Genetic Variation and Genome-Enabled Prediction of White Lupin Frost Resistance in Different Reference Populations
Abstract
1. Introduction
2. Results
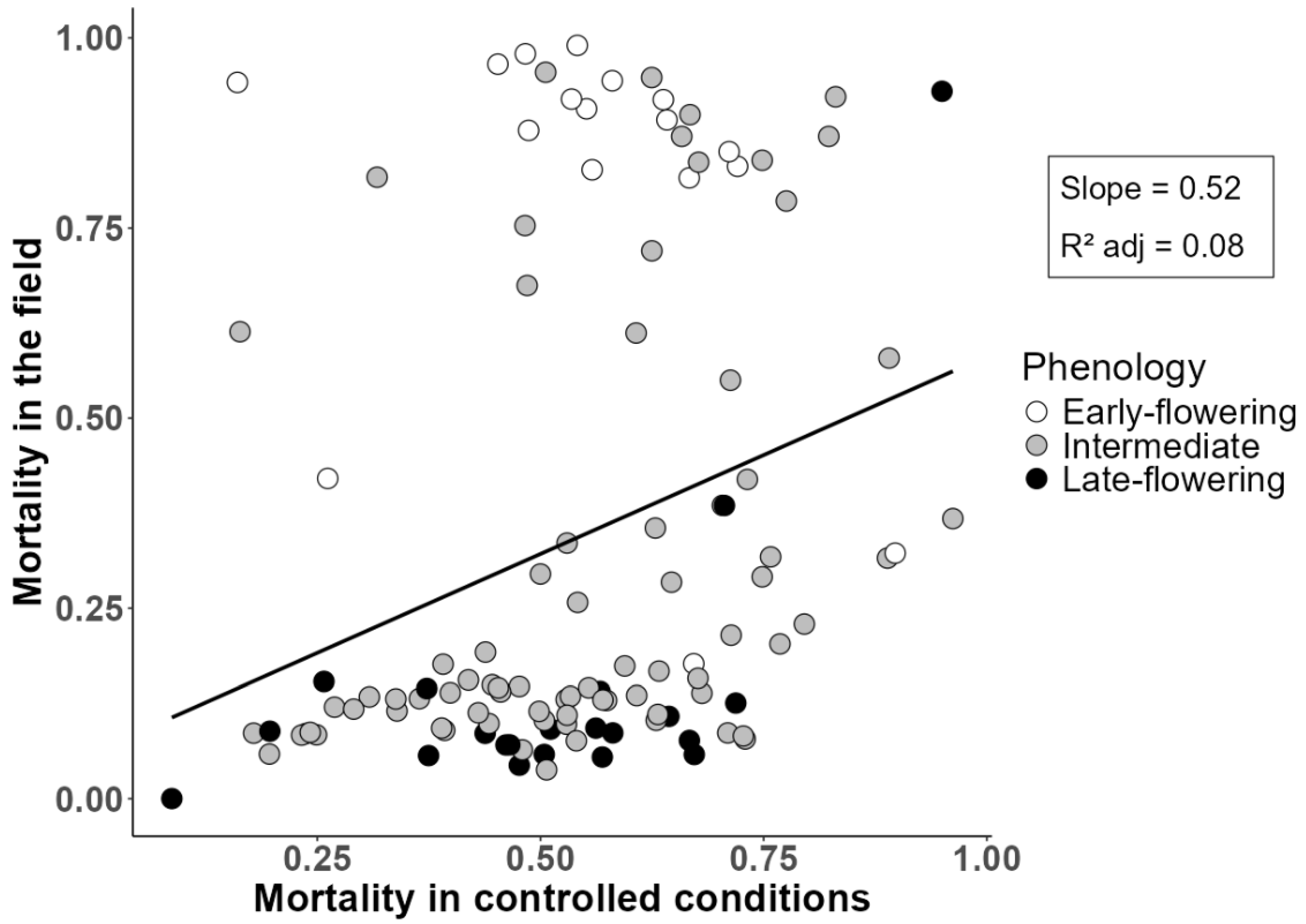
2.1. Frost Resistance Variation and Relationship with Field-Based Winter Mortality
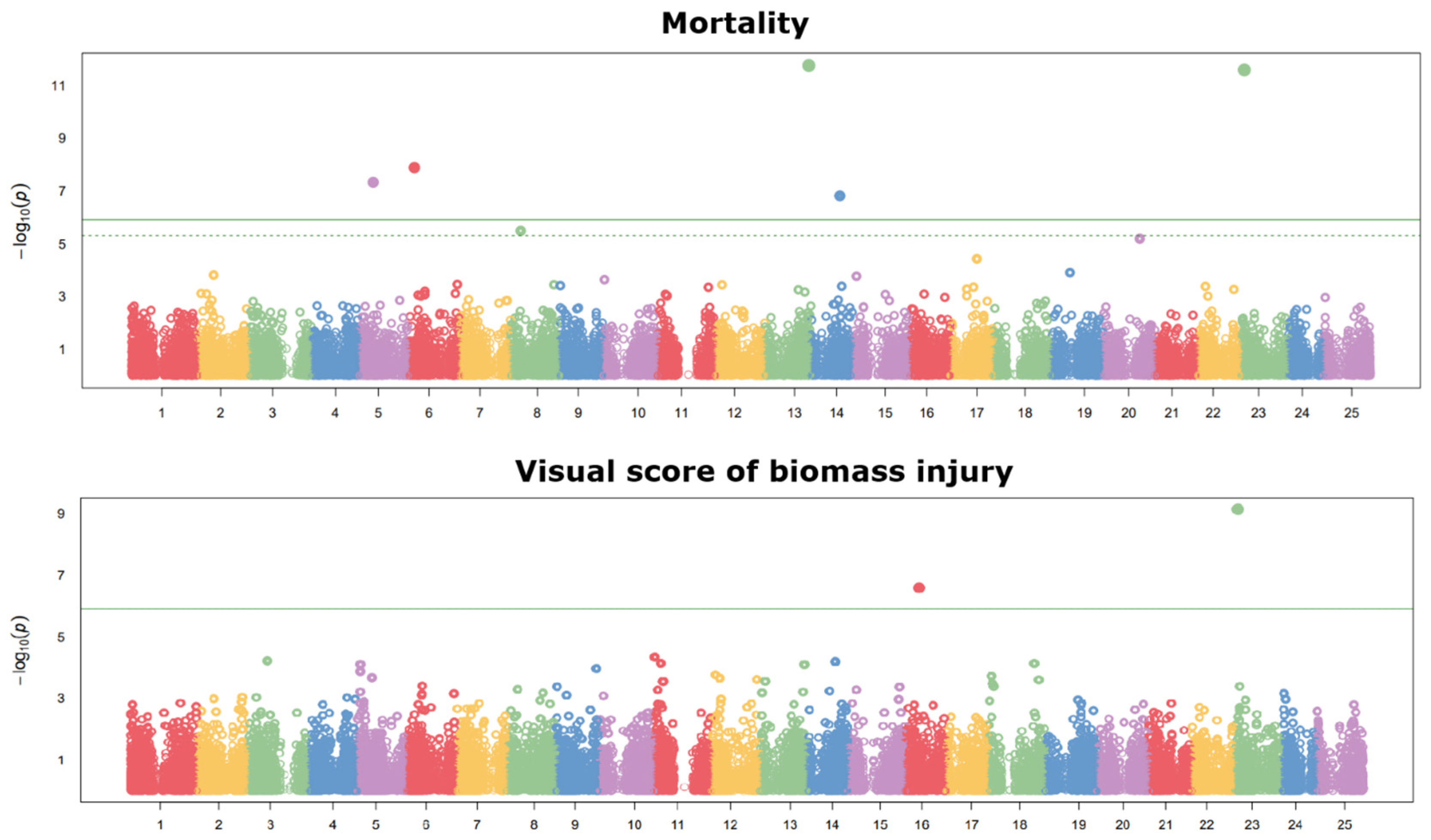
2.2. Analysis of Linkage Disequilibrium Decay and Population Structure, and Genome-Wide Association Study
2.3. Genomic Selection
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Phenotyping
4.3. Phenotypic Data Analysis
4.4. DNA Isolation, GBS Library Construction, and Sequencing
4.5. Genotype SNP Calling Procedures, Data Filtering and Imputation
4.6. Analysis of Linkage Disequilibrium Decay, Population Structure, and Genome-Wide Association Study
4.7. Genomic Selection
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GBS | Genotyping-by-sequencing |
| SNP | Single-Nucleotide Polymorphism |
| FDR | False discovery rate |
| LD | Linkage disequilibrium |
| ANOVA | Analysis of variance |
| BLUE | Best linear unbiased estimate |
| BLUP | Best linear unbiased prediction |
| rrBLUP | Ridge regression best linear unbiased prediction |
| BL | Bayesian Lasso |
References
- Nemecek, T.; von Richthofen, J.S.; Dubois, G.; Casta, P.; Charles, R.; Pahl, H. Environmental impacts of introducing grain legumes into European crop rotations. Eur. J. Agron. 2008, 28, 380–393. [Google Scholar] [CrossRef]
- Watson, C.A.; Reckling, M.; Preissel, S.; Bachinger, J.; Bergkvist, G.; Kuhlman, T.; Lindström, K.; Nemecek, T.; Topp, C.F.E.; Vanhatalo, A.; et al. Grain legume production and use in European agricultural systems. Adv. Agron. 2017, 144, 235–303. [Google Scholar] [CrossRef]
- Boerema, A.; Peeters, A.; Swolfs, S.; Vandevenne, F.; Jacobs, S.; Staes, J.; Meire, P. Soybean trade: Balancing environmental and socio-economic impacts of an intercontinental market. PLoS ONE 2016, 11, e0155222. [Google Scholar] [CrossRef]
- Tadele, Y. White lupin (Lupinus albus) grain, a potential source of protein for ruminants: A review. Res. J. Agric. Environ. Manag. 2015, 4, 180–188. [Google Scholar]
- Annicchiarico, P. Adaptation of cool-season grain legume species across climatically-contrasting environments of southern Europe. Agron. J. 2008, 100, 1647–1654. [Google Scholar] [CrossRef]
- Prusinski, J. White lupin (Lupinus albus L.)—Nutritional and health values in human nutrition—A review. Czech J. Food Sci. 2017, 35, 95–105. [Google Scholar] [CrossRef]
- Boschin, G.; D’Agostina, A.; Annicchiarico, P.; Arnoldi, A. Effect of genotype and environment on fatty acid composition of Lupinus albus L. seed. Food Chem. 2008, 108, 600–606. [Google Scholar] [CrossRef]
- Boukid, F.; Pasqualone, A. Lupine (Lupinus spp.) proteins: Characteristics, safety and food applications. Eur. Food. Res. Technol. 2022, 248, 345–356. [Google Scholar] [CrossRef]
- Quintieri, L.; Nitride, C.; De Angelis, E.; Lamonaca, A.; Pilolli, R.; Russo, F.; Monaci, L. Alternative protein sources and novel foods: Benefits, food applications and safety issues. Nutrients 2023, 15, 1509. [Google Scholar] [CrossRef]
- Gresta, F.; Wink, M.; Prins, U.; Abberton, M.; Capraro, J.; Scarafoni, A.; Hill, G. Lupins in European cropping systems. In Legumes in Cropping Systems; Murphy-Bokern, D., Stoddard, F.L., Watson, C.A., Eds.; CAB International: Wallingford, UK, 2017; pp. 88–108. [Google Scholar]
- Shield, I.F.; Scott, T.; Stevenson, H.J.; Leach, J.E.; Todd, A.D. The causes of over-winter plant losses of autumn-sown white lupins (Lupinus albus) in different regions of the UK over three seasons. J. Agric. Sci. 2000, 135, 173–183. [Google Scholar] [CrossRef]
- López-Bellido, L.; Fuentes, M.; Lhamby, J.C.B.; Castillo, J.E. Growth and yield of white lupin (Lupinus albus) under Mediterranean conditions: Effect of sowing date. Field Crops Res. 1994, 36, 87–94. [Google Scholar] [CrossRef]
- Annicchiarico, P.; Iannucci, A. Winter survival of pea, faba bean and white lupin cultivars in contrasting Italian locations and sowing times, and implications for selection. J. Agric. Sci. 2007, 145, 611–622. [Google Scholar] [CrossRef]
- Francis, J.; Skific, N. Evidence linking rapid Arctic warming to mid-latitude weather patterns. Philos. Trans. R. Soc. A 2015, 373, 20140170. [Google Scholar] [CrossRef]
- Annicchiarico, P.; Harzic, N.; Carroni, A.M. Adaptation, diversity, and exploitation of global white lupin (Lupinus albus L.) landrace genetic resources. Field Crops Res. 2010, 119, 114–124. [Google Scholar] [CrossRef]
- Huyghe, C. White lupin (Lupinus albus L.). Field Crops Res. 1997, 53, 147–160. [Google Scholar] [CrossRef]
- Annicchiarico, P.; Romani, M.; Pecetti, L. White lupin variation for adaptation to severe drought stress. Plant Breed. 2018, 137, 782–789. [Google Scholar] [CrossRef]
- Pecetti, L.; Annicchiarico, P.; Crosta, M.; Notario, T.; Ferrari, B.; Nazzicari, N. White lupin drought tolerance: Genetic variation, trait genetic architecture, and genome-enabled prediction. Int. J. Mol. Sci. 2023, 24, 2351. [Google Scholar] [CrossRef]
- Franguelli, N.; Cavalli, D.; Notario, T.; Pecetti, L.; Annicchiarico, P. Frost tolerance improvement in pea and white lupin by a high-throughput phenotyping platform. Front. Plant Sci. 2024, 15, 1490577. [Google Scholar] [CrossRef]
- Levitt, J. Responses of Plants to Environmental Stresses, 2nd ed.; Academic Press: New York, NY, USA, 1980; Volume 1, pp. 67–346. [Google Scholar]
- Sallam, A.; Martsch, R.; Moursi, Y.S. Genetic variation in morpho-physiological traits associated with frost tolerance in faba bean (Vicia faba L.). Euphytica 2015, 205, 395–408. [Google Scholar] [CrossRef]
- Beck, E.H.; Heim, R.; Hansen, J. Plant resistance to cold stress: Mechanisms and environmental signals triggering frost hardening and dehardening. J. Biosci. 2004, 29, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Junttila, O. Plant adaptation to temperature and photoperiod. Agric. Food Sci. 1996, 5, 251–260. [Google Scholar] [CrossRef]
- Bourion, V.; Lejeune-Hénaut, I.; Munier-Jolain, N.; Salon, C. Cold acclimation of winter and spring peas: Carbon partitioning as affected by light intensity. Eur. J. Agron. 2003, 19, 535–548. [Google Scholar] [CrossRef]
- Arbaoui, M.; Link, W. Effect of hardening on frost tolerance and fatty acid composition of leaves and stems of a set of faba bean (Vicia faba L.) genotypes. Euphytica 2008, 162, 211–219. [Google Scholar] [CrossRef]
- Link, W.; Balko, C.; Stoddard, F.L. Winter hardiness in faba bean: Physiology and breeding. Field Crops Res. 2010, 115, 287–296. [Google Scholar] [CrossRef]
- Papineau, J.; Huyghe, C. Le Lupin Doux Protéagineux; Editions France Agricole: Paris, France, 2004. [Google Scholar]
- Avia, K.; Pilet-Nayel, M.L.; Bahrman, N.; Baranger, A.; Delbreil, B.; Fontaine, V.; Hamon, C.; Hanocq, E.; Niarquin, M.; Sellier, H.; et al. Genetic variability and QTL mapping of freezing tolerance and related traits in Medicago truncatula. Theor. Appl. Genet. 2013, 126, 2353–2366. [Google Scholar] [CrossRef]
- Stoddard, F.L.; Balko, C.; Erskine, W.; Khan, H.R.; Link, W.; Sarker, A. Screening techniques and sources of resistance to abiotic stresses in cool-season food legumes. Euphytica 2006, 147, 167–186. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Książkiewicz, M.; Nazzicari, N.; Yang, H.; Nelson, M.; Renshaw, D.; Rychel-Bielska, S.; Ferrari, B.; Carelli, M.; Tomaszewska, M.; Stawiński, S.; et al. A high-density consensus linkage map of white lupin highlights synteny with narrow-leafed lupin and provides markers tagging key agronomic traits. Sci. Rep. 2017, 7, 15335. [Google Scholar] [CrossRef]
- Annicchiarico, P.; Nazzicari, N.; Ferrari, B.; Harzic, N.; Carroni, A.M.; Romani, M.; Pecetti, L. Genomic prediction of grain yield in contrasting environments for white lupin genetic resources. Mol. Breed. 2019, 39, 142. [Google Scholar] [CrossRef]
- Annicchiarico, P.; de Buck, A.J.; Vlachostergios, D.N.; Heupink, D.; Koskosidis, A.; Nazzicari, N.; Crosta, M. White lupin adaptation to moderately calcareous soils: Phenotypic variation and genome-enabled prediction. Plants 2023, 12, 1139. [Google Scholar] [CrossRef]
- Alkemade, J.A.; Nazzicari, N.; Messmer, M.M.; Annicchiarico, P.; Ferrari, B.; Voegele, R.T.; Finckh, M.R.; Arncken, C.; Hohmann, P. Genome-wide association study reveals white lupin candidate gene involved in anthracnose resistance. Theor. Appl. Genet. 2022, 135, 1011–1024. [Google Scholar] [CrossRef]
- Schwertfirm, G.; Schneider, M.; Haase, F.; Riedel, C.; Lazzaro, M.; Rege-Wehling, B.; Schweizer, G. Genome-wide association study revealed significant SNPs for anthracnose resistance, seed alkaloids and protein content in white lupin. Theor. Appl. Genet. 2024, 137, 155. [Google Scholar] [CrossRef]
- Crosta, M.; Romani, M.; Nazzicari, N.; Ferrari, B.; Annicchiarico, P. Genomic prediction and allele mining of agronomic and morphophysiological traits in pea (Pisum sativum) germplasm collections. Front. Plant Sci. 2023, 14, 1320506. [Google Scholar] [CrossRef] [PubMed]
- Hufnagel, B.; Marques, A.; Soriano, A.; Marquès, L.; Divol, F.; Patrick, D.; Sallet, E.; Mancinotti, D.; Carrere, S.; Marande, W.; et al. High-quality genome sequence of white lupin provides insight into soil exploration and seed quality. Nat. Commun. 2020, 11, 492. [Google Scholar] [CrossRef]
- Sallam, A.; Arbaoui, M.; El-Esawi, M.; Abshire, N.; Martsch, R. Identification and verification of QTL associated with frost tolerance using linkage mapping and GWAS in winter faba bean. Front. Plant Sci. 2016, 7, 1098. [Google Scholar] [CrossRef]
- Lejeune-Hénaut, I.; Hanocq, E.; Béthencourt, L.; Fontaine, V.; Delbreil, B.; Morin, J. The flowering locus Hr colocalizes with a major QTL affecting winter frost tolerance in Pisum sativum L. Theor. Appl. Genet. 2008, 116, 1105–1116. [Google Scholar] [CrossRef]
- Beji, S.; Fontaine, V.; Devaux, R.; Thomas, M.; Negro, S.S.; Bahrman, N.; Siol, M.; Aubert, G.; Burstin, J.; Hilbert, J.L.; et al. Genome-wide association study identifies favorable SNP alleles and candidate genes for frost tolerance in pea. BMC Genom. 2020, 21, 536. [Google Scholar] [CrossRef] [PubMed]
- Zanotto, S.; Ruttink, T.; Pégard, M.; Skøt, L.; Grieder, C.; Kölliker, R.; Ergon, Å. A genome-wide association study of freezing tolerance in red clover (Trifolium pratense L.) germplasm of European origin. Front. Plant Sci. 2023, 14, 1189662. [Google Scholar] [CrossRef]
- Annicchiarico, P.; Osorio, C.; Nazzicari, N.; Ferrari, B.; Barzaghi, S.; Biazzi, E.; Tava, A.; Pecetti, L.; Notario, T.; Romani, M.; et al. Genetic variation and genome-enabled selection of white lupin for key seed quality traits. BMC Genom. 2025, 26, 922. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, K.N.; Buirchell, B.J.; Sweetingham, M.W. Length of vernalization period affects flowering time in three lupin species. Plant Breed. 2012, 131, 631–636. [Google Scholar] [CrossRef]
- Huyghe, C.; Papineau, J. Winter development of autumn sown white lupin: Agronomic and breeding consequences. Agronomie 1990, 10, 709–716. [Google Scholar] [CrossRef]
- Rychel-Bielska, S.; Bielski, W.; Surma, A.; Annicchiarico, P.; Belter, J.; Kozak, B.; Galek, R.; Nathalie, H.; Książkiewicz, M. A GWAS study highlights significant associations between a series of indels in a FLOWERING LOCUS T gene promoter and flowering time in white lupin (Lupinus albus L.). BMC Plant Biol. 2024, 24, 772. [Google Scholar] [CrossRef] [PubMed]
- Homer, A.; Şahin, M.; Küçüközdemir, Ü. Evaluation of pea (Pisum sativum L.) germplasm for winter hardiness in Central Anatolia Turkey using field controlled environment Czech. J. Genet. Plant Breed. 2016, 52, 55–63. [Google Scholar] [CrossRef]
- Auld, D.L.; Ditterline, R.L.; Murray, G.A.; Swensen, J.B. Screening peas for winterhardiness under field and laboratory conditions. Crop Sci. 1983, 23, 85–88. [Google Scholar] [CrossRef]
- Arbaoui, M.; Balko, C.; Link, W. Study of faba bean (Vicia faba L.) winter-hardiness and development of screening methods. Field Crops Res. 2008, 106, 60–67. [Google Scholar] [CrossRef]
- Zanotto, S.; Palmé, A.; Helgadóttir, Á.; Daugstad, K.; Isolahti, M.; Öhlund, L.; Marum, P.; Moen, M.A.; Veteläinen, M.; Rognli, O.A.; et al. Trait characterization of genetic resources reveals useful variation for the improvement of cultivated Nordic red clover. J. Agron. Crop Sci. 2021, 207, 492–503. [Google Scholar] [CrossRef]
- Bateman, G.L. Pathogenicity of fungi associated with winter loss and injury in white lupin. Plant Pathol. 1997, 46, 157–167. [Google Scholar] [CrossRef]
- Hufnagel, B.; Soriano, A.; Taylor, J.; Divol, F.; Kroc, M.; Sanders, H.; Yeheyis, L.; Nelson, M.; Péret, B. Pangenome of white lupin provides insights into the diversity of the species. Plant Biotechnol. J. 2021, 19, 2532–2543. [Google Scholar] [CrossRef]
- Jahed, K.; Saini, A.; Sherif, S. Coping with the cold: Unveiling cryoprotectants, molecular signaling pathways, and strategies for cold stress resilience. Front. Plant Sci. 2023, 14, 1246093. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ding, Y.; Yang, Y.; Song, C.; Wang, B.; Yang, S.; Guo, Y.; Gong, Z. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 2021, 63, 53–78. [Google Scholar] [CrossRef] [PubMed]
- Monroy, A.F.; Sangwan, V.; Dhindsa, R.S. Low temperature signal transduction during cold acclimation: Protein phosphatase 2A as an early target for cold-inactivation. Plant J. 1998, 13, 653–660. [Google Scholar] [CrossRef]
- Jung, W.J.; Jeong, J.H.; Yoon, J.S.; Seo, Y.W. Genome-wide identification of the plant homeodomain-finger family in rye and ScPHD5 functions in cold tolerance and flowering time. Plant Cell Rep. 2024, 43, 142. [Google Scholar] [CrossRef]
- Wu, H.; Lian, B.; Lv, X.; Sun, M.; Wei, F.; An, L.; Li, Y.; Fu, X.; Lu, J.; Ma, L.; et al. Xyloglucan endotransglucosylase-hydrolase 22 positively regulates response to cold stress in upland cotton (Gossypium hirsutum L.). Ind. Crops Prod. 2024, 220, 119273. [Google Scholar] [CrossRef]
- Takahashi, D.; Johnson, K.; Hao, P.; Tuong, T.; Erban, A.; Sampathkumar, A.; Bacic, A.; Livingston, D.; Kopka, J.; Kuroha, T.; et al. Cell wall modification by the xyloglucan endotransglucosylase/hydrolase XTH19 influences freezing tolerance after cold and sub-zero acclimation. Plant Cell Environ. 2020, 44, 915–930. [Google Scholar] [CrossRef]
- Guo, J.; Sun, B.; He, H.; Zhang, Y.; Tian, H.; Wang, B. Current understanding of bHLH transcription factors in plant abiotic stress tolerance. Int. J. Mol. Sci. 2021, 22, 4921. [Google Scholar] [CrossRef] [PubMed]
- Mulet, J.M.; Porcel, R.; Yenush, L. Modulation of potassium transport to increase abiotic stress tolerance in plants. J. Exp. Bot. 2023, 74, 5989–6005. [Google Scholar] [CrossRef]
- Li, G.; Jin, L.; Sheng, S. Genome-Wide Identification of bHLH transcription factor in Medicago sativa in response to cold stress. Genes 2022, 13, 2371. [Google Scholar] [CrossRef] [PubMed]
- Sachdev, S.; Ansari, S.A.; Ansari, M.I.; Fujita, M.; Hasanuzzaman, M. Abiotic stress and reactive oxygen species: Generation, signaling, and defense mechanisms. Antioxidants 2021, 10, 277. [Google Scholar] [CrossRef]
- Song, Q.; Wang, X.; Liu, Y.; Brestic, M.; Yang, X. StLTO1, a lumen thiol oxidoreductase in Solanum tuberosum L., enhances the cold resistance of potato plants. Plant Sci. 2022, 325, 111481. [Google Scholar] [CrossRef] [PubMed]
- Kosmala, A.; Bocian, A.; Rapacz, M.; Jurczyk, B.; Zwierzykowski, Z. Identification of leaf proteins differentially accumulated during cold acclimation between Festuca pratensis plants with distinct levels of frost tolerance. J. Exp. Bot. 2009, 60, 3595–3609. [Google Scholar] [CrossRef]
- Kwon, S.; Kwon, S.I.; Bae, M.; Cho, E.; Park, O. Role of the methionine sulfoxide reductase MsrB3 in cold acclimation in Arabidopsis. Plant Cell Physiol. 2007, 48, 1713–1723. [Google Scholar] [CrossRef]
- Sandve, S.R.; Kosmala, A.; Rudi, H.; Fjellheim, S.; Rapacz, M.; Yamada, T.; Rognli, O.A. Molecular mechanisms underlying frost tolerance in perennial grasses adapted to cold climates. Plant Sci. 2011, 180, 69–77. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, L.; Shi, C.; Zhao, L.; Cui, D.; Chen, F. Identification of proteins using iTRAQ and virus-induced gene silencing reveals three bread wheat proteins involved in the response to combined osmotic-cold Stress. J. Proteome Res. 2018, 17, 2256–2281. [Google Scholar] [CrossRef]
- Kushwaha, H.; Singh, A.; Sopory, S.; Singla-Pareek, S.; Pareek, A. Genome wide expression analysis of CBS domain containing proteins in Arabidopsis thaliana (L.) Heynh and Oryza sativa L. reveals their developmental and stress regulation. BMC Genom. 2009, 10, 200. [Google Scholar] [CrossRef]
- Hao, Q.; Yang, Y.; Shan, Z.; Chen, H.; Zhang, C.; Chen, L.; Yuan, S.; Zhang, X.; Chen, S.; Yang, Z.; et al. Genome-wide investigation and expression profiling under abiotic stresses of a soybean unknown function (DUF21) and cystathionine-β-synthase (CBS) domain-containing protein family. Biochem. Genet. 2021, 59, 83–113. [Google Scholar] [CrossRef]
- Michel, S.; Löschenberger, F.; Hellinger, J.; Strasser, V.; Ametz, C.; Pachler, B.; Sparry, E.; Bürstmayr, H. Improving and maintaining winter hardiness and frost tolerance in bread wheat by genomic selection. Front. Plant Sci. 2019, 10, 1195. [Google Scholar] [CrossRef] [PubMed]
- Erath, W.; Bauer, E.; Fowler, D.B.; Gordillo, A.; Korzun, V.; Ponomareva, M.; Schmidt, M.; Schmiedchen, B.; Wilde, P.; Schön, C.C. Exploring new alleles for frost tolerance in winter rye. Theor. Appl. Genet. 2017, 130, 2151–2164. [Google Scholar] [CrossRef] [PubMed]
- Windhorst, A.; Skovbjerg, C.K.; Angra, D.; O’Sullivan, D.M.; Andersen, S.U.; Link, W. Genome-wide association studies identify promising QTL for freezing tolerance in winter and early spring as a basis for in-depth genetic analysis and implementation in winter faba bean (Vicia faba L.) breeding. bioRxiv 2024. 2024.11.25.625124. [Google Scholar] [CrossRef]
- Buirchell, B.J.; Cowling, W.A. Genetic resources in lupins. In Lupins as Crop Plants. Biology, Production and Utilization; Gladstones, J.S., Atkins, C.A., Hamblin, J., Eds.; CAB International: New York, NY, USA, 1998; pp. 41–66. [Google Scholar]
- Murray, G.A.; Eser, D.; Gusta, L.V.; Eteve, G. Winterhardiness in pea, lentil, faba bean and chickpea. In World Crops: Cool Season Food Legumes. Current Plant Science and Biotechnology in Agriculture; Summerfield, R.J., Ed.; Springer: Dordrecht, The Netherlands, 1988; Volume 5, pp. 831–843. [Google Scholar] [CrossRef]
- DeLacy, I.H.; Basford, K.E.; Cooper, M.; Bull, J.K.; McLaren, C.G. Analysis of multi-environment data—An historical perspective. In Plant Adaptation and Crop Improvement; Cooper, M., Hammer, G.L., Eds.; CAB International: Wallingford, UK, 1996; pp. 39–124. [Google Scholar]
- Nazzicari, N.; Franguelli, N.; Ferrari, B.; Pecetti, L.; Annicchiarico, P. The effect of genome parametrization and SNP marker subsetting on genomic selection in autotetraploid alfalfa. Genes 2024, 15, 449. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Stekhoven, D.J.; Bühlmann, P. MissForest–non-parametric missing value imputation for mixed-type data. Bioinformatics 2012, 28, 112–118. [Google Scholar] [CrossRef]
- Covarrubias-Pazaran, G. Genome-assisted prediction of quantitative traits using the R package sommer. PLoS ONE 2016, 11, e0156744. [Google Scholar] [CrossRef]
- Marroni, F.; Pinosio, S.; Zaina, G.; Fogolari, F.; Felice, N.; Cattonaro, F.; Morgante, M. Nucleotide diversity and linkage disequilibrium in Populus nigra cinnamyl alcohol dehydrogenase (CAD4) gene. Tree Genet. Genomes 2011, 7, 1011–1023. [Google Scholar] [CrossRef]
- Breseghello, F.; Sorrells, M.E. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 2006, 172, 1165–1177. [Google Scholar] [CrossRef]
- Yendle, P.W.; MacFie, H.J. Discriminant principal components analysis. J. Chemom. 1989, 3, 589–600. [Google Scholar] [CrossRef]
- Laurie, C.C.; Doheny, K.F.; Mirel, D.B.; Pugh, E.W.; Bierut, L.J.; Bhangale, T.; Boehm, F.; Caporaso, N.E.; Cornelis, M.C.; Edenberg, H.J.; et al. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet. Epidemiol. 2010, 34, 591–602. [Google Scholar] [CrossRef]
- Jombart, T.; Ahmed, I. Adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.M.; Zhang, Z. BLINK: A package for the next level of genome wide association studies with both individuals and markers in the millions. GigaScience 2019, 8, 154. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Z. GAPIT version 3: Boosting power and accuracy for genomic association and prediction. Genom. Proteom. Bioinform. 2021, 19, 629–640. [Google Scholar] [CrossRef]
- Nazzicari, N.; Biscarini, F. Stacked kinship CNN vs. GBLUP for genomic predictions of additive and complex continuous phenotypes. Sci. Rep. 2022, 12, 19889. [Google Scholar] [CrossRef] [PubMed]
- Endelman, J.B. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 2011, 4, 250–255. [Google Scholar] [CrossRef]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef] [PubMed]
- Habier, D.; Fernando, R.L.; Kizilkaya, K.; Garrick, D.J. Extension of the Bayesian alphabet for genomic selection. BMC Bioinform. 2011, 12, 186. [Google Scholar] [CrossRef]
- Park, T.; Casella, G. The bayesian lasso. J. Am. Stat. Assoc. 2008, 103, 681–686. [Google Scholar] [CrossRef]



| Pool | Material | No. of Genotypes | Mortality | Visual Score | ||
|---|---|---|---|---|---|---|
| Mean a | Range | Mean a | Range | |||
| Winter-type | Cultivar | 4 | 0.21 d | 0.09–0.37 | 5.6 d | 4.8–6.3 |
| Mediterranean-type | Cultivar | 5 | 0.33 cd | 0.13–0.58 | 6.2 cd | 4.4–7.5 |
| Greece | Landrace | 12 | 0.44 bc | 0.08–0.70 | 7.0 bc | 5.3–8.6 |
| Spain | Landrace | 11 | 0.48 bc | 0.17–0.70 | 7.1 bc | 5.7–8.3 |
| Madeira and Canaries | Landrace | 10 | 0.51 bc | 0.27–0.73 | 7.3 b | 6.2–8.5 |
| Azores | Landrace | 11 | 0.51 bc | 0.18–0.80 | 7.2 b | 5.1–8.7 |
| West Asia | Landrace | 12 | 0.52 ab | 0.26–0.76 | 7.3 b | 5.9–8.8 |
| Turkey | Landrace | 12 | 0.53 ab | 0.16–0.73 | 7.5 b | 6.1–8.9 |
| Italy | Landrace | 14 | 0.54 ab | 0.26–0.75 | 7.5 ab | 5.7–8.7 |
| Portugal | Landrace | 10 | 0.55 ab | 0.25–0.76 | 7.7 ab | 6.6–8.7 |
| Egypt | Landrace | 14 | 0.56 ab | 0.16–0.82 | 7.7 ab | 5.2–9.0 |
| East Africa | Landrace | 10 | 0.57 ab | 0.21–0.95 | 7.6 ab | 5.5–9.4 |
| Spring-type | Cultivar | 8 | 0.63 ab | 0.12–0.90 | 7.9 ab | 4.9–9.5 |
| Maghreb | Landrace | 11 | 0.71 a | 0.45–0.96 | 8.4 a | 7.4–9.7 |
| LSD (p < 0.05) | 0.16 | 0.9 | ||||
| Landrace | Mortality | Visual Score | ||
|---|---|---|---|---|
| Parent Value | Progeny Value | Parent Value | Progeny Value | |
| Gr56 | 0.24 | 0.49 | 6.0 | 7.6 |
| La646 | 0.39 | 0.57 | 6.6 | 8.0 |
| La246 | 0.36 | 0.66 | 6.4 | 8.5 |
| LAP123 | 0.72 | 0.77 | 8.6 | 8.9 |
| LSD (p < 0.05) | 0.34 | 0.06 | 1.5 | 0.6 |
| SNP | Population | Trait | Candidate Gene | Putative Protein | Putative Role |
|---|---|---|---|---|---|
| Chr02_14306413 | 2 | Mortality, visual score | Chr02g0156401 | Metallo-dependent phosphatase | Cold signal regulation |
| Chr02_14306413 | 2 | Mortality, visual score | Chr02g0156391 | Multi antimicrobial extrusion protein | |
| Chr02_14306413 | 2 | Mortality, visual score | Chr02g0156411 | UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase | |
| Chr04_7632627 | 2 | Mortality | Chr04g0256531 | Hydrolase | Stabilization of cell wall |
| Chr04_7632627 | 2 | Mortality | Chr04g0256541 | Potassium transporter | Cryoprotection and osmoprotection |
| Chr05_4820341 | 1 | Mortality | Chr05g0219341 | CBS domain-containing protein (CDCPs) | |
| Chr05_4820341 | 1 | Mortality | Chr05g0219331 | Polyadenylate binding protein, human types 1, 2, 3, 4 | |
| Chr06_1878682 | 1 | Mortality | Chr06g0163651 | Oxidoreductase | Enhancement of ROS scavenging |
| Chr06_1878682 | 1 | Mortality | Chr06g0163661 | mRNA splicing factor SYF2 | |
| Chr08_3511620 | 1 | Mortality | Chr08g0234251 | Cellulose synthase (UDP-forming) chromatin regulator PHD family | Gene expression regulation |
| Chr08_3511620 | 1 | Mortality | Chr08g0234241 | QWRF family protein | |
| Chr13_15386976 | 1 | Mortality | Chr13g0303311 | Ribosomal protein S4/S9 | Ribosomal biogenesis |
| Chr13_15386976 | 1 | Mortality | Chr13g0303301 | Transcription factor bHLH family | Cryoprotection and osmoprotection |
| Chr14_10127694 | 1 | Mortality | Chr14g0368501 | Leucine-rich repeat domain, L domain-containing protein | Primary cold sensor |
| Chr14_10127694 | 1 | Mortality | Chr14g0368511 | Plus-end-directed kinesin ATPase | |
| Chr16_5032297 | 1 | Visual score | Chr16g0384801 | Kinase RLK-Pelle-URK-1 family | Primary cold sensor |
| Chr19_17886074 | 2 | Mortality | Chr19g0139891 | Vacuolar protein sorting-associated protein | |
| Chr19_17886074 | 2 | Mortality | Chr19g0139901 | Methionine--tRNA ligase | |
| Chr19_17886074 | 2 | Mortality | Chr19g0139881 | Phosphatase 4 core regulatory subunit R2 | |
| Chr21_1050253 | 2 | Mortality | Chr21g0306351 | Serine/threonine phosphatase, protein kinase CMGC-GSKL family | Cold signal regulation |
| Chr23_1146188 | 1 | Mortality, visual score | - | - |
| Trait | Training Set | Validation Set | Model | Predictive Abilities |
|---|---|---|---|---|
| Mortality | Population 1 | Population 1 | rrBLUP | 0.414 |
| Mortality | Population 2 | Population 2 | Bayesian Lasso | 0.672 |
| Mortality | Population 1 | Population 2 | Bayesian Lasso | 0.393 |
| Mortality | Population 2 | Population 1 | Bayesian Lasso | 0.255 |
| Visual score | Population 1 | Population 1 | rrBLUP | 0.376 |
| Visual score | Population 2 | Population 2 | Bayesian Lasso | 0.678 |
| Visual score | Population 1 | Population 2 | Bayesian Lasso | 0.386 |
| Visual score | Population 2 | Population 1 | Bayesian Lasso | 0.232 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Franguelli, N.; Cavalli, D.; Nazzicari, N.; Pecetti, L.; Notario, T.; Annicchiarico, P. Genetic Variation and Genome-Enabled Prediction of White Lupin Frost Resistance in Different Reference Populations. Int. J. Mol. Sci. 2025, 26, 10224. https://doi.org/10.3390/ijms262010224
Franguelli N, Cavalli D, Nazzicari N, Pecetti L, Notario T, Annicchiarico P. Genetic Variation and Genome-Enabled Prediction of White Lupin Frost Resistance in Different Reference Populations. International Journal of Molecular Sciences. 2025; 26(20):10224. https://doi.org/10.3390/ijms262010224
Chicago/Turabian StyleFranguelli, Nicolò, Daniele Cavalli, Nelson Nazzicari, Luciano Pecetti, Tommaso Notario, and Paolo Annicchiarico. 2025. "Genetic Variation and Genome-Enabled Prediction of White Lupin Frost Resistance in Different Reference Populations" International Journal of Molecular Sciences 26, no. 20: 10224. https://doi.org/10.3390/ijms262010224
APA StyleFranguelli, N., Cavalli, D., Nazzicari, N., Pecetti, L., Notario, T., & Annicchiarico, P. (2025). Genetic Variation and Genome-Enabled Prediction of White Lupin Frost Resistance in Different Reference Populations. International Journal of Molecular Sciences, 26(20), 10224. https://doi.org/10.3390/ijms262010224








