Decomposition Analysis of Theoretical Raman Spectra for Efficient Interpretation of Experimental Spectra of Thin-Film Functional Materials
Abstract
1. Introduction
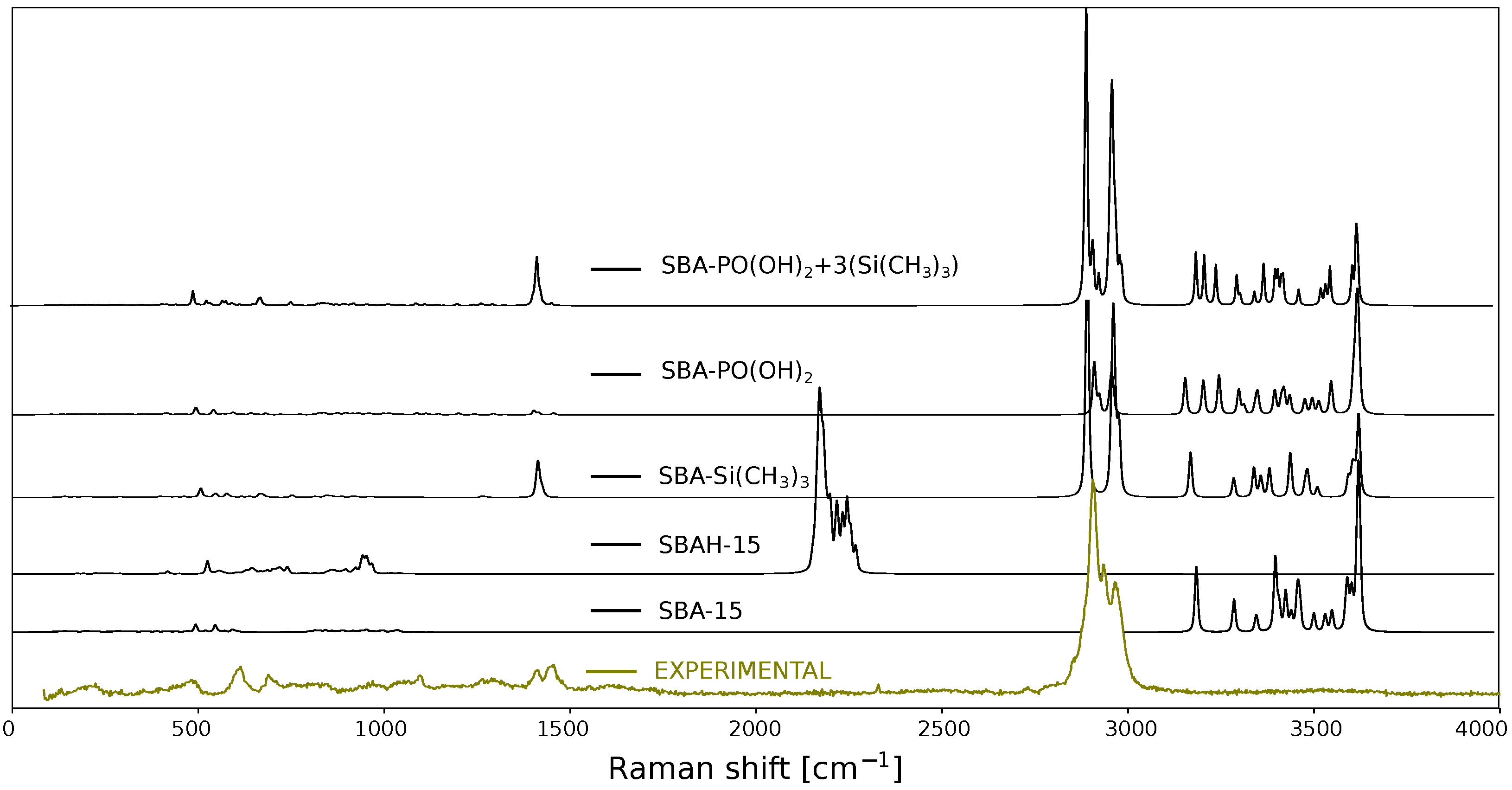
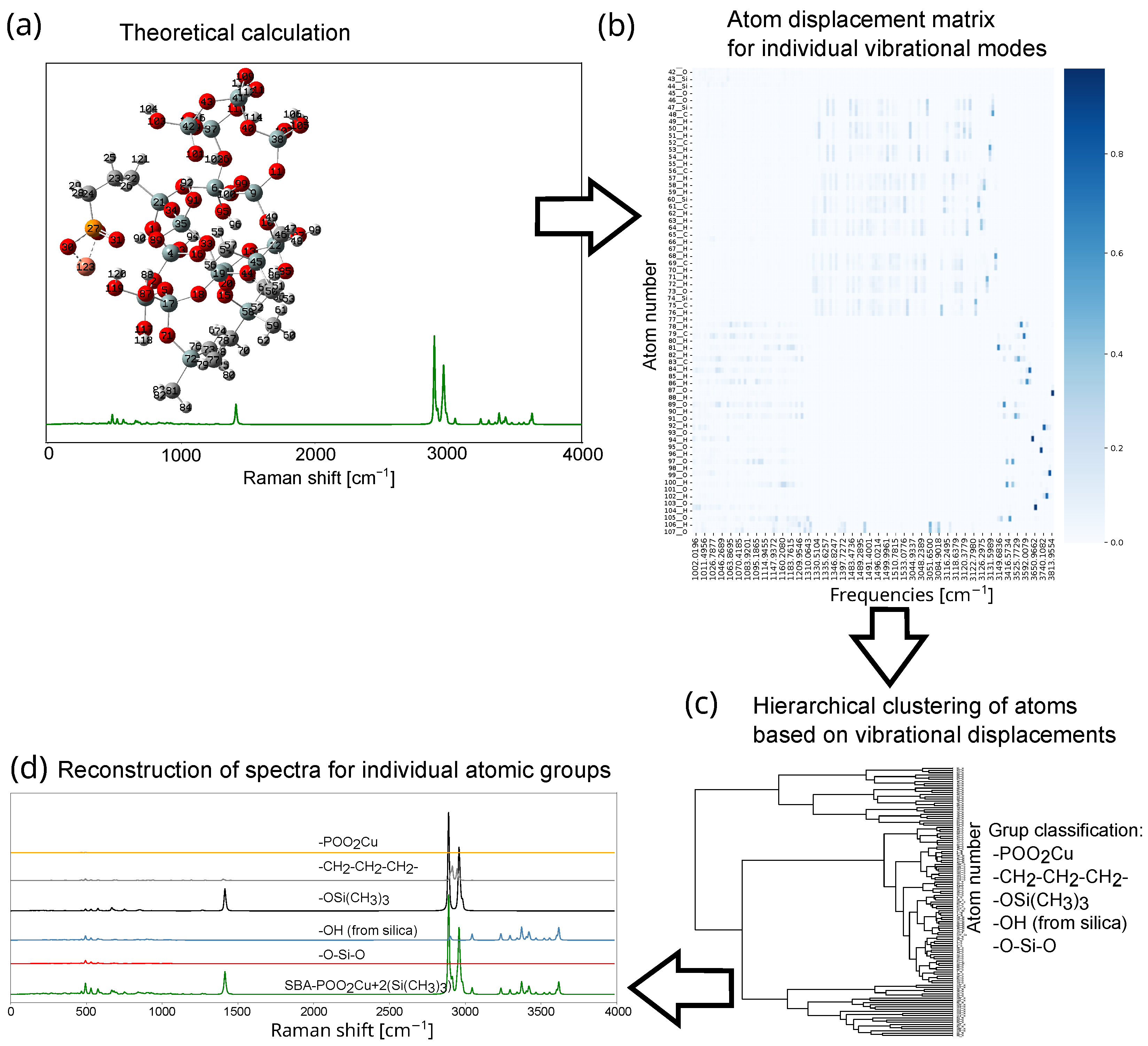
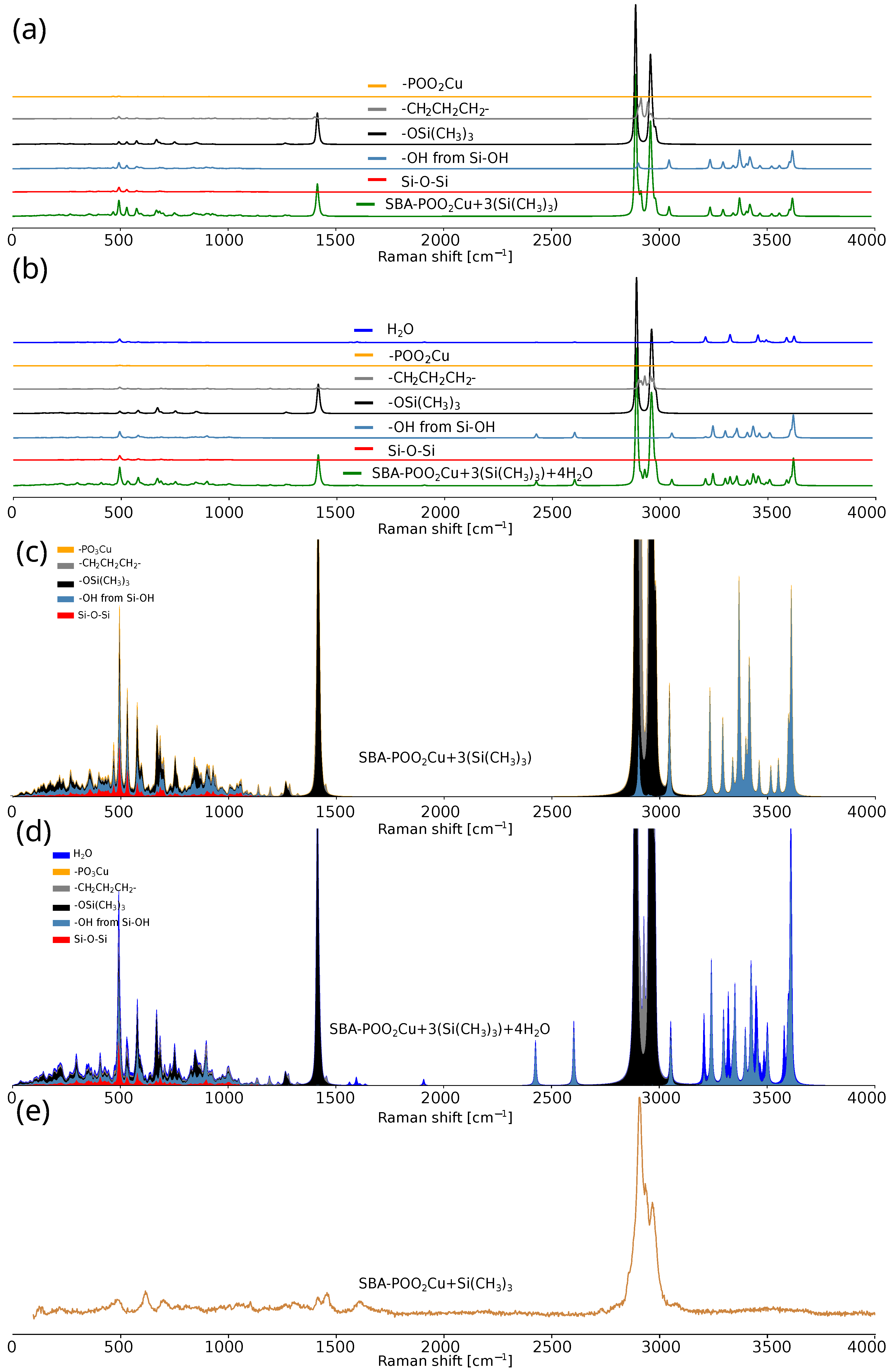
2. Results and Discussion
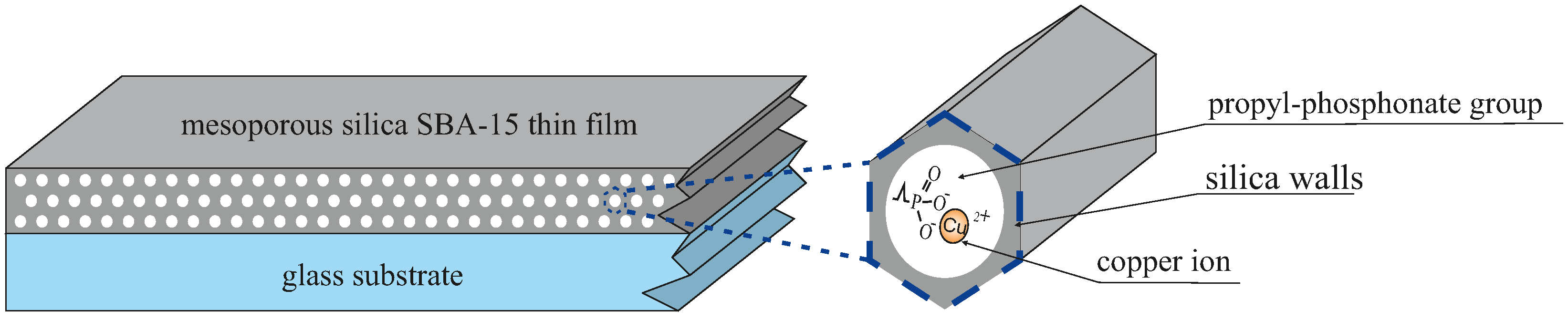
- SBA-PO(OH)2+3(Si(CH3)3) consists of four silica rings forming both planar and three-dimensional structures, three TMS groups (O-Si(CH3)3) substituting hydroxyl groups, one hydroxyl group, and one -CH2CH2CH2PO(OH)2 group, and 17 Si-H hydrogen atoms. This model consists of 124 atoms.
- SBAH-PO(OH)2+3(Si(CH3)3) is similar to SBA-PO(OH)2+3(Si(CH3)3) but lacks Si-H groups, replacing them with Si-OH. This model consists of 107 atoms.
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| B3LYP | Becke, 3-parameter, Lee–Yang–Parr |
| D3(BJ) | Grimme’s dispersion and the Becke–Johnson damping parameter |
| DFT | Density functional theory |
| HWHM | Half-width at half-maximum |
| LLM | Large language model |
| NA | Numerical aperture |
| SBA-15 | Santa Barbara Amorphous-15 |
| SERS | Surface-Enhanced Raman Spectroscopy |
| TEOS | Tetraethyl orthosilicate |
| TEM | Transmission electron microscopy |
| TERS | Tip-Enhanced Raman Scattering |
| TMS | Trimetylosilan |
| PPTES | Phosphonate propyl diethyl triethoxysilane |
| P123 | (Poly(ethylene glycol)20-poly(propylene glycol)70-poly(ethylene glycol)20 |
References
- Ashkenasy, G.; Cahen, D.; Cohen, R.; Shanzer, A.; Vilan, A. Molecular engineering of semiconductor surfaces and devices. Accounts Chem. Res. 2002, 35, 121–128. [Google Scholar] [CrossRef]
- Fagan, P.J.; Ward, M.D.; Calabrese, J.C. Molecular engineering of solid-state materials: Organometallic building blocks. J. Am. Chem. Soc. 1989, 111, 1698–1719. [Google Scholar] [CrossRef]
- Sanchez, C.; Boissiere, C.; Cassaignon, S.; Chanéac, C.; Durupthy, O.; Faustini, M.; Grosso, D.; Laberty-Robert, C.; Nicole, L.; Portehault, D.; et al. Molecular engineering of functional inorganic and hybrid materials. Chem. Mater. 2014, 26, 221–238. [Google Scholar] [CrossRef]
- Nasrollahzadeh, M.; Sajadi, S.M.; Sajjadi, M.; Issaabadi, Z. An introduction to nanotechnology. In Interface Science and Technology; Elsevier: Amsterdam, The Netherlands, 2019; Volume 28, pp. 1–27. [Google Scholar]
- Gharpure, K.M.; Wu, S.Y.; Li, C.; Lopez-Berestein, G.; Sood, A.K. Nanotechnology: Future of oncotherapy. Clin. Cancer Res. 2015, 21, 3121–3130. [Google Scholar] [CrossRef] [PubMed]
- McNeil, S.E. Nanotechnology for the biologist. J. Leukoc. Biol. 2005, 78, 585–594. [Google Scholar] [CrossRef]
- Emerich, D.F.; Thanos, C.G. Nanotechnology and medicine. Expert Opin. Biol. Ther. 2003, 3, 655–663. [Google Scholar] [CrossRef]
- Laskowska, M.; Kowalczyk, P.; Karczmarska, A.; Pogoda, K.; Zubko, M.; Laskowski, Ł. Functionalised Mesoporous Silica Thin Films as ROS-Generating Antimicrobial Coatings. Int. J. Mol. Sci. 2025, 26, 7154. [Google Scholar] [CrossRef]
- Rojo, L.; García-Fernández, L.; Aguilar, M.R.; Vázquez-Lasa, B. Antimicrobial polymeric biomaterials based on synthetic, nanotechnology, and biotechnological approaches. Curr. Opin. Biotechnol. 2022, 76, 102752. [Google Scholar] [CrossRef]
- Yin, Y.; Talapin, D. The chemistry of functional nanomaterials. Chem. Soc. Rev. 2013, 42, 2484–2487. [Google Scholar] [CrossRef]
- Sattler, K.D. Handbook of Nanophysics: Functional Nanomaterials; CRC Press: Boca Raton, FL, USA, 2010. [Google Scholar]
- Giraldo, L.; López, B.; Pérez, L.; Urrego, S.; Sierra, L.; Mesa, M. Mesoporous silica applications. In Proceedings of the Macromolecular Symposia, Buzios, State of Rio De Janeiro, Brazil, 16–20 April 2007; Wiley Online Library: Hoboken, NJ, USA, 2007; Volume 258, pp. 129–141. [Google Scholar]
- Kumar, S.; Malik, M.; Purohit, R. Synthesis methods of mesoporous silica materials. Mater. Today Proc. 2017, 4, 350–357. [Google Scholar] [CrossRef]
- Boissière, C.; Martines, M.A.; Tokumoto, M.; Larbot, A.; Prouzet, E. Mechanisms of pore size control in MSU-X mesoporous silica. Chem. Mater. 2003, 15, 509–515. [Google Scholar] [CrossRef]
- Kishor, R.; Ghoshal, A.K. APTES grafted ordered mesoporous silica KIT-6 for CO2 adsorption. Chem. Eng. J. 2015, 262, 882–890. [Google Scholar] [CrossRef]
- Henning, L.M.; Cubas, D.D.; Colmenares, M.G.; Schmidt, J.; Bekheet, M.F.; Pauw, B.R.; Gurlo, A.; Simon, U. High specific surface area ordered mesoporous silica COK-12 with tailored pore size. Microporous Mesoporous Mater. 2019, 280, 133–143. [Google Scholar] [CrossRef]
- Małecka, M.A.; Kraszkiewicz, P.; Matus, K.; Laskowska, M.; Doskocz, M. A novel method for the synthesis of bubble-like mesoporous silica—The influence of dopants. Mater. Chem. Phys. 2025, 333, 130405. [Google Scholar] [CrossRef]
- Karczmarska, A.; Zieliński, P.M.; Laskowski, Ł.; Prusik, K.; Pawlik, K.; Laskowska, M. Mesoporous Silica-Carbon Composites with Enhanced Conductivity: Analysis of Powder and Thin Film Forms. Materials 2024, 17, 6274. [Google Scholar] [CrossRef]
- El Houbbadi, S.; Laskowska, M.; Walcarius, A.; Doskocz, M.; Maximenko, A.; Olejniczak, Z.; Laskowski, Ł. Revealing the molecular structure of copper phosphonate groups anchored inside SBA-15 silica channels: Theoretical and experimental study. Appl. Surf. Sci. 2024, 669, 160425. [Google Scholar] [CrossRef]
- Laskowski, Ł.; Laskowska, M.; Walcarius, A.; Doskocz, M.; Vila, N.; Karczmarska, A.; Pawlik, P.; Goraus, J.; Balin, K.; Dulski, M. Synthesis and characterization of SBA-15 silica containing cyclam inside pores for capturing iron chloride: Analysis of interactions between ferrous chloride and cyclam in a system with strongly dispersed functional groups on the SiO2 surface. Microporous Mesoporous Mater. 2024, 378, 113243. [Google Scholar] [CrossRef]
- Kraszkiewicz, P.; Małecka, M.; Miśta, W. Sintering-resistant and highly active Au/SBA-15 catalyst for carbon monoxide oxidation. Microporous Mesoporous Mater. 2022, 346, 112338. [Google Scholar] [CrossRef]
- Yang, W.; Shirazian, S.; Soltani, R.; Zare, M.H. Bio-originated mesosilicate SBA-15: Synthesis, characterization, and application for heavy metal removal. Npj Clean Water 2024, 7, 49. [Google Scholar] [CrossRef]
- Díaz, A.; López, T.; Manjarrez, J.; Basaldella, E.; Martínez-Blanes, J.; Odriozola, J. Growth of hydroxyapatite in a biocompatible mesoporous ordered silica. Acta Biomaterialia 2006, 2, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Rehman, F.; Khan, A.J.; Sama, Z.U.; Alobaid, H.M.; Gilani, M.A.; Safi, S.Z.; Muhammad, N.; Rahim, A.; Ali, A.; Guo, J.; et al. Surface engineered mesoporous silica carriers for the controlled delivery of anticancer drug 5-fluorouracil: Computational approach for the drug-carrier interactions using density functional theory. Front. Pharmacol. 2023, 14, 1146562. [Google Scholar] [CrossRef]
- Eivazzadeh-Keihan, R.; Chenab, K.K.; Taheri-Ledari, R.; Mosafer, J.; Hashemi, S.M.; Mokhtarzadeh, A.; Maleki, A.; Hamblin, M.R. Recent advances in the application of mesoporous silica-based nanomaterials for bone tissue engineering. Mater. Sci. Eng. C 2020, 107, 110267. [Google Scholar] [CrossRef]
- Huang, J.; Liu, H.B.; Wang, J. Functionalized mesoporous silica as a fluorescence sensor for selective detection of Hg2+ in aqueous medium. Spectrochim. Acta Part A: Mol. Biomol. Spectrosc. 2021, 246, 118974. [Google Scholar] [CrossRef]
- Yuan, S.; Wang, M.; Liu, J.; Guo, B. Recent advances of SBA-15-based composites as the heterogeneous catalysts in water decontamination: A mini-review. J. Environ. Manag. 2020, 254, 109787. [Google Scholar] [CrossRef]
- Sharma, R.K.; Kaushik, B.; Yadav, S.; Rana, P.; Rana, P.; Solanki, K.; Rawat, D. Ingeniously designed Silica nanostructures as an exceptional support: Opportunities, potential challenges and future prospects for viable degradation of pesticides. J. Environ. Manag. 2022, 301, 113821. [Google Scholar] [CrossRef] [PubMed]
- Feliczak-Guzik, A.; Wawrzyńczak, A.; Nowak, I. Photocatalysis by Mixed Oxides Containing Niobium, Vanadium, Silica, or Tin. Catalysts 2025, 15, 118. [Google Scholar] [CrossRef]
- Ganji, S.; Kola, R.; Gullapelli, K.; Martha, R. Cu-Ag/SBA-15 nano catalysts for the control of microorganisms in water. Discov. Nano 2025, 20, 16. [Google Scholar] [CrossRef] [PubMed]
- Matthews, T.; Seroka, N.S.; Khotseng, L. Potential application of green synthesised SBA-15 from biomass wastes as a catalyst for sustainable biodiesel production. Discov. Appl. Sci. 2024, 6, 525. [Google Scholar] [CrossRef]
- Luján Ferreira, M.; Pedernera, M.; Esperanza Adrover, M. Enhanced CO2 capture by functionalization of SBA-15 with APTES and l-lysine. Chem. Eng. J. 2024, 498, 155431. [Google Scholar] [CrossRef]
- Lin, D.H.; Jiang, Y.X.; Wang, Y.; Sun, S.G. Silver Nanoparticles Confined in SBA-15 Mesoporous Silica and the Application as a Sensor for Detecting Hydrogen Peroxide. J. Nanomater. 2008, 2008, 473791. [Google Scholar] [CrossRef]
- Laskowski, Ł.; Laskowska, M.; Vila, N.; Schabikowski, M.; Walcarius, A. Mesoporous Silica-Based Materials for Electronics-Oriented Applications. Molecules 2019, 24, 2395. [Google Scholar] [CrossRef]
- Deryło-Marczewska, A.; Zienkiewicz-Strzałka, M.; Skrzypczyńska, K.; Świątkowski, A.; Kuśmierek, K. Evaluation of the SBA-15 materials ability to accumulation of 4-chlorophenol on carbon paste electrode. Adsorption 2016, 22, 801–812. [Google Scholar] [CrossRef]
- Liang, B.; Zhu, P.; Gu, J.; Yuan, W.; Xiao, B.; Hu, H.; Rao, M. Advancing Adsorption and Separation with Modified SBA-15: A Comprehensive Review and Future Perspectives. Molecules 2024, 29, 3543. [Google Scholar] [CrossRef]
- Laskowska, M.; Kityk, I.; Pastukh, O.; Dulski, M.; Zubko, M.; Jedryka, J.; Cpałka, K.; Zieliński, P.M.; Laskowski, Ł. Nanocomposite for photonics—Nickel pyrophosphate nanocrystals synthesised in silica nanoreactors. Microporous Mesoporous Mater. 2020, 306, 110435. [Google Scholar] [CrossRef]
- Wang, M.; Xia, Y.; Qiu, J.; Ren, X. Carbon quantum dots embedded mesoporous silica for rapid fluorescent detection of acidic gas. Spectrochim. Acta Part A: Mol. Biomol. Spectrosc. 2019, 206, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Poryvaev, A.S.; Gjuzi, E.; Polyukhov, D.M.; Hoffmann, F.; Fröba, M.; Fedin, M.V. Blatter-Radical-Grafted Mesoporous Silica as Prospective Nanoplatform for Spin Manipulation at Ambient Conditions. Angew. Chem. Int. Ed. 2021, 60, 8683–8688. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.F.; Hu, Z.B.; Shao, C.; Ouyang, Z.; Wang, Z.; Song, Y. Molecular Spin Qubits Impregnated in a Hexagonal Self-Ordered Mesoporous Silica. Chem. Mater. 2022, 34, 8427–8436. [Google Scholar] [CrossRef]
- Laskowski, Ł.; Laskowska, M.; Jelonkiewicz, J.; Boullanger, A. Molecular approach to hopfield neural network. In Proceedings of the Artificial Intelligence and Soft Computing: 14th International Conference, ICAISC 2015, Zakopane, Poland, 14–18 June 2015; Proceedings, Part I 14; Springer: Heidelberg, Germany, 2015; pp. 72–78. [Google Scholar]
- Jaafar, A.H.; Shao, L.; Dai, P.; Zhang, T.; Han, Y.; Beanland, R.; Kemp, N.T.; Bartlett, P.N.; Hector, A.L.; Huang, R. 3D-structured mesoporous silica memristors for neuromorphic switching and reservoir computing. Nanoscale 2022, 14, 17170–17181. [Google Scholar] [CrossRef]
- Sigle, M.; Rohlfing, A.K.; Kenny, M.; Scheuermann, S.; Sun, N.; Graeßner, U.; Haug, V.; Sudmann, J.; Seitz, C.M.; Heinzmann, D.; et al. Translating genomic tools to Raman spectroscopy analysis enables high-dimensional tissue characterization on molecular resolution. Nat. Commun. 2023, 14, 5799. [Google Scholar] [CrossRef]
- Laskowska, M.; Laskowski, L.; Jelonkiewicz, J. SBA-15 mesoporous silica activated by metal ions—Verification of molecular structure on the basis of Raman spectroscopy supported by numerical simulations. J. Mol. Struct. 2015, 1100, 21–26. [Google Scholar] [CrossRef]
- Laskowski, L.; Laskowska, M. Functionalization of SBA-15 mesoporous silica by Cu-phosphonate units: Probing of synthesis route. J. Solid State Chem. 2014, 220, 221–226. [Google Scholar] [CrossRef]
- Szymanski, N.J.; Rendy, B.; Fei, Y.; Kumar, R.E.; He, T.; Milsted, D.; McDermott, M.J.; Gallant, M.; Cubuk, E.D.; Merchant, A.; et al. An autonomous laboratory for the accelerated synthesis of novel materials. Nature 2023, 624, 86–91. [Google Scholar] [CrossRef]
- Xie, Y.; Sattari, K.; Zhang, C.; Lin, J. Toward autonomous laboratories: Convergence of artificial intelligence and experimental automation. Prog. Mater. Sci. 2023, 132, 101043. [Google Scholar] [CrossRef]
- Lo, S.; Baird, S.G.; Schrier, J.; Blaiszik, B.; Carson, N.; Foster, I.; Aguilar-Granda, A.; Kalinin, S.V.; Maruyama, B.; Politi, M.; et al. Review of low-cost self-driving laboratories in chemistry and materials science: The “frugal twin” concept. Digit. Discov. 2024, 3, 842–868. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian, 16th ed.; Gaussian Inc.: Wallingford, UK, 2016. [Google Scholar]
- Marchand, N.; Lienard, P.; Siehl, H.; Izato, H. Applications of molecular simulation software SCIGRESS in industry and university. Fujitsu Sci. Tech. J. 2014, 50, 46–51. [Google Scholar]
- Bursch, M.; Mewes, J.M.; Hansen, A.; Grimme, S. Best-Practice DFT Protocols for Basic Molecular Computational Chemistry**. Angew. Chem. Int. Ed. 2022, 61, e202205735. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, D.; Xia, Y.; Huang, Y.P.; Lin, X.; Han, X.; Ni, N.; Wang, Z.; Yu, F.; Yang, L.; et al. Artificial Intelligence Enhanced Molecular Simulations. J. Chem. Theory Comput. 2023, 19, 4338–4350. [Google Scholar] [CrossRef] [PubMed]
- Dral, P.O. AI in computational chemistry through the lens of a decade-long journey. Chem. Commun. 2024, 60, 3240–3258. [Google Scholar] [CrossRef]
- Kebabsa, A.; Maurel, F.; Brémond, Ł. Boosting the Modeling of Infrared and Raman Spectra of Bulk Phase Chromophores with Machine Learning. J. Chem. Theory Comput. 2024, 20, 7009–7015. [Google Scholar] [CrossRef]
- Lappala, A. The next revolution in computational simulations: Harnessing AI and quantum computing in molecular dynamics. Curr. Opin. Struct. Biol. 2024, 89, 102919. [Google Scholar] [CrossRef]
- Xu, N.; Rosander, P.; Schäfer, C.; Lindgren, E.; Österbacka, N.; Fang, M.; Chen, W.; He, Y.; Fan, Z.; Erhart, P. Tensorial Properties via the Neuroevolution Potential Framework: Fast Simulation of Infrared and Raman Spectra. J. Chem. Theory Comput. 2024, 20, 3273–3284. [Google Scholar] [CrossRef]
- Eckwert, J.; Ahmed, R.A.; Kopp, W.A.; Leonhard, K. Simulation of Raman-Spectra of water using machine learning potentials. Chem. Phys. 2025, 595, 112698. [Google Scholar] [CrossRef]
- Li, Y.; Lee, D.K.J.; Cai, P.; Zhang, Z.; Gorai, P.; Canepa, P. A database of computed Raman spectra of inorganic compounds with accurate hybrid functionals. Sci. Data 2024, 11, 105. [Google Scholar] [CrossRef] [PubMed]
- Landau, L.; Lifshitz, E.; Reichl, L.E. Statistical Physics; Part 1; Butterworth-Heinemann: Oxford, UK, 1981. [Google Scholar]
- Brown, B.P.; Stein, R.A.; Meiler, J.; Mchaourab, H.S. Approximating Projections of Conformational Boltzmann Distributions with AlphaFold2 Predictions: Opportunities and Limitations. J. Chem. Theory Comput. 2024, 20, 1434–1447. [Google Scholar] [CrossRef] [PubMed]
- Pasterny, K.; Wrzalik, R.; Kupka, T.; Pasterna, G. Theoretical and experimental vibrational studies on liquid thiophene and its acetonitrile solution. J. Mol. Struct. 2002, 614, 297–304. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, D.; Zhao, Z.; Chen, Y.; Lan, J. A DFT study of the structural units in SBA-15 mesoporous molecular sieve. Comput. Theor. Chem. 2011, 963, 403–411. [Google Scholar] [CrossRef]
- Hanrahan, M.P.; Fought, E.L.; Windus, T.L.; Wheeler, L.M.; Anderson, N.C.; Neale, N.R.; Rossini, A.J. Characterization of Silicon Nanocrystal Surfaces by Multidimensional Solid-State NMR Spectroscopy. Chem. Mater. 2017, 29, 10339–10351. [Google Scholar] [CrossRef]
- Venetos, M.C.; Wen, M.; Persson, K.A. Machine Learning Full NMR Chemical Shift Tensors of Silicon Oxides with Equivariant Graph Neural Networks. J. Phys. Chem. A 2023, 127, 2388–2398. [Google Scholar] [CrossRef] [PubMed]
- Kaur, N.; Gupta, S.; Goel, N. Enantioselective synthesis of sulfoxide using an SBA-15 supported vanadia catalyst: A computational elucidation using a QM/MM approach. Phys. Chem. Chem. Phys. 2017, 19, 25059–25070, Correction in Phys. Chem. Chem. Phys. 2018, 20, 1293–1293.. [Google Scholar] [CrossRef]
- Laskowski, L.; Laskowska, M.; Jelonkiewicz, J.; Galkowski, T.; Pawlik, P.; Piech, H.; Doskocz, M. Iron Doped SBA-15 Mesoporous Silica Studied by Mossbauer Spectroscopy. J. Nanomater. 2016, 2016, 160425. [Google Scholar] [CrossRef]
- Katari, M.; Nicol, E.; Steinmetz, V.; van der Rest, G.; Carmichael, D.; Frison, G. Improved Infrared Spectra Prediction by DFT from a New Experimental Database. Chem. A Eur. J. 2017, 23, 8414–8423. [Google Scholar] [CrossRef]
- Jacobsen, R.L.; Johnson, R.D.; Irikura, K.K.; Kacker, R.N. Anharmonic Vibrational Frequency Calculations Are Not Worthwhile for Small Basis Sets. J. Chem. Theory Comput. 2013, 9, 951–954. [Google Scholar] [CrossRef] [PubMed]
- Cornaton, Y.; Ringholm, M.; Louant, O.; Ruud, K. Analytic calculations of anharmonic infrared and Raman vibrational spectra. Phys. Chem. Chem. Phys. 2016, 18, 4201–4215. [Google Scholar] [CrossRef]
- Hermsen, A.; Hertel, F.; Wilbert, D.; Mayer, C.; Jaeger, M. MD and DFT Calculations to Analyze Raman and SERS Spectra of Paraquat—From Computer Aided Spectra Interpretation to Pesticide Identification. Appl. Res. 2024, 4, e2400182. [Google Scholar] [CrossRef]
- Demtröder, W. Laser Spectroscopy; Springer: Berlin/Heidelberg, Germany, 1973; Volume 2. [Google Scholar]
- Bi, X.; Czajkowsky, D.M.; Shao, Z.; Ye, J. Digital colloid-enhanced Raman spectroscopy by single-molecule counting. Nature 2024, 628, 771–775. [Google Scholar] [CrossRef]
- Dennington, R.; Keith, T.A.; Millam, J.M. GaussView, 6th ed.; Semichem Inc.: Shawnee Mission, KS, USA, 2019. [Google Scholar]
- Parker, S.F. The Analysis of Vibrational Spectra: Past, Present and Future. ChemPlusChem 2024, 90, e202400461. [Google Scholar] [CrossRef]
- Teixeira, F.; Cordeiro, M.N.D.S. Improving Vibrational Mode Interpretation Using Bayesian Regression. J. Chem. Theory Comput. 2018, 15, 456–470. [Google Scholar] [CrossRef] [PubMed]
- Oenen, K.; Dinu, D.F.; Liedl, K.R. Determining internal coordinate sets for optimal representation of molecular vibration. J. Chem. Phys. 2024, 160, 014104. [Google Scholar] [CrossRef]
- Kamiya, K.; Sugawara, Y.; Umeyama, H. Algorithm for normal mode analysis with general internal coordinates. J. Comput. Chem. 2003, 24, 826–841. [Google Scholar] [CrossRef] [PubMed]
- Kraka, E.; Quintano, M.; La Force, H.W.; Antonio, J.J.; Freindorf, M. The Local Vibrational Mode Theory and Its Place in the Vibrational Spectroscopy Arena. J. Phys. Chem. A 2022, 126, 8781–8798. [Google Scholar] [CrossRef]
- Wilson, E.; Decius, J.; Cross, P. Molecular Vibrations: The Theory of Infrared and Raman Vibrational Spectra; Dover Books on Chemistry Series; Dover Publications: Mineola, NY, USA, 1980. [Google Scholar]
- Xing, J.; Takeuchi, K.; Kamei, K.; Nakamuro, T.; Harano, K.; Nakamura, E. Atomic-number (Z)-correlated atomic sizes for deciphering electron microscopic molecular images. Proc. Natl. Acad. Sci. USA 2022, 119, e2114432119. [Google Scholar] [CrossRef]
- Dubey, A.; Jauhri, A.; Pandey, A.; Kadian, A.; Al-Dahle, A.; Letman, A.; Mathur, A.; Schelten, A.; Yang, A.; Fan, A.; et al. The Llama 3 Herd of Models. arXiv 2024. [Google Scholar] [CrossRef]
- Laskowski, L.; Laskowska, M.; Jelonkiewicz, J.; Dulski, M.; Wojtyniak, M.; Fitta, M.; Balanda, M. SBA-15 mesoporous silica free-standing thin films containing copper ions bounded via propyl phosphonate units-preparation and characterization. J. Solid State Chem. 2016, 241, 143–151. [Google Scholar] [CrossRef]
- Corriu, R.; Mehdi, A.; Reyé, C. Nanoporous materials: A good opportunity for nanosciences. J. Organomet. Chem. 2004, 689, 4437–4450. [Google Scholar] [CrossRef]
- Doskocz, M.; Kubas, K.; Frąckowiak, A.; Gancarz, R. NMR and ab initio studies of Mg2+, Ca2+, Zn2+, Cu2+ alizarin complexes. Polyhedron 2009, 28, 2201–2205. [Google Scholar] [CrossRef]
- Persson, I. Structure and size of complete hydration shells of metal ions and inorganic anions in aqueous solution. Dalton Trans. 2024, 53, 15517–15538. [Google Scholar] [CrossRef] [PubMed]
- Becke, A.D. Density-functional thermochemistry. I. The effect of the exchange-only gradient correction. J. Chem. Phys. 1992, 96, 2155–2160. [Google Scholar] [CrossRef]
- Stephens, P.J.; Devlin, F.J.; Chabalowski, C.F.; Frisch, M.J. Ab Initio Calculation of Vibrational Absorption and Circular Dichroism Spectra Using Density Functional Force Fields. J. Phys. Chem. 1994, 98, 11623–11627. [Google Scholar] [CrossRef]
- Grimme, S.; Ehrlich, S.; Goerigk, L. Effect of the damping function in dispersion corrected density functional theory. J. Comput. Chem. 2011, 32, 1456–1465. [Google Scholar] [CrossRef]
- Millman, K.J.; Aivazis, M. Python for scientists and engineers. Comput. Sci. Eng. 2011, 13, 9–12. [Google Scholar] [CrossRef]
- Bressert, E. SciPy and NumPy: An Overview for Developers; O’Reilly Media: Sebastopol, CA, USA, 2012. [Google Scholar]
- McKinney, W. pandas: A foundational Python library for data analysis and statistics. Python High Perform. Sci. Comput. 2011, 14, 1–9. [Google Scholar]
- Perkel, J.M. Why Jupyter is data scientists’ computational notebook of choice. Nature 2018, 563, 145–146. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doskocz, M.; Laskowski, Ł.; Kujawski, J.; Karczmarska, A.; Cpałka, K.; Lipiec, E.; Laskowska, M. Decomposition Analysis of Theoretical Raman Spectra for Efficient Interpretation of Experimental Spectra of Thin-Film Functional Materials. Int. J. Mol. Sci. 2025, 26, 10237. https://doi.org/10.3390/ijms262010237
Doskocz M, Laskowski Ł, Kujawski J, Karczmarska A, Cpałka K, Lipiec E, Laskowska M. Decomposition Analysis of Theoretical Raman Spectra for Efficient Interpretation of Experimental Spectra of Thin-Film Functional Materials. International Journal of Molecular Sciences. 2025; 26(20):10237. https://doi.org/10.3390/ijms262010237
Chicago/Turabian StyleDoskocz, Marek, Łukasz Laskowski, Jacek Kujawski, Agnieszka Karczmarska, Krzysztof Cpałka, Ewelina Lipiec, and Magdalena Laskowska. 2025. "Decomposition Analysis of Theoretical Raman Spectra for Efficient Interpretation of Experimental Spectra of Thin-Film Functional Materials" International Journal of Molecular Sciences 26, no. 20: 10237. https://doi.org/10.3390/ijms262010237
APA StyleDoskocz, M., Laskowski, Ł., Kujawski, J., Karczmarska, A., Cpałka, K., Lipiec, E., & Laskowska, M. (2025). Decomposition Analysis of Theoretical Raman Spectra for Efficient Interpretation of Experimental Spectra of Thin-Film Functional Materials. International Journal of Molecular Sciences, 26(20), 10237. https://doi.org/10.3390/ijms262010237







