Regulatory Mechanisms and Functional Roles of Readthrough Transcripts in Tumorigenesis
Abstract
1. Introduction
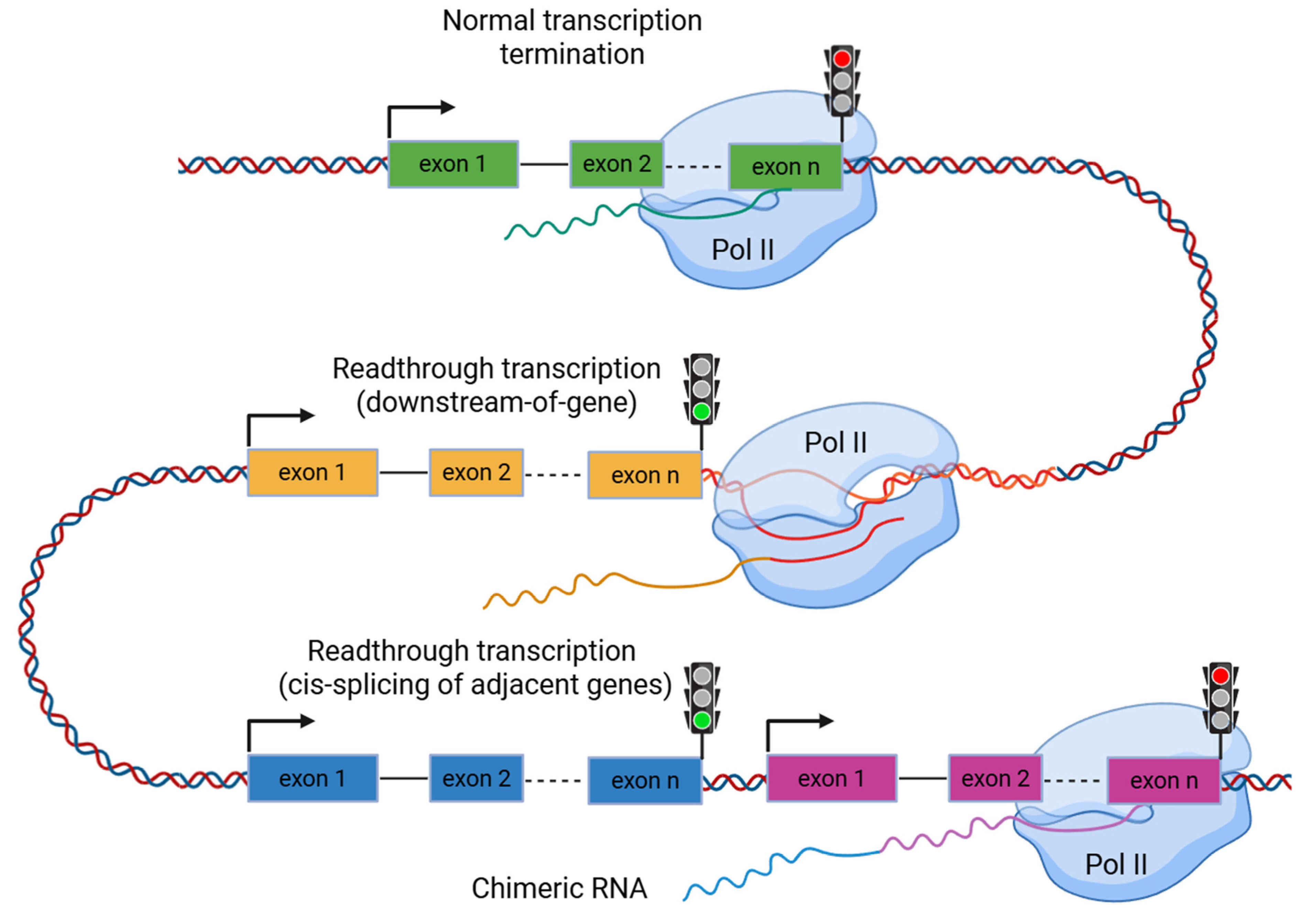
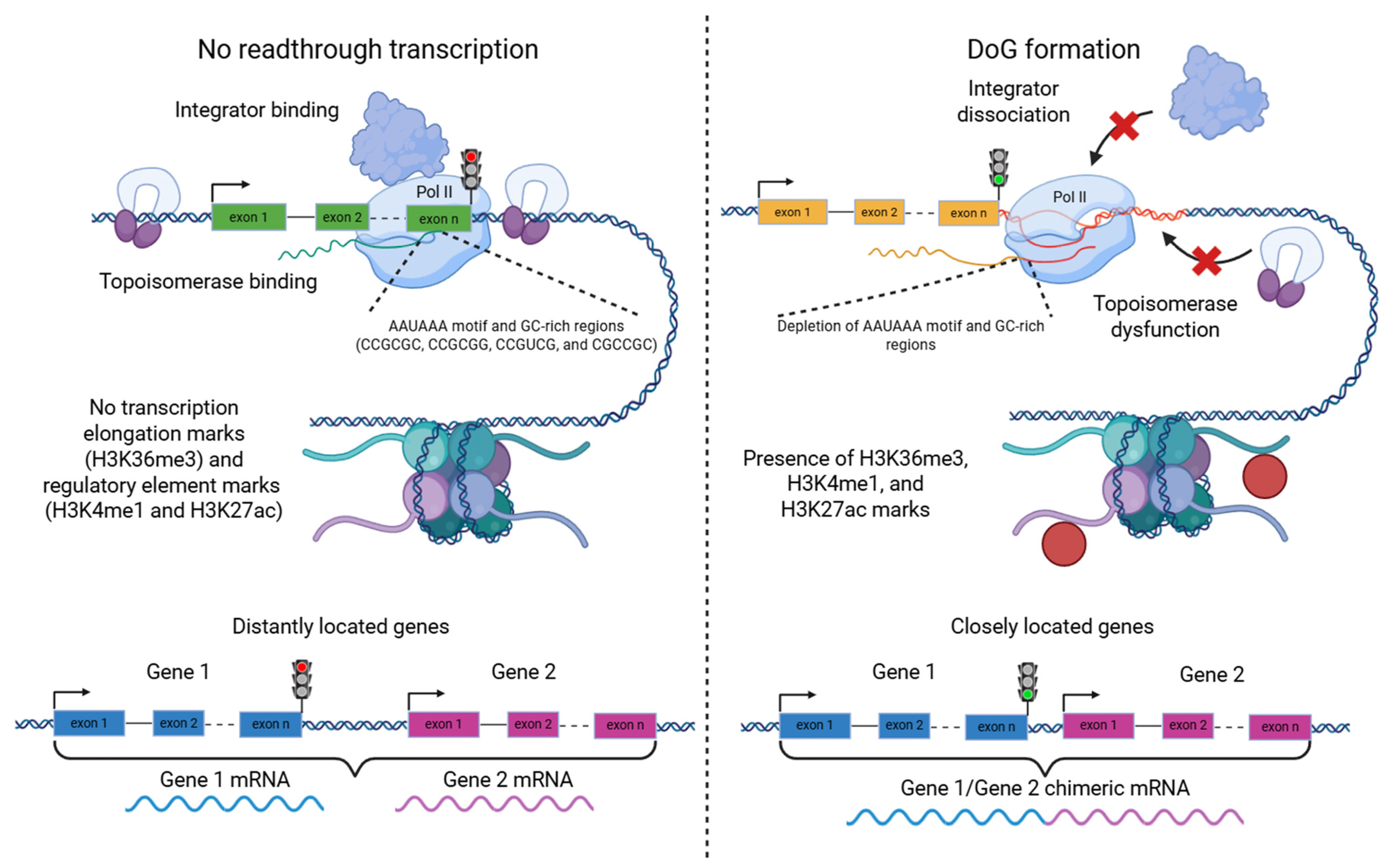
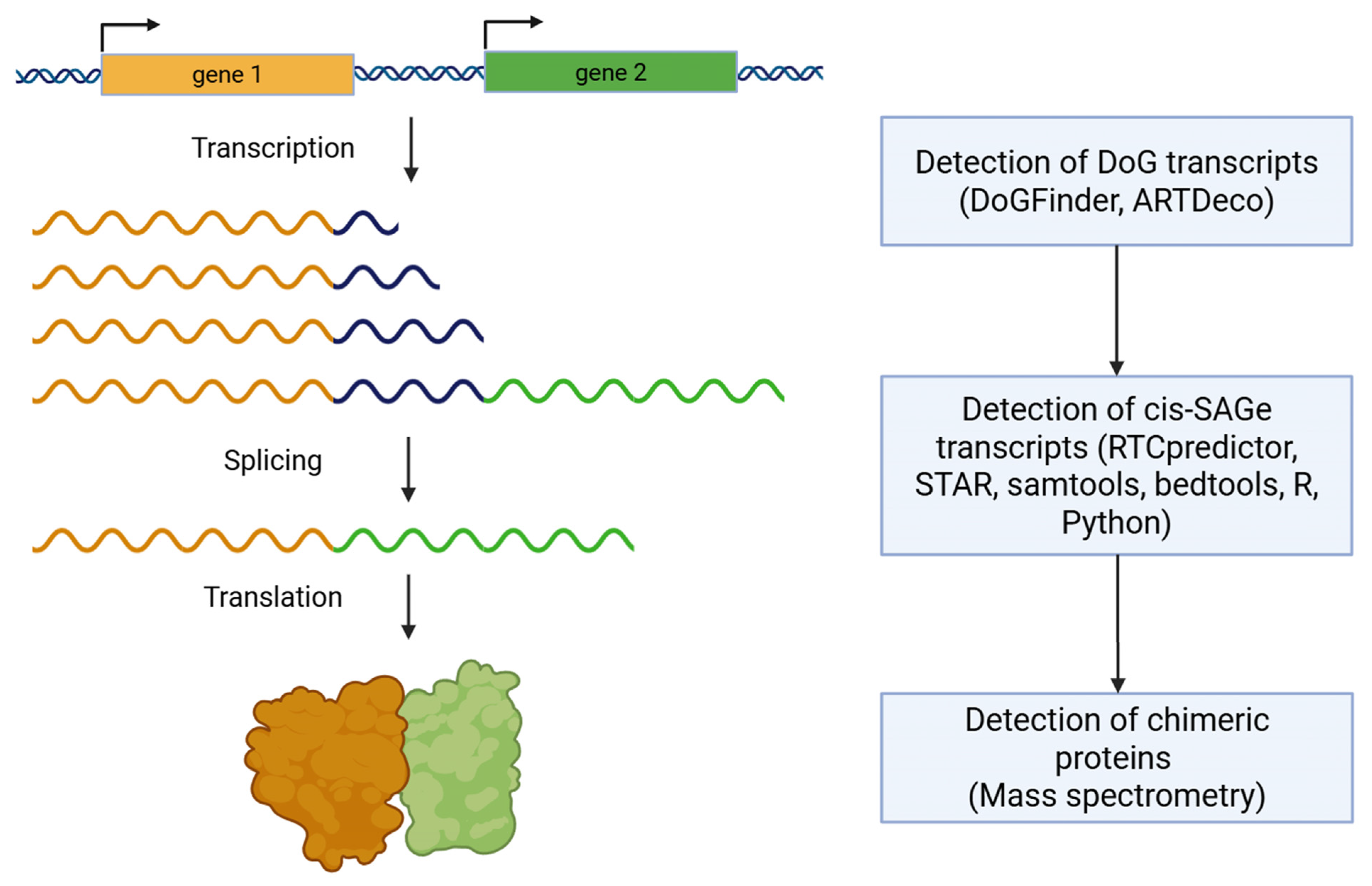
2. Biological Mechanism of Readthrough Transcription
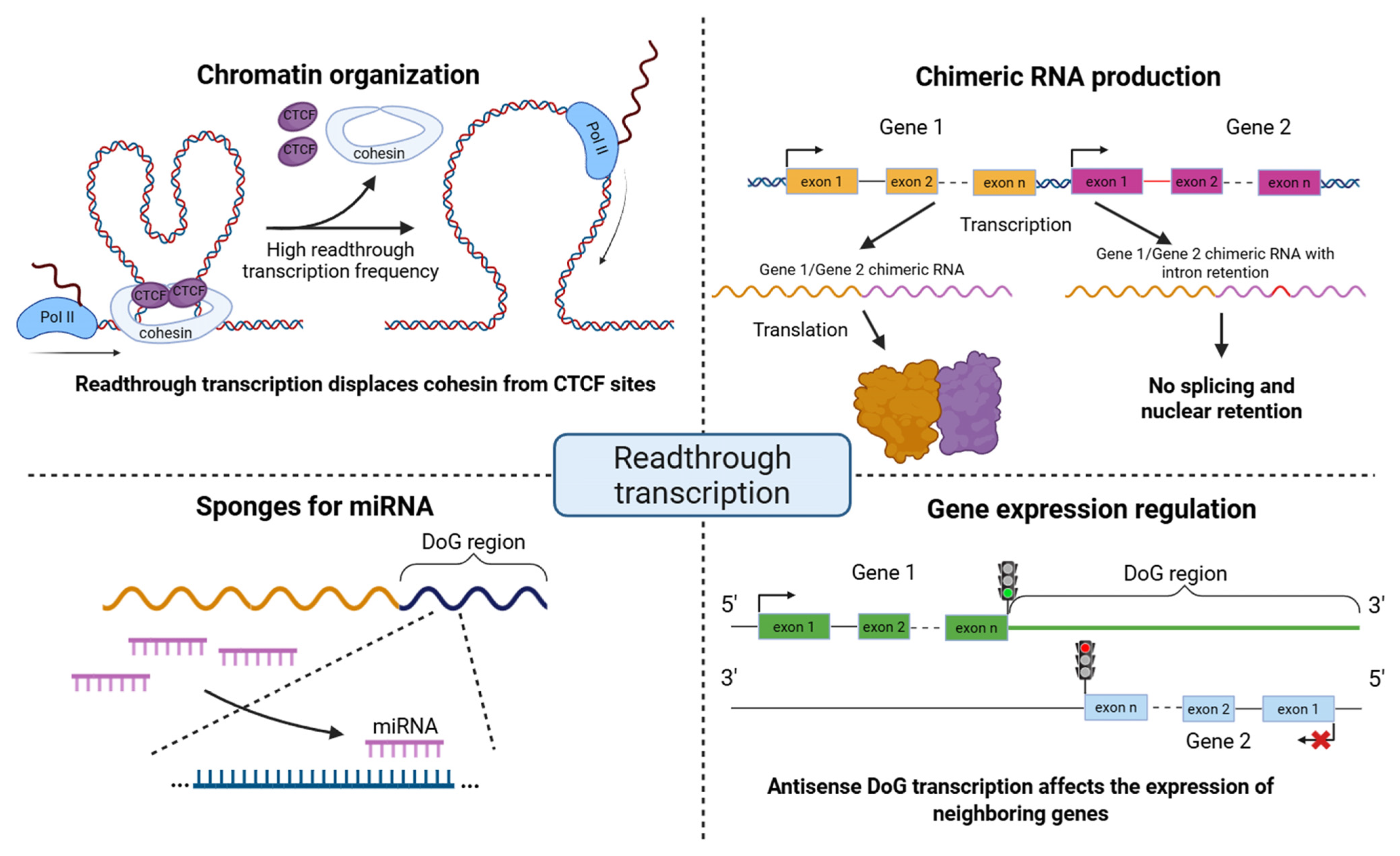
3. Functional Roles of Readthrough Transcription
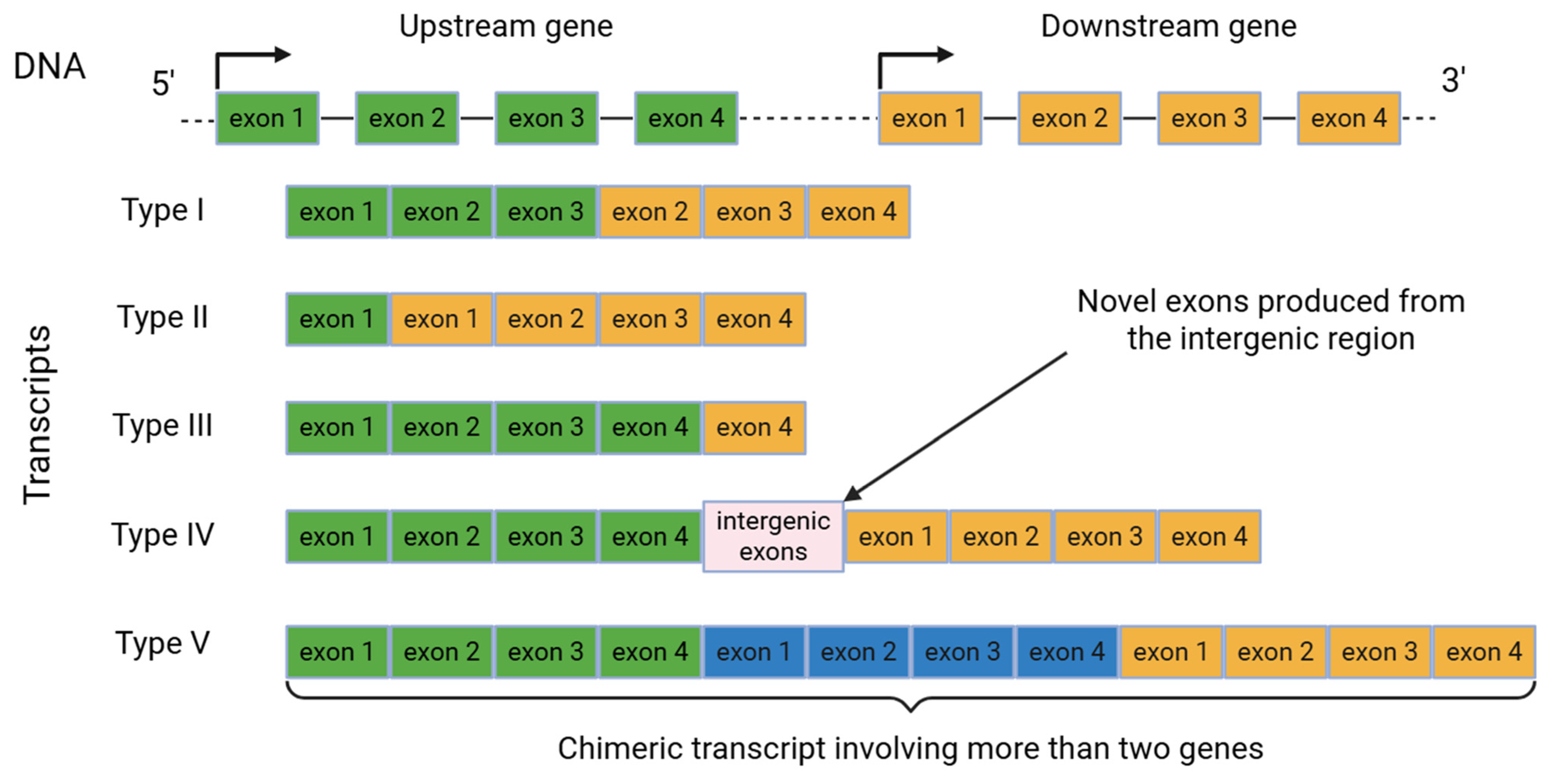
4. Readthrough Transcription Events in Cancer Progression
| cis-SAGes (Upstream Gene–Downstream Gene) | Tumor Type | Reference |
|---|---|---|
| SLC45A3-ELK4 | prostate cancer | [18,24,37,48,49] |
| BCL2L2-PABPN1 | bladder cancer, glioblastoma | [42,47] |
| RRM2-C2orf48 | colorectal cancer | [44] |
| METTL21B-TSFM | colorectal cancer | [44] |
| SF3A2-AMH | colorectal cancer | [44] |
| TMEM189-UBE2V1 | colorectal cancer | [45] |
| POLA2-CDC42EP2 | gastrointestinal stromal tumors | [50] |
| C8orf42-FBXO25 | gastrointestinal stromal tumors | [50] |
| STX16-NPEPL1 | gastrointestinal stromal tumors | [50] |
| RPL17-C18orf32 | gastric cancer | [51] |
| PRR5-ARHGAP8 | gastric cancer | [51] |
| PHOSPHO2-KLHL23 | gastric cancer | [51] |
| MSMB-NCOA4 | prostate cancer | [24] |
| D2HGDH-GAL3ST2 | prostate cancer | [38] |
| SCNN1A-TNFRSF1A | breast cancer | [40] |
| CTSD-IFITM10 | breast cancer | [40] |
| LHX6-NDUFA8 | cervical cancer | [41] |
| SLC2A11-MIF | cervical cancer | [41] |
| TWE-PRIL | T- and B-lymphoma | [52] |
| HNRNPA1L2-SUGT1 | bladder cancer | [43] |
| CHFR-GOLGA3 | bladder cancer | [42] |
| CTSC-RAB38 | kidney cancer | [7] |
| KDSR-BCL2 | kidney cancer | [7] |
| KLK4-KRSP1 | kidney cancer | [46] |
| TSNAX-DISC1 | endometrial cancer | [53] |
| INS-IGF2 | non-small-cell lung cancer | [54] |
| DUS4L-BCAP29 | prostate cancer, gastric cancer | [39] |
| JMJD7-PLA2G4B | human head and neck squamous cell carcinoma | [55] |
| NFATC3-PLA2G15 | T-acute lymphoblastic leukemia | [56] |
5. DoG and Cis-SAGe Detection
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chwalenia, K.; Qin, F.; Singh, S.; Tangtrongstittikul, P.; Li, H. Connections between Transcription Downstream of Genes and Cis-SAGe Chimeric RNA. Genes 2017, 8, 338. [Google Scholar] [CrossRef] [PubMed]
- Vilborg, A.; Sabath, N.; Wiesel, Y.; Nathans, J.; Levy-Adam, F.; Yario, T.A.; Steitz, J.A.; Shalgi, R. Comparative Analysis Reveals Genomic Features of Stress-Induced Transcriptional Readthrough. Proc. Natl. Acad. Sci. USA 2017, 114, E8362–E8371. [Google Scholar] [CrossRef]
- Babiceanu, M.; Qin, F.; Xie, Z.; Jia, Y.; Lopez, K.; Janus, N.; Facemire, L.; Kumar, S.; Pang, Y.; Qi, Y.; et al. Recurrent Chimeric Fusion RNAs in Non-Cancer Tissues and Cells. Nucleic Acids Res. 2016, 44, 2859–2872. [Google Scholar] [CrossRef]
- Caldas, P.; Luz, M.; Baseggio, S.; Andrade, R.; Sobral, D.; Grosso, A.R. Transcription Readthrough Is Prevalent in Healthy Human Tissues and Associated with Inherent Genomic Features. Commun. Biol. 2024, 7, 100. [Google Scholar] [CrossRef] [PubMed]
- Bartz, J.; Jung, H.; Wasiluk, K.; Zhang, L.; Dong, X. Progress in Discovering Transcriptional Noise in Aging. Int. J. Mol. Sci. 2023, 24, 3701. [Google Scholar] [CrossRef] [PubMed]
- Abe, K.; Maunze, B.; Lopez, P.-A.; Xu, J.; Muhammad, N.; Yang, G.-Y.; Katz, D.; Liu, Y.; Lauberth, S.M. Downstream-of-Gene (DoG) Transcripts Contribute to an Imbalance in the Cancer Cell Transcriptome. Sci. Adv. 2024, 10, eadh9613. [Google Scholar] [CrossRef]
- Grosso, A.R.; Leite, A.P.; Carvalho, S.; Matos, M.R.; Martins, F.B.; Vítor, A.C.; Desterro, J.M.P.; Carmo-Fonseca, M.; de Almeida, S.F. Pervasive Transcription Read-through Promotes Aberrant Expression of Oncogenes and RNA Chimeras in Renal Carcinoma. eLife 2015, 4, e09214. [Google Scholar] [CrossRef]
- Sorokin, M.; Lyadov, V.; Suntsova, M.; Garipov, M.; Semenova, A.; Popova, N.; Guguchkin, E.; Heydarov, R.; Zolotovskaia, M.; Zhao, X.; et al. Detection of Fusion Events by RNA Sequencing in FFPE versus Freshly Frozen Colorectal Cancer Tissue Samples. Front. Mol. Biosci. 2025, 11, 1448792. [Google Scholar] [CrossRef]
- Buzdin, A.A.; Heydarov, R.N.; Golounina, O.O.; Suntsova, M.V.; Matrosova, A.V.; Bondarenko, E.V.; Roumiantsev, S.A.; Sorokin, M.I.; Kholodenko, R.V.; Kholodenko, I.V.; et al. Transcriptome-Wide Analysis of Pituitary and Ectopic Adrenocorticotropic Hormone-Secreting Tumors. Cancers 2025, 17, 658. [Google Scholar] [CrossRef]
- Rosa-Mercado, N.A.; Zimmer, J.T.; Apostolidi, M.; Rinehart, J.; Simon, M.D.; Steitz, J.A. Hyperosmotic Stress Alters the RNA Polymerase II Interactome and Induces Readthrough Transcription despite Widespread Transcriptional Repression. Mol. Cell 2021, 81, 502–513.e4. [Google Scholar] [CrossRef]
- Dasilva, L.F.; Blumenthal, E.; Beckedorff, F.; Cingaram, P.R.; Gomes Dos Santos, H.; Edupuganti, R.R.; Zhang, A.; Dokaneheifard, S.; Aoi, Y.; Yue, J.; et al. Integrator Enforces the Fidelity of Transcriptional Termination at Protein-Coding Genes. Sci. Adv. 2021, 7, eabe3393. [Google Scholar] [CrossRef] [PubMed]
- Vilborg, A.; Passarelli, M.C.; Yario, T.A.; Tycowski, K.T.; Steitz, J.A. Widespread Inducible Transcription Downstream of Human Genes. Mol. Cell 2015, 59, 449–461. [Google Scholar] [CrossRef]
- Fianu, I.; Ochmann, M.; Walshe, J.L.; Dybkov, O.; Cruz, J.N.; Urlaub, H.; Cramer, P. Structural Basis of Integrator-Dependent RNA Polymerase II Termination. Nature 2024, 629, 219–227. [Google Scholar] [CrossRef]
- Liu, H.; Heller-Trulli, D.; Moore, C.L. Targeting the mRNA Endonuclease CPSF73 Inhibits Breast Cancer Cell Migration, Invasion, and Self-Renewal. iScience 2022, 25, 104804. [Google Scholar] [CrossRef]
- Chwalenia, K.; Qin, F.; Singh, S.; Li, H. A Cell-Based Splicing Reporter System to Identify Regulators of Cis-Splicing between Adjacent Genes. Nucleic Acids Res. 2019, 47, e24. [Google Scholar] [CrossRef]
- Yao, J.; Ding, D.; Li, X.; Shen, T.; Fu, H.; Zhong, H.; Wei, G.; Ni, T. Prevalent Intron Retention Fine-Tunes Gene Expression and Contributes to Cellular Senescence. Aging Cell 2020, 19, e13276. [Google Scholar] [CrossRef]
- Qin, F.; Song, Z.; Babiceanu, M.; Song, Y.; Facemire, L.; Singh, R.; Adli, M.; Li, H. Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate Cells. PLoS Genet. 2015, 11, e1005001. [Google Scholar] [CrossRef]
- Zhang, Y.; Gong, M.; Yuan, H.; Park, H.G.; Frierson, H.F.; Li, H. Chimeric Transcript Generated by Cis-Splicing of Adjacent Genes Regulates Prostate Cancer Cell Proliferation. Cancer Discov. 2012, 2, 598–607. [Google Scholar] [CrossRef] [PubMed]
- Hennig, T.; Michalski, M.; Rutkowski, A.J.; Djakovic, L.; Whisnant, A.W.; Friedl, M.-S.; Jha, B.A.; Baptista, M.A.P.; L’Hernault, A.; Erhard, F.; et al. HSV-1-Induced Disruption of Transcription Termination Resembles a Cellular Stress Response but Selectively Increases Chromatin Accessibility Downstream of Genes. PLoS Pathog. 2018, 14, e1006954. [Google Scholar] [CrossRef]
- Heinz, S.; Texari, L.; Hayes, M.G.B.; Urbanowski, M.; Chang, M.W.; Givarkes, N.; Rialdi, A.; White, K.M.; Albrecht, R.A.; Pache, L.; et al. Transcription Elongation Can Affect Genome 3D Structure. Cell 2018, 174, 1522–1536.e22. [Google Scholar] [CrossRef]
- Alkan, A.H.; Akgül, B. Endogenous miRNA Sponges. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2022; Volume 2257, pp. 91–104. [Google Scholar] [CrossRef]
- Hadar, S.; Meller, A.; Saida, N.; Shalgi, R. Stress-Induced Transcriptional Readthrough into Neighboring Genes Is Linked to Intron Retention. iScience 2022, 25, 105543. [Google Scholar] [CrossRef]
- Jia, Y.; Xie, Z.; Li, H. Intergenically Spliced Chimeric RNAs in Cancer. Trends Cancer 2016, 2, 475–484. [Google Scholar] [CrossRef]
- Nacu, S.; Yuan, W.; Kan, Z.; Bhatt, D.; Rivers, C.S.; Stinson, J.; Peters, B.A.; Modrusan, Z.; Jung, K.; Seshagiri, S.; et al. Deep RNA Sequencing Analysis of Readthrough Gene Fusions in Human Prostate Adenocarcinoma and Reference Samples. BMC Med. Genom. 2011, 4, 11. [Google Scholar] [CrossRef]
- Lu, G.; Wu, J.; Zhao, G.; Wang, Z.; Chen, W.; Mu, S. Abundant and Broad Expression of Transcription-Induced Chimeras and Protein Products in Mammalian Genomes. Biochem. Biophys. Res. Commun. 2016, 470, 759–765. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Han, Y.; Zellmer, L.; Yang, W.; Guan, Z.; Yu, W.; Huang, H.; Liao, D.J. It Is Imperative to Establish a Pellucid Definition of Chimeric RNA and to Clear Up a Lot of Confusion in the Relevant Research. Int. J. Mol. Sci. 2017, 18, 714. [Google Scholar] [CrossRef]
- Vidal, A.F. Read-through Circular RNAs Reveal the Plasticity of RNA Processing Mechanisms in Human Cells. RNA Biol. 2020, 17, 1823–1826. [Google Scholar] [CrossRef] [PubMed]
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.-M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881.e13. [Google Scholar] [CrossRef]
- Chen, Z.; Huang, L.; Wang, K.; Zhang, L.; Zhong, X.; Yan, Z.; Liu, B.; Zhu, P. rtcisE2F Promotes the Self-Renewal and Metastasis of Liver Tumor-Initiating Cells via N6-Methyladenosine-Dependent E2F3/E2F6 mRNA Stability. Sci. China Life Sci. 2022, 65, 1840–1854. [Google Scholar] [CrossRef]
- Morgan, M.; Shiekhattar, R.; Shilatifard, A.; Lauberth, S.M. It’s a DoG-Eat-DoG World-Altered Transcriptional Mechanisms Drive Downstream-of-Gene (DoG) Transcript Production. Mol. Cell 2022, 82, 1981–1991. [Google Scholar] [CrossRef] [PubMed]
- Alpert, T.; Straube, K.; Oesterreich, F.C.; Herzel, L.; Neugebauer, K.M. Widespread Transcriptional Readthrough Caused by Nab2 Depletion Leads to Chimeric Transcripts with Retained Introns. Cell Rep. 2020, 33, 108496. [Google Scholar] [CrossRef]
- Herzel, L.; Straube, K.; Neugebauer, K.M. Long-Read Sequencing of Nascent RNA Reveals Coupling among RNA Processing Events. Genome Res. 2018, 28, 1008–1019. [Google Scholar] [CrossRef]
- Rutkowski, A.J.; Erhard, F.; L’Hernault, A.; Bonfert, T.; Schilhabel, M.; Crump, C.; Rosenstiel, P.; Efstathiou, S.; Zimmer, R.; Friedel, C.C.; et al. Widespread Disruption of Host Transcription Termination in HSV-1 Infection. Nat. Commun. 2015, 6, 7126. [Google Scholar] [CrossRef]
- Mei, Y.; Cheng, Z.; Lu, Y.; Wu, S.; Chen, X. Comprehensive Resource for Transcription Readthrough Events in Healthy Human Tissues. Sci. Data 2025, 12, 1176. [Google Scholar] [CrossRef]
- Stoeger, T.; Grant, R.A.; McQuattie-Pimentel, A.C.; Anekalla, K.R.; Liu, S.S.; Tejedor-Navarro, H.; Singer, B.D.; Abdala-Valencia, H.; Schwake, M.; Tetreault, M.-P.; et al. Aging Is Associated with a Systemic Length-Associated Transcriptome Imbalance. Nat. Aging 2022, 2, 1191–1206. [Google Scholar] [CrossRef]
- He, H.; Wang, Y.; Ye, P.; Yi, D.; Cheng, Y.; Tang, H.; Zhu, Z.; Wang, X.; Jin, S. Long Noncoding RNA ZFPM2-AS1 Acts as a miRNA Sponge and Promotes Cell Invasion through Regulation of miR-139/GDF10 in Hepatocellular Carcinoma. J. Exp. Clin. Cancer Res. 2020, 39, 159. [Google Scholar] [CrossRef]
- Rickman, D.S.; Pflueger, D.; Moss, B.; VanDoren, V.E.; Chen, C.X.; de la Taille, A.; Kuefer, R.; Tewari, A.K.; Setlur, S.R.; Demichelis, F.; et al. SLC45A3-ELK4 Is a Novel and Frequent Erythroblast Transformation-Specific Fusion Transcript in Prostate Cancer. Cancer Res. 2009, 69, 2734–2738. [Google Scholar] [CrossRef]
- Qin, F.; Song, Z.; Chang, M.; Song, Y.; Frierson, H.; Li, H. Recurrent Cis-SAGe Chimeric RNA, D2HGDH-GAL3ST2, in Prostate Cancer. Cancer Lett. 2016, 380, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Qin, F.; Liu, A.; Li, H. Recurrent Fusion RNA DUS4L-BCAP29 in Non-Cancer Human Tissues and Cells. Oncotarget 2017, 8, 31415–31423. [Google Scholar] [CrossRef] [PubMed]
- Varley, K.E.; Gertz, J.; Roberts, B.S.; Davis, N.S.; Bowling, K.M.; Kirby, M.K.; Nesmith, A.S.; Oliver, P.G.; Grizzle, W.E.; Forero, A.; et al. Recurrent Read-through Fusion Transcripts in Breast Cancer. Breast Cancer Res. Treat. 2014, 146, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Yang, S.; Singh, S.; Qin, F.; Kumar, S.; Wang, L.; Ma, D.; Li, H. The Landscape and Implications of Chimeric RNAs in Cervical Cancer. eBioMedicine 2018, 37, 158–167. [Google Scholar] [CrossRef]
- Zhu, D.; Singh, S.; Chen, X.; Zheng, Z.; Huang, J.; Lin, T.; Li, H. The Landscape of Chimeric RNAs in Bladder Urothelial Carcinoma. Int. J. Biochem. Cell Biol. 2019, 110, 50–58. [Google Scholar] [CrossRef]
- Wu, H.; Singh, S.; Shi, X.; Xie, Z.; Lin, E.; Li, X.; Li, H. Functional Heritage: The Evolution of Chimeric RNA into a Gene. RNA Biol. 2019, 17, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Singh, S.; Xie, Z.; Li, X.; Li, H. Landscape Characterization of Chimeric RNAs in Colorectal Cancer. Cancer Lett. 2020, 489, 56–65. [Google Scholar] [CrossRef]
- Thomson, T.M.; Lozano, J.J.; Loukili, N.; Carrió, R.; Serras, F.; Cormand, B.; Valeri, M.; Díaz, V.M.; Abril, J.; Burset, M.; et al. Fusion of the Human Gene for the Polyubiquitination Coeffector UEV1 with Kua, a Newly Identified Gene. Genome Res. 2000, 10, 1743–1756. [Google Scholar] [CrossRef]
- Pflueger, D.; Mittmann, C.; Dehler, S.; Rubin, M.A.; Moch, H.; Schraml, P. Functional Characterization of BC039389-GATM and KLK4-KRSP1 Chimeric Read-through Transcripts Which Are up-Regulated in Renal Cell Cancer. BMC Genom. 2015, 16, 247. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, D.; Han, X.; Guo, X.; Cao, Y.; Xia, Y.; Gao, D. Novel Read-through Fusion Transcript Bcl2l2-Pabpn1 in Glioblastoma Cells. J. Cell. Mol. Med. 2022, 26, 4686–4697. [Google Scholar] [CrossRef]
- Kumar-Sinha, C.; Kalyana-Sundaram, S.; Chinnaiyan, A.M. SLC45A3-ELK4 Chimera in Prostate Cancer: Spotlight on Cis-Splicing. Cancer Discov. 2012, 2, 582–585. [Google Scholar] [CrossRef]
- Qin, F.; Zhang, Y.; Liu, J.; Li, H. SLC45A3-ELK4 Functions as a Long Non-Coding Chimeric RNA. Cancer Lett. 2017, 404, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.; Yun, H.; Sun, C.-H.; Park, I.; Lee, S.; Kwon, J.; Do, I.; Hong, M.E.; Van Vrancken, M.; Lee, J.; et al. Integrated Genomic Analyses Identify Frequent Gene Fusion Events and VHL Inactivation in Gastrointestinal Stromal Tumors. Oncotarget 2016, 7, 6538–6551. [Google Scholar] [CrossRef]
- Choi, E.-S.; Lee, H.; Lee, C.-H.; Goh, S.-H. Overexpression of KLHL23 Protein from Read-through Transcription of PHOSPHO2-KLHL23 in Gastric Cancer Increases Cell Proliferation. FEBS Open Bio 2016, 6, 1155–1164. [Google Scholar] [CrossRef] [PubMed]
- Kolfschoten, G.M.; Pradet-Balade, B.; Hahne, M.; Medema, J.P. TWE-PRIL; a Fusion Protein of TWEAK and APRIL. Biochem. Pharmacol. 2003, 66, 1427–1432. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zheng, J.; Li, H.; Deng, J.; Hu, M.; Wu, H.; Li, W.; Li, F.; Lan, X.; Lu, J.; et al. Identification of Chimeric TSNAX-DISC1 Resulting from Intergenic Splicing in Endometrial Carcinoma through High-Throughput RNA Sequencing. Carcinogenesis 2014, 35, 2687–2697. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Lin, Z.; Li, C.; Wang, Y.; Yang, L.; Zou, B.; Chen, J.; Li, J.; Feng, D.; Song, Z.; et al. lncINS-IGF2 Promotes Cell Proliferation and Migration by Promoting G1/S Transition in Lung Cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033818823029. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, Y.; Li, J.; Chang, I.; Wang, C.-Y. A Novel Read-through Transcript JMJD7-PLA2G4B Regulates Head and Neck Squamous Cell Carcinoma Cell Proliferation and Survival. Oncotarget 2017, 8, 1972–1982. [Google Scholar] [CrossRef]
- Bond, J.; Tran Quang, C.; Hypolite, G.; Belhocine, M.; Bergon, A.; Cordonnier, G.; Ghysdael, J.; Macintyre, E.; Boissel, N.; Spicuglia, S.; et al. Novel Intergenically Spliced Chimera, NFATC3-PLA2G15, Is Associated with Aggressive T-ALL Biology and Outcome. Mol. Cancer Res. MCR 2018, 16, 470–475. [Google Scholar] [CrossRef]
- Chai, Y.; Liu, J.-L.; Zhang, S.; Li, N.; Xu, D.-Q.; Liu, W.-J.; Fu, R.-J.; Tang, Y.-P. The Effective Combination Therapies with Irinotecan for Colorectal Cancer. Front. Pharmacol. 2024, 15, 1356708. [Google Scholar] [CrossRef]
- Gupta, A.; De Jesus-Acosta, A.; Zheng, L.; Lee, V.; Kamel, I.; Le, D.; Pishvaian, M.; Laheru, D. Clinical Outcomes of Liposomal Irinotecan in Advanced Pancreatic Adenocarcinoma Patients Previously Treated with Conventional Irinotecan-Based Chemotherapy: A Real-World Study. Front. Oncol. 2023, 13, 1250136. [Google Scholar] [CrossRef] [PubMed]
- Tate, S.; Nishikimi, K.; Matsuoka, A.; Otsuka, S.; Kato, K.; Takahashi, Y.; Shozu, M. Tailored-Dose Chemotherapy with Gemcitabine and Irinotecan in Patients with Platinum-Refractory/Resistant Ovarian or Primary Peritoneal Cancer: A Phase II Trial. J. Gynecol. Oncol. 2021, 32, e8. [Google Scholar] [CrossRef]
- Seto, Z.; Takata, N.; Murayama, N.; Tokui, K.; Okazawa, S.; Kambara, K.; Imanishi, S.; Miwa, T.; Hayashi, R.; Matsui, S.; et al. Irinotecan Monotherapy as Third- or Further-Line Treatment for Patients with Small Cell Lung Cancer. Tumori 2021, 107, 536–541. [Google Scholar] [CrossRef]
- Reyhanoglu, G.; Smith, T. Irinotecan. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Cameron, D.P.; Grosser, J.; Ladigan, S.; Kuzin, V.; Iliopoulou, E.; Wiegard, A.; Benredjem, H.; Jackson, K.; Liffers, S.T.; Lueong, S.; et al. Coinhibition of Topoisomerase 1 and BRD4-Mediated Pause Release Selectively Kills Pancreatic Cancer via Readthrough Transcription. Sci. Adv. 2023, 9, eadg5109. [Google Scholar] [CrossRef]
- Khobta, A.; Ferri, F.; Lotito, L.; Montecucco, A.; Rossi, R.; Capranico, G. Early Effects of Topoisomerase I Inhibition on RNA Polymerase II along Transcribed Genes in Human Cells. J. Mol. Biol. 2006, 357, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Cilloni, D.; Saglio, G. Molecular Pathways: BCR-ABL. Clin. Cancer Res. 2012, 18, 930–937. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.V.; Robinson, G.W.; Gauvain, K.; Basu, E.M.; Macy, M.E.; Maese, L.; Whipple, N.S.; Sabnis, A.J.; Foster, J.H.; Shusterman, S.; et al. Entrectinib in Children and Young Adults with Solid or Primary CNS Tumors Harboring NTRK, ROS1, or ALK Aberrations (STARTRK-NG). Neuro-Oncology 2022, 24, 1776–1789. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, M.; Rabushko, E.; Rozenberg, J.M.; Mohammad, T.; Seryakov, A.; Sekacheva, M.; Buzdin, A. Clinically Relevant Fusion Oncogenes: Detection and Practical Implications. Ther. Adv. Med. Oncol. 2022, 14, 175883592211441. [Google Scholar] [CrossRef]
- Zakharova, G.; Suntsova, M.; Rabushko, E.; Mohammad, T.; Drobyshev, A.; Seryakov, A.; Poddubskaya, E.; Moisseev, A.; Smirnova, A.; Sorokin, M.; et al. A New Approach of Detecting ALK Fusion Oncogenes by RNA Sequencing Exon Coverage Analysis. Cancers 2024, 16, 3851. [Google Scholar] [CrossRef]
- Marín-Aguilera, M.; Reig, Ò.; Milà-Guasch, M.; Font, A.; Domènech, M.; Rodríguez-Vida, A.; Carles, J.; Suárez, C.; Del Alba, A.G.; Jiménez, N.; et al. The Influence of Treatment Sequence in the Prognostic Value of TMPRSS2-ERG as Biomarker of Taxane Resistance in Castration-Resistant Prostate Cancer. Int. J. Cancer 2019, 145, 1970–1981. [Google Scholar] [CrossRef]
- Errani, C.; Zhang, L.; Shao, S.Y.; Hajdu, M.; Singer, S.; Maki, R.G.; Healey, J.H.; Antonescu, C.R. A Novel WWTR1-CAMTA1 Gene Fusion Is a Consistent Abnormality in Epithelioid Hemangioendothelioma of Different Anatomic Sites. Genes. Chromosomes Cancer 2011, 50, 644–653. [Google Scholar] [CrossRef]
- Rabushko, E.; Sorokin, M.; Suntsova, M.; Seryakov, A.P.; Kuzmin, D.V.; Poddubskaya, E.; Buzdin, A.A. Experimentally Deduced Criteria for Detection of Clinically Relevant Fusion 3′ Oncogenes from FFPE Bulk RNA Sequencing Data. Biomedicines 2022, 10, 1866. [Google Scholar] [CrossRef]
- Cruz-Rico, G.; Avilés-Salas, A.; Segura-González, M.; Espinosa-García, A.M.; Ramírez-Tirado, L.A.; Morales-Oyarvide, V.; Rojas-Marín, C.; Cardona, A.-F.; Arrieta, O. Diagnosis of EML4-ALK Translocation with FISH, Immunohistochemistry, and Real-Time Polymerase Chain Reaction in Patients with Non-Small Cell Lung Cancer. Am. J. Clin. Oncol. 2017, 40, 631–638. [Google Scholar] [CrossRef]
- Ali, J.; Khan, S.A.; Rauf, S.-E.-; Ayyub, M.; Ali, N.; Afridi, N.K. Comparative Analysis of Fluorescence In Situ Hybridization and Real Time Polymerase Chain Reaction in Diagnosis of Chronic Myeloid Leukemia. J. Coll. Physicians Surg. Pak. JCPSP 2017, 27, 26–29. [Google Scholar]
- Singh, S.; Li, H. Comparative Study of Bioinformatic Tools for the Identification of Chimeric RNAs from RNA Sequencing. RNA Biol. 2021, 18, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Roth, S.J.; Heinz, S.; Benner, C. ARTDeco: Automatic Readthrough Transcription Detection. BMC Bioinform. 2020, 21, 214. [Google Scholar] [CrossRef] [PubMed]
- Wiesel, Y.; Sabath, N.; Shalgi, R. DoGFinder: A Software for the Discovery and Quantification of Readthrough Transcripts from RNA-Seq. BMC Genom. 2018, 19, 597. [Google Scholar] [CrossRef]
- Melnick, M.; Gonzales, P.; Cabral, J.; Allen, M.A.; Dowell, R.D.; Link, C.D. Heat Shock in C. Elegans Induces Downstream of Gene Transcription and Accumulation of Double-Stranded RNA. PLoS ONE 2019, 14, e0206715. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, H.; Zhang, D.; Chen, R.; Luo, J. Readon: A Novel Algorithm to Identify Read-through Transcripts with Long-Read Sequencing Data. Bioinformatics 2024, 40, btae336. [Google Scholar] [CrossRef]
- Singh, S.; Shi, X.; Haddox, S.; Elfman, J.; Ahmad, S.B.; Lynch, S.; Manley, T.; Piczak, C.; Phung, C.; Sun, Y.; et al. RTCpredictor: Identification of Read-through Chimeric RNAs from RNA Sequencing Data. Brief. Bioinform. 2024, 25, bbae251. [Google Scholar] [CrossRef]
- Modestov, A.; Buzdin, A.; Prassolov, V. Omics Wise Analysis of RNA Splicing. In Handbook of Translational Transcriptomics; Elsevier: Amsterdam, The Netherlands, 2025; pp. 213–229. ISBN 978-0-443-19110-7. [Google Scholar]
- Musatov, I.; Sorokin, M.; Buzdin, A. Detection of Fusion Transcripts by RNA-Sequencing Data. In Handbook of Translational Transcriptomics; Elsevier: Amsterdam, The Netherlands, 2025; pp. 463–487. ISBN 978-0-443-19110-7. [Google Scholar]
- Logsdon, G.A.; Vollger, M.R.; Eichler, E.E. Long-Read Human Genome Sequencing and Its Applications. Nat. Rev. Genet. 2020, 21, 597–614. [Google Scholar] [CrossRef]
- Modestov, A.; Buzdin, A.; Suntsova, M. Unveiling RNA Editing by ADAR and APOBEC Protein Gene Families. Front. Biosci.-Landmark 2025, 30, 26298. [Google Scholar] [CrossRef]
- Balamurali, D.; Gorohovski, A.; Detroja, R.; Palande, V.; Raviv-Shay, D.; Frenkel-Morgenstern, M. ChiTaRS 5.0: The Comprehensive Database of Chimeric Transcripts Matched with Druggable Fusions and 3D Chromatin Maps. Nucleic Acids Res. 2020, 48, D825–D834. [Google Scholar] [CrossRef]
- Barresi, V.; Cosentini, I.; Scuderi, C.; Napoli, S.; Di Bella, V.; Spampinato, G.; Condorelli, D.F. Fusion Transcripts of Adjacent Genes: New Insights into the World of Human Complex Transcripts in Cancer. Int. J. Mol. Sci. 2019, 20, 5252. [Google Scholar] [CrossRef]
- Kim, P.; Tan, H.; Liu, J.; Lee, H.; Jung, H.; Kumar, H.; Zhou, X. FusionGDB 2.0: Fusion Gene Annotation Updates Aided by Deep Learning. Nucleic Acids Res. 2022, 50, D1221–D1230. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.E.; Jang, I.; Kim, S.; Cho, S.; Kim, D.; Kim, K.; Kim, J.; Hwang, J.; Kim, S.; Kim, J.; et al. ChimerDB 4.0: An Updated and Expanded Database of Fusion Genes. Nucleic Acids Res. 2020, 48, D817–D824. [Google Scholar] [CrossRef] [PubMed]
- DSouza, D.; Bik, L.; Giwa, O.; Cohen, S.; Barazany, H.L.; Siegal, T.; Frenkel-Morgenstern, M. ChiTaRS 8.0: The Comprehensive Database of Chimeric Transcripts and RNA-Seq Data with Applications in Liquid Biopsy. Nucleic Acids Res. 2024, 53, D1302–D1312. [Google Scholar] [CrossRef] [PubMed]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, B.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) Project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Modestov, A.; Zakharova, G.; Poddubskaya, E.; Buzdin, A. Regulatory Mechanisms and Functional Roles of Readthrough Transcripts in Tumorigenesis. Int. J. Mol. Sci. 2025, 26, 9975. https://doi.org/10.3390/ijms26209975
Modestov A, Zakharova G, Poddubskaya E, Buzdin A. Regulatory Mechanisms and Functional Roles of Readthrough Transcripts in Tumorigenesis. International Journal of Molecular Sciences. 2025; 26(20):9975. https://doi.org/10.3390/ijms26209975
Chicago/Turabian StyleModestov, Alexander, Galina Zakharova, Elena Poddubskaya, and Anton Buzdin. 2025. "Regulatory Mechanisms and Functional Roles of Readthrough Transcripts in Tumorigenesis" International Journal of Molecular Sciences 26, no. 20: 9975. https://doi.org/10.3390/ijms26209975
APA StyleModestov, A., Zakharova, G., Poddubskaya, E., & Buzdin, A. (2025). Regulatory Mechanisms and Functional Roles of Readthrough Transcripts in Tumorigenesis. International Journal of Molecular Sciences, 26(20), 9975. https://doi.org/10.3390/ijms26209975






