1. Introduction
The human microbiome, comprising diverse microorganisms that exist predominantly as complex, structured communities known as biofilms, is a critical factor in maintaining health and influencing disease within the host’s internal environment and on its external surfaces [
1,
2]. These polymicrobial associations, composed of various types of microorganisms such as bacteria, viruses, archae, micromycete, and eukarya, colonize different human organs and tissues, forming what is known as the ‘microbiome’ [
3]. The microbiome constitutes a complex ecosystem that can have a wide range of impacts on human health, which can be neutral, beneficial, such as vitamin synthesis and fiber degradation, or detrimental, such as infections and carcinogenesis [
4,
5,
6]. While bacterial communities represent the most diverse and abundant component [
7], the overall composition and stability of this ecosystem are crucial for host wellbeing.
The alteration in the composition of microbiota that occurs during dysbiosis frequently involves a reduction in microbial diversity, an increased prevalence of pathogenic microbes, and disruption of normal polymicrobial relationships [
8]. This dysbiosis has been linked to the pathogenesis of various health problems, including acute and chronic inflammation, infections, allergies, and the development of tumors, both benign and malignant [
8,
9,
10]. Consequently, scientific research has explored the role of microbes in tumor formation at various anatomical sites such as the intestines, lung, skin and others [
11,
12]. However, its role in breast pathology is only beginning to be understood.
Following this, the results of the Human Microbiome Project’s sequencing efforts revealed significant interindividual variability in microbial community diversity and abundance [
4]. A multitude of factors are known to influence the formation of microbiomes and the incidence of dysbiosis [
13], encompassing previous infections, inflammatory disorders [
8], dietary habits, chemical exposures [
14], and genetic predispositions [
4,
15].
Breast pathology is one of the most urgent public health problems, and breast cancer (BC), the most common type of cancer among women, is a significant socio-economic issue. According to the World Health Organization (WHO), 2.3 million women were diagnosed with breast cancer in 2022 [
16]. The burden of this disease is substantial, profoundly impacting patients’ quality of life and posing high global costs [
17,
18].
In contrast, benign breast pathology, despite its high frequency, has received comparatively less scientific and clinical attention. Research shows that breast disease is mostly benign, although accurate data on its prevalence is not available [
19,
20]. In the presence of breast symptoms, breast cancer is detected only in 3–6% of cases. Consequently, there is a significant lack of evidence-based approaches for managing patients with benign breast conditions, since the main focus has been on optimizing diagnosis and treatment for breast cancer [
21].
Notwithstanding these emerging associations, significant gaps persist in our mechanistic understanding of the breast microbiome’s role in pathogenesis. New studies suggest a link between certain microorganisms in breast tissue and the development of benign breast dysplasia. This finding is promising for the advancement of diagnostics and treatment methods. Critical unresolved questions include the precise taxonomic and functional definition of a ‘eubiotic’ breast microbiome; the causal mechanisms by which specific microbial consortia or their metabolites initiate or promote benign proliferative changes; the potential for microbial-host immune system crosstalk to modulate disease progression; and the translational applicability of microbial profiles as biomarkers for diagnostics, risk stratification, or novel therapeutic targets.
Research on the microbiome’s role in breast pathology is still in its early stages. Most studies focus on dysbiosis in breast tissues, such as the skin, nipple-areola complex secretions, ducts, and glandular fibrous tissues. Additionally, intestinal flora are also being investigated. This review aims to synthesize the current understanding of how human microbiomes might contribute to the development of benign breast diseases (BBDs), with a specific emphasis on identifying these fundamental knowledge gaps and outlining future research directions to address them.
2. Pathways of Mammary Gland Microbiota Formation and Mechanisms of Its Interaction with the Host Organism
Breast tissue obtained under aseptic conditions from patients without clinical signs of a local infectious process possesses its own unique microbiota [
22,
23,
24]. A significant diversity of microorganisms is specific to breast tissue and distinct from the microbiota of the skin and other body sites [
11,
22,
24,
25]. Furthermore, the mammary gland (MG) is considered an organ with a low microbial biomass compared to other body areas [
9].
2.1. Origin of the Mammary Gland Microbiota
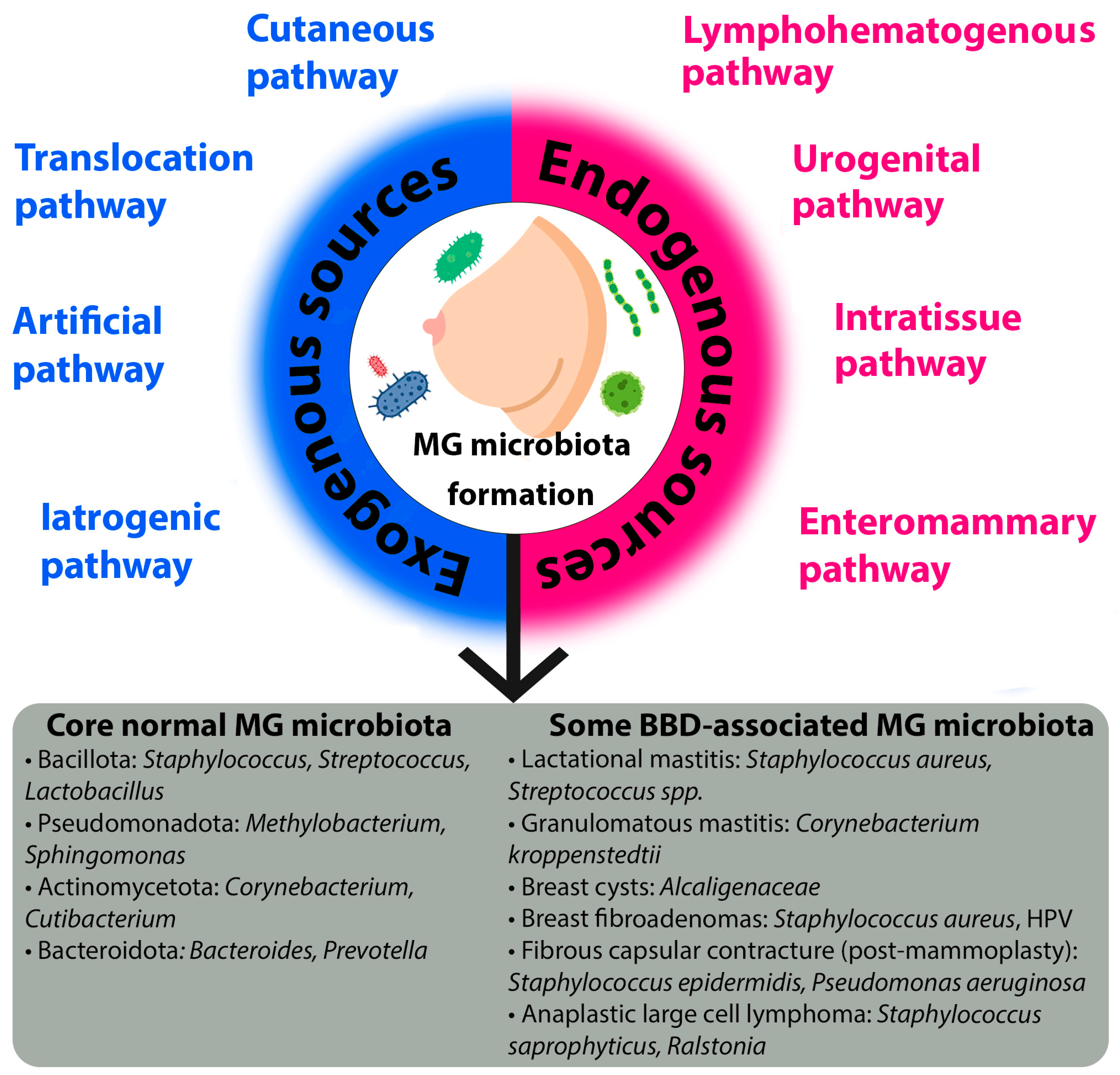
The microbiota of breast tissue is formed through the influx of microorganisms via the nipple-areolar complex [
23,
26], translocation from the gastrointestinal tract and oral cavity [
27], the urinary [
28] and reproductive systems [
22], the skin surface and other sources [
29]. The unique MG microbiota is established via distinct exogenous and endogenous pathways, each characterized by specific mechanisms and influenced by key factors (
Figure 1).
Exogenous sources refer to the introduction of microorganisms from the external environment. The cutaneous pathway is a primary route, whereby microbes from the skin surface translocate to the breast tissue. Although the microbiota of the MG and the overlying skin share species diversity, they remain distinct communities [
24,
29]. This pathway is significantly influenced by environmental factors (e.g., climate, geography, occupation, living conditions), lifestyle habits, particularly hygiene practices, and host physiology, such as a compromised skin barrier function. The evidence for this pathway is strong, primarily based on comparative 16S rRNA sequencing studies that show overlapping but distinct taxonomic profiles between skin and breast tissue samples [
24,
29].
Another critical exogenous route is the translocation pathway, which involves the retrograde inoculation of microbes via the lactiferous ducts [
30,
31,
32,
33,
34,
35]. This pathway is highly dependent on breastfeeding practices, including the type of feeding (direct breastfeeding, pumping, or formula), the mode of delivery (if breastfeeding), and the sex of the infant. Support for this pathway is moderate and indirect, largely inferred from studies detecting oral-associated bacteria in breast milk and the observed dynamic changes in milk microbiota in response to infant feeding patterns [
32,
35].
Medical interventions constitute two additional exogenous pathways. The artificial pathway describes the direct introduction of microbiota during procedures such as breast biopsies or surgery [
36]. Conversely, the iatrogenic pathway refers to alterations in the existing microbiota induced by medical treatments, most notably the use of antibiotics or probiotics. Evidence for these exogenous pathways is currently limited and primarily hypothetical, based on analogous findings from other body sites, though this represents a critical area for future controlled studies.
Endogenous sources involve the translocation of microorganisms from within the host organism. The most well-studied is the enteromammary pathway, a sophisticated mechanism where gut and oral microbiota are transported to the MG via immune cells, such as CD18+ cells and dendritic cells [
27,
29,
33,
37,
38,
39,
40]. This pathway creates a direct link between the gut and the breast, and its efficiency is influenced by the composition of the gut microbiota (e.g., states of dysbiosis associated with high body mass index (BMI)) and host diet.
The urogenital pathway proposes that microbiota from the urogenital tract may serve as a source for MG colonization [
9], although the specific mechanisms are less defined. Furthermore, the local tissue environment, or intratissue pathway, dictated by the MG tissue structure (specifically the ratio of glandular/fibrous to adipose tissue) [
41], shapes a unique ecological niche that determines which microbial communities can survive and thrive. Finally, the lymphohematogenous pathway involves the systemic dissemination of microbes to the breast through the lymphatic system and bloodstream [
35], representing a potential route for bacteria from distant sites of infection or colonization.
In conclusion, the establishment of the mammary gland microbiota is a dynamic process governed by a multitude of exogenous and endogenous pathways. Understanding the intricate interplay between these routes and their modulating factors is crucial for elucidating the role of the microbiome in breast health and disease.
2.2. Microbiota of Unaltered Mammary Gland Tissues
The composition of the microbial community in unaltered mammary gland tissues has been characterized through diversity analyses. These analyses reveal that while breast tissue harbors a greater number of bacterial species (alpha-diversity) than skin, the communities are distinct in their composition (beta-diversity), primarily due to differences in rare or less abundant species [
24,
42]. The resulting foundational profile is dominated by the species listed in
Table 1 and
Figure 1.
The higher prevalence of the phyla Pseudomonadota (Proteobacteria) and Bacillota (Firmicutes) compared to other taxonomic groups may be due to the affinity of these microorganisms for the fatty acid-rich environment of breast tissues [
4]. Disruption of polymicrobial interactions leading to dysbiosis may contribute to the development of breast diseases [
43].
Table 1.
Representatives of the bacterial community identified in unaltered female breast tissue.
Table 1.
Representatives of the bacterial community identified in unaltered female breast tissue.
| Phylum | Family | Genus | Species | References |
|---|
| Pseudomonadota (Proteobacteria) | Sphingomonadaceae | - | - | [44] |
| Methylobacteriaceae | Methylobacterium | - | [4,9] |
| Burkholderiaceae | Ralstonia | - | [4,44,45] |
| Sphingomonadaceae | Sphingomonas | yanoikuyae | [4] |
| - | [22,24,44] |
| Pseudomonadaceae | Pseudomonas | - | [22,44,46] |
| Comamonadaceae | - | - | [22] |
| Enterobacteriaceae | - | - | [22] |
| Moraxellaceae | Acinetobacter | - | [22] |
| Pasteurellaceae | Haemophilus | - | [46] |
| Neisseriaceae | Neisseria | - | [46] |
| - | - | - | [4,9,22,26,45] |
| Bacillota (Firmicutes) | Veillonellaceae | Veillonella | - | [46] |
| Staphylococcaceae | Staphylococcus | - | [22,44] |
| Streptococcaceae | Lactococcus | - | [44] |
| Streptococcus | - | [44] |
| Clostridiaceae | Clostridium | - | [44] |
| Lactobacillaceae | Lactobacillus | - | [44] |
| Listeriaceae | Listeria | welshimeri | [22] |
| - | - | - | [4,9,22,26,45,47] |
| Actinomycetota (Actinobacteria) | Corynebacteriaceae | Corynebacterium | - | [44] |
| Micrococcaceae | Micrococcus | - | [4] |
| Propionibacteriaceae | Propionibacterium | - | [22,44] |
| - | - | - | [4,26] |
| Bacteroidota (Bacteroidetes) | Bacteroidaceae | Bacteroides | - | [9] |
| Prevotellaceae | Prevotella | - | [22,44] |
| - | - | - | [9,26,44] |
In recent years, several studies have clarified the role of viruses and fungi (micromycetes) in the human microbiome [
48]. For instance, DNA from human papillomavirus (HPV), Epstein–Barr virus (EBV), human cytomegalovirus (HCMV), herpes simplex virus (HSV), and human herpesvirus type 8 (HHV-8) has been detected in breast tissue both under normal conditions and with pathological changes [
49,
50]. Given that viruses are obligate parasites, it is difficult to distinguish a separate infectious process in the MG from long-term viral persistence in breast tissue cells with potential carcinogenicity—in this case, one can refer to the MG virome. The predominant focus on the bacterial fraction of the human microbiome in most studies has led to an incomplete understanding of the role of micromycetes in polymicrobial interactions. Research into the role of fungal communities (the mycobiome) is complicated by the low fungal biomass in breast tissues compared to the bacterial component. According to published studies,
Candida albicans and
Saccharomyces are identified in normal breast tissue structure, while
Malassezia,
Davidiella,
Sistotrema, and
Penicillium are found in breast milk during lactation [
6,
30]. However, their specific role of viruses and fungi in the pathogenesis of BBD remains largely speculative.
2.3. The Microbiota-Human Organism Interaction
The MG microbiota interacts with the host organism by regulating immune, metabolic, transcriptional, and epigenetic processes through the production of enzymes and other biologically active substances [
4,
35,
41,
51,
52,
53,
54].
Bacterial metabolites (e.g., short-chain fatty acids, acetate, butyrate, pyruvate, formate, amines (cadaverine), bile acid derivatives (lithocholic acid, deoxycholic acid), indole, etc.) can influence processes of cell growth, apoptosis, epithelial–mesenchymal transition, anti-tumor immunological surveillance, and also exert cytotoxic and genotoxic effects [
55,
56]. Dysbiosis leads to changes in the bacterial metabolite profile [
56,
57]. Some bacterial metabolites and their mediated effects are presented in
Table 2.
Furthermore, microorganisms inhabiting remote areas of the body can influence each other through metabolites and immune factors [
12,
40]. These interactions can impact the onset and development of pathological conditions, particularly breast diseases [
58,
59,
60].
2.4. Mechanisms of Microbiota-Induced Breast Pathogenesis
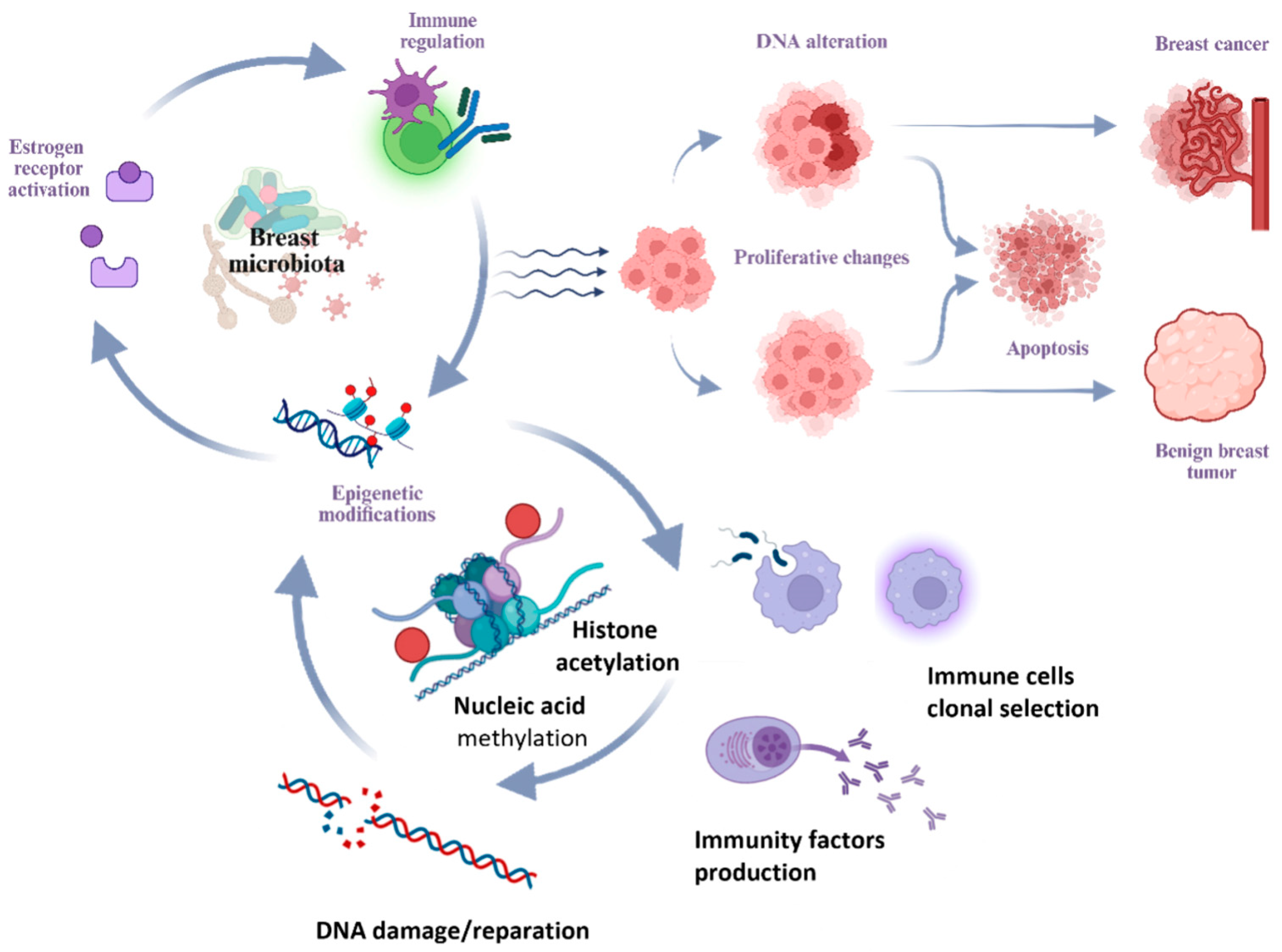
Several hypotheses have been proposed regarding the ability of microorganisms to cause DNA damage, modulate estrogen metabolism, shape the immune microenvironment of the breast—inducing chronic inflammation and local immunosuppression—and potentiate proliferative processes (
Figure 2) [
11,
61,
62,
63].
One of the primary mechanisms involves the regulation of estrogen metabolism. Elevated estrogen levels are closely associated with the activation of estrogen receptors, promoting proliferative processes in breast tissue in BBD and BC [
11,
64,
65]. The gut microbiota regulates processes of enterohepatic circulation of active estrogen forms [
66,
67]. and the synthesis of estrogen-like substances [
57,
60,
61]. The collection of microorganisms influencing systemic levels of estrogen and its metabolites constitutes the concept of the estrobolome [
68,
69]. The role of local estrogen levels in breast tissue in BC should also be noted [
9].
Beyond hormonal influence, the microbiota exerts a significant effect through local immunomodulation and the maintenance of chronic inflammation. Available research data indicate that commensal microflora of the gastrointestinal tract possesses immunomodulatory properties. The gut microbiota can maintain chronic inflammation by altering the balance between proliferation and apoptosis and triggering unregulated innate and adaptive immune responses. Immunoglobulin A (IgA), which maintains mucosal barrier integrity, is involved in recognizing and regulating the composition of the gut microbiota [
23,
57,
70].
A further mechanism by which the microbiota may influence breast tissue is through epigenetic modifications. Previous studies have shown that the microbiota can influence processes of epigenetic regulation [
71]. Epigenetic mechanisms include DNA methylation, acetylation and deacetylation of histone proteins, and modification of non-coding RNAs and microRNAs. Epigenetic modifications lead to enhanced or attenuated cell growth and regulation of cellular signaling pathways. Therefore, studying epigenetics as one of the mechanisms by which the microbiota influences pathological states of the breast appears promising [
40].
3. Microbiota Composition and Diversity in BBD
3.1. General Characteristics of BBD
BBD is among the most common breast diseases and includes conditions varying by clinical, morphological, and etiological criteria. These conditions are based on an imbalance between the epithelial and connective tissue components due to specific features of proliferation and regression processes in breast tissue. Given the heterogeneity of benign pathology, a number of terms are used to denote it: BBD, mastopathy, fibrocystic mastopathy, fibrocystic changes, fibrocystic disease, dyshormonal hyperplasia of the mammary glands, fibroadenomatosis, etc. The prevalence of benign breast changes in the female population is 50% or higher. Considering the prevalence and the increased risk of developing BC with certain types of benign changes, the issues of improving diagnosis, differential diagnosis, and treatment are relevant [
72,
73].
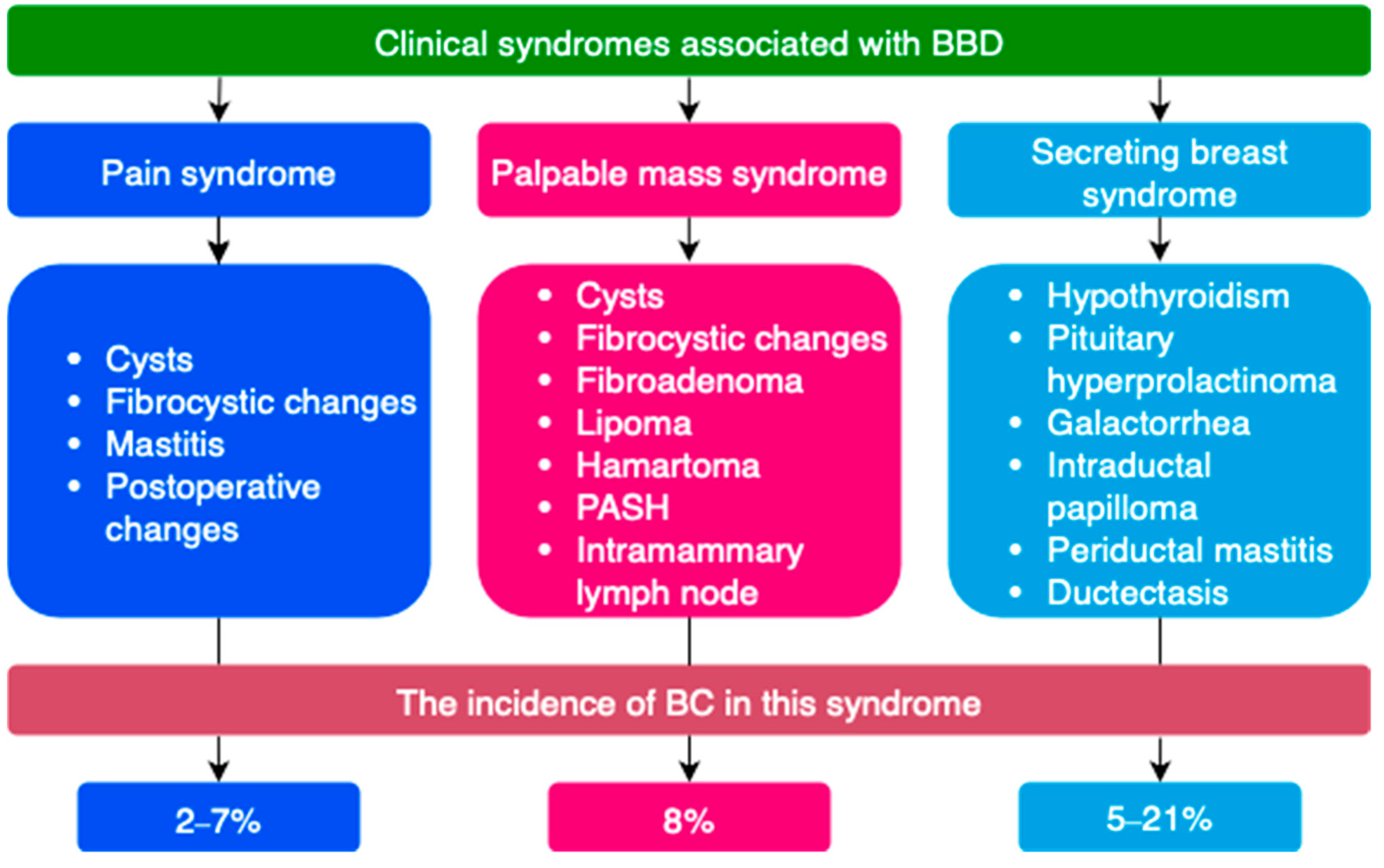
BBD is most often classified by clinical manifestations, degree of proliferative changes, type of pathomorphological changes on biopsy, etc. (
Figure 3 and
Figure 4) [
21,
72,
74,
75,
76,
77]. Breast tissue changes on histological examination are classified from B1 to B5 depending on the degree of suspicion for BC [
21,
78].
A special group of diseases consists of lesions of uncertain malignant potential (B3). Some proliferative breast conditions may be associated with the presence of DCIS, invasive BC, or be markers of an increased risk of developing BC [
57,
79,
80]. Such conditions are classified as lesions of uncertain malignant potential (B3) on histological examination [
21]. These include atypical ductal hyperplasia (ADH), flat epithelial atypia (FEA), classical lobular neoplasia (LN, ALH, LCIS), as well as lesions of heterogeneous structure with a risk of incomplete sampling during biopsy—intraductal papilloma with/without atypia, radial scar, and complex sclerosing lesion [
21].
3.2. General Characteristics of Microbiota in BBD
The main task of clinical mammology is the differential diagnosis of benign and malignant breast neoplasms. Consequently, research is primarily aimed at identifying the association between changes in the microbiota of breast tissues and gut flora in BC, while studies assessing the features of the microbiota in BBD have been published in limited numbers to date. Most of them are aimed precisely at comparing and identifying similarities and differences in the tissue microbiota of unaltered breast tissues and tumor formations (if present) and gut microflora in the absence of breast pathology, BC, and BBD [
24,
43,
60].
Studies of taxonomic profiles show that the bacterial microbiota of breast tissue in BC and BBD is similar, dominated by representatives of the phyla Bacteroidota (Bacteroidetes), Bacillota (Firmicutes), Pseudomonadota (Proteobacteria), and the phylum Actinomycetota (Actinobacteria) [
11,
24,
43]. However, more granular analysis reveals taxon-specific differences that may hold diagnostic potential. In unaltered adjacent tissues in BC, a significantly greater abundance of certain bacterial taxa was found, including the genus
Bacillus,
Staphylococcus, and members of the family Enterobacteriaceae, compared to patients with benign breast tumors and healthy women [
43]. Moreover, the relative abundance of Pseudomonadota (Proteobacteria) in benign diseases was found to be significantly lower than in malignant ones [
11]. While these findings are descriptively valuable, their utility as standalone diagnostic biomarkers is currently limited by small cohort sizes and methodological variability. The critical question of whether these microbial signatures are drivers of pathogenesis or merely secondary consequences of the altered tumor microenvironment remains unresolved.
In BBD, a low level of bacteria influencing DNA damage processes is noted, which may be a possible factor preventing malignant transformation [
43]. This observation suggests a potential functional role for the microbiota in disease progression beyond mere taxonomic composition. Taxonomic profiles in benign tumors are more similar to profiles of normal adjacent tissue in women with malignant tumors than to profiles of tissues from healthy patients [
43]. This similarity could indicate a shared microenvironmental niche or an early, ‘field effect’ change that predisposes tissue to neoplasia, warranting investigation into whether specific microbial consortia can predict the risk of malignant transformation in BBD. Beta-diversity assessment shows that the microbial community in unaltered breast tissue adjacent to invasive cancer differs from that in women with benign diseases, mainly due to rare and/or less abundant species. Alpha-diversity analysis revealed no significant differences [
4,
24]. The diagnostic value of these beta-diversity shifts, particularly the role of low-abundance taxa, is a promising but unvalidated area for future research aimed at developing non-invasive microbial biomarkers for differential diagnosis.
Metabolic pathways mediated by the tissue microbiota also differ between BC and BBD. According to KEGG (Kyoto Encyclopedia of Genes and Genomes) PATHWAY, benign tissues exhibit elevated metabolism of cysteine and methionine, glycosyltransferases, and fatty acid biosynthesis, while the microbiota of malignant tissues demonstrates reduced inositol phosphate metabolism [
24]. Bacterial lipopolysaccharides are often present in many human solid tumors [
21,
60].
A number of viruses possess oncogenic potential by initiating malignant transformation of epithelial cells, prolonging the cell cycle, activating cell proliferation, and preventing apoptosis [
35]. The phenomenon of the influence of androgens and estrogens on the replicative activity of some viruses makes viruses a possible etiological factor in the development of a number of hormone-sensitive tumors and conditions, particularly of the breast.
For example, HPV can integrate into the cell genome and mediate oncogenic transformation via E6 and E7 proteins. HPV DNA was identified in 20.3% of malignant neoplasms and 35% of benign neoplasms of the breast [
49]. It was also detected in 27.3% of biopsy samples of unaltered breast tissue. It was not found in biopsy material of in situ neoplasms or borderline lesions. HCMV can modulate the tumor immune microenvironment and stimulate tumor cell growth. In breast tissues, HCMV genetic material is more frequently detected in malignant neoplasms than in normal tissue. Very low levels of EBV DNA are detected in breast tissues in BC by quantitative PCR. Thus, the role of EBV and HCMV in the development of breast pathology, primarily breast cancer, remains debatable [
21].
Colonization of epithelial tissues and cells by some species (tropism) of micromycetes may indicate a potential role of fungi in breast diseases [
51]. Available studies only highlight the association of micromycetes with malignant neoplasms. Thus, micromycetes have been associated with 35 cancer types, yet their role in carcinogenesis remains unexplored [
48,
51]. In the context of breast cancer (BC), the most significant fungal-bacterial-immune interactions have been identified, particularly involving representatives of the genera
Aspergillus,
Malassezia, and
Cladosporium (
Cladosporium sphaerospermum) [
81]. By analogy, it is plausible that similar polymicrobial crosstalk could occur in the inflammatory milieu of BBD, such as duct ectasia or mastitis, but this remains a hypothesis awaiting clinical validation.
Fungi remain understudied but important commensals/opportunistic pathogens that shape unique host immune responses. Research has demonstrated that specific fungi like
Malassezia contribute to oncogenesis through mechanisms including pro-inflammatory cytokine induction and complement activation, collectively establishing an immunosuppressive tumor niche [
81]. By extension, it is plausible that fungal colonization of the breast ductal system may similarly influence local immunodynamics in BBD. For instance, persistent fungal antigens could potentially sustain granulomatous inflammation in conditions such as idiopathic granulomatous mastitis. Given the possibility of symbiotic and antagonistic interactions (physical, biochemical) between bacteria and fungi, further research is required to assess the role of fungi in the polymicrobial interaction of the tumor environment. If a distinct mycobiota is associated with BBD, it could open new avenues for management, such as the use of antifungal or immunomodulatory therapies in refractory cases, similar to approaches used in other fungal-driven inflammatory conditions. Defining the breast mycobiota could ultimately help identify patients with specific microbiological risk profiles for disease progression, but this potential remains entirely speculative without foundational data.
3.3. Features of Gut Microbiota in Breast Pathology
The role of gut dysbiosis in the development of BBD is also currently debatable. In patients with malignant breast tumors, the species diversity of the gut microbiota is lower but more homogeneous than in patients with benign tumors [
59]. It should be noted that in a number of studies on the composition of gut microbiota in BBD and BC, no statistical differences were observed in the assessment of α- and β-diversity [
60].
When comparing patients with BBD and healthy individuals, no differences in α-diversity indices were found [
57].
Furthermore, assessment by beta-diversity noticeably differed among the three groups (BC/BBD/normal). These results indicate an altered composition of the gut microbiota in healthy women, BC patients, and BBD patients [
57].
An increase in the number of representatives of the genera
Clostridium,
Faecalibacterium,
Lachnospira,
Romboutsia,
Fusicatenibacter,
Xylophilus,
Arcanobacterium,
Escherichia,
Peptoniphilus,
Coprobacillus,
Lactobacillus,
Porphyromonas, and the family Erysipelotrichaceae in the gut microbiota of patients with benign tumors has been reported, along with a decrease in the abundance of the genera
Collinsella,
Alistipes,
Megamonas,
Butyricimonas,
Acidaminococcus,
Asaccharobacter,
Tissierella, and
Cloacibacillus [
57,
58,
60].
The metabolic pathways of the gut microbiota in patients with malignant tumors differed significantly from those in patients with benign neoplasms [
60]. In patients with malignant tumors, unlike BBD, marked activation of lipopolysaccharide biosynthesis pathways was noted. Conversely, KEGG analysis revealed significant activation of sporulation in patients with benign tumors [
60,
82].
3.4. Mammary Gland Microbiota in Specific Types of BBD
Current scientific data do not cover the entire spectrum of microbiota changes across all types of BBD. Research is focused mainly, as noted earlier, on fundamental differences in microbiota (gut and breast tissue) in malignant and benign breast conditions in general. Below, a number of specific benign breast pathologies and their association with the composition of tissue and gut microbiota are considered. Summary data on associated and protective microorganisms for various BBDs are presented in
Table 3 and
Figure 1.
3.4.1. Breast Cysts
Among representatives of the gut microbiota, the family Alcaligenaceae is associated with an increased risk of developing breast cysts, while the genera
Eubacterium ruminantium and
Lactococcus are associated with a reduced risk [
23,
57]. Cysts can be complicated by secondary infection with clinical and radiological signs of a complicated cyst (cyst with inflammation); see the
Section 3.4.7.
HPV was detected in 40.0% of biopsies taken from patients with fibrocystic mastopathy [
83].
3.4.2. Breast Fibroadenomas
Staphylococcus aureus is an important factor causing mutation of the MED12 gene, which may contribute to the development of breast fibroadenoma and uterine leiomyoma [
57,
84]. Among benign neoplasms, HPV DNA was identified in 38.9% of histological material from fibroadenomas [
49].
3.4.3. Lactational Mastitis
Lactational mastitis is the most common type of mastitis, accounting for 33% of all breast diseases [
85]. In 90% of cases, the etiological agent is Staphylococcus aureus, less frequently coagulase-negative
Staphylococcus,
Streptococcus,
Pseudomonas aeruginosa, and
Escherichia coli [
21]. The presence of representatives of the genera
Anaerofilum and
Anaerotruncus in the gut microbiota is associated with cases of lactational mastitis, while the genus
Butyricimonas and the orders
Coriobacteriales,
Pasteurellales, and
Verrucomicrobiales had a negative association [
23]. In this regard, probiotics are used in the treatment of lactational mastitis and are potentially effective for chronic and subclinical forms of mastitis as an alternative to antibacterial therapy [
57].
3.4.4. Non-Lactational Mastitis
Non-lactational mastitis accounts for up to 3% of all benign breast diseases [
85]. Given the clinical picture and difficulties in diagnosis and treatment, non-lactational mastitis can cause distress in some women. Non-lactational mastitis occurs at any age, although it more often affects young and middle-aged women [
85]. The pathological forms of this disease are diverse, including duct ectasia (MDE), periductal mastitis (PDM), and granulomatous lobular mastitis (GLM) [
85].
The influence of the MG microbiota on the development of inflammatory breast diseases has been studied [
29,
86,
87]. Representatives of the gut flora, for example, the family Prevotellaceae, are associated with inflammatory breast changes [
23]. The breast tissue microbiota in patients with non-lactational mastitis demonstrates differences in the composition of microbial communities compared to healthy patients. In patients with non-lactational mastitis, representatives of bacterial communities inhabiting the gut, particularly the genera
Ruminococcus,
Coprococcus, and
Clostridium, were identified in breast tissue [
85]. Non-lactational mastitis can also be associated with autoimmune reactions [
85].
3.4.5. Granulomatous Mastitis
Granulomatous mastitis is a rare inflammatory breast disease in women of reproductive age [
29,
88]. The etiology of this condition is unknown; nevertheless, a significant role is assigned to local dyshormonal changes, hyperprolactinemia, autoimmune reactions, and infectious agents [
21,
85]. A role for the genus
Corynebacterium, particularly the species
Corynebacterium kroppenstedtii, has been identified in the pathogenesis of granulomatous inflammation [
85,
87]. Other taxa encountered in granulomatous mastitis include representatives of the genera
Pseudomonas,
Brevundimonas,
Stenotrophomonas,
Acinetobacter, and
Aspergillus [
29].
3.4.6. Ductal Changes
Periductal mastitis is an inflammatory disease of the subareolar lactiferous ducts, with a prevalence of up to 9% outside the lactation period [
89]. In nipple discharge, bacterial flora is detected in 50% of cases, while against the background of duct ectasia, it is detected in 62% of cases [
85]. This may be associated with structural changes in the duct wall during a persistent inflammatory process against the background of combined infections caused by representatives of the genera
Enterococcus,
Streptococcus, and
Bacteroides [
85,
90].
3.4.7. Purulent-Septic Changes of the Breast
In abscesses associated with non-lactational mastitis, the most frequent bacterial strains were coagulase-negative staphylococci and peptostreptococci,
Staphylococcus aureus (50% of cases MRSA) [
91], including as part of combined bacterial infection [
21,
85]. Certain diseases associated with impaired skin barrier function, such as atopic dermatitis, promote contamination of deep skin layers and underlying tissues, causing the development of an infectious process, including breast abscesses [
29]. Representatives of the genera
Corynebacterium and
Pseudomonas (
Pseudomonas aeruginosa) are often associated with infections of the skin and underlying soft tissues (abscesses, phlegmons, fistulas, etc.) [
92,
93]. These skin microorganisms are capable of metabolizing fatty acids and are considered potential pathobionts in breast tissues [
29].
3.4.8. Fibrous Capsular Contracture
The transfer of microorganisms during surgery via surgical instruments can lead to opportunistic subclinical infection. Staphylococci,
Cutibacterium acnes,
Pseudomonas aeruginosa, Staphylococcus lugdunensis, Staphylococcus hominis,
Staphylococcus epidermidis,
Sphingomonas paucimobilis, and
Aeromonas salmonicida, which are found on the skin of healthy individuals, are also frequently identified in infections associated with surgical interventions [
36,
94,
95]. The frequency of infectious-inflammatory complications in breast surgeries ranges from 3 to 15%. Despite prophylactic antibiotic use and adherence to aseptic and antiseptic principles, positive culture results (breast tissue) were found in 20.4% of cases [
96].
Augmentation mammoplasty is a common breast surgery and is associated with the development of capsular contracture [
97]. Up to 56% of capsular contracture cases are associated with the detection of bacterial flora on the surface of implants or in the fibrous capsule, particularly
Staphylococcus epidermidis [
28,
98]. Furthermore, species isolated from the structure of the fibrous capsule included
Escherichia coli,
Diaphorobacter nitroreducens,
Cutibacterium acnes,
Staphylococcus aureus, and
Staphylococcus spp. [
97,
99]. The formation of bacterial biofilms on the implant surface promotes resistance to antibiotic therapy and allows microorganisms to escape immune surveillance, also complicating the assessment of the species composition of the bacterial flora [
97].
3.4.9. Anaplastic Large Cell Lymphoma
In addition to capsular contracture, bacterial colonization and biofilm formation are linked to breast implant-associated anaplastic large cell lymphoma (BIA-ALCL) [
100]. The species
Staphylococcus saprophyticus and representatives of the genus
Ralstonia are the most frequently detected microorganisms in BIA-ALCL, both on the side of interest and in the contralateral breast [
97,
101].
4. Breast Microbiota in Men
Before puberty, breast tissue is identical in both sexes. During puberty, boys experience transient proliferation of the milk ducts and stroma (due to estrogen stimulation), followed by involution of these structures (with increasing testosterone levels) [
46,
102]. The male breast does not develop terminal ductal lobular units due to the absence of progesterone [
102]. The spectrum of pathology in the male breast is limited and related to the gender-specific histological structure of the organ [
46].
Most hyperplastic processes in the male breast are benign, etiologically and pathogenetically similar to changes in the female breast. Male breast cancer (1 per 100,000 in Europe with a peak at 71 years) is rare, accounting for approximately 1% of breast pathologies [
46,
102].
Benign Conditions of the Male Breast [
46,
102]:
Developmental anomalies (amastia, polymastia, nipple inversion, athelia, polythelia, etc.);
Inflammatory and reactive changes (mastitis, abscess, Mondor’s disease, etc.);
Ductal changes (duct ectasia, intraductal papilloma, etc.);
Systemic diseases/symptoms of systemic diseases (diabetic mastopathy, gynecomastia);
Benign neoplasms (lipoma, angiolipoma, cavernous hemangioma, myofibroblastoma, epidermal cysts, pseudoangiomatous stromal hyperplasia (PASH), hamartoma, etc.);
Traumatic and post-traumatic changes (hematoma, fat necrosis).
Gynecomastia is the most common pathological benign condition of the male breast. The prevalence of gynecomastia is high, especially in the neonatal period (60–90%), during puberty (48–64%), in the reproductive period (up to 30%), and in the elderly (60% after 70 years). The development of gynecomastia is often associated with transient physiological changes, endocrine disorders, systemic diseases, drug therapy, and can also develop idiopathically [
103]. The development of glandular tissue creates a morphological substrate for the development of other breast pathologies.
A critical barrier in this field is the paucity of data concerning the microbiota of the male breast. The existing literature is exceptionally limited and precludes definitive conclusions.
A critical barrier in this field is the paucity of data concerning the microbiota of the male breast. The existing literature is exceptionally limited and precludes definitive conclusions. While the concept of a sexually dimorphic breast microbiome—a microgenderome—has been proposed for both cancerous and histologically normal tissues, the evidence base for male-specific characterization is nascent. A single study has reported that histologically normal male breast tissue exhibits greater microbial richness (alpha-diversity) and distinct community composition (beta-diversity) compared to female tissue, noting a predominance of Bacteroidaceae, Caulobacteraceae, Comamonadaceae, Enterococcaceae, Microbacteriaceae, Peptoniphilaceae families, and the genera
Brevundimonas,
Clavibacter,
Comamonas, and
Rhodococcus [
46]. However, these findings are derived from a limited sample set and must be considered preliminary.
Consequently, there exists a substantial research gap in the comprehensive characterization of the male breast microbiome and its potential role in health and disease states, such as gynecomastia and male breast cancer. Present clinical paradigms for diagnosing and managing breast pathologies in men remain largely extrapolated from studies conducted in female populations. Therefore, a targeted investigation of the male breast microbiota is warranted to elucidate its pathophysiological significance and to determine whether microbial dysbiosis contributes to disease etiology in a sex-specific manner.
5. Limitations in Studying the Microbiota in BBD
This narrative review of available scientific data on the microbiota in BBD reveals a field in its early stages, characterized by several significant challenges. These limitations can be broadly categorized into methodological and analytical, clinical, and conceptual hurdles that must be addressed to advance the field.
5.1. Methodological and Analytical Limitations
The technical study of the breast microbiome presents unique difficulties. Foremost among these is the inherently low microbial biomass of breast tissue, which amplifies the risk of contamination during sampling and sequencing and complicates data analysis and interpretation [
21]. This issue is exacerbated by a lack of standardized, cross-sectional studies and unified protocols. Existing research often involves small, heterogeneous patient cohorts and employs varied methods for DNA extraction, sequencing, and bioinformatic analysis, making meta-analysis and direct comparison of findings across studies unreliable [
31]. Furthermore, the current characterization of the breast microbiome is incomplete, with a pronounced lack of high-quality data on the virome and mycobiome, focusing almost exclusively on bacterial communities.
5.2. Clinical and Cohort-Related Limitations
From a clinical perspective, obtaining appropriate samples for research is a major obstacle. There is a critical paucity of baseline data on the ‘healthy’ breast microbiome due to the obvious inaccessibility of truly normal breast tissue from healthy individuals. Consequently, studies often rely on adjacent histologically normal tissue from cancer patients as controls, which may not represent a genuine healthy state and could introduce bias. This is compounded by a dominant research focus on malignant breast neoplasms, which has left the BBD microbiota comparatively neglected. The diagnostic category of ‘lesions of uncertain malignant potential (B3)’ introduces additional complexity, as the ambiguous nature of these lesions makes it difficult to define clean and distinct patient cohorts for microbiological studies.
5.3. Conceptual and Interpretative Limitations
Beyond technical and clinical issues, there are profound conceptual challenges. The most significant is the inability to infer causality from current data. It remains unknown whether observed microbial dysbiosis is a causative factor in the pathogenesis of BBD, a consequence of the altered tissue microenvironment, or merely a bystander effect [
46]. This is complicated by the high degree of inter-individual variability in the microbiota and the multifactorial etiology of BBD, making it difficult to disentangle the specific contribution of microbes from other genetic, hormonal, and environmental factors. Finally, while the potential for microbial signatures to serve as clinical biomarkers is a compelling goal, the transition from correlative observations to validated, actionable diagnostic or therapeutic tools for personalized medicine remains a considerable challenge [
60,
104].
In conclusion, addressing these multifaceted limitations is paramount for building a robust scientific foundation. Future research must prioritize standardized protocols, larger prospective cohorts, and innovative analytical methods to move from descriptive association to mechanistic understanding, ultimately unlocking the potential of the microbiome for the prevention, diagnosis, and treatment of BBD.
6. Materials and Methods
This narrative review was conducted by systematically searching the PubMed, Scopus, and Google Scholar databases for literature published between January 2000 and June 2025. The search utilized a comprehensive set of keywords and their combinations, including ‘breast microbiota’, ‘mammary gland microbiota’, ‘breast tumor microbiota’, ‘benign breast diseases’, ‘benign breast lesions’, ‘breast tissue microbiota’, and ‘human mammary gland microbiota’. Duplicate records were removed, and the search was restricted to titles and abstracts focusing on microbiological findings in human breast tissue, milk, or associated bodily fluids (e.g., urine, nipple aspirate fluid) in the context of both benign and malignant breast conditions to provide a comparative context. The initial search yielded a total of 2815 studies. After screening titles and abstracts for relevance to the mammary gland microbiota and benign breast diseases, 215 full-text articles were assessed for eligibility. Studies were excluded if they focused solely on cancer without a benign disease comparison, were purely methodological, or involved animal models. Ultimately, a final set of 85 studies was included in the synthesis. While not a systematic review, this approach aimed to comprehensively cover the current state of knowledge on the features of the microbiome in patients with benign breast pathology.
7. Conclusions
Accumulating evidence underscores a compelling association between microbial dysbiosis and the pathogenesis of breast diseases. This review synthesizes current understanding of the MG microbiota—a distinct community formed through exogenous (cutaneous, retrograde translocation) and endogenous (enteromammary, hematogenous) pathways—and its intricate interactions with the host via regulation of estrogen metabolism, immunomodulation, and epigenetic mechanisms.
A central and unresolved question is whether the observed microbial shifts in BBD are a cause or a consequence of the disease. While the low microbial biomass and correlative nature of most studies preclude definitive conclusions, several lines of evidence suggest a potential causative role for specific taxa. For instance, the consistent association of Corynebacterium kroppenstedtii with the lipid-rich microenvironment of granulomatous mastitis and the established pathogenicity of Staphylococcus aureus in acute lactational mastitis point to microbes that may actively drive pathology.
Looking ahead, the translation of microbiota research into clinical practice for BBD represents a key frontier. Within the next 5–10 years, we anticipate that validated microbial signatures could augment traditional diagnostics. Specifically, the relative abundance of Pseudomonadota or the presence of a Corynebacterium kroppenstedtii-dominant profile in biopsy samples or ductal lavage fluid could serve as a molecular tool to differentiate inflammatory BBD subtypes or stratify the risk of malignant transformation in proliferative benign conditions. This could lead to the development of non-invasive microbial risk scores to guide patient management.
To realize this clinical potential and move beyond correlation to causation, future research on BBD must be unequivocally directed toward a multi-omics and mechanistic framework. The critical next steps should include longitudinal cohort studies specifically tracking microbial dynamics across different BBD subtypes—from initial development through potential progression or resolution. This should be coupled with integrated meta-omics approaches to define functional relationships between microbial communities and BBD pathogenesis, particularly focusing on inflammatory processes in conditions like granulomatous mastitis and proliferative changes in benign tumors. Furthermore, developing BBD-specific experimental models, including patient-derived organoids from fibroadenoma and duct ectasia tissues, will be crucial for validating the functional role of candidate microbes and testing potential microbiota-modulating therapies.
Ultimately, advancing this field necessitates strengthened interdisciplinary collaboration across microbiology, oncology, and immunology to translate these fundamental insights into tangible benefits for patients with benign breast diseases.
Author Contributions
Conceptualization, Y.V.Z., M.I.K. and L.I.K.; methodology, M.I.K., Y.V.Z. and L.I.K.; writing—original draft preparation, N.I.U.; writing—review and editing, M.I.K., Y.V.Z., L.I.K. and A.B.A.; visualization, N.I.U., M.I.K. and Y.V.Z.; supervision, M.I.K., L.I.K., A.B.A. and Y.V.Z. All authors have read and agreed to the published version of the manuscript.
Funding
The work was carried out within the framework of the Russian Science Foundation, grant No. 25-18-00901.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable.
Conflicts of Interest
Yury V. Zhernov was employed by the company Fomin Clinic. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| BBD | benign breast disease |
| BC | breast cancer |
| EBV | Epstein–Barr virus |
| HCMV | human cytomegalovirus |
| HHV-8 | human herpesvirus type 8 |
| HPV | human papillomavirus |
| HSV | herpes simplex virus |
| MG | mammary gland |
| MRSA | methicillin-resistant Staphylococcus aureus |
References
- Swidsinski, A.; Amann, R.; Guschin, A.; Swidsinski, S.; Loening-Baucke, V.; Mendling, W.; Sobel, J.D.; Lamont, R.F.; Vaneechoutte, M.; Baptista, P.V.; et al. Polymicrobial consortia in the pathogenesis of biofilm vaginosis visualized by FISH. Historic review outlining the basic principles of the polymicrobial infection theory. Microbes Infect. 2024, 26, 105403. [Google Scholar] [CrossRef] [PubMed]
- Cerca, N.; Vaneechoutte, M.; Guschin, A.; Swidsinski, A. Polymicrobial infections and biofilms in women’s health: Gahro Expert Group Meeting Report. Res. Microbiol. 2017, 168, 902–904. [Google Scholar] [CrossRef] [PubMed]
- NIH HMP Working Group; Peterson, J.; Garges, S.; Giovanni, M.; McInnes, P.; Wang, L.; Schloss, J.A.; Bonazzi, V.; McEwen, J.E.; Wetterstrand, K.A.; et al. The NIH Human Microbiome Project. Genome Res. 2009, 19, 2317–2323. [Google Scholar] [CrossRef]
- D’Afonseca, V.; Muñoz, E.V.; Leal, A.L.; Soto, P.M.A.S.; Parra-Cid, C. Implications of the microbiome and metabolic intermediaries produced by bacteria in breast cancer. Genet. Mol. Biol. 2024, 47 (Suppl. S1), e20230316. [Google Scholar] [CrossRef]
- El-Sayed, A.; Aleya, L.; Kamel, M. Microbiota’s role in health and diseases. Environ. Sci. Pollut. Res. Int. 2021, 28, 36967–36983. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Zhou, Y.; Feng, Y.; Sun, T.; Xu, J. Tumor-related fungi and crosstalk with gut fungi in the tumor microenvironment. Cancer Biol. Med. 2024, 21, 977–994. [Google Scholar] [CrossRef]
- Manos, J. The human microbiome in disease and pathology. Apmis 2022, 130, 690–705. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, Q.; Huo, X.; Liu, Z.; Da, M.; Yuan, M.; Zhao, Y.; Shen, G. Impact of intestinal dysbiosis on breast cancer metastasis and progression. Front. Oncol. 2022, 12, 1037831. [Google Scholar] [CrossRef]
- Wang, H.; Altemus, J.; Niazi, F.; Green, H.; Calhoun, B.C.; Sturgis, C.; Grobmyer, S.R.; Eng, C. Breast tissue, oral and urinary microbiomes in breast cancer. Oncotarget 2017, 8, 88122–88138. [Google Scholar] [CrossRef] [PubMed]
- Alhamwe, A.; López, J.F.; Zhernov, Y.; von Strandmann, E.P.; Karaulov, A.; Kolahian, S.; Geßner, R.; Renz, H. Impact of local human microbiota on the allergic diseases: Organ-organ interaction. Pediatr. Allergy Immunol. Off. Publ. Eur. Soc. Pediatr. Allergy Immunol. 2023, 34, e13976. [Google Scholar] [CrossRef]
- Samkari, A.A.; Alsulami, M.; Bataweel, L.; Altaifi, R.; Altaifi, A.; Saleem, A.M.; Farsi, A.H.; Iskanderani, O.; Akeel, N.Y.; Malibary, N.H.; et al. Body Microbiota and Its Relationship with Benign and Malignant Breast Tumors: A Systematic Review. Cureus 2022, 14, e25473b. [Google Scholar] [CrossRef]
- Kovács, P.; Csonka, T.; Kovács, T.; Sári, Z.; Ujlaki, G.; Sipos, A.; Karányi, Z.; Szeőcs, D.; Hegedűs, C.; Uray, K.; et al. Lithocholic Acid, a Metabolite of the Microbiome, Increases Oxidative Stress in Breast Cancer. Cancers 2019, 11, 1255. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.; Thaiss, C.A.; Zeevi, D.; Dohnalová, L.; Zilberman-Schapira, G.; Mahdi, J.A.; David, E.; Savidor, A.; Korem, T.; Herzig, Y.; et al. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell 2015, 163, 1428–1443. [Google Scholar] [CrossRef]
- Sonnenburg, E.D.; Smits, S.A.; Tikhonov, M.; Higginbottom, S.K.; Wingreen, N.S.; Sonnenburg, J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nature 2016, 529, 212–215. [Google Scholar] [CrossRef] [PubMed]
- Stappenbeck, T.S.; Virgin, H.W. Accounting for reciprocal host–microbiome interactions in experimental science. Nature 2016, 534, 191–199. [Google Scholar] [CrossRef]
- World Health Organization. Breast Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer (accessed on 12 November 2024).
- Fernandez-Rodriguez, E.J.; Taboada-Taboada, R.; Garcia-Martin, A.; Sanchez-Gomez, C.; Saez-Gutierrez, S.; Rihuete-Galve, M.I.; Fonseca-Sánchez, E. Study on the additional financial burden of breast cancer disease on cancer patients and their families. Financial toxicity in cancer. Front. Public Health 2024, 12, 1324334. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Cao, Z.; Prettner, K.; Kuhn, M.; Yang, J.; Jiao, L.; Wang, Z.; Li, W.; Geldsetzer, P.; Bärnighausen, T.; et al. Estimates and Projections of the Global Economic Cost of 29 Cancers in 204 Countries and Territories from 2020 to 2050. JAMA Oncol. 2023, 9, 465. [Google Scholar] [CrossRef]
- van de Voort, E.M.; Struik, G.M.; van Streun, S.P.; Verhoef, C.; Groot, C.A.U.-D.; Klem, T.M. Hospital costs and cosmetic outcome of benign and high-risk breast lesions managed by vacuum-assisted excision versus surgical excision. Br. J. Radiol. 2022, 95, 20220117. [Google Scholar] [CrossRef]
- Zhu, L.; Zeng, X.; Jiang, S.; Ruan, S.; Ma, H.; Li, Y.; Ye, C.; Dong, J. Prevalence of breast fibroadenoma in healthy physical examination population in Guangdong province of China: A cross-sectional study. BMJ Open 2022, 12, e057080. [Google Scholar] [CrossRef]
- Stachs, A.; Stubert, J.; Reimer, T.; Hartmann, S. Benign Breast Disease in Women. Dtsch. Arztebl. Int. 2019, 116, 565–574. [Google Scholar] [CrossRef]
- Urbaniak, C.; Cummins, J.; Brackstone, M.; Macklaim, J.M.; Gloor, G.B.; Baban, C.K.; Scott, L.; O'Hanlon, D.M.; Burton, J.P.; Francis, K.P.; et al. Microbiota of Human Breast Tissue. Appl. Environ. Microbiol. 2014, 80, 3007–3014. [Google Scholar] [CrossRef]
- Wang, X.; Gao, H.; Zeng, Y.; Chen, J. Exploring the relationship between gut microbiota and breast diseases using Mendelian randomization analysis. Front. Med. 2024, 11, 298. [Google Scholar] [CrossRef]
- Hieken, T.J.; Chen, J.; Hoskin, T.L.; Walther-Antonio, M.; Johnson, S.; Ramaker, S.; Xiao, J.; Radisky, D.C.; Knutson, K.L.; Kalari, K.R.; et al. The Microbiome of Aseptically Collected Human Breast Tissue in Benign and Malignant Disease. Sci. Rep. 2016, 6, 30751. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Tian, T.; Wei, Z.; Shih, N.; Feldman, M.D.; Peck, K.N.; DeMichele, A.M.; Alwine, J.C.; Robertson, E.S. Distinct microbial signatures associated with different breast cancer types. Front. Microbiol. 2018, 9, 951. [Google Scholar] [CrossRef]
- Xuan, C.; Shamonki, J.M.; Chung, A.; Dinome, M.L.; Chung, M.; Sieling, P.A.; Lee, D.J. Microbial Dysbiosis Is Associated with Human Breast Cancer. PLoS ONE 2014, 9, e83744. [Google Scholar] [CrossRef]
- Rodríguez, J.M. The Origin of Human Milk Bacteria: Is There a Bacterial Entero-Mammary Pathway during Late Pregnancy and Lactation? Adv. Nutr. 2014, 5, 779–784. [Google Scholar] [CrossRef]
- An, J.; Yang, J.; Lee, W.-H.; Kim, J.B.; Yang, Y.; Kim, J.; Kim, H.; Paek, S.H.; Lee, J.W.; Woo, J.; et al. Diagnostic Kit of breast cancer via urine microbiome. Eur. J. Surg. Oncol. 2020, 46, e33. [Google Scholar] [CrossRef]
- Wang, K.; Nakano, K.; Naderi, N.; Bajaj-Elliott, M.; Mosahebi, A. Is the skin microbiota a modifiable risk factor for breast disease?: A systematic review. Breast 2021, 59, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Boix-Amorós, A.; Puente-Sánchez, F.; du Toit, E.; Linderborg, K.M.; Zhang, Y.; Yang, B.; Salminen, S.; Isolauri, E.; Tamames, J.; Mira, A.; et al. Mycobiome Profiles in Breast Milk from Healthy Women Depend on Mode of Delivery, Geographic Location, and Interaction with Bacteria. Appl. Environ. Microbiol. 2019, 85, e02994-18. [Google Scholar] [CrossRef]
- Laborda-Illanes, A.; Sanchez-Alcoholado, L.; Dominguez-Recio, M.E.; Jimenez-Rodriguez, B.; Lavado, R.; Comino-Méndez, I.; Alba, E.; Queipo-Ortuño, M.I. Breast and Gut Microbiota Action Mechanisms in Breast Cancer Pathogenesis and Treatment. Cancers 2020, 12, 2465. [Google Scholar] [CrossRef]
- Gay, M.; Koleva, P.; Slupsky, C.; Toit, E.; Eggesbo, M.; Johnson, C.; Wegienka, G.; Shimojo, N.; Campbell, D.; Prescott, S.; et al. In Vivo LactoActive Study Investigators. Worldwide variation in human milk metabolome: Indicators of breast physiology and maternal lifestyle? Nutrients 2018, 10, 1151. [Google Scholar] [CrossRef] [PubMed]
- Moossavi, S.; Sepehri, S.; Robertson, B.; Bode, L.; Goruk, S.; Field, C.J.; Lix, L.M.; de Souza, R.J.; Becker, A.B.; Mandhane, P.J.; et al. Composition and Variation of the Human Milk Microbiota Are Influenced by Maternal and Early-Life Factors. Cell Host Microbe 2019, 25, 324–335.e4. [Google Scholar] [CrossRef]
- Cabrera-Rubio, R.; Collado, M.C.; Laitinen, K.; Salminen, S.; Isolauri, E.; Mira, A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am. J. Clin. Nutr. 2012, 96, 544–551. [Google Scholar] [CrossRef]
- Ye, Z.; Gao, L.; Guo, Z.; Wang, Q. Oral and intestinal flora translocation and tumor development. J. Cancer Res. Ther. 2025, 21, 323–333. [Google Scholar] [CrossRef]
- Owens, C.D.; Stoessel, K. Surgical site infections: Epidemiology, microbiology and prevention. J. Hosp. Infect. 2008, 70, 3–10. [Google Scholar] [CrossRef]
- Zhong, Z.; Tang, H.; Shen, T.; Ma, X.; Zhao, F.; Kwok, L.Y.; Sun, Z.; Bilige, M.; Zhang, H. Bifidobacterium animalis subslactis Probio-M8 undergoes host adaptive evolution by glcU mutation and translocates to the infant’s gut via oral-/entero-mammary routes through lactation. Microbiome 2022, 10, 197. [Google Scholar] [CrossRef]
- Soto-Pantoja, D.R.; Gaber, M.; Arnone, A.A.; Bronson, S.M.; Cruz-Diaz, N.; Wilson, A.S.; Clear, K.Y.J.; Ramirez, M.U.; Kucera, G.L.; Levine, E.A.; et al. Diet Alters Entero-Mammary Signaling to Regulate the Breast Microbiome and Tumorigenesis. Cancer Res. 2021, 81, 3890–3904. [Google Scholar] [CrossRef] [PubMed]
- Angelopoulou, A.; Field, D.; Ryan, C.A.; Stanton, C.; Hill, C.; Ross, R.P. The microbiology and treatment of human mastitis. Med. Microbiol. Immunol. 2018, 207, 83–94. [Google Scholar] [CrossRef]
- Wu, H.; Ganguly, S.; Tollefsbol, T.O. Modulating Microbiota as a New Strategy for Breast Cancer Prevention and Treatment. Microorganisms 2022, 10, 1727. [Google Scholar] [CrossRef] [PubMed]
- Hieken, T.J.; Chen, J.; Chen, B.; Johnson, S.; Hoskin, T.L.; Degnim, A.C.; Walther-Antonio, M.R.; Chia, N. The breast tissue microbiome, stroma, immune cells and breast cancer. Neoplasia 2022, 27, 100786. [Google Scholar] [CrossRef]
- Chen, J.; Bittinger, K.; Charlson, E.S.; Hoffmann, C.; Lewis, J.; Wu, G.D.; Collman, R.G.; Bushman, F.D.; Li, H. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 2012, 28, 2106–2113. [Google Scholar] [CrossRef]
- Urbaniak, C.; Gloor, G.B.; Brackstone, M.; Scott, L.; Tangney, M.; Reid, G. The Microbiota of Breast Tissue and Its Association with Breast Cancer. Appl. Environ. Microbiol. 2016, 82, 5039–5048. [Google Scholar] [CrossRef]
- Chan, A.A.; Bashir, M.; Rivas, M.N.; Duvall, K.; Sieling, P.A.; Pieber, T.R.; Vaishampayan, P.A.; Love, S.M.; Lee, D.J. Characterization of the microbiome of nipple aspirate fluid of breast cancer survivors. Sci. Rep. 2016, 6, 28061. [Google Scholar] [CrossRef]
- Costantini, L.; Magno, S.; Albanese, D.; Donati, C.; Molinari, R.; Filippone, A.; Masetti, R.; Merendino, N. Characterization of human breast tissue microbiota from core needle biopsies through the analysis of multi hypervariable 16S-rRNA gene regions. Sci. Rep. 2018, 8, 16893. [Google Scholar] [CrossRef]
- Niccolai, E.; Baldi, S.; Nannini, G.; Gensini, F.; Papi, L.; Vezzosi, V.; Bianchi, S.; Orzalesi, L.; Ramazzotti, M.; Amedei, A. Breast cancer: The first comparative evaluation of oncobiome composition between males and females. Biol. Sex Differ. 2023, 14, 37. [Google Scholar] [CrossRef]
- Thompson, K.J.; Ingle, J.N.; Tang, X.; Chia, N.; Jeraldo, P.R.; Walther-Antonio, M.R.; Kandimalla, K.K.; Johnson, S.; Yao, J.Z.; Harrington, S.C.; et al. A comprehensive analysis of breast cancer microbiota and host gene expression. PLoS ONE 2017, 12, e0188873. [Google Scholar] [CrossRef] [PubMed]
- Narunsky-Haziza, L.; Sepich-Poore, G.D.; Livyatan, I.; Asraf, O.; Martino, C.; Nejman, D.; Gavert, N.; Stajich, J.E.; Amit, G.; González, A.; et al. Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell 2022, 185, 3789–3806.e17. [Google Scholar] [CrossRef] [PubMed]
- Maldonado-Rodríguez, E.; Hernández-Barrales, M.; Reyes-López, A.; Godina-González, S.; Gallegos-Flores, P.I.; Esparza-Ibarra, E.L.; González-Curiel, I.E.; Aguayo-Rojas, J.; López-Saucedo, A.; Mendoza-Almanza, G.; et al. Presence of Human Papillomavirus DNA in Malignant Neoplasia and Non-Malignant Breast Disease. Curr. Issues Mol. Biol. 2022, 44, 3648–3665. [Google Scholar] [CrossRef] [PubMed]
- Lawson, J.S.; Günzburg, W.H.; Whitaker, N.J. Viruses and human breast cancer. Future Microbiol. 2006, 1, 33–51. [Google Scholar] [CrossRef]
- Guglietta, S.; Li, X.; Saxena, D. Role of Fungi in Tumorigenesis: Promises and Challenges. Annu. Rev. Pathol. 2025, 20, 459–482. [Google Scholar] [CrossRef]
- Levy, M.; Thaiss, C.A.; Elinav, E. Metagenomic cross-talk: The regulatory interplay between immunogenomics and the microbiome. Genome Med. 2015, 7, 120. [Google Scholar] [CrossRef] [PubMed]
- Kovács, T.; Mikó, E.; Ujlaki, G.; Sári, Z.; Bai, P. The Microbiome as a Component of the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1225, 137–153. [Google Scholar] [CrossRef] [PubMed]
- Luu, M.; Visekruna, A. Microbial metabolites: Novel therapeutic tools for boosting cancer therapies. Trends Cell Biol. 2021, 31, 873–875. [Google Scholar] [CrossRef]
- Kovács, T.; Mikó, E.; Vida, A.; Sebő, É.; Toth, J.; Csonka, T.; Boratkó, A.; Ujlaki, G.; Lente, G.; Kovács, P.; et al. Cadaverine, a metabolite of the microbiome, reduces breast cancer aggressiveness through trace amino acid receptors. Sci. Rep. 2019, 9, 1300. [Google Scholar] [CrossRef]
- Kovács, T.; Mikó, E.; Ujlaki, G.; Yousef, H.; Csontos, V.; Uray, K.; Bai, P. The involvement of oncobiosis and bacterial metabolite signaling in metastasis formation in breast cancer. Cancer Metastasis Rev. 2021, 40, 1223–1249. [Google Scholar] [CrossRef]
- Ma, Z.; Qu, M.; Wang, X. Analysis of Gut Microbiota in Patients with Breast Cancer and Benign Breast Lesions. Pol. J. Microbiol. 2022, 71, 217–226. [Google Scholar] [CrossRef]
- Zhu, J.; Liao, M.; Yao, Z.; Liang, W.; Li, Q.; Liu, J.; Yang, H.; Ji, Y.; Wei, W.; Tan, A.; et al. Breast cancer in postmenopausal women is associated with an altered gut metagenome. Microbiome 2018, 6, 136. [Google Scholar] [CrossRef]
- Goedert, J.J.; Jones, G.; Hua, X.; Xu, X.; Yu, G.; Flores, R.; Falk, R.T.; Gail, M.H.; Shi, J.; Ravel, J.; et al. Investigation of the Association Between the Fecal Microbiota and Breast Cancer in Postmenopausal Women: A Population-Based Case-Control Pilot Study. J. Natl. Cancer Inst. 2015, 107, djv147. [Google Scholar] [CrossRef]
- Yang, P.; Wang, Z.; Peng, Q.; Lian, W.; Chen, D. Comparison of the Gut Microbiota in Patients with Benign and Malignant Breast Tumors: A Pilot Study. Evol. Bioinform. 2021, 17, 11769343211057573. [Google Scholar] [CrossRef]
- Parida, S.; Sharma, D. The Microbiome–Estrogen Connection and Breast Cancer Risk. Cells 2019, 8, 1642. [Google Scholar] [CrossRef] [PubMed]
- Fernández, M.F.; Reina-Pérez, I.; Astorga, J.M.; Rodríguez-Carrillo, A.; Plaza-Díaz, J.; Fontana, L. Breast Cancer and Its Relationship with the Microbiota. Int. J. Environ. Res. Public Health 2018, 15, 1747. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 2013, 13, 800–812. [Google Scholar] [CrossRef]
- Al-Shami, K.; Awadi, S.; Khamees, A.; Alsheikh, A.M.; Al-Sharif, S.; Ala’ Bereshy, R.; Al-Eitan, S.F.; Banikhaled, S.H.; Al-Qudimat, A.R.; Al-Zoubi, R.M.; et al. Estrogens and the risk of breast cancer: A narrative review of literature. Heliyon 2023, 9, e20224. [Google Scholar] [CrossRef]
- Doisneau-Sixou, S.F.; Sergio, C.M.; Carroll, J.S.; Hui, R.; Musgrove, E.A.; Sutherland, R.L. Estrogen and antiestrogen regulation of cell cycle progression in breast cancer cells. Endocr. Relat. Cancer 2003, 10, 179–186. [Google Scholar] [CrossRef]
- Mikó, E.; Kovács, T.; Sebő, É.; Tóth, J.; Csonka, T.; Ujlaki, G.; Sipos, A.; Szabó, J.; Méhes, G.; Bai, P. Microbiome—Microbial Metabolome—Cancer Cell Interactions in Breast Cancer—Familiar, but Unexplored. Cells 2019, 8, 293. [Google Scholar] [CrossRef]
- Deng, X.; Yang, H.; Tian, L.; Ling, J.; Ruan, H.; Ge, A.; Liu, L.; Fan, H. Bibliometric analysis of global research trends between gut microbiota and breast cancer: From 2013 to 2023. Front. Microbiol. 2024, 15, 1393422. [Google Scholar] [CrossRef]
- Flores, R.; Shi, J.; Fuhrman, B.; Xu, X.; Veenstra, T.D.; Gail, M.H.; Gajer, P.; Ravel, J.; Goedert, J.J. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: A cross-sectional study. J. Transl. Med. 2012, 10, 253. [Google Scholar] [CrossRef]
- Fuhrman, B.J.; Feigelson, H.S.; Flores, R.; Gail, M.H.; Xu, X.; Ravel, J.; Goedert, J.J. Associations of the fecal microbiome with urinary estrogens and estrogen metabolites in postmenopausal women. J. Clin. Endocrinol. Metab. 2014, 99, 4632–4640. [Google Scholar] [CrossRef]
- Peterson, D.A.; McNulty, N.P.; Guruge, J.L.; Gordon, J.I. IgA Response to Symbiotic Bacteria as a Mediator of Gut Homeostasis. Cell Host Microbe 2007, 2, 328–339. [Google Scholar] [CrossRef]
- Haque, S.; Raina, R.; Afroze, N.; Hussain, A.; Alsulimani, A.; Singh, V.; Mishra, B.N.; Kaul, S.; Kharwar, R.N. Microbial dysbiosis and epigenetics modulation in cancer development—A chemopreventive approach. Semin. Cancer Biol. 2022, 86, 666–681. [Google Scholar] [CrossRef]
- Dyrstad, S.W.; Yan, Y.; Fowler, A.M.; Colditz, G.A. Breast cancer risk associated with benign breast disease: Systematic review and meta-analysis. Breast Cancer Res. Treat. 2015, 149, 569–575. [Google Scholar] [CrossRef]
- Onstad, M.; Stuckey, A. Benign Breast Disorders. Obstet. Gynecol. Clin. N. Am. 2013, 40, 459–473. [Google Scholar] [CrossRef]
- Iddon, J.; Dixon, J.M. Mastalgia. BMJ 2013, 347, f3213. [Google Scholar] [CrossRef]
- Barton, M.B.; Elmore, J.G.; Fletcher, S.W. Breast Symptoms among Women Enrolled in a Health Maintenance Organization: Frequency, Evaluation, and Outcome. Ann. Intern. Med. 1999, 130, 651–657. [Google Scholar] [CrossRef]
- Expert Panel on Breast Imaging; Lee, S.J.; Trikha, S.; Moy, L.; Baron, P.; diFlorio, R.M.; Green, E.D.; Heller, S.L.; Holbrook, A.I.; Lewin, A.A.; et al. ACR Appropriateness Criteria® Evaluation of Nipple Discharge. J. Am. Coll. Radiol. 2017, 14, S138–S153. [Google Scholar] [CrossRef]
- Dupont, S.C.; Boughey, J.C.; Jimenez, R.E.; Hoskin, T.L.; Hieken, T.J. Frequency of diagnosis of cancer or high-risk lesion at operation for pathologic nipple discharge. Surgery 2015, 158, 988–995. [Google Scholar] [CrossRef]
- Ellis, I.O.; Humphreys, S.; Michell, M.; Pinder, S.E.; Wells, C.A.; Zakhour, H.D. UK National Coordinating Commmittee for Breast Screening Pathology; European Commission Working Group on Breast Screening Pathology. Best Practice No 179. Guidelines for breast needle core biopsy handling and reporting in breast screening assessment. J. Clin. Pathol. 2004, 57, 897–902. [Google Scholar] [CrossRef]
- Worsham, M.J.; Raju, U.; Lu, M.; Kapke, A.; Botttrell, A.; Cheng, J.; Shah, V.; Savera, A.; Wolman, S.R. Risk factors for breast cancer from benign breast disease in a diverse population. Breast Cancer Res. Treat. 2009, 118, 1–7. [Google Scholar] [CrossRef]
- Johansson, A.; Christakou, A.E.; Iftimi, A.; Eriksson, M.; Tapia, J.; Skoog, L.; Benz, C.C.; Rodriguez-Wallberg, K.A.; Hall, P.; Czene, K.; et al. Characterization of Benign Breast Diseases and Association with Age, Hormonal Factors, and Family History of Breast Cancer Among Women in Sweden. JAMA Netw. Open 2021, 4, e2114716. [Google Scholar] [CrossRef]
- Banerjee, S.; Wei, Z.; Tian, T.; Bose, D.; Shih, N.N.C.; Feldman, M.D.; Khoury, T.; De Michele, A.; Robertson, E.S. Prognostic correlations with the microbiome of breast cancer subtypes. Cell Death Dis. 2021, 12, 831. [Google Scholar] [CrossRef]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef]
- Swamy, N.; Rohilla, M.; Raichandani, S.; Bryant-Smith, G. Epidemiology of male breast diseases: A 10-year institutional review. Clin. Imaging 2021, 72, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Bullerdiek, J.; Rommel, B. Factors targeting MED12 to drive tumorigenesis? F1000Research 2018, 7, 359. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wu, J.; Liang, Z.; Mo, C.; Qi, T.; Liang, S.; Lian, T.; Qiu, R.; Yu, X.; Tang, X.; et al. Interactions between the breast tissue microbiota and host gene regulation in nonpuerperal mastitis. Microbes Infect. 2022, 24, 104904. [Google Scholar] [CrossRef] [PubMed]
- Park, S.M.; Choi, W.S.; Yoon, Y.; Jung, G.H.; Lee, C.K.; Ahn, S.H.; Wonsuck, Y.; Yoo, Y. Breast abscess caused by Staphylococcus aureus in 2 adolescent girls with atopic dermatitis. Korean J. Pediatr. 2018, 61, 200. [Google Scholar] [CrossRef]
- Wang, J.; Xu, H.; Li, Z.; Li, F.; Yang, Y.; Yu, X.; Jiang, D.; Xing, L.; Sun, H.; Shao, M. Pathogens in patients with granulomatous lobular mastitis. Int. J. Infect. Dis. 2019, 81, 123–127. [Google Scholar] [CrossRef]
- Yu, H.J.; Deng, H.; Ma, J.; Huang, S.J.; Yang, J.M.; Huang, Y.F.; Mu, X.P.; Zhang, L.; Wang, Q. Clinical metagenomic analysis of bacterial communities in breast abscesses of granulomatous mastitis. Int. J. Infect. Dis. 2016, 53, 30–33. [Google Scholar] [CrossRef]
- Boakes, E.; Woods, A.; Johnson, N.; Kadoglou, N. Breast Infection: A Review of Diagnosis and Management Practices. Eur. J. Breast Health 2018, 14, 136. [Google Scholar] [CrossRef]
- Liu, L.; Zhou, F.; Wang, P.; Yu, L.; Ma, Z.; Li, Y.; Gao, D.; Zhang, Q.; Li, L.; Yu, Z. Periductal Mastitis: An Inflammatory Disease Related to Bacterial Infection and Consequent Immune Responses? Mediat. Inflamm. 2017, 2017, 5309081. [Google Scholar] [CrossRef]
- Moazzez, A. Breast Abscess Bacteriologic Features in the Era of Community-Acquired Methicillin-Resistant Staphylococcus aureus Epidemics. Arch. Surg. 2007, 142, 881. [Google Scholar] [CrossRef]
- Wu, D.C.; Chan, W.W.; Metelitsa, A.I.; Fiorillo, L.; Lin, A.N. Pseudomonas Skin Infection. Am. J. Clin. Dermatol. 2011, 12, 157–169. [Google Scholar] [CrossRef]
- Rudresh, S.M. Non Diphtheritic Corynebacteria: An Emerging Nosocomial Pathogen in Skin and Soft Tissue Infection. J. Clin. Diagn. Res. 2015, 9, DC19. [Google Scholar] [CrossRef]
- Torrens, C.; Marí, R.; Alier, A.; Puig, L.; Santana, F.; Corvec, S. Cutibacterium acnes in primary reverse shoulder arthroplasty: From skin to deep layers. J. Shoulder Elb. Surg. 2019, 28, 839–846. [Google Scholar] [CrossRef]
- Fazli, M.; Bjarnsholt, T.; Kirketerp-Møller, K.; Jørgensen, B.; Andersen, A.S.; Krogfelt, K.A.; Givskov, M.; Tolker-Nielsen, T. Nonrandom Distribution of Pseudomonas aeruginosa and Staphylococcus aureus in Chronic Wounds. J. Clin. Microbiol. 2009, 47, 4084–4089. [Google Scholar] [CrossRef]
- Stachon, H.; Amoroso, V.; Urban, C.; Bioni, P.; Spautz, C.; Lima, R.S.; Anselmi, K.; Kuroda, F.; Rabinovich, I.; Alvarez, T.; et al. Intraoperative Assessment of Endogenous Microbiota in the Breast. Rev. Bras. Ginecol. Obs. 2021, 43, 759–764. [Google Scholar] [CrossRef]
- Cook, J.; Holmes, C.J.; Wixtrom, R.; Newman, M.I.; Pozner, J.N. Characterizing the Microbiome of the Contracted Breast Capsule Using Next Generation Sequencing. Aesthetic Surg. J. 2021, 41, 440–447. [Google Scholar] [CrossRef]
- Carvajal, J.; Carvajal, M.; Hernández, G. Back to Basics: Could the Preoperative Skin Antiseptic Agent Help Prevent Biofilm-Related Capsular Contracture? Aesthetic Surg. J. 2019, 39, 848–859. [Google Scholar] [CrossRef]
- Bachour, Y.; Poort, L.; Verweij, S.P.; van Selms, G.; Winters, H.A.H.; Ritt, M.J.P.F.; Niessen, F.B.; Budding, A.E. PCR Characterization of Microbiota on Contracted and Non-Contracted Breast Capsules. Aesthetic Plast. Surg. 2019, 43, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Johani, K.; Almatroudi, A.; Vickery, K.; Van Natta, B.; Kadin, M.E.; Brody, G.; Clemens, M.; Cheah, C.Y.; Lade, S.; et al. Bacterial Biofilm Infection Detected in Breast Implant–Associated Anaplastic Large-Cell Lymphoma. Plast. Reconstr. Surg. 2016, 137, 1659–1669. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.N.; Walker, J.N.; Hanson, B.M.; Pinkner, C.L.; Simar, S.R.; Pinkner, J.S.; Parikh, R.; Clemens, M.W.; Hultgren, S.J.; Myckatyn, T.M. Insights into the Microbiome of Breast Implants and Periprosthetic Tissue in Breast Implant-Associated Anaplastic Large Cell Lymphoma. Sci. Rep. 2019, 9, 10393. [Google Scholar] [CrossRef] [PubMed]
- Charlot, M.; Béatrix, O.; Chateau, F.; Dubuisson, J.; Golfier, F.; Valette, P.J.; Réty, F. Pathologies of the male breast. Diagn. Interv. Imaging 2013, 94, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Firmin-Lefebvre, D.; Misery, L. Pathologie du sein de l’homme Male breast diseases. Ann. Dermatol. Venereol. 2013, 140, 436–443. [Google Scholar] [CrossRef] [PubMed]
- Chadha, J.; Nandi, D.; Atri, Y.; Nag, A. Significance of human microbiome in breast cancer: Tale of an invisible and an invincible. Semin. Cancer Biol. 2021, 70, 112–127. [Google Scholar] [CrossRef] [PubMed]
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).