Overexpression of Abiotic Stress-Responsive SsCor413-1 Gene Enhances Salt and Drought Tolerance in Sugarcane (Saccharum spp. Hybrid)
Abstract
1. Introduction
2. Results
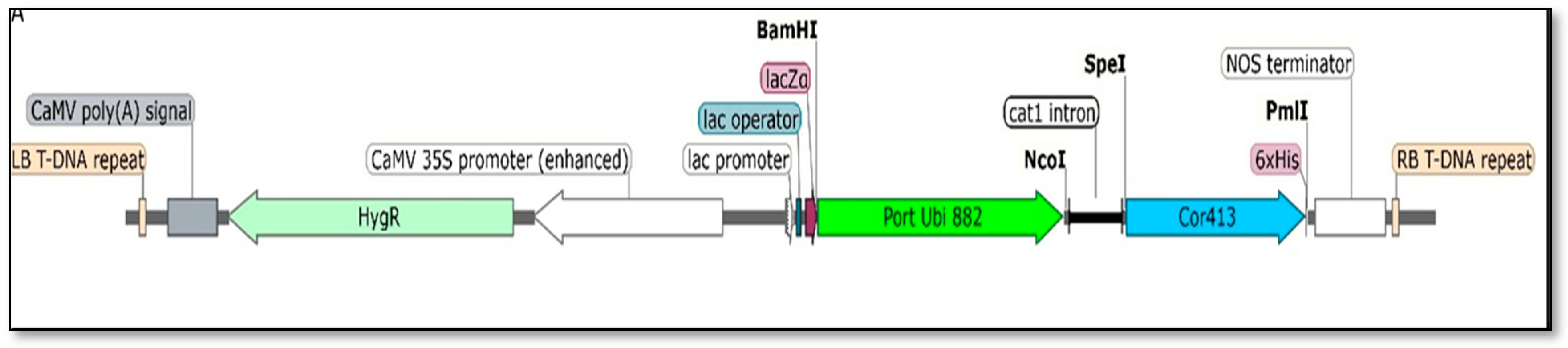
2.1. Isolation of Cor413-1 Gene from S. spontaneum and Cloning into Plant Expression Vector
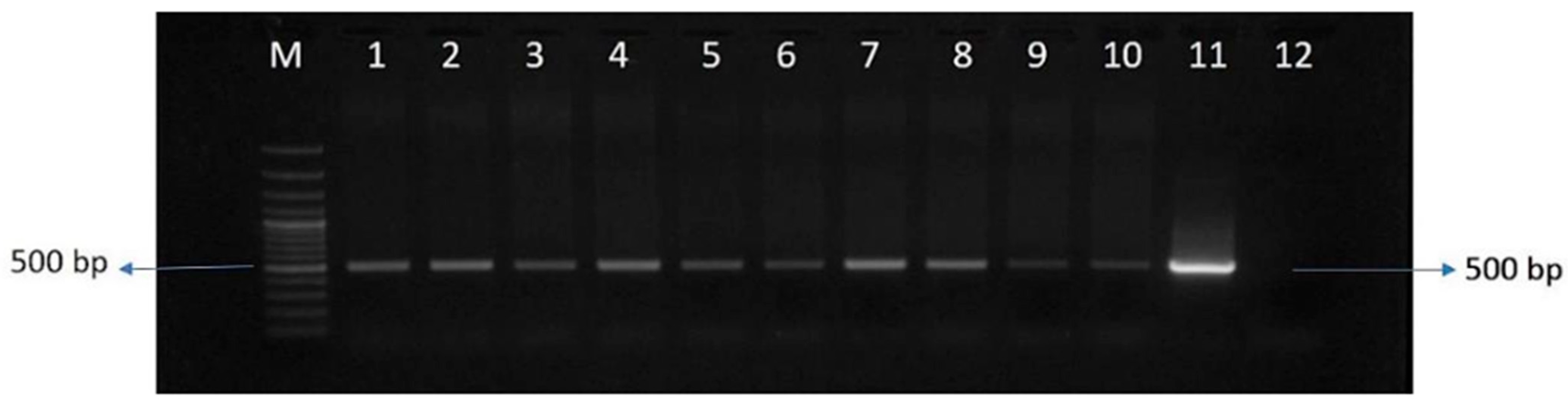
2.2. Generation of Cor413-1 Gene Transgenic Lines
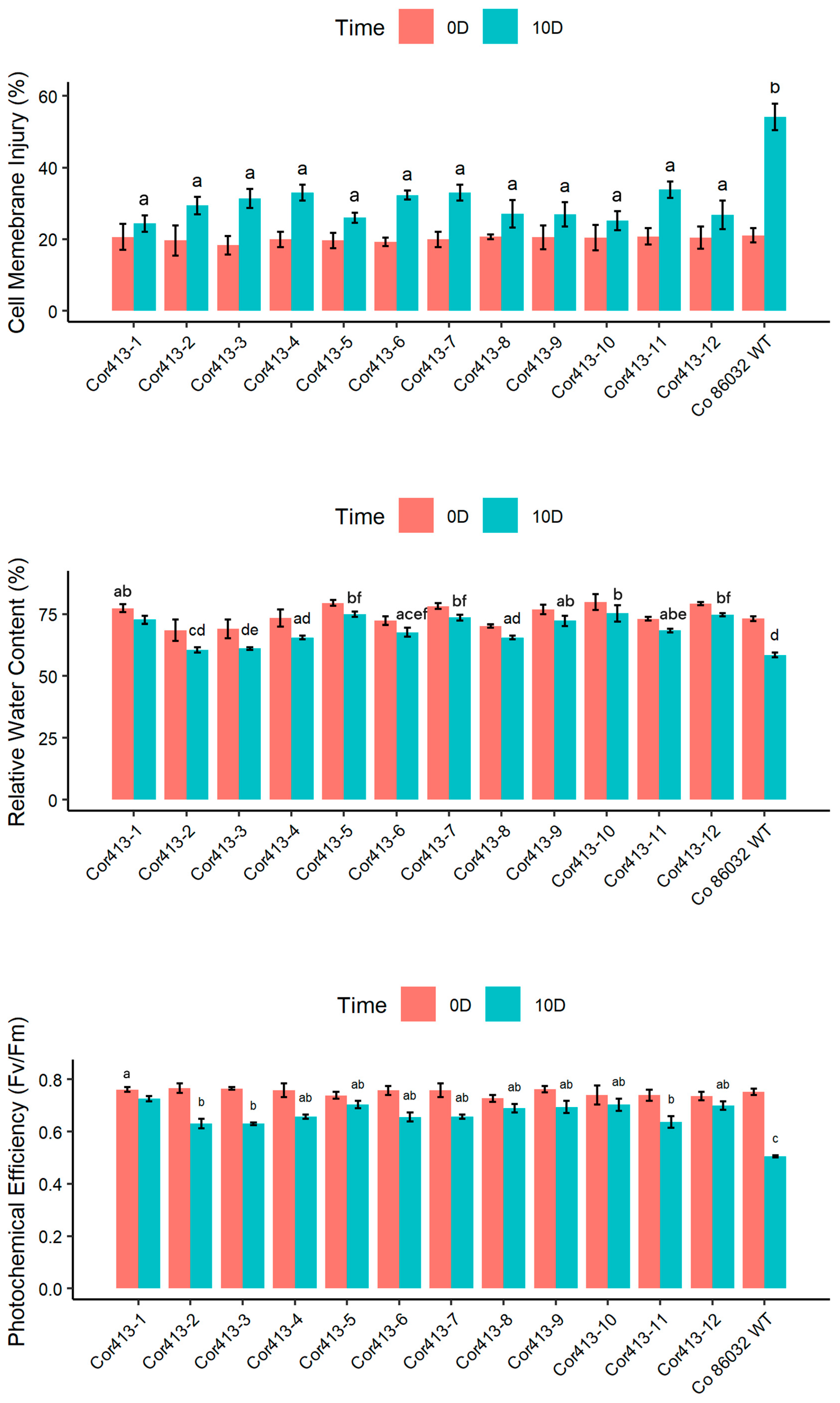
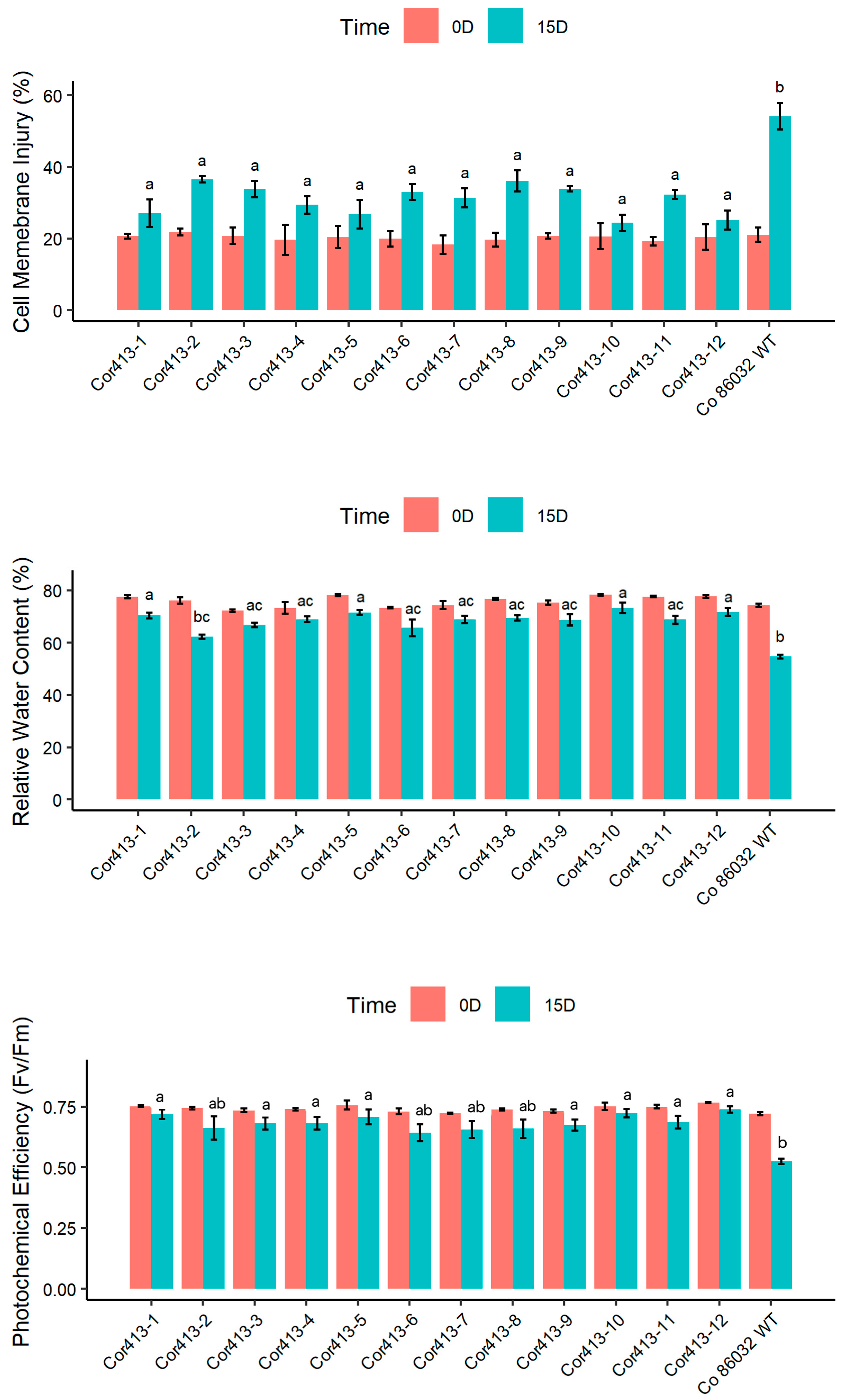
2.3. Evaluation of Physiological Traits During Drought and Salinity Stress Exposure
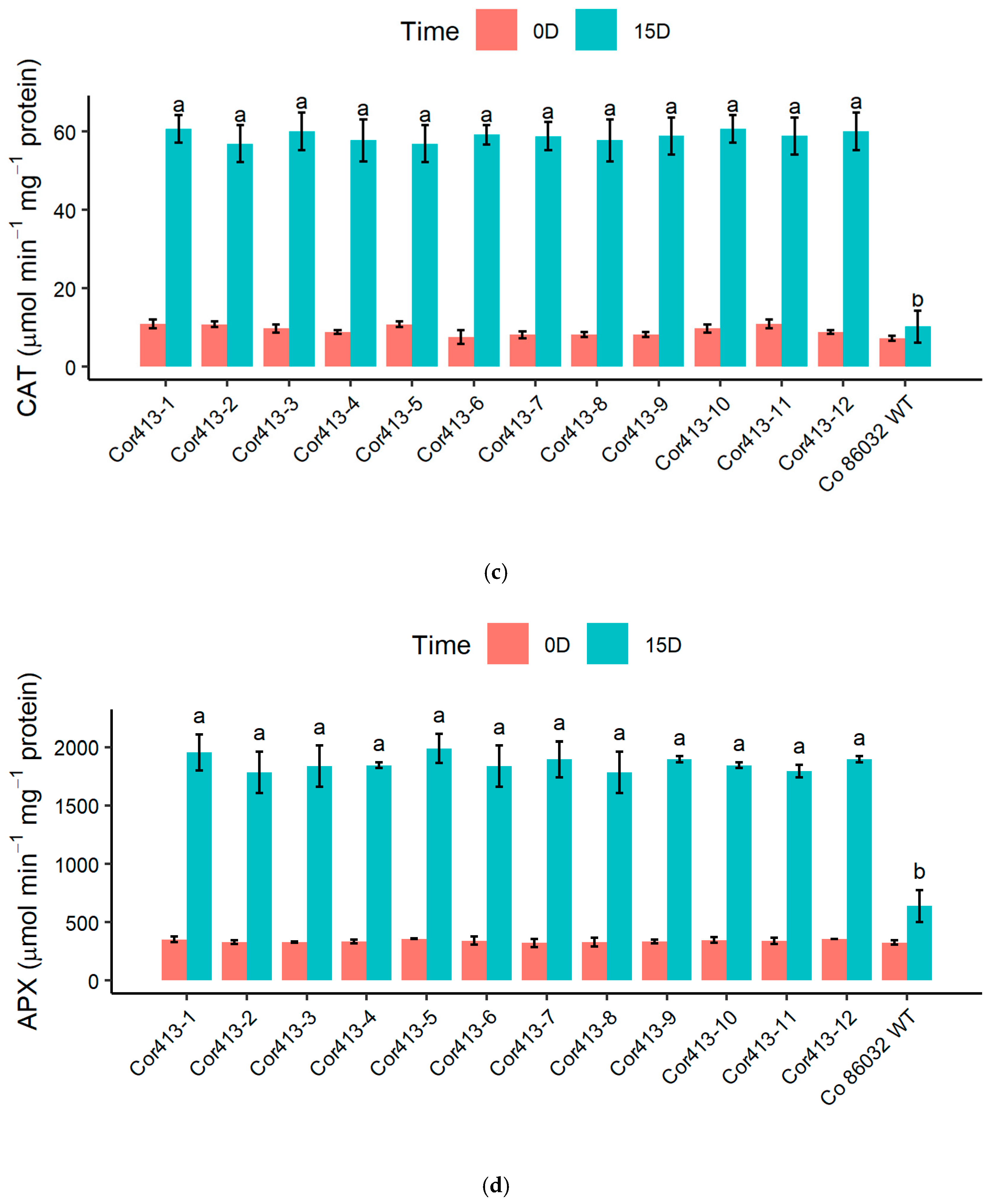
2.4. Evaluation of Stress-Induced Biochemical Changes
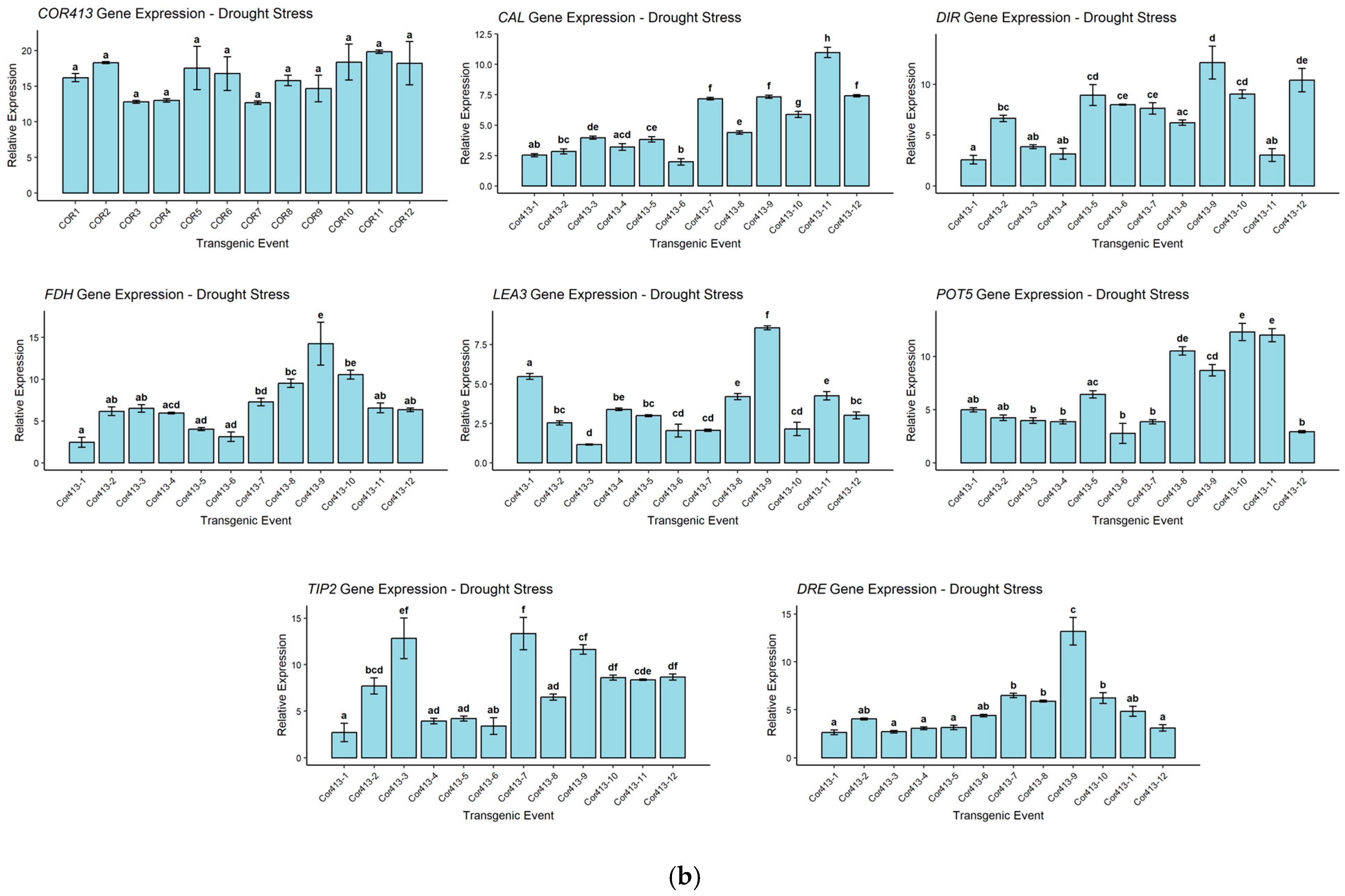
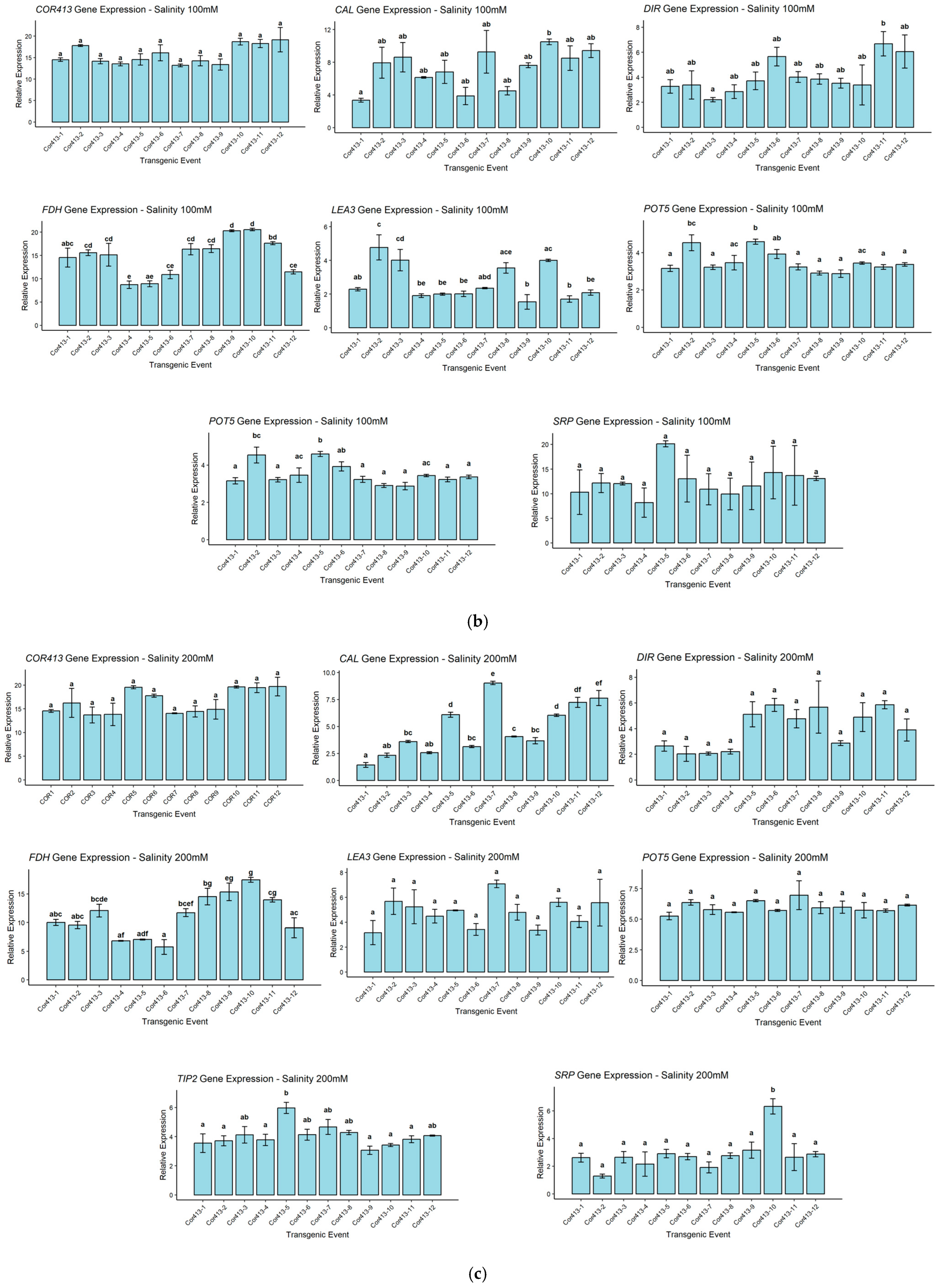
2.5. Expression Analysis of Stress-Inducible Gene in SsCor413-1 Gene Transgenic Lines Under Drought and Salinity Using qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Cor413-1 Gene Isolation and Vector Construction
4.2. Generation of Sugarcane Transgenic Lines and Selection of Putative Events
4.3. Stress Treatments and Assessment of Transgenic Events in V1 Generation
4.4. Physiological and Biochemical Parameter Evaluation
4.5. Gene Expression Studies and Transgene Copy Number Integration Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| COR | Cold-Regulated |
| SOD | Superoxide Dismutase |
| CAT | Catalase |
| APX | Ascorbate Peroxidase |
| MDA | Malondialdehyde |
| HSP70 | Heat Shock Protein 70 |
| LEA3 | Late Embryogenesis Abundant 3 |
| DREB2 | Dehydration-Responsive Element-Binding Protein 2 |
| DIR | Dirigent |
| TIP2 | Tonoplast Intrinsic Protein |
| FAD | Fatty Acid Dehydrogenase |
| HPTII | Hygromycin Phosphotransferase |
References
- Hoang, N.V.; Furtado, A.; Botha, F.C.; Simmons, B.A.; Henry, R.J. Potential for Genetic Improvement of Sugarcane as a Source of Biomass for Biofuels. Front. Bioeng. Biotechnol. 2015, 3, 182. [Google Scholar] [CrossRef] [PubMed]
- Statista Area of Cultivation for Sugarcane in India FY 2005–2025. Available online: https://www.statista.com/statistics/765883/india-area-of-cultivation-for-sugarcane/ (accessed on 30 September 2025).
- Marconi, T.G.; Costa, E.A.; Miranda, H.R.; Mancini, M.C.; Cardoso-Silva, C.B.; Oliveira, K.M.; Pinto, L.R.; Mollinari, M.; Garcia, A.A.; Souza, A.P. Functional Markers for Gene Mapping and Genetic Diversity Studies in Sugarcane. BMC Res. Notes 2011, 4, 264. [Google Scholar] [CrossRef] [PubMed]
- Narayan, J.A.; Chakravarthi, M.; Nerkar, G.; Manoj, V.; Dharshini, S.; Subramonian, N.; Premachandran, M.; Kumar, R.A.; Surendar, K.K.; Hemaprabha, G. Overexpression of Expansin EaEXPA1, a Cell Wall Loosening Protein Enhances Drought Tolerance in Sugarcane. Ind. Crops Prod. 2021, 159, 113035. [Google Scholar] [CrossRef]
- Nerkar, G.; Thorat, A.; Sheelavantmath, S.; Kassa, H.B.; Devarumath, R. Genetic Transformation of Sugarcane and Field Performance of Transgenic Sugarcane. In Biotechnologies of Crop Improvement; Springer: Berlin/Heidelberg, Germany, 2018; Volume 2, pp. 207–226. [Google Scholar]
- Amalraj, V.; Balasundaram, N. Status of sugar-cane genetic resources in India. Plant Genet. Resour. Newsl. 2006, 148, 1–6. [Google Scholar]
- Fukuhara, S.; Terajima, Y.; Irei, S.; Sakaigaichi, T.; Ujihara, K.; Sugimoto, A.; Matsuoka, M. Identification and Characterization of Intergeneric Hybrid of Commercial Sugarcane (Saccharum spp. Hybrid) and Erianthus arundinaceus (Retz.) Jeswiet. Euphytica 2013, 189, 321–327. [Google Scholar] [CrossRef]
- Healey, A.; Garsmeur, O.; Lovell, J.; Shengquiang, S.; Sreedasyam, A.; Jenkins, J.; Plott, C.; Piperidis, N.; Pompidor, N.; Llaca, V. The Complex Polyploid Genome Architecture of Sugarcane. Nature 2024, 628, 804–810. [Google Scholar] [CrossRef]
- Thomashow, M.F. Plant Cold Acclimation: Freezing Tolerance Genes and Regulatory Mechanisms. Annu. Rev. Plant Bio. 1999, 50, 571–599. [Google Scholar] [CrossRef]
- Ma, X.; Wang, G.; Zhao, W.; Yang, M.; Ma, N.; Kong, F.; Dong, X.; Meng, Q. SlCOR413IM1: A Novel Cold-Regulation Gene from Tomato, Enhances Drought Stress Tolerance in Tobacco. J. Plant Physiol. 2017, 216, 88–99. [Google Scholar] [CrossRef]
- Paul, S.; Roychoudhury, A. Transgenic Plants for Improved Salinity and Drought Tolerance. In Biotechnologies of Crop Improvement, Volume 2: Transgenic Approaches; Springer: Berlin/Heidelberg, Germany, 2018; pp. 141–181. [Google Scholar]
- Zhang, C.; Li, C.; Liu, J.; Lv, Y.; Yu, C.; Li, H.; Zhao, T.; Liu, B. The OsABF1 Transcription Factor Improves Drought Tolerance by Activating the Transcription of COR413-TM1 in Rice. J. Exp. Bot. 2017, 68, 4695–4707. [Google Scholar] [CrossRef]
- Mahajan, S.; Tuteja, N. Cold, Salinity and Drought Stresses: An Overview. Arch. Biochem. Biophys. 2005, 444, 139–158. [Google Scholar] [CrossRef]
- Manna, M.; Thakur, T.; Chirom, O.; Mandlik, R.; Deshmukh, R.; Salvi, P. Transcription Factors as Key Molecular Target to Strengthen the Drought Stress Tolerance in Plants. Physiol. Plant. 2021, 172, 847–868. [Google Scholar] [CrossRef]
- Mei, W.; Lepeng, W.; Xiangxue, Y.; Jingyi, Z.; Zhijia, T.; Xiaohong, L.; Guoping, W.; Li, Z.; Xinyong, G. Enhancing Cold and Drought Tolerance in Cotton: A Protective Role of SikCOR413PM1. BMC Plant Biol. 2023, 23, 577. [Google Scholar] [CrossRef]
- Salehi-Lisar, S.Y.; Bakhshayeshan-Agdam, H. Drought Stress in Plants: Causes, Consequences, and Tolerance. In Drought Stress Tolerance in Plants, Vol 1: Physiology and Biochemistry; Hossain, M.A., Wani, S.H., Bhattacharjee, S., Burritt, D.J., Tran, L.-S.P., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 1–16. ISBN 978-3-319-28899-4. [Google Scholar]
- Raza, M.A.; Sohail, H.; Hassan, M.A.; Sajad, S.; Xing, Y.; Song, J. Cold Stress in Brassica Vegetables: Morpho-Physiological and Molecular Responses Underlying Adaptive Mechanism. Sci. Horticult. 2024, 329, 113002. [Google Scholar] [CrossRef]
- Long, S.; Yan, F.; Yang, L.; Sun, Z.; Wei, S. Responses of Manila Grass (Zoysia matrella) to Chilling Stress: From Transcriptomics to Physiology. PLoS ONE 2020, 15, e0235972. [Google Scholar] [CrossRef]
- Amin, I.; Rasool, S.; Mir, M.A.; Wani, W.; Masoodi, K.Z.; Ahmad, P. Ion Homeostasis for Salinity Tolerance in Plants: A Molecular Approach. Physiol. Plant. 2021, 171, 578–594. [Google Scholar] [CrossRef] [PubMed]
- Appunu, C.; Krishna, S.S.; Chandar, S.H.; Valarmathi, R.; Suresha, G.S.; Sreenivasa, V.; Malarvizhi, A.; Manickavasagam, M.; Arun, M.; Kumar, R.A. Overexpression of EaALDH7, an Aldehyde Dehydrogenase Gene from Erianthus arundinaceus Enhances Salinity Tolerance in Transgenic Sugarcane (Saccharum spp. Hybrid). Plant Sci. 2024, 348, 112206. [Google Scholar] [CrossRef]
- Augustine, S.M.; Ashwin Narayan, J.; Syamaladevi, D.P.; Appunu, C.; Chakravarthi, M.; Ravichandran, V.; Tuteja, N.; Subramonian, N. Overexpression of EaDREB2 and Pyramiding of EaDREB2 with the Pea DNA Helicase Gene (PDH45) Enhance Drought and Salinity Tolerance in Sugarcane (Saccharum spp. Hybrid). Plant Cell Rep. 2015, 34, 247–263. [Google Scholar] [CrossRef] [PubMed]
- Chinnaswamy, A.; Sakthivel, S.K.; Channappa, M.; Ramanathan, V.; Shivalingamurthy, S.G.; Peter, S.C.; Kumar, R.; Kumar, R.A.; Dhansu, P.; Meena, M.R.; et al. Overexpression of an NF-YB Gene Family Member, EaNF-YB2, Enhances Drought Tolerance in Sugarcane (Saccharum Spp. Hybrid). BMC Plant Biol. 2024, 24, 1246. [Google Scholar] [CrossRef] [PubMed]
- Chinnaswamy, A.; Harish Chandar, S.R.; Ramanathan, V.; Chennappa, M.; Sakthivel, S.K.; Arthanari, M.; Thangavel, S.; Raja, A.K.; Devarumath, R.; Vijayrao, S.K.; et al. Ectopic Expression of Choline Oxidase (codA) Gene from Arthrobacter globiformis Confers Drought Stress Tolerance in Transgenic Sugarcane. 3 Biotech 2024, 14, 309. [Google Scholar] [CrossRef]
- Breton, G.; Danyluk, J.; Charron, J.-B.F.; Sarhan, F. Expression Profiling and Bioinformatic Analyses of a Novel Stress-Regulated Multispanning Transmembrane Protein Family from Cereals and Arabidopsis. Plant Physiol. 2003, 132, 64–74. [Google Scholar] [CrossRef]
- Wang, J.; Zuo, K.-J.; Qin, J.; Zhang, L.; Su, L.; Liu, J.; Ling, H.; Zhao, J.-Y.; Cao, Y.-F.; Tang, K.-X. Isolation and Bioinformatics Analyses of a COR413-like Gene from Gossypium barbadense. Acta Physiol. Plant. 2007, 29, 1–9. [Google Scholar] [CrossRef]
- Dharshini, S.; Manoj, V.; Suresha, G.; Narayan, J.A.; Padmanabhan, T.S.; Kumar, R.; Meena, M.R.; Manickavasagam, M.; Ram, B.; Appunu, C. Isolation and Characterization of Nuclear Localized Abiotic Stress Responsive Cold Regulated Gene 413 (SsCor413) from Saccharum spontaneum. Plant Mol. Biol. Rep. 2020, 38, 628–640. [Google Scholar] [CrossRef]
- Saravanan, S.; Kumar, K.; Raveendran, M.; Sudhakar, D.; Arul, L.; Kokiladevi, E. Genetic Engineering of Sugarcane for Drought and Salt Tolerant Transgenic Plants Expressing the BcZAT12 Gene. Int. J. Curr. Microbiol. App. Sci. 2018, 7, 1594–1613. [Google Scholar] [CrossRef]
- Mbambalala, N.; Panda, S.K.; van der Vyver, C. Overexpression of AtBBX29 Improves Drought Tolerance by Maintaining Photosynthesis and Enhancing the Antioxidant and Osmolyte Capacity of Sugarcane Plants. Plant Mol. Biol. Rep. 2021, 39, 419–433. [Google Scholar] [CrossRef]
- Masoabi, M.; Burger, N.F.V.; Botha, A.-M.; Le Roux, M.L.; Vlok, M.; Snyman, S.; Van der Vyver, C. Overexpression of the Small Ubiquitin-Like Modifier Protease OTS1 Gene Enhances Drought Tolerance in Sugarcane (Saccharum spp. Hybrid). Plant Biol. 2023, 25, 1121–1141. [Google Scholar] [CrossRef] [PubMed]
- Haghpanah, M.; Hashemipetroudi, S.; Arzani, A.; Araniti, F. Drought Tolerance in Plants: Physiological and Molecular Responses. Plants 2024, 13, 2962. [Google Scholar] [CrossRef] [PubMed]
- Stewart, R.R.; Bewley, J.D. Lipid Peroxidation Associated with Accelerated Aging of Soybean Axes. Plant Physiol. 1980, 65, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Tanentzap, F.M.; Stempel, A.; Ryser, P. Reliability of Leaf Relative Water Content (RWC) Measurements after Storage: Consequences for in Situ Measurements. Botany 2015, 93, 535–541. [Google Scholar] [CrossRef]
- Jongdee, B.; Fukai, S.; Cooper, M. Leaf Water Potential and Osmotic Adjustment as Physiological Traits to Improve Drought Tolerance in Rice. Field Crops Res. 2002, 76, 153–163. [Google Scholar] [CrossRef]
- Sheldon, A.R.; Dalal, R.C.; Kirchhof, G.; Kopittke, P.M.; Menzies, N.W. The Effect of Salinity on Plant-Available Water. Plant Soil 2017, 418, 477–491. [Google Scholar] [CrossRef]
- Molinari, H.B.C.; Marur, C.J.; Daros, E.; De Campos, M.K.F.; De Carvalho, J.F.R.P.; Filho, J.C.B.; Pereira, L.F.P.; Vieira, L.G.E. Evaluation of the Stress-inducible Production of Proline in Transgenic Sugarcane (Saccharum spp.): Osmotic Adjustment, Chlorophyll Fluorescence and Oxidative Stress. Physiol. Plant. 2007, 130, 218–229. [Google Scholar] [CrossRef]
- Sharma, D.K.; Andersen, S.B.; Ottosen, C.-O.; Rosenqvist, E. Wheat Cultivars Selected for High Fv/Fm under Heat Stress Maintain High Photosynthesis, Total Chlorophyll, Stomatal Conductance, Transpiration and Dry Matter. Physiol. Plant. 2015, 153, 284–298. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Li, J.; Fan, Y.; Xu, S.; Zhang, Z. High Temperature Effects on Photosynthesis, PSII Functionality and Antioxidant Activity of Two Festuca Arundinacea Cultivars with Different Heat Susceptibility. Bot. Stud. 2006, 47, 61–69. [Google Scholar]
- An, D.; Zhao, B.; Liu, Y.; Xu, Z.; Kong, R.; Yan, C.; Su, J. Simulation of Photosynthetic Quantum Efficiency and Energy Distribution Analysis Reveals Differential Drought Response Strategies in Two (Drought-Resistant and -Susceptible) Sugarcane Cultivars. Plants 2023, 12, 1042. [Google Scholar] [CrossRef]
- Del Rio, D.; Stewart, A.J.; Pellegrini, N. A Review of Recent Studies on Malondialdehyde as Toxic Molecule and Biological Marker of Oxidative Stress. Nutr. Metab. Cardiovasc. Dis. 2005, 15, 316–328. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, M.; Guo, J.; Wang, Y.; Min, D.; Jiang, Q.; Ji, H.; Huang, C.; Wei, W.; Xu, H. Overexpression of Soybean DREB1 Enhances Drought Stress Tolerance of Transgenic Wheat in the Field. J. Exp. Bot. 2020, 71, 1842–1857. [Google Scholar] [CrossRef] [PubMed]
- van Beek, C.R.; Guzha, T.; Kopana, N.; van der Westhuizen, C.S.; Panda, S.K.; van der Vyver, C. The SlNAC2 Transcription Factor from Tomato Confers Tolerance to Drought Stress in Transgenic Tobacco Plants. Physiol. Mol. Biol. Plants 2021, 27, 907–921. [Google Scholar] [CrossRef]
- Zhang, P.; Yuan, Z.; Wei, L.; Qiu, X.; Wang, G.; Liu, Z.; Fu, J.; Cao, L.; Wang, T. Overexpression of ZmPP2C55 Positively Enhances Tolerance to Drought Stress in Transgenic Maize Plants. Plant Sci. 2022, 314, 111127. [Google Scholar] [CrossRef]
- Leonowicz, G.; Trzebuniak, K.F.; Zimak-Piekarczyk, P.; Ślesak, I.; Mysliwa-Kurdziel, B. The Activity of Superoxide Dismutases (SODs) at the Early Stages of Wheat Deetiolation. PLoS ONE 2018, 13, e0194678. [Google Scholar] [CrossRef]
- Cheng, Z.; Zhang, X.; Zhao, K.; Zhou, B.; Jiang, T. Ectopic Expression of a Poplar Gene NAC13 Confers Enhanced Tolerance to Salinity Stress in Transgenic Nicotiana tabacum. J. Plant Res. 2020, 133, 727–737. [Google Scholar] [CrossRef]
- Caverzan, A.; Passaia, G.; Rosa, S.B.; Ribeiro, C.W.; Lazzarotto, F.; Margis-Pinheiro, M. Plant Responses to Stresses: Role of Ascorbate Peroxidase in the Antioxidant Protection. Genet. Mol. Biol. 2012, 35, 1011–1019. [Google Scholar] [CrossRef]
- Shafi, A.; Chauhan, R.; Gill, T.; Swarnkar, M.K.; Sreenivasulu, Y.; Kumar, S.; Kumar, N.; Shankar, R.; Ahuja, P.S.; Singh, A.K. Expression of SOD and APX Genes Positively Regulates Secondary Cell Wall Biosynthesis and Promotes Plant Growth and Yield in Arabidopsis under Salt Stress. Plant Mol. Biol. 2015, 87, 615–631. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Liu, P.; Yuan, G.; Jia, J.; Li, X.; Qi, D.; Chen, S.; Ma, T.; Liu, G.; Cheng, L. New Insights on Drought Stress Response by Global Investigation of Gene Expression Changes in Sheepgrass (Leymus chinensis). Front. Plant Sci. 2016, 7, 954. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Roychoudhury, A. Group II Late Embryogenesis Abundant (LEA) Proteins: Structural and Functional Aspects in Plant Abiotic Stress. Plant Growth Regul. 2016, 79, 1–17. [Google Scholar] [CrossRef]
- Feng, Z.-J.; Liu, N.; Zhang, G.-W.; Niu, F.-G.; Xu, S.-C.; Gong, Y.-M. Investigation of the AQP Family in Soybean and the Promoter Activity of TIP2;6 in Heat Stress and Hormone Responses. Int. J. Mol. Sci. 2019, 20, 262. [Google Scholar] [CrossRef] [PubMed]
- Xiao, R.; Zou, Y.; Guo, X.; Li, H.; Lu, H. Fatty Acid Desaturases (FADs) Modulate Multiple Lipid Metabolism Pathways to Improve Plant Resistance. Mol. Bio. Rep. 2022, 49, 9997–10011. [Google Scholar] [CrossRef]
- Paniagua, C.; Bilkova, A.; Jackson, P.; Dabravolski, S.; Riber, W.; Didi, V.; Houser, J.; Gigli-Bisceglia, N.; Wimmerova, M.; Budínská, E.; et al. Dirigent Proteins in Plants: Modulating Cell Wall Metabolism during Abiotic and Biotic Stress Exposure. J. Exp. Bot. 2017, 68, 3287–3301. [Google Scholar] [CrossRef]
- Manoj, V.M.; Anunanthini, P.; Swathik, P.C.; Dharshini, S.; Ashwin Narayan, J.; Manickavasagam, M.; Sathishkumar, R.; Suresha, G.S.; Hemaprabha, G.; Ram, B. Comparative Analysis of Glyoxalase Pathway Genes in Erianthus arundinaceus and Commercial Sugarcane Hybrid under Salinity and Drought Conditions. BMC Genom. 2019, 19, 986. [Google Scholar] [CrossRef]
- Selvarajan, D.; Mohan, C.; Dhandapani, V.; Nerkar, G.; Jayanarayanan, A.N.; Vadakkancherry Mohanan, M.; Murugan, N.; Kaur, L.; Chennappa, M.; Kumar, R. Differential Gene Expression Profiling through Transcriptome Approach of Saccharum spontaneum L. under Low Temperature Stress Reveals Genes Potentially Involved in Cold Acclimation. 3 Biotech 2018, 8, 195. [Google Scholar] [CrossRef]
- Dharshini, S.; Hoang, N.V.; Mahadevaiah, C.; Padmanabhan, T.S.; Alagarasan, G.; Suresha, G.; Kumar, R.; Meena, M.R.; Ram, B.; Appunu, C. Root Transcriptome Analysis of Saccharum Spontaneum Uncovers Key Genes and Pathways in Response to Low-Temperature Stress. Environ. Exp. Bot. 2020, 171, 103935. [Google Scholar] [CrossRef]
- Philip, A.; Syamaladevi, D.P.; Chakravarthi, M.; Gopinath, K.; Subramonian, N. 5′ Regulatory Region of Ubiquitin 2 Gene from Porteresia Coarctata Makes Efficient Promoters for Transgene Expression in Monocots and Dicots. Plant Cell Rep. 2013, 32, 1199–1210. [Google Scholar] [CrossRef]
- Narayan, J.A.; Manoj, V.; Nerkar, G.; Chakravarthi, M.; Dharshini, S.; Subramonian, N.; Premachandran, M.; Valarmathi, R.; Kumar, R.A.; Gomathi, R. Transgenic Sugarcane with Higher Levels of BRK1 Showed Improved Drought Tolerance. Plant Cell Rep. 2023, 42, 1611–1628. [Google Scholar] [CrossRef]
- Bower, R.; Birch, R.G. Transgenic Sugarcane Plants via Microprojectile Bombardment. Plant J. 1992, 2, 409–416. [Google Scholar] [CrossRef]
- Christy, L.A.; Arvinth, S.; Saravanakumar, M.; Kanchana, M.; Mukunthan, N.; Srikanth, J.; Thomas, G.; Subramonian, N. Engineering Sugarcane Cultivars with Bovine Pancreatic Trypsin Inhibitor (Aprotinin) Gene for Protection against Top Borer (Scirpophaga excerptalis Walker). Plant Cell Rep. 2009, 28, 175–184. [Google Scholar] [CrossRef]
- Doyle, J. DNA Protocols for Plants. In Molecular Techniques in Taxonomy; Springer: Berlin/Heidelberg, Germany, 1991; pp. 283–293. [Google Scholar]
- Mohanan, M.V.; Pushpanathan, A.; Padmanabhan, S.; Sasikumar, T.; Jayanarayanan, A.N.; Selvarajan, D.; Ramalingam, S.; Ram, B.; Chinnaswamy, A. Overexpression of Glyoxalase III Gene in Transgenic Sugarcane Confers Enhanced Performance under Salinity Stress. J. Plant Res. 2021, 134, 1083–1094. [Google Scholar] [CrossRef]
- Augustine, S.M.; Narayan, J.A.; Syamaladevi, D.P.; Appunu, C.; Chakravarthi, M.; Ravichandran, V.; Subramonian, N. Erianthus Arundinaceus HSP70 (EaHSP70) Overexpression Increases Drought and Salinity Tolerance in Sugarcane (Saccharum spp. Hybrid). Plant Sci. 2015, 232, 23–34. [Google Scholar] [CrossRef]
- Boaretto, L.F.; Carvalho, G.; Borgo, L.; Creste, S.; Landell, M.G.; Mazzafera, P.; Azevedo, R.A. Water Stress Reveals Differential Antioxidant Responses of Tolerant and Non-Tolerant Sugarcane Genotypes. Plant Physiol. Biochem. 2014, 74, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Giannopolitis, C.N.; Ries, S.K. Superoxide Dismutases: I. Occurrence in Higher Plants. Plant Physiol. 1977, 59, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Aebi, H. Catalase in Vitro. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1984; Volume 105, pp. 121–126. ISBN 0076-6879. [Google Scholar]
- Nakano, Y.; Asada, K. Hydrogen Peroxide Is Scavenged by Ascorbate-Specific Peroxidase in Spinach Chloroplasts. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar] [CrossRef]
- Casu, R.E.; Selivanova, A.; Perroux, J.M. High-Throughput Assessment of Transgene Copy Number in Sugarcane Using Real-Time Quantitative PCR. Plant Cell Rep. 2012, 31, 167–177. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

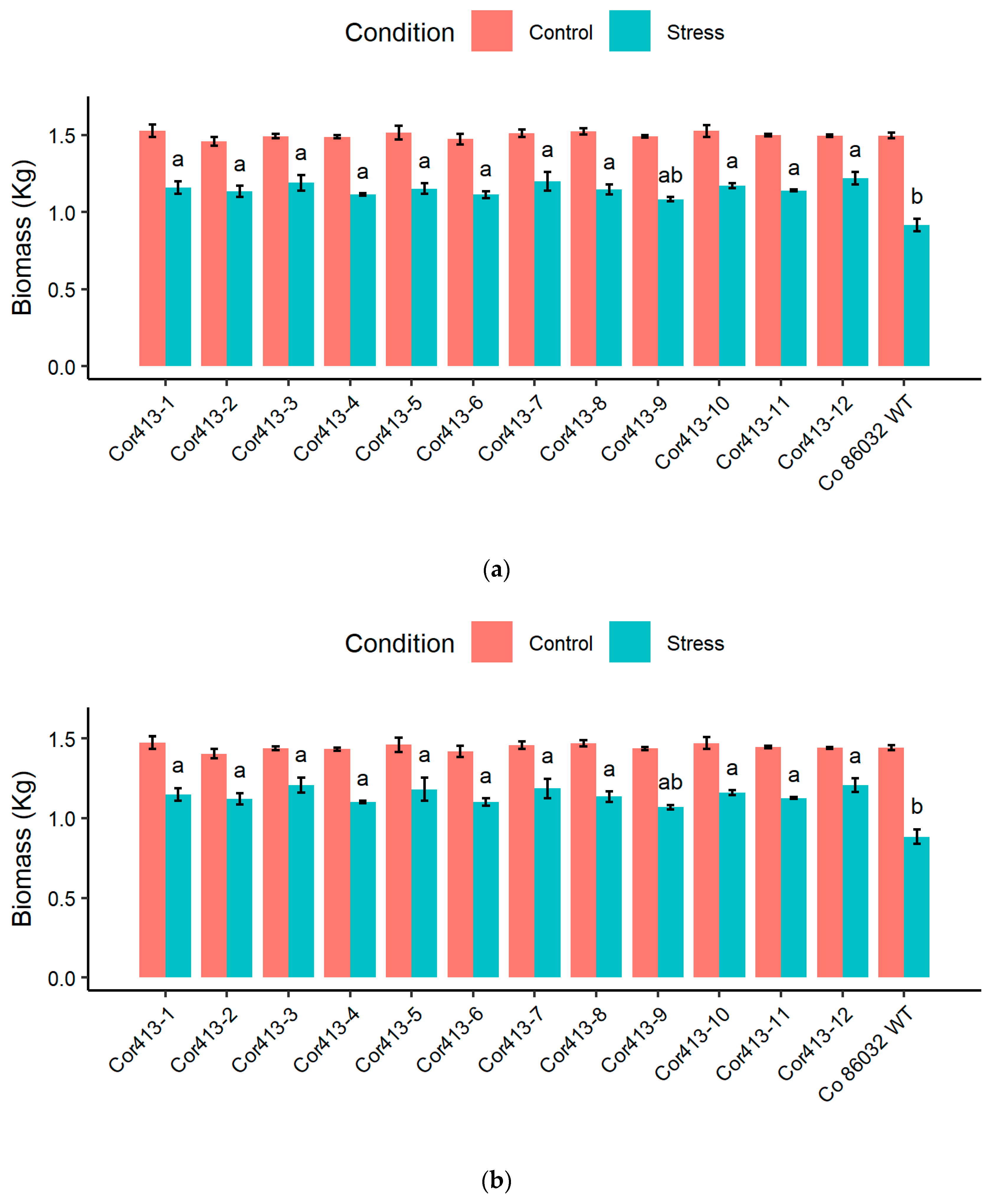
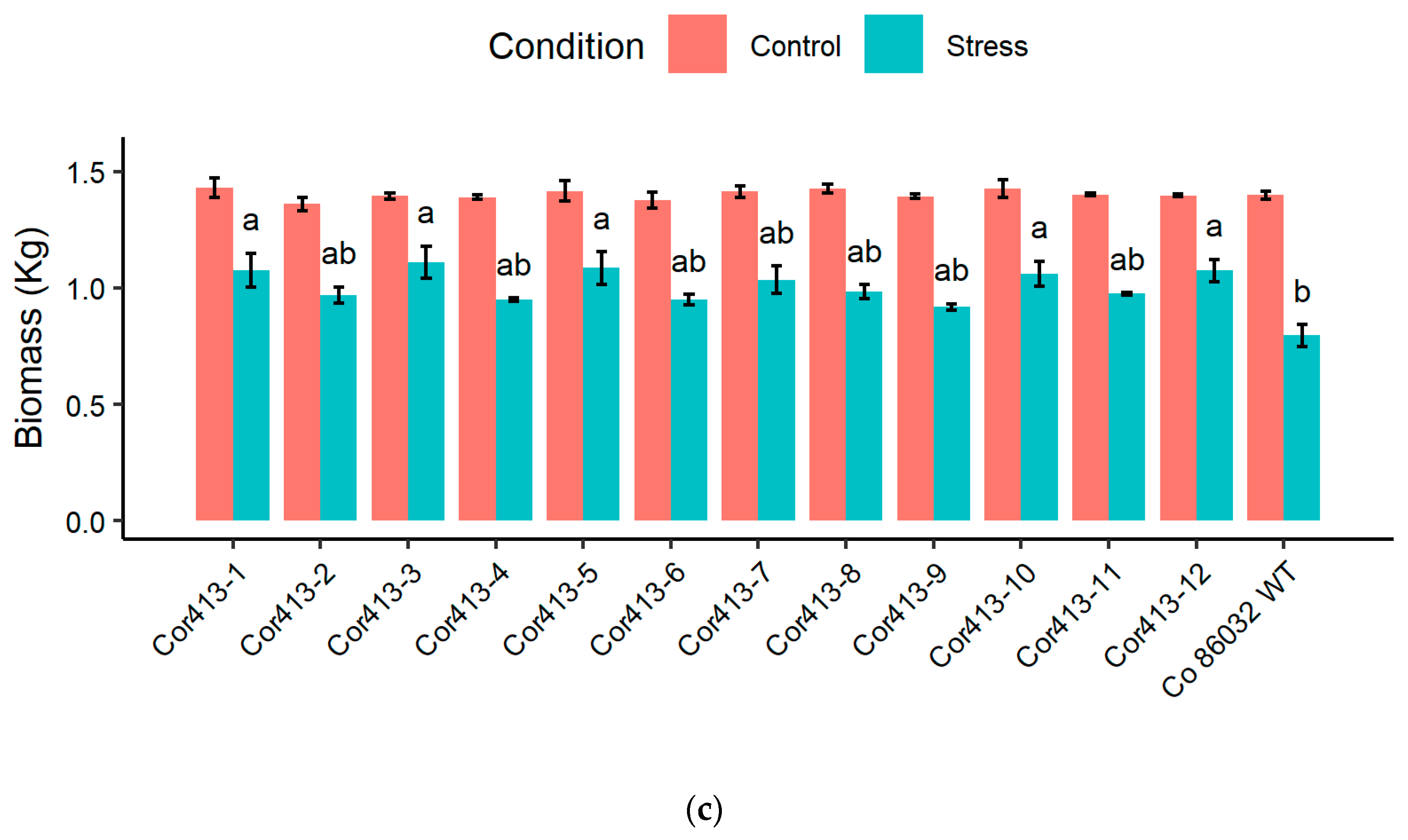
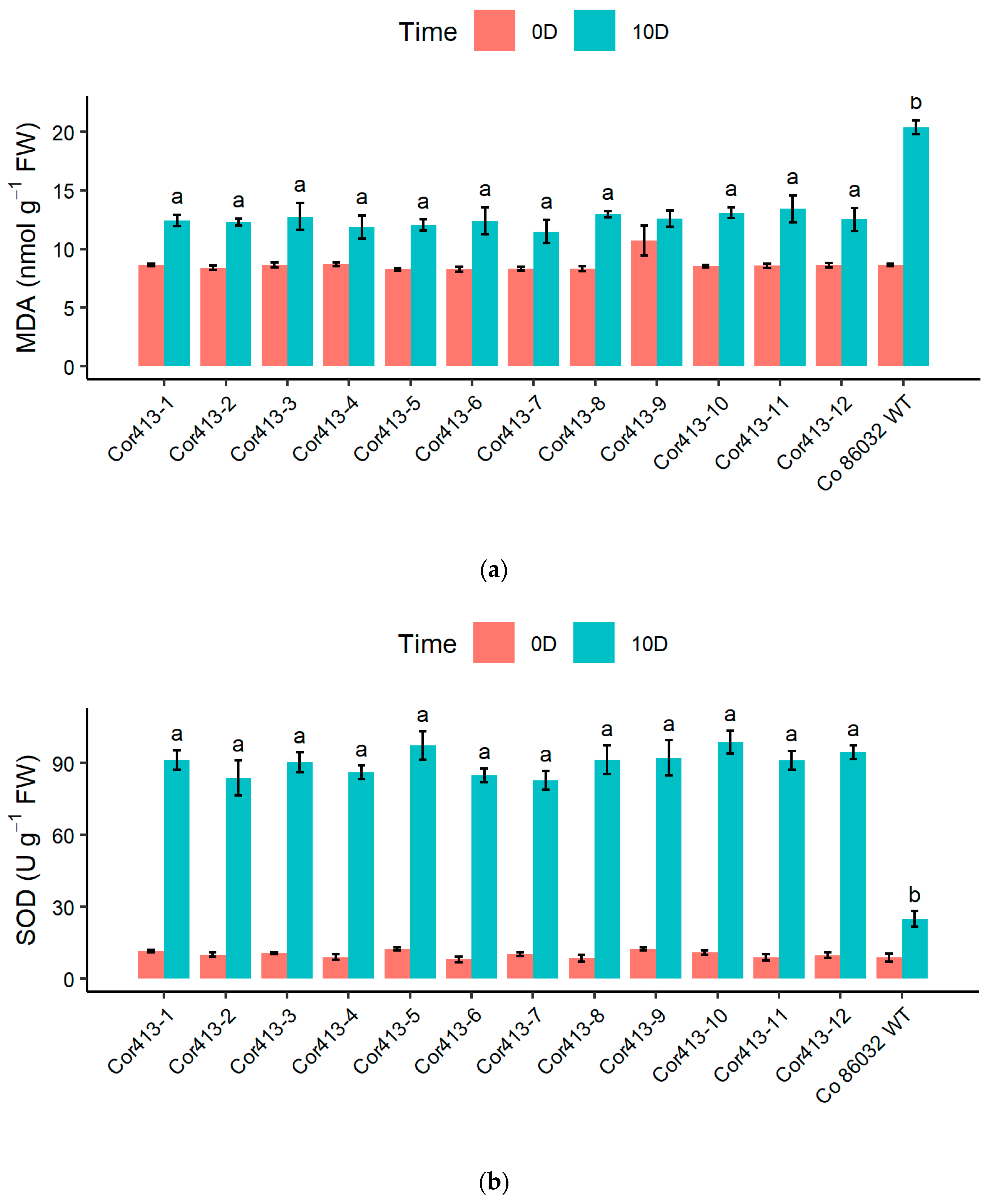

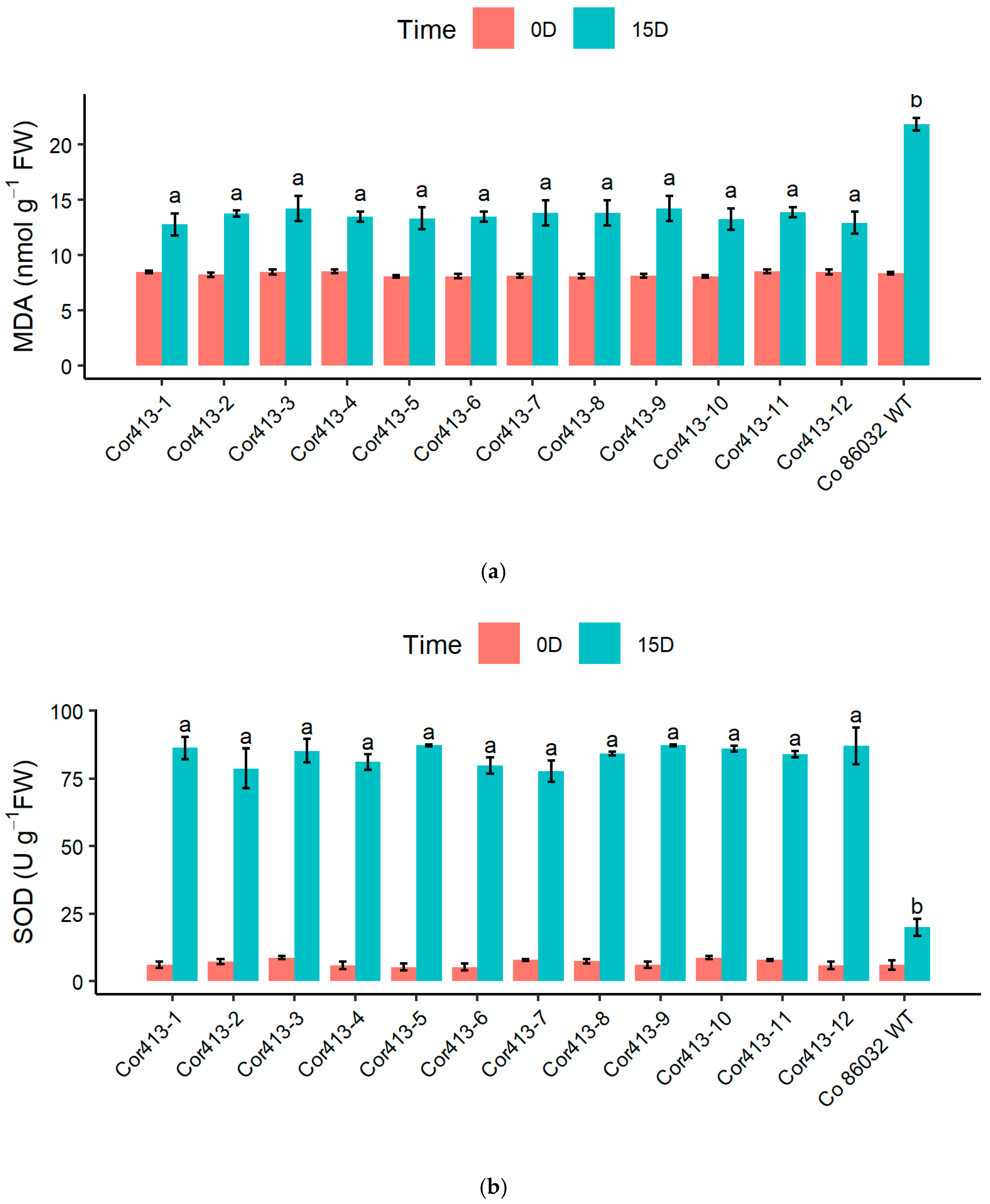


| Sl. No | Transgenic Events | Average_100 mM | Average_200 mM |
|---|---|---|---|
| 1. | Cor413-1 | 2 | 2 |
| 2. | Cor413-2 | 1 | 0.5 |
| 3. | Cor413-3 | 2 | 2 |
| 4. | Cor413-4 | 1 | 1 |
| 5. | Cor413-5 | 1 | 2 |
| 6. | Cor413-6 | 1 | 2 |
| 7. | Cor413-7 | 1 | 0.5 |
| 8. | Cor413-8 | 1 | 1 |
| 9. | Cor413-9 | 1 | 1 |
| 10. | Cor413-10 | 1 | 0.5 |
| 11. | Cor413-11 | 1 | 2 |
| 12. | Cor413-12 | 1 | 2 |
| 13. | Co 86032 | 3 | 5 |
| Sl. No | Transgenic Events | Gene Copy Number |
|---|---|---|
| 1. | Cor413-1 | 2 |
| 2. | Cor413-2 | 1 |
| 3. | Cor413-3 | 3 |
| 4. | Cor413-4 | 2 |
| 5. | Cor413-5 | 2 |
| 6. | Cor413-6 | 3 |
| 7. | Cor413-7 | 2 |
| 8. | Cor413-8 | 3 |
| 9. | Cor413-9 | 2 |
| 10. | Cor413-10 | 1 |
| 11. | Cor413-11 | 1 |
| 12. | Cor413-12 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dharshini, S.; Swathi, T.; Lekshmi, L.A.; Surya Krishna, S.; Harish Chandar, S.R.; Manoj, V.M.; Ashwin Narayan, J.; Sarath Padmanabhan, T.S.; Valarmathi, R.; Kumar, R.A.; et al. Overexpression of Abiotic Stress-Responsive SsCor413-1 Gene Enhances Salt and Drought Tolerance in Sugarcane (Saccharum spp. Hybrid). Int. J. Mol. Sci. 2025, 26, 9868. https://doi.org/10.3390/ijms26209868
Dharshini S, Swathi T, Lekshmi LA, Surya Krishna S, Harish Chandar SR, Manoj VM, Ashwin Narayan J, Sarath Padmanabhan TS, Valarmathi R, Kumar RA, et al. Overexpression of Abiotic Stress-Responsive SsCor413-1 Gene Enhances Salt and Drought Tolerance in Sugarcane (Saccharum spp. Hybrid). International Journal of Molecular Sciences. 2025; 26(20):9868. https://doi.org/10.3390/ijms26209868
Chicago/Turabian StyleDharshini, Selvarajan, Thangavel Swathi, L. Ananda Lekshmi, Sakthivel Surya Krishna, S. R. Harish Chandar, Vadakkenchery Mohanan Manoj, Jayanarayanan Ashwin Narayan, Thelakat Sasikumar Sarath Padmanabhan, Ramanathan Valarmathi, Raja Arun Kumar, and et al. 2025. "Overexpression of Abiotic Stress-Responsive SsCor413-1 Gene Enhances Salt and Drought Tolerance in Sugarcane (Saccharum spp. Hybrid)" International Journal of Molecular Sciences 26, no. 20: 9868. https://doi.org/10.3390/ijms26209868
APA StyleDharshini, S., Swathi, T., Lekshmi, L. A., Surya Krishna, S., Harish Chandar, S. R., Manoj, V. M., Ashwin Narayan, J., Sarath Padmanabhan, T. S., Valarmathi, R., Kumar, R. A., Boominathan, P., & Appunu, C. (2025). Overexpression of Abiotic Stress-Responsive SsCor413-1 Gene Enhances Salt and Drought Tolerance in Sugarcane (Saccharum spp. Hybrid). International Journal of Molecular Sciences, 26(20), 9868. https://doi.org/10.3390/ijms26209868





