From Transcription Factors Dysregulation to Malignancy: In Silico Reconstruction of Cancer’s Foundational Drivers—The Eternity Triangle
Abstract
1. Introduction
2. Results
2.1. TF Families Identified as Interactors in Oncogene Promoters
- E2F Family
- MYC Family
- KLF Family
- FOXO Family
- SP Family
- GATA Family
- STAT Family
- IRF Family
- NF-κB family
- SOX and HOX Family
- SMAD Family
- Sex hormone receptor families
- ETS Family
- ZNF Family
- PRDM Family
- Retinoic receptor Family
- EGR Family
- MEF2 Family
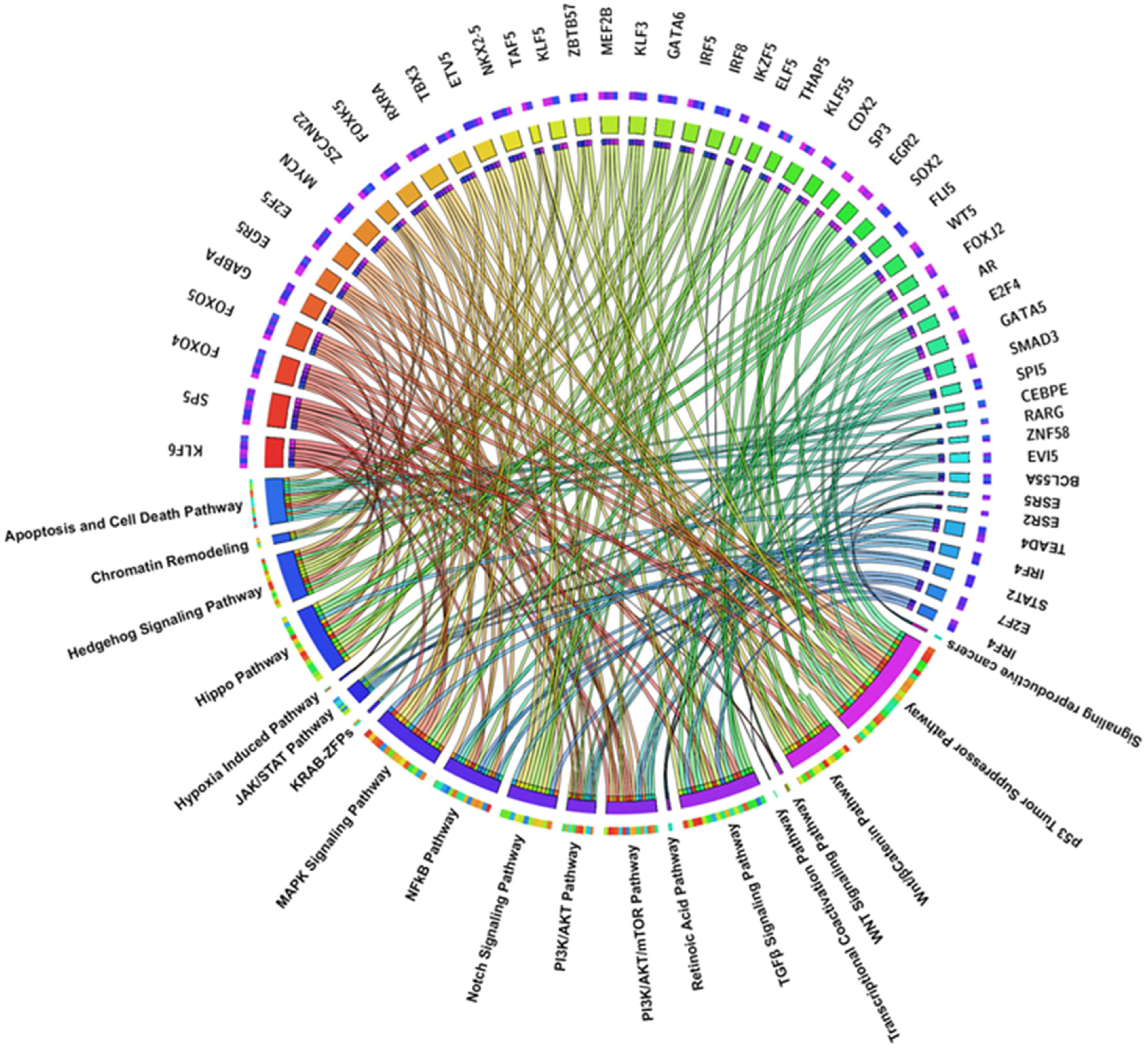
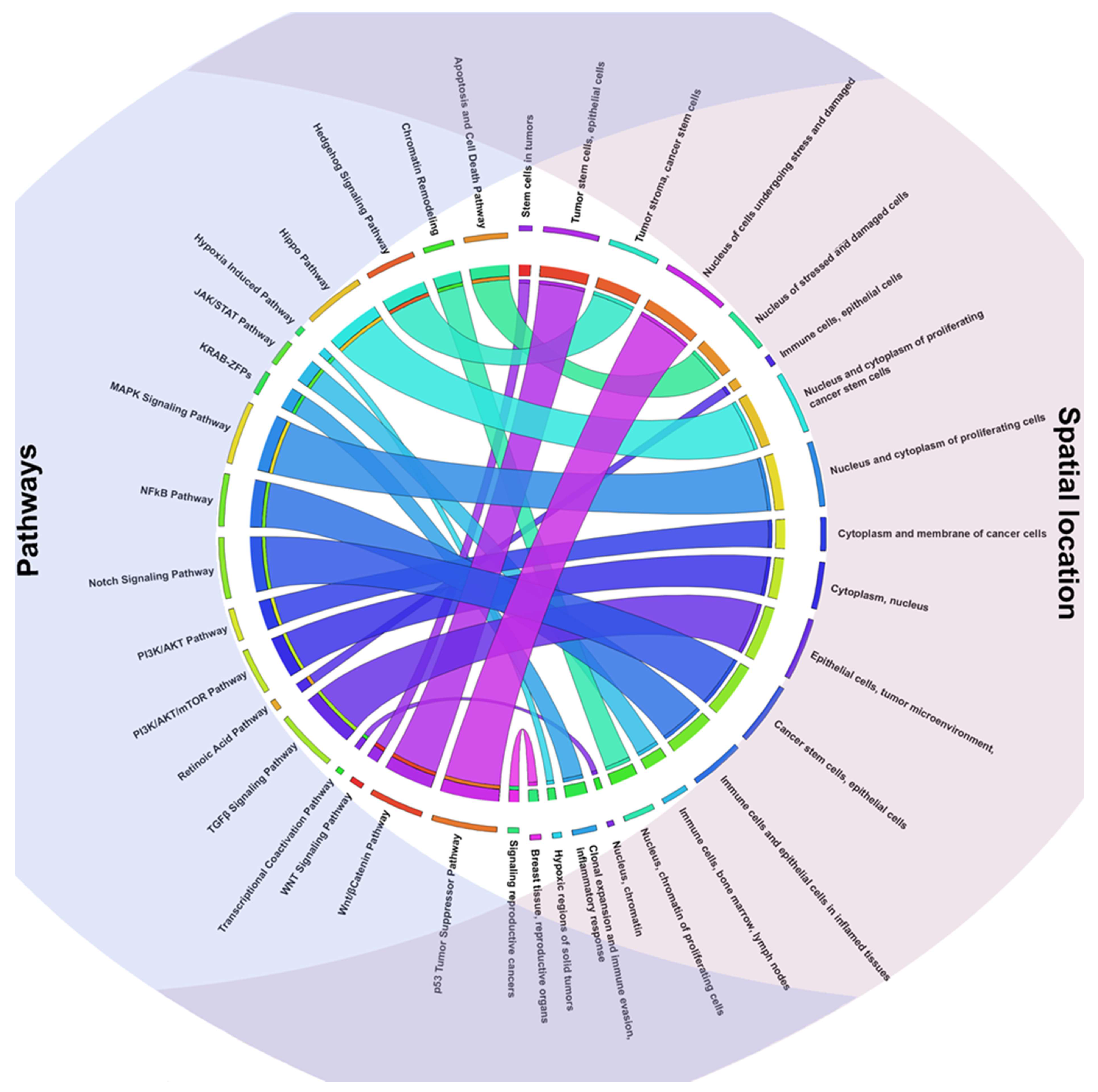
2.2. TFs Associated Signaling Pathways and Their Role in Tumorigenesis
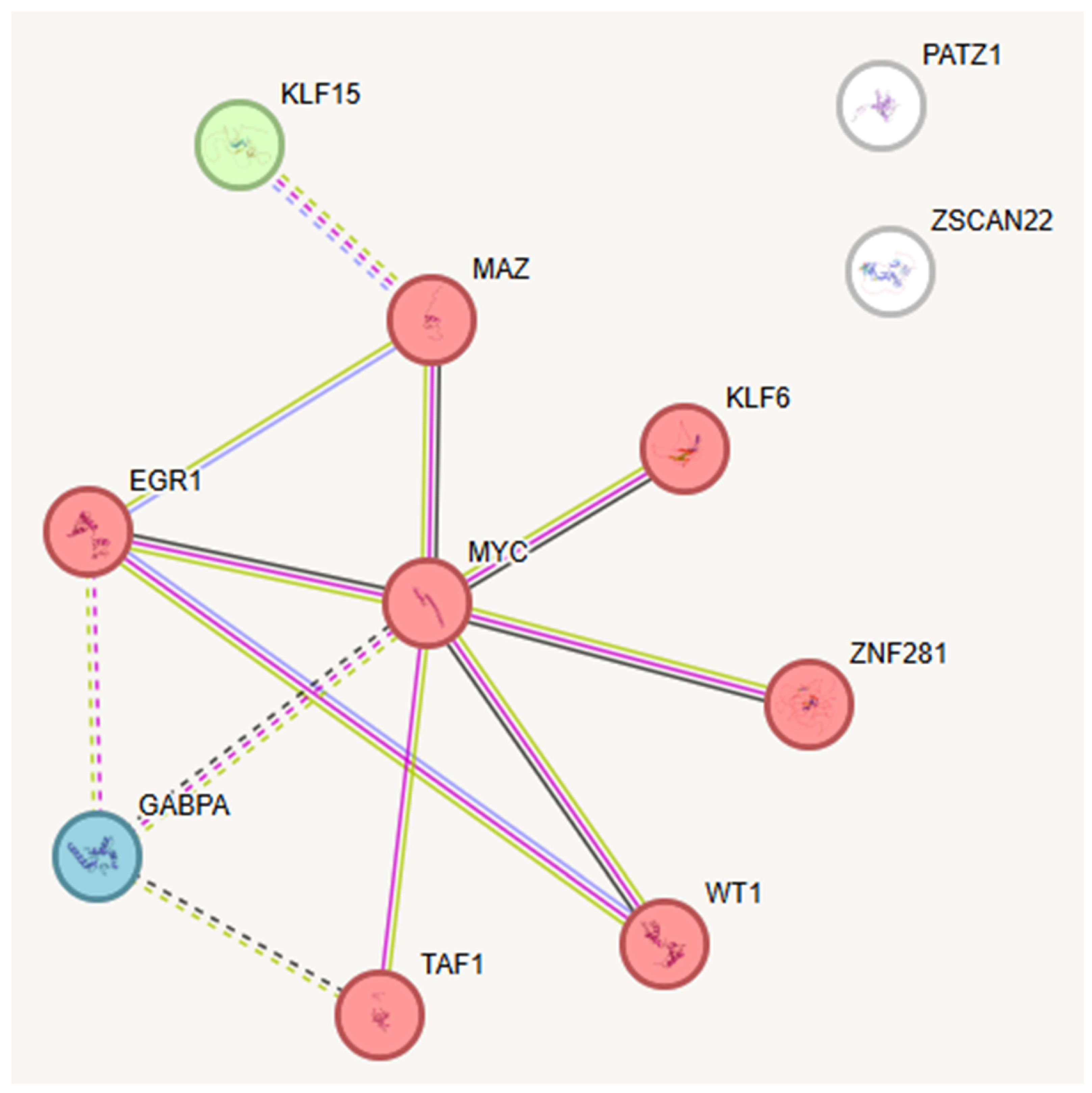
2.3. Key TFs as Regulatory Hubs in Oncogene Promoter Dysregulation
3. Discussion
4. Materials and Methods
4.1. Data Collection and Gene Promoter Analysis
4.2. Data Comparison and Reporting

5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ocsenas, O.; Reimand, J. Chromatin Accessibility of Primary Human Cancers Ties Regional Mutational Processes and Signatures with Tissues of Origin. PLoS Comput. Biol. 2022, 18, e1010393. [Google Scholar] [CrossRef]
- Saxena, S.; Zou, L. Hallmarks of DNA Replication Stress. Mol. Cell 2022, 82, 2298–2314. [Google Scholar] [CrossRef]
- Curti, L.; Campaner, S. MYC-Induced Replicative Stress: A Double-Edged Sword for Cancer Development and Treatment. Int. J. Mol. Sci. 2021, 22, 6168. [Google Scholar] [CrossRef] [PubMed]
- García-Gutiérrez, L.; Delgado, M.D.; León, J. MYC Oncogene Contributions to Release of Cell Cycle Brakes. Genes 2019, 10, 244. [Google Scholar] [CrossRef]
- Kim, U.; Shin, H.Y. Genomic Mutations of the STAT5 Transcription Factor Are Associated with Human Cancer and Immune Diseases. Int. J. Mol. Sci. 2022, 23, 11297. [Google Scholar] [CrossRef]
- Yang, H.; Lan, L. Transcription-coupled DNA Repair Protects Genome Stability upon Oxidative Stress-derived DNA Strand Breaks. FEBS Lett. 2024, 599, 168–176. [Google Scholar] [CrossRef]
- Zhu, W.; Zhang, Y.; Sahay, H.; Wasserman, H.; Afek, A.; Williams, J.; Shaltz, S.; Johnson, C.; Pinheiro, K.; MacAlpine, D.M.; et al. DNA Mutagenesis Driven by Transcription Factor Competition with Mismatch Repair. Cell 2025. Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Lindström, M.S.; Bartek, J.; Maya-Mendoza, A. P53 at the Crossroad of DNA Replication and Ribosome Biogenesis Stress Pathways. Cell Death Differ. 2022, 29, 972–982. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Dong, W.; Zhang, P.; Zhang, T.; Ma, L.; Qu, M.; Ma, X.; Zhou, X.; He, Q. Comprehensive Analysis of Regulatory Factors and Immune-Associated Patterns to Decipher Common and BRCA1/2 Mutation-Type-Specific Critical Regulation in Breast Cancer. Front. Cell Dev. Biol. 2021, 9, 750897. [Google Scholar] [CrossRef]
- Pellarin, I.; Dall’Acqua, A.; Favero, A.; Segatto, I.; Rossi, V.; Crestan, N.; Karimbayli, J.; Belletti, B.; Baldassarre, G. Cyclin-Dependent Protein Kinases and Cell Cycle Regulation in Biology and Disease. Signal Transduct. Target. Ther. 2025, 10, 11. [Google Scholar] [CrossRef]
- Fouad, S.; Hauton, D.; D’Angiolella, V. E2F1: Cause and Consequence of DNA Replication Stress. Front. Mol. Biosci. 2021, 7, 599332. [Google Scholar] [CrossRef]
- Vélez-Cruz, R.; Johnson, D. The Retinoblastoma (RB) Tumor Suppressor: Pushing Back against Genome Instability on Multiple Fronts. Int. J. Mol. Sci. 2017, 18, 1776. [Google Scholar] [CrossRef]
- Peterson, A.F.; Ingram, K.; Huang, E.J.; Parksong, J.; McKenney, C.; Bever, G.S.; Regot, S. Systematic Analysis of the MAPK Signaling Network Reveals MAP3K-Driven Control of Cell Fate. Cell Syst. 2022, 13, 885–894.e4. [Google Scholar] [CrossRef]
- Fernández-Medarde, A.; De Las Rivas, J.; Santos, E. 40 Years of RAS—A Historic Overview. Genes 2021, 12, 681. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, F.S.; Ahmadi, A.; Kesharwani, P.; Hosseini, H.; Sahebkar, A. Regulatory Effects of Statins on Akt Signaling for Prevention of Cancers. Cell Signal. 2024, 120, 111213. [Google Scholar] [CrossRef] [PubMed]
- Niehrs, C.; Luke, B. Regulatory R-Loops as Facilitators of Gene Expression and Genome Stability. Nat. Rev. Mol. Cell Biol. 2020, 21, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Kant, R.; Manne, R.K.; Anas, M.; Penugurti, V.; Chen, T.; Pan, B.-S.; Hsu, C.-C.; Lin, H.-K. Deregulated Transcription Factors in Cancer Cell Metabolisms and Reprogramming. Semin. Cancer Biol. 2022, 86, 1158–1174. [Google Scholar] [CrossRef]
- Alhmoud, J.F.; Woolley, J.F.; Al Moustafa, A.-E.; Malki, M.I. DNA Damage/Repair Management in Cancers. Cancers 2020, 12, 1050. [Google Scholar] [CrossRef]
- Chen, Y.; Liang, R.; Li, Y.; Jiang, L.; Ma, D.; Luo, Q.; Song, G. Chromatin Accessibility: Biological Functions, Molecular Mechanisms and Therapeutic Application. Signal Transduct. Target. Ther. 2024, 9, 340. [Google Scholar] [CrossRef]
- Cadet, J.; Wagner, J.R. DNA Base Damage by Reactive Oxygen Species, Oxidizing Agents, and UV Radiation. Cold Spring Harb. Perspect. Biol. 2013, 5, a012559. [Google Scholar] [CrossRef]
- Juul, R.I.; Nielsen, M.M.; Juul, M.; Feuerbach, L.; Pedersen, J.S. The landscape and driver potential of site-specific hotspots across cancer genomes. npj Genom. Med. 2021, 6, 33. [Google Scholar] [CrossRef]
- Makova, K.D.; Hardison, R.C. The Effects of Chromatin Organization on Variation in Mutation Rates in the Genome. Nat. Rev. Genet. 2015, 16, 213–223. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data With or Without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Selvam, K.; Sivapragasam, S.; Poon, G.M.K.; Wyrick, J.J. Detecting Recurrent Passenger Mutations in Melanoma by Targeted UV Damage Sequencing. Nat. Commun. 2023, 14, 2702. [Google Scholar] [CrossRef]
- Bailey, C.G.; Gupta, S.; Metierre, C.; Amarasekera, P.M.S.; O’Young, P.; Kyaw, W.; Laletin, T.; Francis, H.; Semaan, C.; Sharifi Tabar, M.; et al. Structure–Function Relationships Explain CTCF Zinc Finger Mutation Phenotypes in Cancer. Cell Mol. Life Sci. 2021, 78, 7519–7536. [Google Scholar] [CrossRef] [PubMed]
- Dehingia, B.; Milewska, M.; Janowski, M.; Pękowska, A. CTCF Shapes Chromatin Structure and Gene Expression in Health and Disease. EMBO Rep. 2022, 23, e55146. [Google Scholar] [CrossRef]
- Gandini, S.; Zanna, I.; De Angelis, S.; Palli, D.; Raimondi, S.; Ribero, S.; Masala, G.; Suppa, M.; Bellerba, F.; Corso, F.; et al. TERT Promoter Mutations and Melanoma Survival: A Comprehensive Literature Review and Meta-Analysis. Crit. Rev. Oncol. Hematol. 2021, 160, 103288. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Larsson, C.; Xu, D. Mechanisms Underlying the Activation of TERT Transcription and Telomerase Activity in Human Cancer: Old Actors and New Players. Oncogene 2019, 38, 6172–6183. [Google Scholar] [CrossRef]
- Gupta, S.; Vanderbilt, C.M.; Lin, Y.-T.; Benhamida, J.K.; Jungbluth, A.A.; Rana, S.; Momeni-Boroujeni, A.; Chang, J.C.; Mcfarlane, T.; Salazar, P.; et al. A Pan-Cancer Study of Somatic TERT Promoter Mutations and Amplification in 30,773 Tumors Profiled by Clinical Genomic Sequencing. J. Mol. Diagn. 2021, 23, 253–263. [Google Scholar] [CrossRef]
- Gundem, G.; Perez-Llamas, C.; Jene-Sanz, A.; Kedzierska, A.; Islam, A.; Deu-Pons, J.; Furney, S.J.; Lopez-Bigas, N. IntOGen: Integration and Data Mining of Multidimensional Oncogenomic Data. Nat. Methods 2010, 7, 92–93. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-Z.; Tsai, S.-Y.; Leone, G. Emerging Roles of E2Fs in Cancer: An Exit from Cell Cycle Control. Nat. Rev. Cancer 2009, 9, 785–797. [Google Scholar] [CrossRef]
- Bartolucci, D.; Montemurro, L.; Raieli, S.; Lampis, S.; Pession, A.; Hrelia, P.; Tonelli, R. MYCN Impact on High-Risk Neuroblastoma: From Diagnosis and Prognosis to Targeted Treatment. Cancers 2022, 14, 4421. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Dong, T.; Wu, Z.; Zhu, D.; Gu, H. The Effects of MYC on Tumor Immunity and Immunotherapy. Cell Death Discov. 2023, 9, 103. [Google Scholar] [CrossRef]
- Zhang, Y.; Yao, C.; Ju, Z.; Jiao, D.; Hu, D.; Qi, L.; Liu, S.; Wu, X.; Zhao, C. Krüppel-like Factors in Tumors: Key Regulators and Therapeutic Avenues. Front. Oncol. 2023, 13, 1080720. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Zheng, Q.-K.; Ma, R.-J.; Ma, C.; Sun, Z.-G.; Zhang, N. Krüppel-Like Factor 6 Splice Variant 1: An Oncogenic Transcription Factor Involved in the Progression of Multiple Malignant Tumors. Front. Cell Dev. Biol. 2021, 9, 661731. [Google Scholar] [CrossRef]
- McConnell, B.B.; Yang, V.W. Mammalian Krüppel-Like Factors in Health and Diseases. Physiol. Rev. 2010, 90, 1337–1381. [Google Scholar] [CrossRef]
- Brown, A.K.; Webb, A.E. Regulation of FOXO Factors in Mammalian Cells. In Current Topics in Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 165–192. [Google Scholar]
- Jiramongkol, Y.; Lam, E.W.-F. FOXO Transcription Factor Family in Cancer and Metastasis. Cancer Metastasis Rev. 2020, 39, 681–709. [Google Scholar] [CrossRef]
- Mei, W.; Mei, B.; Chang, J.; Liu, Y.; Zhou, Y.; Zhu, N.; Hu, M. Role and Regulation of FOXO3a: New Insights into Breast Cancer Therapy. Front. Pharmacol. 2024, 15, 1346745. [Google Scholar] [CrossRef]
- Safe, S.; Abbruzzese, J.; Abdelrahim, M.; Hedrick, E. Specificity Protein Transcription Factors and Cancer: Opportunities for Drug Development. Cancer Prev. Res. 2018, 11, 371–382. [Google Scholar] [CrossRef]
- Gao, Y.; Gan, K.; Liu, K.; Xu, B.; Chen, M. SP1 Expression and the Clinicopathological Features of Tumors: A Meta-Analysis and Bioinformatics Analysis. Pathol. Oncol. Res. 2021, 27, 581998. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Blobel, G.A. GATA Transcription Factors and Cancer. Genes Cancer 2010, 1, 1178–1188. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, W.; Huang, Y.; Luo, S.; Tang, X.; Yi, Q. Association of GATA3 Expression in Triple-Positive Breast Cancer with Overall Survival and Immune Cell Infiltration. Sci. Rep. 2024, 14, 17795. [Google Scholar] [CrossRef]
- Arnal-Estapé, A.; Cai, W.L.; Albert, A.E.; Zhao, M.; Stevens, L.E.; López-Giráldez, F.; Patel, K.D.; Tyagi, S.; Schmitt, E.M.; Westbrook, T.F.; et al. Tumor Progression and Chromatin Landscape of Lung Cancer Are Regulated by the Lineage Factor GATA6. Oncogene 2020, 39, 3726–3737. [Google Scholar] [CrossRef]
- Alikarami, F.; Xie, H.M.; Riedel, S.S.; Goodrow, H.T.; Barrett, D.R.; Mahdavi, L.; Lenard, A.; Chen, C.; Yamauchi, T.; Danis, E.; et al. GATA2 Links Stemness to Chemotherapy Resistance in Acute Myeloid Leukemia. Blood 2025, 145, 2179–2195. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Pardoll, D.; Jove, R. STATs in Cancer Inflammation and Immunity: A Leading Role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef] [PubMed]
- Witalisz-Siepracka, A.; Klein, K.; Zdársky, B.; Stoiber, D. The Multifaceted Role of STAT3 in NK-Cell Tumor Surveillance. Front. Immunol. 2022, 13, 947568. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, Y.; Zhang, N.; Xian, Y.; Tang, Y.; Ye, J.; Reza, F.; He, G.; Wen, X.; Jiang, X. The Multiple Roles of Interferon Regulatory Factor Family in Health and Disease. Signal Transduct. Target. Ther. 2024, 9, 282. [Google Scholar] [CrossRef] [PubMed]
- Savitsky, D.; Tamura, T.; Yanai, H.; Taniguchi, T. Regulation of Immunity and Oncogenesis by the IRF Transcription Factor Family. Cancer Immunol. Immunother. 2010, 59, 489–510. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhong, Z.; Karin, M. NF-κB: A Double-Edged Sword Controlling Inflammation. Biomedicines 2022, 10, 1250. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-κB Signaling in Inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Mao, H.; Zhao, X.; Sun, S. NF-κB in Inflammation and Cancer. Cell Mol. Immunol. 2025, 22, 811–839. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Jin, Y.; Chen, X.; Ye, X.; Shen, X.; Lin, M.; Zeng, C.; Zhou, T.; Zhang, J. NF-κB in Biology and Targeted Therapy: New Insights and Translational Implications. Signal Transduct. Target. Ther. 2024, 9, 53. [Google Scholar] [CrossRef]
- Zhang, S.; Xiong, X.; Sun, Y. Functional Characterization of SOX2 as an Anticancer Target. Signal Transduct. Target. Ther. 2020, 5, 135. [Google Scholar] [CrossRef]
- Shenoy, U.S.; Adiga, D.; Alhedyan, F.; Kabekkodu, S.P.; Radhakrishnan, R. HOXA9 Transcription Factor Is a Double-Edged Sword: From Development to Cancer Progression. Cancer Metastasis Rev. 2023, 43, 709–728. [Google Scholar] [CrossRef]
- Mamun, M.A.; Mannoor, K.; Cao, J.; Qadri, F.; Song, X. SOX2 in Cancer Stemness: Tumor Malignancy and Therapeutic Potentials. J. Mol. Cell Biol. 2018, 12, 85–98. [Google Scholar] [CrossRef]
- Shenoy, U.S.; Basavarajappa, D.S.; Kabekkodu, S.P.; Radhakrishnan, R. Pan-cancer exploration of oncogenic and clinical impacts revealed that HOXA9 is a diagnostic indicator of tumorigenesis. Clin. Exp. Med. 2024, 24, 134. [Google Scholar] [CrossRef]
- Deng, Z.; Fan, T.; Xiao, C.; Tian, H.; Zheng, Y.; Li, C.; He, J. TGF-β Signaling in Health, Disease and Therapeutics. Signal Transduct. Target. Ther. 2024, 9, 61. [Google Scholar] [CrossRef]
- Principe, D.R.; Underwood, P.W.; Kumar, S.; Timbers, K.E.; Koch, R.M.; Trevino, J.G.; Munshi, H.G.; Rana, A. Loss of SMAD4 Is Associated With Poor Tumor Immunogenicity and Reduced PD-L1 Expression in Pancreatic Cancer. Front. Oncol. 2022, 12, 806963. [Google Scholar] [CrossRef] [PubMed]
- Boye, A.; Osei, S.A.; Brah, A.S. Therapeutic Prospects of Sex Hormone Receptor Signaling in Hormone-Responsive Cancers. Biomed. Pharmacother. 2024, 180, 117473. [Google Scholar] [CrossRef]
- Li, C.; Cheng, D.; Li, P. Androgen Receptor Dynamics in Prostate Cancer: From Disease Progression to Treatment Resistance. Front. Oncol. 2025, 15, 1542811. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.T.; Gou, X.; Seker, S.; Ellis, M.J. ESR1 Alterations and Metastasis in Estrogen Receptor Positive Breast Cancer. J. Cancer Metastasis Treat. 2019, 5, 38. [Google Scholar] [CrossRef]
- A Fry, E.; Inoue, K. Aberrant Expression of ETS1 and ETS2 Proteins in Cancer. Cancer Rep. Rev. 2018, 2, 1000151. [Google Scholar] [CrossRef]
- Wang, S.; Wan, L.; Zhang, X.; Fang, H.; Zhang, M.; Li, F.; Yan, D. ETS-1 in Tumor Immunology: Implications for Novel Anti-Cancer Strategies. Front. Immunol. 2025, 16, 1526368. [Google Scholar] [CrossRef]
- Zhao, J.; Wen, D.; Zhang, S.; Jiang, H.; Di, X. The Role of Zinc Finger Proteins in Malignant Tumors. FASEB J. 2023, 37, e23157. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Xu, G.; Hong, H.; Zhang, J.; Cui, Z.; Yu, Z. The Zinc Finger Protein560(ZNF560) Functions as a Novel Oncogenic Gene in Osteosarcoma. Sci. Rep. 2025, 15, 79. [Google Scholar] [CrossRef]
- Casamassimi, A.; Rienzo, M.; Di Zazzo, E.; Sorrentino, A.; Fiore, D.; Proto, M.C.; Moncharmont, B.; Gazzerro, P.; Bifulco, M.; Abbondanza, C. Multifaceted Role of PRDM Proteins in Human Cancer. Int. J. Mol. Sci. 2020, 21, 2648. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, L.; You, W.; Xu, J.; Dai, J.; Hua, D.; Zhang, R.; Yao, F.; Zhou, S.; Huang, W.; et al. PRDM1/BLIMP1 Induces Cancer Immune Evasion by Modulating the USP22-SPI1-PD-L1 Axis in Hepatocellular Carcinoma Cells. Nat. Commun. 2022, 13, 7677. [Google Scholar] [CrossRef]
- Schmidt, C.; Cohen, S.; Gudenas, B.L.; Husain, S.; Carlson, A.; Westelman, S.; Wang, L.; Phillips, J.J.; Northcott, P.A.; Weiss, W.A.; et al. PRDM6 Promotes Medulloblastoma by Repressing Chromatin Accessibility and Altering Gene Expression. Sci. Rep. 2024, 14, 16074. [Google Scholar] [CrossRef] [PubMed]
- Duong, V.; Rochette-Egly, C. The Molecular Physiology of Nuclear Retinoic Acid Receptors. Health Dis. Biochim. Biophys. Acta—Mol. Basis Dis. 2011, 1812, 1023–1031. [Google Scholar] [CrossRef]
- di Martino, O.; Welch, J.S. Retinoic Acid Receptors in Acute Myeloid Leukemia Therapy. Cancers 2019, 11, 1915. [Google Scholar] [CrossRef]
- Hunsu, V.O.; Facey, C.O.B.; Fields, J.Z.; Boman, B.M. Retinoids as Chemo-Preventive and Molecular-Targeted Anti-Cancer Therapies. Int. J. Mol. Sci. 2021, 22, 7731. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Wang, R.; Zhang, W.; Li, Y.; Wang, Y.; Wang, H.; Li, X.; Song, J. Multifaceted Regulatory Mechanisms of the EGR Family in Tumours and Prospects for Therapeutic Applications (Review). Int. J. Mol. Med. 2025, 56, 113. [Google Scholar] [CrossRef]
- Wang, B.; Guo, H.; Yu, H.; Chen, Y.; Xu, H.; Zhao, G. The Role of the Transcription Factor EGR1 in Cancer. Front. Oncol. 2021, 11, 642547. [Google Scholar] [CrossRef]
- Di Giorgio, E.; Hancock, W.W.; Brancolini, C. MEF2 and the Tumorigenic Process, Hic Sunt Leones. Biochim. Biophys. Acta—Rev. Cancer 2018, 1870, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Ou, W.-C.; Fang, L.; Tian, C.-W.; Xiong, Y. Myocyte Enhancer Factor 2A Plays a Central Role in the Regulatory Networks of Cellular Physiopathology. Aging Dis. 2022, 14, 331–349. [Google Scholar] [CrossRef]
- Zhang, M.; Truscott, J.; Davie, J. Loss of MEF2D Expression Inhibits Differentiation and Contributes to Oncogenesis in Rhabdomyosarcoma Cells. Mol. Cancer 2013, 12, 150. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.I.; Young, R.A. Transcriptional Regulation and Its Misregulation in Disease. Cell 2013, 152, 1237–1251. [Google Scholar] [CrossRef]
- Tufail, M.; Jiang, C.-H.; Li, N. Wnt Signaling in Cancer: From Biomarkers to Targeted Therapies and Clinical Translation. Mol. Cancer 2025, 24, 107. [Google Scholar] [CrossRef]
- Song, P.; Gao, Z.; Bao, Y.; Chen, L.; Huang, Y.; Liu, Y.; Dong, Q.; Wei, X. Wnt/β-Catenin Signaling Pathway in Carcinogenesis and Cancer Therapy. J. Hematol. Oncol. 2024, 17, 46. [Google Scholar] [CrossRef]
- Peri, S.S.; Narayanaa, Y.K.; Hubert, T.D.; Rajaraman, R.; Arfuso, F.; Sundaram, S.; Archana, B.; Warrier, S.; Dharmarajan, A.; Perumalsamy, L.R. Navigating Tumour Microenvironment and Wnt Signalling Crosstalk: Implications for Advanced Cancer Therapeutics. Cancers 2023, 15, 5847. [Google Scholar] [CrossRef]
- Williams, A.B.; Schumacher, B. P53 in the DNA-Damage-Repair Process. Cold Spring Harb. Perspect. Med. 2016, 6, a026070. [Google Scholar] [CrossRef]
- Song, B.; Yang, P.; Zhang, S. Cell Fate Regulation Governed by P53: Friends or Reversible Foes in Cancer Therapy. Cancer Commun. 2024, 44, 297–360. [Google Scholar] [CrossRef]
- Dhanasekaran, R.; Deutzmann, A.; Mahauad-Fernandez, W.D.; Hansen, A.S.; Gouw, A.M.; Felsher, D.W. The MYC Oncogene —The Grand Orchestrator of Cancer Growth and Immune Evasion. Nat. Rev. Clin. Oncol. 2021, 19, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Doha, Z.O.; Sears, R.C. Unraveling MYC’s Role in Orchestrating Tumor Intrinsic and Tumor Microenvironment Interactions Driving Tumorigenesis and Drug Resistance. Pathophysiology 2023, 30, 400–419. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Du, Y.; Nie, R.; Wang, S.; Wang, H.; Li, P. Notch Signaling in Cancers: Mechanism and Potential Therapy. Front. Cell Dev. Biol. 2025, 13, 1542967. [Google Scholar] [CrossRef]
- Trindade, A.; Duarte, A. Notch Signaling Function in the Angiocrine Regulation of Tumor Development. Cells 2020, 9, 2467. [Google Scholar] [CrossRef]
- Zhang, T.; Ma, C.; Zhang, Z.; Zhang, H.; Hu, H. NF-κB Signaling in Inflammation and Cancer. MedComm 2021, 2, 618–653. [Google Scholar] [CrossRef]
- Lu, H.; Ouyang, W.; Huang, C. Inflammation, a key event in cancer development. Mol. Cancer Res. 2006, 4, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Fu, V.; Plouffe, S.W.; Guan, K.-L. The Hippo Pathway in Organ Development, Homeostasis, and Regeneration. Curr. Opin. Cell Biol. 2017, 49, 99–107. [Google Scholar] [CrossRef]
- Calses, P.C.; Crawford, J.J.; Lill, J.R.; Dey, A. Hippo Pathway in Cancer: Aberrant Regulation and Therapeutic Opportunities. Trends Cancer 2019, 5, 297–307. [Google Scholar] [CrossRef]
- Glaviano, A.; Foo, A.S.C.; Lam, H.Y.; Yap, K.C.H.; Jacot, W.; Jones, R.H.; Eng, H.; Nair, M.G.; Makvandi, P.; Geoerger, B.; et al. PI3K/AKT/mTOR Signaling Transduction Pathway and Targeted Therapies in Cancer. Mol. Cancer 2023, 22, 138. [Google Scholar] [CrossRef]
- Moghbeli, M. PI3K/AKT pathway as a pivotal regulator of epithelial-mesenchymal transition in lung tumor cells. Cancer Cell Int. 2024, 24, 165. [Google Scholar] [CrossRef]
- Yu, L.; Wei, J.; Liu, P. Attacking the PI3K/Akt/mTOR Signaling Pathway for Targeted Therapeutic Treatment in Human Cancer. Semin. Cancer Biol. 2022, 85, 69–94. [Google Scholar] [CrossRef]
- Bahar, M.E.; Kim, H.J.; Kim, D.R. Targeting the RAS/RAF/MAPK Pathway for Cancer Therapy: From Mechanism to Clinical Studies. Signal Transduct. Target. Ther. 2023, 8, 455. [Google Scholar] [CrossRef]
- Liu, S.; Ren, J.; ten Dijke, P. Targeting TGFβ Signal Transduction for Cancer Therapy. Signal Transduct. Target. Ther. 2021, 6, 8. [Google Scholar] [CrossRef]
- Xu, J.; Lamouille, S.; Derynck, R. TGF-β-Induced Epithelial to Mesenchymal Transition. Cell Res. 2009, 19, 156–172. [Google Scholar] [CrossRef] [PubMed]
- Pickup, M.; Novitskiy, S.; Moses, H.L. The Roles of TGFβ in the Tumour Microenvironment. Nat. Rev. Cancer 2013, 13, 788–799. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Liao, C.; Zhang, Q. Hypoxia-Driven Effects in Cancer: Characterization, Mechanisms, and Therapeutic Implications. Cells 2021, 10, 678. [Google Scholar] [CrossRef] [PubMed]
- Shaikat, A.H.; Azad, S.M.A.K.; Tamim, M.A.R.; Ullah, M.S.; Amin, M.N.; Sabbir, M.K.; Tarun, M.T.I.; Mostaq, M.S.; Sabrin, S.; Mahmud, M.Z.; et al. Investigating Hypoxia-Inducible Factor Signaling in Cancer: Mechanisms, Clinical Implications, Targeted Therapeutic Strategies, and Resistance. Cancer Pathog. Ther. 2025. Online ahead of print. [Google Scholar] [CrossRef]
- Qian, S.; Wei, Z.; Yang, W.; Huang, J.; Yang, Y.; Wang, J. The role of BCL-2 family proteins in regulating apoptosis and cancer therapy. Front. Oncol. 2022, 12, 985363. [Google Scholar] [CrossRef]
- Xu, X.-C. Tumor-Suppressive Activity of Retinoic Acid Receptor-β in Cancer. Cancer Lett. 2007, 253, 14–24. [Google Scholar] [CrossRef]
- Chen, Y.; Tong, X.; Lu, R.; Zhang, Z.; Ma, T. All-Trans Retinoic Acid in Hematologic Disorders: Not Just Acute Promyelocytic Leukemia. Front. Pharmacol. 2024, 15, 1404092. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Kim, H.; Baek, S.H. Unraveling the Physiological Roles of Retinoic Acid Receptor-Related Orphan Receptor α. Exp. Mol. Med. 2021, 53, 1278–1286. [Google Scholar] [CrossRef]
- Chen, P.; Li, B.; Ou-Yang, L. Role of Estrogen Receptors in Health and Disease. Front. Endocrinol. 2022, 13, 839005. [Google Scholar] [CrossRef]
- Han, R.; Gu, S.; Zhang, Y.; Luo, A.; Jing, X.; Zhao, L.; Zhao, X.; Zhang, L. Estrogen Promotes Progression of Hormone-Dependent Breast Cancer through CCL2-CCR2 Axis by Upregulation of Twist via PI3K/AKT/NF-κB Signaling. Sci. Rep. 2018, 8, 9575. [Google Scholar] [CrossRef] [PubMed]
- Cong, G.; Zhu, X.; Chen, X.R.; Chen, H.; Chong, W. Mechanisms and Therapeutic Potential of the Hedgehog Signaling Pathway in Cancer. Cell Death Discov. 2025, 11, 40. [Google Scholar] [CrossRef] [PubMed]
- Sari, I.N.; Phi, L.T.H.; Jun, N.; Wijaya, Y.T.; Lee, S.; Kwon, H.Y. Hedgehog Signaling in Cancer: A Prospective Therapeutic Target for Eradicating Cancer Stem Cells. Cells 2018, 7, 208. [Google Scholar] [CrossRef]
- Xue, C.; Yao, Q.; Gu, X.; Shi, Q.; Yuan, X.; Chu, Q.; Bao, Z.; Lu, J.; Li, L. Evolving Cognition of the JAK-STAT Signaling Pathway: Autoimmune Disorders and Cancer. Signal Transduct. Target. Ther. 2023, 8, 204. [Google Scholar] [CrossRef]
- Sabaawy, H.E.; Ryan, B.M.; Khiabanian, H.; Pine, S.R. JAK/STAT of All Trades: Linking Inflammation with Cancer Development, Tumor Progression and Therapy Resistance. Carcinogenesis 2021, 42, 1411–1419. [Google Scholar] [CrossRef]
- Gopinathan, G.; Diekwisch, T.G.H. Epigenetics and Early Development. J. Dev. Biol. 2022, 10, 26. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, S.; Zhang, X.; Du, Y.; Ni, T.; Hao, S. Crosstalk between Metabolic and Epigenetic Modifications during Cell Carcinogenesis. iScience 2024, 27, 111359. [Google Scholar] [CrossRef] [PubMed]
- Spiegelman, B.M.; Heinrich, R. Biological Control through Regulated Transcriptional Coactivators. Cell 2004, 119, 157–167. [Google Scholar] [CrossRef]
- Sobocińska, J.; Molenda, S.; Machnik, M.; Oleksiewicz, U. KRAB-ZFP Transcriptional Regulators Acting as Oncogenes and Tumor Suppressors: An Overview. Int. J. Mol. Sci. 2021, 22, 2212. [Google Scholar] [CrossRef] [PubMed]
- Martins, F.; Rosspopoff, O.; Carlevaro-Fita, J.; Forey, R.; Offner, S.; Planet, E.; Pulver, C.; Pak, H.; Huber, F.; Michaux, J.; et al. A Cluster of Evolutionarily Recent KRAB Zinc Finger Proteins Protects Cancer Cells from Replicative Stress–Induced Inflammation. Cancer Res. 2024, 84, 808–826. [Google Scholar] [CrossRef] [PubMed]
- Crombie, E.M.; Cleverley, K.; Timmers, H.T.M.; Fisher, E.M.C. The Roles of TAF1 in Neuroscience and Beyond. R. Soc. Open Sci. 2024, 11, 240790. [Google Scholar] [CrossRef]
- Li, H.-H.; Li, A.G.; Sheppard, H.M.; Liu, X. Phosphorylation on Thr-55 by TAF1 Mediates Degradation of P53. Mol. Cell 2004, 13, 867–878. [Google Scholar] [CrossRef]
- Zhang, J.; Li, R.; Zhang, B.; Cui, X. TAF1 Promotes NSCLC Cell Epithelial-Mesenchymal Transition by Transcriptionally Activating TGFβ1. Biochem. Biophys. Res. Commun. 2022, 636, 113–118. [Google Scholar] [CrossRef]
- Tavassoli, P.; Wafa, L.A.; Cheng, H.; Zoubeidi, A.; Fazli, L.; Gleave, M.; Snoek, R.; Rennie, P.S. TAF1 Differentially Enhances Androgen Receptor Transcriptional Activity via Its N-Terminal Kinase and Ubiquitin-Activating and -Conjugating Domains. Mol. Endocrinol. 2010, 24, 696–708. [Google Scholar] [CrossRef]
- Sánchez-Ramos, J.; Pozo-Molina, G.; Garrido, E. The Expression of the Transcription Factor TAF1 Is Modified by the HPV16 E2 Protein. Acta Virol. 2020, 64, 375–379. [Google Scholar] [CrossRef]
- Zhou, L.; Yao, Q.; Ma, L.; Li, H.; Chen, J. TAF1 Inhibitor Bay-299 Induces Cell Death in Acute Myeloid Leukemia. Transl. Cancer Res. 2021, 10, 5307–5318. [Google Scholar] [CrossRef]
- Wan, C.; Keany, M.P.; Dong, H.; Al-Alem, L.F.; Pandya, U.M.; Lazo, S.; Boehnke, K.; Lynch, K.N.; Xu, R.; Zarrella, D.T.; et al. Enhanced Efficacy of Simultaneous PD-1 and PD-L1 Immune Checkpoint Blockade in High-Grade Serous Ovarian Cancer. Cancer Res. 2021, 81, 158–173. [Google Scholar] [CrossRef] [PubMed]
- Nian, Q.; Lin, Y.; Zeng, J.; Zhang, Y.; Liu, R. Multifaceted Functions of the Wilms Tumor 1 Protein: From Its Expression in Various Malignancies to Targeted Therapy. Transl. Oncol. 2025, 52, 102237. [Google Scholar] [CrossRef]
- Syafruddin, S.E.; Mohtar, M.A.; Wan Mohamad Nazarie, W.F.; Low, T.Y. Two Sides of the Same Coin: The Roles of KLF6 in Physiology and Pathophysiology. Biomolecules 2020, 10, 1378. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Li, J.; Zhang, G.; Fang, H.; Du, Y.; Liang, Y. KLF15 Transcriptionally Activates LINC00689 to Inhibit Colorectal Cancer Development. Commun. Biol. 2024, 7, 130. [Google Scholar] [CrossRef]
- Hahn, S.; Hermeking, H. ZNF281/ZBP-99: A New Player in Epithelial–Mesenchymal Transition, Stemness, and Cancer. J. Mol. Med. 2014, 92, 571–581. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhang, C.; Zhang, X.; Wang, S.; Guo, T.; Yin, Y.; Zhang, H.; Li, Z.; Si, Y.; Lu, Y.; et al. ZNF281 Inhibits Mitochondrial Biogenesis to Facilitate Metastasis of Hepatocellular Carcinoma. Cell Death Discov. 2023, 9, 396. [Google Scholar] [CrossRef]
- Hahn, S.; Jackstadt, R.; Siemens, H.; Hünten, S.; Hermeking, H. SNAIL and miR-34a Feed-Forward Regulation of ZNF281/ZBP99 Promotes Epithelial-Mesenchymal Transition. EMBO J. 2013, 32, 3079–3095. [Google Scholar] [CrossRef]
- Adamson, E.D.; Mercola, D. Egr1 transcription factor: Multiple roles in prostate tumor cell growth and survival. Tumor Biol. 2002, 23, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ameri, A.H.; Wang, S.; Jansson, K.H.; Casey, O.M.; Yang, Q.; Beshiri, M.L.; Fang, L.; Lake, R.G.; Agarwal, S.; et al. EGR1 Regulates Angiogenic and Osteoclastogenic Factors in Prostate Cancer and Promotes Metastasis. Oncogene 2019, 38, 6241–6255. [Google Scholar] [CrossRef]
- Ma, Z.; Gao, X.; Shuai, Y.; Wu, X.; Yan, Y.; Xing, X.; Ji, J. EGR1-mediated Linc01503 Promotes Cell Cycle Progression and Tumorigenesis in Gastric Cancer. Cell Prolif. 2020, 54, 12922. [Google Scholar] [CrossRef] [PubMed]
- Baron, V.; Adamson, E.D.; Calogero, A.; Ragona, G.; Mercola, D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFbeta1, PTEN, p53, and fibronectin. Cancer Gene Ther. 2006, 13, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Calogero, A.; Arcella, A.; De Gregorio, G.; Porcellini, A.; Mercola, D.; Liu, C.; Lombari, V.; Zani, M.; Giannini, G.; Gagliardi, F.M.; et al. The early growth response gene EGR-1 behaves as a suppressor gene that is down-regulated independent of ARF/Mdm2 but not p53 alterations in fresh human gliomas. Clin. Cancer Res. 2001, 7, 2788–2796. [Google Scholar]
- Ma, H.; Ow, J.R.; Tan, B.C.P.; Goh, Z.; Feng, B.; Loh, Y.H.; Fedele, M.; Li, H.; Wu, Q. The Dosage of Patz1 Modulates Reprogramming Process. Sci. Rep. 2014, 4, 7519. [Google Scholar] [CrossRef]
- Tian, X.; Sun, D.; Zhang, Y.; Zhao, S.; Xiong, H.; Fang, J. Zinc Finger Protein 278, a Potential Oncogene in Human Colorectal Cancer. Acta Biochim. Biophys. Sin. 2008, 40, 289–296. [Google Scholar] [CrossRef]
- Yang, W.-L.; Ravatn, R.; Kudoh, K.; Alabanza, L.; Chin, K.-V. Interaction of the Regulatory Subunit of the cAMP-Dependent Protein Kinase with PATZ1 (ZNF278). Biochem. Biophys. Res. Commun. 2010, 391, 1318–1323. [Google Scholar] [CrossRef]
- Fedele, M.; Franco, R.; Salvatore, G.; Paronetto, M.; Barbagallo, F.; Pero, R.; Chiariotti, L.; Sette, C.; Tramontano, D.; Chieffi, G.; et al. PATZ1 Gene Has a Critical Role in the Spermatogenesis and Testicular Tumours. J. Pathol. 2008, 215, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Hatzi, K.; Melnick, A. Breaking bad in the germinal center: How deregulation of BCL6 contributes to lymphomagenesis. Trends Mol. Med. 2014, 20, 343–352. [Google Scholar] [CrossRef]
- Zheng, C.; Wu, H.; Jin, S.; Li, D.; Tan, S.; Zhu, X. Roles of Myc-associated Zinc Finger Protein in Malignant Tumors. Asia-Pac. J. Clin. Oncol. 2022, 18, 506–514. [Google Scholar] [CrossRef]
- Triner, D.; Castillo, C.; Hakim, J.B.; Xue, X.; Greenson, J.K.; Nuñez, G.; Chen, G.Y.; Colacino, J.A.; Shah, Y.M. Myc-Associated Zinc Finger Protein Regulates the Proinflammatory Response in Colitis and Colon Cancer via STAT3 Signaling. Mol. Cell Biol. 2018, 38, e00386-18. [Google Scholar] [CrossRef] [PubMed]
- Ni, G.; Sun, Y.; Jia, H.; Xiahou, Z.; Li, Y.; Zhao, F.; Zang, H. MAZ-Mediated Tumor Progression and Immune Evasion in Hormone Receptor-Positive Breast Cancer: Targeting Tumor Microenvironment and PCLAF+ Subtype-Specific Therapy. Transl. Oncol. 2025, 52, 102280. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Ni, Q.; Wu, N.; Xie, T.; Yun, F.; Zhang, X.; Gao, L.; Gai, Y.; Li, E.; Yi, X.; et al. Molecular Mechanisms of MAZ Targeting Up-Regulation of NDUFS3 Expression to Promote Malignant Progression in Melanoma. Commun. Biol. 2024, 7, 1491. [Google Scholar] [CrossRef]
- Li, X.; Han, M.; Zhang, H.; Liu, F.; Pan, Y.; Zhu, J.; Liao, Z.; Chen, X.; Zhang, B. Structures and biological functions of zinc finger proteins and their roles in hepatocellular carcinoma. Biomark. Res. 2022, 10, 2. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, H.; Kornblau, S.M.; Graber, D.A.; Zhang, N.; Matthews, J.A.; Wang, M.; Weber, D.M.; Thomas, S.K.; Shah, J.J.; et al. Evidence of a Role for the Novel Zinc-Finger Transcription Factor ZKSCAN3 in Modulating Cyclin D2 Expression in Multiple Myeloma. Oncogene 2010, 30, 1329–1340. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Luo, Y.; Fu, S.; He, L.; Pan, G.; Fan, D.; Wen, Q.; Fan, Y. Zinc Finger and SCAN Domain-Containing Protein 18 Is a Potential DNA Methylation-Modified Tumor Suppressor and Biomarker in Breast Cancer. Front. Endocrinol. 2023, 14, 1095604. [Google Scholar] [CrossRef]
- Ristevski, S.; O’Leary, D.A.; Thornell, A.P.; Owen, M.J.; Kola, I.; Hertzog, P.J. The ETS Transcription Factor GABPα Is Essential for Early Embryogenesis. Mol. Cell. Biol. 2004, 24, 5844–5849. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Wang, M.; Zhang, C.; Zhang, Y. The Transcription Factor GABPA Is a Master Regulator of Naive Pluripotency. Nat. Cell Biol. 2025, 27, 48–58. [Google Scholar] [CrossRef]
- Rosmarin, A. GA-Binding Protein Transcription Factor: A Review of GABP as an Integrator of Intracellular Signaling and Protein–Protein Interactions. Blood Cells Mol. Dis. 2004, 32, 143–154. [Google Scholar] [CrossRef]
- Beumer, J.; Clevers, H. Hallmarks of Stemness in Mammalian Tissues. Cell Stem Cell 2024, 31, 7–24. [Google Scholar] [CrossRef]
- Ciwinska, M.; Messal, H.A.; Hristova, H.R.; Lutz, C.; Bornes, L.; Chalkiadakis, T.; Harkes, R.; Langedijk, N.S.M.; Hutten, S.J.; Menezes, R.X.; et al. Mechanisms That Clear Mutations Drive Field Cancerization in Mammary Tissue. Nature 2024, 633, 198–206. [Google Scholar] [CrossRef]
- Bunz, F. Passengers, Drivers, and "Goners". Int. J. Cancer 2024, 155, 1696–1698. [Google Scholar] [CrossRef]
- Chetta, M.; Cammarota, A.L.; De Marco, M.; Bukvic, N.; Marzullo, L.; Rosati, A. The Continuous Adaptive Challenge Played by Arboviruses: An In Silico Approach to Identify a Possible Interplay between Conserved Viral RNA Sequences and Host RNA Binding Proteins (RBPs). Int. J. Mol. Sci. 2023, 24, 11051. [Google Scholar] [CrossRef]
- Chetta, M.; Tarsitano, M.; Oro, M.; Rivieccio, M.; Bukvic, N. An in Silico Pipeline Approach Uncovers a Potentially Intricate Network Involving Spike SARS-CoV-2 RNA, RNA Vaccines, Host RNA-Binding Proteins (RBPs), and Host miRNAs at the Cellular Level. J. Genet. Eng. Biotechnol. 2022, 20, 129. [Google Scholar] [CrossRef]
- Gonzalez-Perez, A.; Perez-Llamas, C.; Deu-Pons, J.; Tamborero, D.; Schroeder, M.P.; Jene-Sanz, A.; Santos, A.; Lopez-Bigas, N. IntOGen-mutations identifies cancer drivers across tumor types. Nat. Methods 2013, 10, 1081–1082. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Potter, S.C.; Clarke, L.; Curwen, V.; Keenan, S.; Mongin, E.; Searle, S.M.; Stabenau, A.; Storey, R.; Clamp, M. The Ensembl analysis pipeline. Genome Res. 2004, 14, 934–941. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Machanick, P.; Bailey, T.L. MEME-ChIP: Motif Analysis of Large DNA Datasets. Bioinformatics 2011, 27, 1696–1697. [Google Scholar] [CrossRef] [PubMed]
- Kulakovskiy, I.V.; Medvedeva, Y.A.; Schaefer, U.; Kasianov, A.S.; Vorontsov, I.E.; Bajic, V.B.; Makeev, V.J. HOCOMOCO: A Comprehensive Collection of Human Transcription Factor Binding Sites Models. Nucleic Acids Res. 2012, 41, D195–D202. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein–Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2018, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
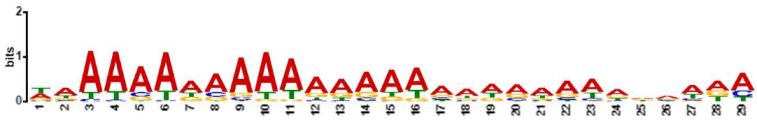
| MEME-ChIP Motif (Sequence Logos) | E-Value | Human TFs |
|---|---|---|
 | 3.4 × 10−143 | ZNF770, IKZF1, WT1, ZNF263, KLF6, ZSC22, ZN281, ZNF250, TEAD4, GFI1, EGR1, PATZ1, ELF5, ZNF341, GFI1B, ZNF467, MAZ, KLF15, ZNF322, TAF1, GABPA, SP3, SP1, TFAP4, DUX4 |
 | 6.3 × 10−140 | AIRE, ALX1, ANDR, BC11A, CDX1, CDX2, ELF3, EVI1, FOXJ2, FOXJ3, FOXK1, FOXO1, FOXO4, FOXQ1, GATA3, GATA6, HNF6, IRF1, IRF2, IRF3, IRF4, IRF8, LEF1, LHX3, MEF2A, MEF2B, MEF2D, NFAC1, NKX61, NR2E3, PIT1, PO2F1, PRDM6, SOX2, SOX4, SOX5, SPI1, SPIB, SRF, SRY, STAT2, TCF7, Z354A, ZFP28, ZFP82, ZIM3, ZN394, ZNF260, ZNF354A, ZNF394, ZNF85, |
 | 6.6 × 10−168 | THAP1, TEAD1, RARG, ZSC31, ESR2, ESR1, CEBPE, ZNF257, ZNF18, MYB, TBX3, ETV5, NKX21, TAF1, SMCA5, NKX25 |
 | 3.8 × 10−153 | PATZ1, SP2, SP1, SP3, ZN467, WT1, MAZ, VEZF1, KLF15, ZN341, EGR1, KLF6, ZN263, KLF3, SP4, EGR2, ZBT17, ZN770, ZN281, PTF1A, TAF1, E2F6, KLF9, RXRA, THAP1, E2F1, ASCL1, KLF1, FLI1, E2F4, MYOD1, KLF12, E2F7, COT1, MBD2, MXI1, MYOG, SRBP2, USF2, TFDP1, GABPA, NR1H4, SALL4, RFX1, ZSC22, ZN335, CTCFL, MYCN |
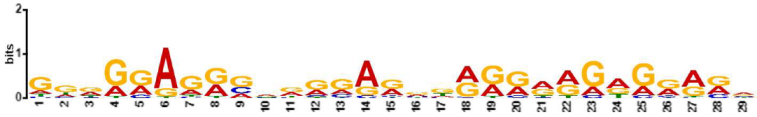 | 4.6 × 10−141 | MAZ, ZN467, VEZF1, ZN263, FLI1, WT1, ZN341, PATZ1, KLF15, IRF3, ZBT17, RXRA, BC11A, ETS2, E2F6, E2F1, ETV5, SPI1, SPIB, ELF5, SP3, EGR2, PTF1A, SP4, TFDP1, ZN281, GATA2, E2F4, E2F7, SP2, OLIG2, KLF6, TBX3, SP1, ETV2, SALL4, KLF3, IRF4, ERG, E2F3, SOX2, EGR1, SMAD3, GATA1, ZN770, NR1D1, TAL1, ETV4, ETS1, IRF8, PRDM6, ZFP82, COT1, RARA, SOX4, ELF3, PRDM1, NFAC1, FOXO1, MZF1, ZSC22, GABPA, ZNF41, IRF2, ZN418, TBX21, TAF1, SMAD2, ZN816, NKX25, IRF1, ZN586, ZN768, ELF2, NFIC, SRBP2 |
| TF Family | Representative Members | Main Functions | Oncogenic/Tumor Suppressor Role | Associated Cancers/Implications |
|---|---|---|---|---|
| E2F Family | E2F1, E2F3, E2F4, E2F6, E2F7 | Regulate genes essential for G1→S phase transition in the cell cycle | Frequently oncogenic when deregulated; loss of regulation leads to uncontrolled proliferation | Retinoblastoma, breast cancer, multiple carcinomas |
| MYC Family | MYCN | Controls proliferation, metabolism, and interaction with PI3K/AKT and CDKs | Oncogenic; amplification linked to poor prognosis | Neuroblastoma, lung carcinoma, lymphoma, breast cancer |
| KLF Family | KLF1, KLF3, KLF6, KLF12, KLF15 | Regulate apoptosis, oxidative stress, angiogenesis, and proliferation | KLF6 as tumor suppressor; KLF3 and KLF15 can be oncogenic | Prostate, liver, colon cancers |
| FOXO Family | FOXO1, FOXO3, FOXO4 | Control apoptosis, DNA repair, and stress response | Tumor suppressors; often inactivated via AKT phosphorylation | Skin, liver, lung, prostate cancers |
| SP Family | SP1, SP3 | Regulate genes for proliferation, DNA synthesis, stress response | Oncogenic when overexpressed | Breast cancer, chemoresistance |
| GATA Family | GATA2, GATA3, GATA6 | Control EMT, migration, metastasis, immune regulation | Context-dependent; can promote or suppress tumors | Breast cancer, lung adenocarcinoma, AML |
| STAT Family | STAT3 | Regulate proliferation, survival, immune evasion | Oncogenic; promotes NK cell evasion | Breast, lung, melanoma |
| IRF Family | IRF1, IRF8 | Regulate apoptosis, immune activation | Tumor suppressors in immunity | Various cancers; loss promotes immune suppression |
| NF-κB Family | NFAC1, NF-κB1 | Promote inflammation, proliferation, metastasis | Oncogenic when persistently active | Skin, colon, breast cancers |
| SOX Family | SOX2 | Maintain stemness, plasticity, therapy resistance | Oncogenic in stem cell maintenance | Lung carcinoma, glioblastoma |
| HOX Family | HOXA9 | Regulate differentiation, proliferation | Oncogenic in leukemia and solid tumors | Leukemia, various solid tumors |
| SMAD Family | SMAD4 | TGF-β signaling, apoptosis, differentiation | Tumor suppressor; loss promotes immune evasion | Pancreatic cancer, others |
| Sex Hormone Receptors | AR, ERα, ERβ | Regulate hormone-dependent growth | Oncogenic in deregulated states | Prostate, breast, ovarian, endometrial cancers |
| ETS Family | ETS1, ETS2 | Regulate proliferation, migration, angiogenesis | Oncogenic | Multiple cancers, immune modulation |
| ZNF Family | ZNF18, ZNF250, ZNF560 | Gene regulation, tumor behavior modulation | Oncogenic when overexpressed | Osteosarcoma |
| PRDM Family | PRDM1, PRDM6 | Epigenetic regulation, gene repression | Oncogenic; immune evasion | HCC, medulloblastoma |
| Retinoic Receptor Family | RARA, RARG | Regulate differentiation, proliferation, apoptosis | Disrupted function oncogenic | APL, skin, lung cancers |
| EGR Family | EGR1, EGR2 | Control proliferation, apoptosis, immune response | Dual role; tumor suppressor or oncogenic depending on context | Multiple cancers, therapy resistance |
| MEF2 Family | MEF2A, MEF2B, MEF2D | Regulate differentiation, metabolism, angiogenesis | Oncogenic when dysregulated | Sarcomas, solid tumors |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cammarota, A.L.; Carrizzo, A.; De Marco, M.; Bukvic, N.; Romano, F.J.; Rosati, A.; Chetta, M. From Transcription Factors Dysregulation to Malignancy: In Silico Reconstruction of Cancer’s Foundational Drivers—The Eternity Triangle. Int. J. Mol. Sci. 2025, 26, 9933. https://doi.org/10.3390/ijms26209933
Cammarota AL, Carrizzo A, De Marco M, Bukvic N, Romano FJ, Rosati A, Chetta M. From Transcription Factors Dysregulation to Malignancy: In Silico Reconstruction of Cancer’s Foundational Drivers—The Eternity Triangle. International Journal of Molecular Sciences. 2025; 26(20):9933. https://doi.org/10.3390/ijms26209933
Chicago/Turabian StyleCammarota, Anna Lisa, Albino Carrizzo, Margot De Marco, Nenad Bukvic, Francesco Jacopo Romano, Alessandra Rosati, and Massimiliano Chetta. 2025. "From Transcription Factors Dysregulation to Malignancy: In Silico Reconstruction of Cancer’s Foundational Drivers—The Eternity Triangle" International Journal of Molecular Sciences 26, no. 20: 9933. https://doi.org/10.3390/ijms26209933
APA StyleCammarota, A. L., Carrizzo, A., De Marco, M., Bukvic, N., Romano, F. J., Rosati, A., & Chetta, M. (2025). From Transcription Factors Dysregulation to Malignancy: In Silico Reconstruction of Cancer’s Foundational Drivers—The Eternity Triangle. International Journal of Molecular Sciences, 26(20), 9933. https://doi.org/10.3390/ijms26209933










