | 0.23 |
| Gln | CAA * | 0.08 | Cys | UGU *** | 1.04 | Arg | CGU *** | 0.9 | Asp | GAU * | 0.16 | Arg | CGU *** | 1.14 |
| Gly | GGU *** | 0.89 | Gln | CAA * | 0.12 | Cys | UGU *** | 0.5 | Cys | UGU *** | 0.6 | Asp | GAU * | 0.19 |
| His | CAU * | 0.18 | Gly | GGU *** | 0.79 | Gly | GGU ** | 0.36 | Gln | CAA * | 0.27 | Cys | UGU ** | 0.42 |
| Ile | AUU * | 0.27 | Ile | AUU ** | 0.42 | Leu | CUA *** | 0.67 | Gly | GGU *** | 1 | Gln | CAA * | 0.1 |
| Leu | UUA *** | 1.56 | Leu | CUU * | 0.1 | Leu | UUA *** | 0.6 | His | CAU * | 0.11 | Gly | GGU *** | 1.06 |
| Lys | AAA *** | 0.6 | Leu | UUA *** | 1.07 | Lys | AAA * | 0.09 | Ile | AUU ** | 0.44 | His | CAU * | 0.15 |
| Phe | UUU * | 0.14 | Lys | AAA * | 0.26 | Pro | CCU *** | 1.09 | Leu | CUA * | 0.18 | Ile | AUU ** | 0.31 |
| Pro | CCU *** | 1.22 | Pro | CCA * | 0.18 | Ser | UCU *** | 1.19 | Leu | UUA *** | 1.16 | Leu | CUU * | 0.21 |
| Ser | AGU * | 0.24 | Pro | CCU *** | 0.97 | Thr | ACU *** | 0.57 | Pro | CCU *** | 0.84 | Leu | UUA *** | 0.97 |
| Ser | UCU *** | 1.25 | Ser | AGU * | 0.14 | Val | GUA * | 0.24 | Ser | AGU ** | 0.43 | Pro | CCU *** | 0.84 |
| Thr | ACA * | 0.12 | Ser | UCU *** | 0.96 | Ser | UCU *** | 0.78 | Ser | AGU * | 0.18 | |||
| Thr | ACU *** | 0.78 | Thr | ACA * | 0.24 | Thr | ACA * | 0.29 | Ser | UCU *** | 0.78 | |||
| Val | GUU ** | 0.39 | Thr | ACU *** | 0.55 | Thr | ACU ** | 0.3 | Thr | ACU * | 0.1 | |||
| Val | GUA * | 0.26 | Tyr | UAU * | 0.08 | Tyr | UAU * | 0.2 | ||||||
| Val | GUA * | 0.17 | Val | GUA *** | 0.65 | |||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, T.; Zhang, Y.; Du, W.; Wang, Z. Comparative Genomics of Triticum, Secale, and Triticale: Codon Usage Bias in Chloroplast Genomes and Its Implications for Evolution and Genetic Engineering. Int. J. Mol. Sci. 2025, 26, 10266. https://doi.org/10.3390/ijms262110266
Tian T, Zhang Y, Du W, Wang Z. Comparative Genomics of Triticum, Secale, and Triticale: Codon Usage Bias in Chloroplast Genomes and Its Implications for Evolution and Genetic Engineering. International Journal of Molecular Sciences. 2025; 26(21):10266. https://doi.org/10.3390/ijms262110266
Chicago/Turabian StyleTian, Tian, Yinxia Zhang, Wenhua Du, and Zhijun Wang. 2025. "Comparative Genomics of Triticum, Secale, and Triticale: Codon Usage Bias in Chloroplast Genomes and Its Implications for Evolution and Genetic Engineering" International Journal of Molecular Sciences 26, no. 21: 10266. https://doi.org/10.3390/ijms262110266
APA StyleTian, T., Zhang, Y., Du, W., & Wang, Z. (2025). Comparative Genomics of Triticum, Secale, and Triticale: Codon Usage Bias in Chloroplast Genomes and Its Implications for Evolution and Genetic Engineering. International Journal of Molecular Sciences, 26(21), 10266. https://doi.org/10.3390/ijms262110266





