Selective Endocytosis-Mediated Omicron S1-RBD Internalization Revealed by Reconstitution of ACE2-S1-RBD Interaction on Micropatterned Membrane Substrates
Abstract
1. Introduction
2. Results
2.1. Reconstitution of ACE2-S1-RBD Interaction in a Hybrid Live Cell–SLB Platform
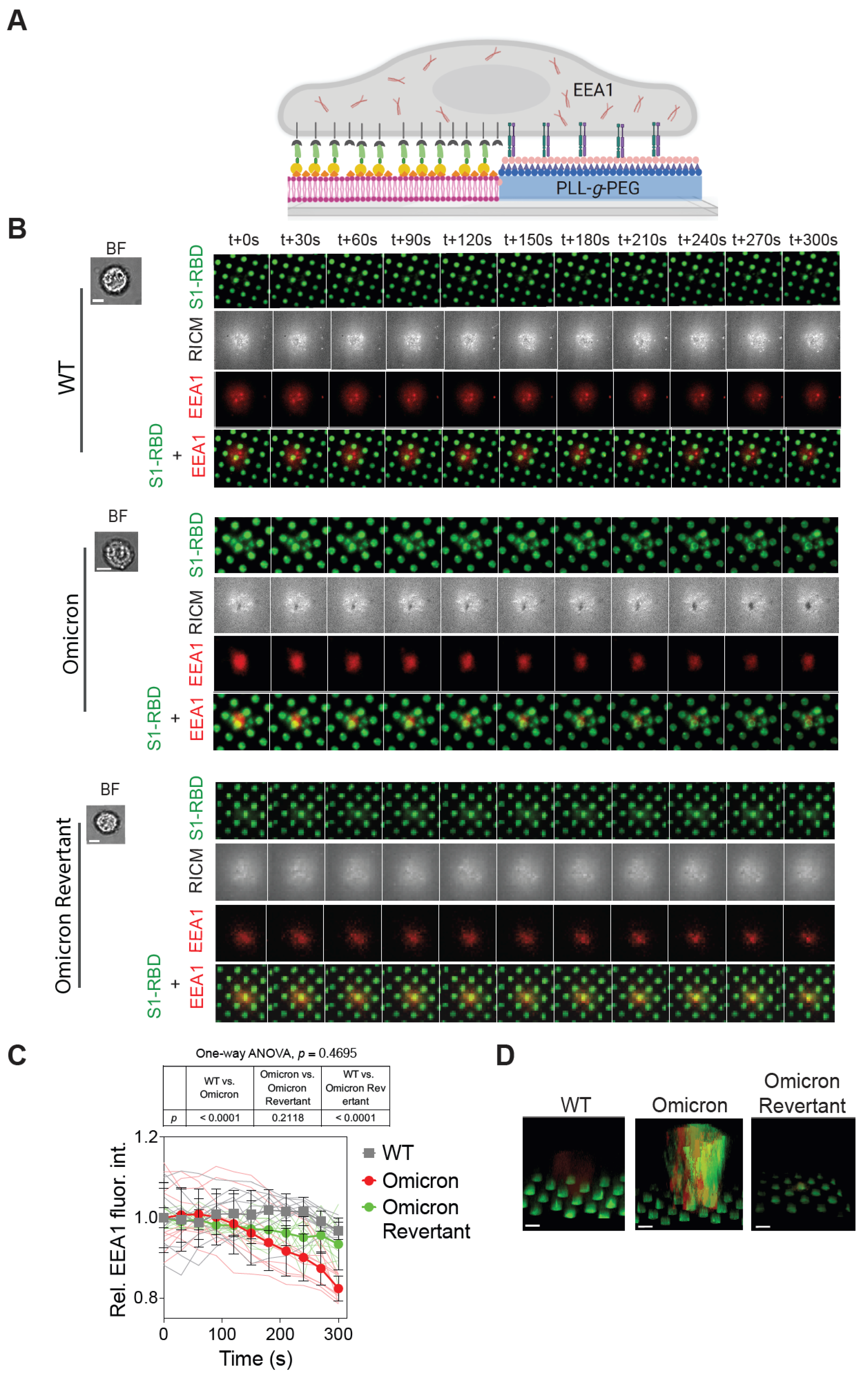
2.2. Significant Depletion of Omicron S1-RBD- from SLB Corral upon Interaction with A549 Cells
2.3. Omicron S1-RBD Depletion from SLB Corrals Is Mediated by the Host Cell Endocytic Machinery
2.4. Pitstop 2 Abrogates, While Blebbistatin Partially Reduces, Omicron S1-RBD Depletion from SLBs
3. Discussion
4. Materials and Methods
4.1. Plasmid Design
4.2. Protein Expression and Purification
4.3. Supported Lipid Bilayer Preparation
4.4. Micropatterned Supported Lipid Bilayer Preparation
4.5. Functionalization of SLB with S1-RBD
4.6. Cell Culture and Preparation of Cells for Live Cell Experiments
4.7. Assay for Monitoring the ACE2-Mediated Spike Interaction and Internalization on Bilayer
4.8. Monitoring the Effect of Inhibiting Endocytosis and Actin in Spike Internalization
4.9. Microscopy and Image Analysis
4.10. Data Analysis and Figure Preparation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Markov, P.V.; Ghafari, M.; Beer, M.; Lythgoe, K.; Simmonds, P.; Stilianakis, N.I.; Katzourakis, A. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 361–379. [Google Scholar] [CrossRef]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.-f.; Xu, W.; Liu, S.-w. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Hoter, A.; Naim, H.Y. Biochemical Characterization of SARS-CoV-2 Spike RBD Mutations and Their Impact on ACE2 Receptor Binding. Front. Mol. Biosci. 2022, 9, 893843. [Google Scholar] [CrossRef]
- Borkotoky, S.; Dey, D.; Hazarika, Z. Interactions of angiotensin-converting enzyme-2 (ACE2) and SARS-CoV-2 spike receptor-binding domain (RBD): A structural perspective. Mol. Biol. Rep. 2023, 50, 2713–2721. [Google Scholar] [CrossRef]
- Carabelli, A.M.; Peacock, T.P.; Thorne, L.G.; Harvey, W.T.; Hughes, J.; de Silva, T.I.; Peacock, S.J.; Barclay, W.S.; de Silva, T.I.; Towers, G.J.; et al. SARS-CoV-2 variant biology: Immune escape, transmission and fitness. Nat. Rev. Microbiol. 2023, 21, 162–177. [Google Scholar] [CrossRef]
- Chang, X.; Liu, X.; Martina, B.; Zeltins, A.; Augusto, G.; Vogel, M.; Mohsen, M.O.; Speiser, D.E.; Bachmann, M.F. Vaccination using mutated receptor binding domains of SARS-CoV-2: Evidence for partial immune escape but not serotype formation. Front. Immunol. 2023, 14, 1114396. [Google Scholar] [CrossRef]
- Wang, Q.; Iketani, S.; Li, Z.; Liu, L.; Guo, Y.; Huang, Y.; Bowen, A.D.; Liu, M.; Wang, M.; Yu, J.; et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 2023, 186, 279–286.e8. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Yisimayi, A.; Jian, F.; Song, W.; Xiao, T.; Wang, L.; Du, S.; Wang, J.; Li, Q.; Chen, X.; et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature 2022, 608, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Alkhalil, E.; Jarad, B.; Saleh, T.; Aldarwish, W.; Joujeh, D. BioNTech/Pfizer (BNT162b2) COVID-19 mRNA vaccine: Manufacturing, immunogenicity, efficacy and safety. Prospect. Pharm. Sci. 2025, 23, 31–46. [Google Scholar] [CrossRef]
- Ahmed, W.S.; Philip, A.M.; Biswas, K.H. Decreased Interfacial Dynamics Caused by the N501Y Mutation in the SARS-CoV-2 S1 Spike:ACE2 Complex. Front. Mol. Biosci. 2022, 9, 846996. [Google Scholar] [CrossRef]
- Philip, A.M.; Ahmed, W.S.; Biswas, K.H. Reversal of the unique Q493R mutation increases the affinity of Omicron S1-RBD for ACE2. Comput. Struct. Biotechnol. J. 2023, 21, 1966–1977. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Wu, L.; Xu, Z.; Wang, X.; Xie, Y.; Chai, Y.; Zheng, A.; Zhou, J.; Qiao, S.; Huang, M.; et al. An updated atlas of antibody evasion by SARS-CoV-2 Omicron sub-variants including BQ.1.1 and XBB. Cell Rep. Med. 2023, 4, 100991. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Huang, Z.; Guo, Y.; Guo, H.; Jian, L.; Xiao, J.; Yao, X.; Yu, H.; Cheng, T.; Zhang, Y.; et al. Evolving spike mutations in SARS-CoV-2 Omicron variants facilitate evasion from breakthrough infection-acquired antibodies. Cell Discov. 2023, 9, 86. [Google Scholar] [CrossRef]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Bayati, A.; Kumar, R.; Francis, V.; McPherson, P.S. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J. Biol. Chem. 2021, 296, 100306. [Google Scholar] [CrossRef]
- Nejat, R.; Torshizi, M.F.; Najafi, D.J. S Protein, ACE2 and Host Cell Proteases in SARS-CoV-2 Cell Entry and Infectivity; Is Soluble ACE2 a Two Blade Sword? A Narrative Review. Vaccines 2023, 11, 204. [Google Scholar] [CrossRef]
- Zhou, Y.-Q.; Wang, K.; Wang, X.-Y.; Cui, H.-Y.; Zhao, Y.; Zhu, P.; Chen, Z.-N. SARS-CoV-2 pseudovirus enters the host cells through spike protein-CD147 in an Arf6-dependent manner. Emerg. Microbes Infect. 2022, 11, 1135–1144. [Google Scholar] [CrossRef]
- Miao, L.; Yan, C.; Chen, Y.; Zhou, W.; Zhou, X.; Qiao, Q.; Xu, Z. SIM imaging resolves endocytosis of SARS-CoV-2 spike RBD in living cells. Cell Chem. Biol. 2023, 30, 248–260.e4. [Google Scholar] [CrossRef]
- Reider, A.; Wendland, B. Endocytic adaptors—Social networking at the plasma membrane. J. Cell Sci. 2011, 124, 1613–1622. [Google Scholar] [CrossRef] [PubMed]
- Kamentseva, R.; Kosheverova, V.; Kharchenko, M.; Zlobina, M.; Salova, A.; Belyaeva, T.; Nikolsky, N.; Kornilova, E. Functional cycle of EEA1-positive early endosome: Direct evidence for pre-existing compartment of degradative pathway. PLoS ONE 2020, 15, e0232532. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Qiu, Z.; Hou, Y.; Deng, X.; Xu, W.; Zheng, T.; Wu, P.; Xie, S.; Bian, W.; Zhang, C.; et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021, 31, 126–140. [Google Scholar] [CrossRef]
- Singh, A.; Soler, J.A.; Lauer, J.; Grill, S.W.; Jahnel, M.; Zerial, M.; Thutupalli, S. Two-component molecular motor driven by a GTPase cycle. Nat. Phys. 2023, 19, 1185–1192. [Google Scholar] [CrossRef]
- Meng, B.; Abdullahi, A.; Ferreira, I.A.T.M.; Goonawardane, N.; Saito, A.; Kimura, I.; Yamasoba, D.; Gerber, P.P.; Fatihi, S.; Rathore, S.; et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 2022, 603, 706–714. [Google Scholar] [CrossRef]
- Groves, J.T.; Boxer, S.G. Electric field-induced concentration gradients in planar supported bilayers. Biophys. J. 1995, 69, 1972–1975. [Google Scholar] [CrossRef] [PubMed]
- Groves, J.T.; Wulfing, C.; Boxer, S.G. Electrical manipulation of glycan-phosphatidyl inositol-tethered proteins in planar supported bilayers. Biophys. J. 1996, 71, 2716–2723. [Google Scholar] [CrossRef] [PubMed]
- Groves, J.T.; Ulman, N.; Boxer, S.G. Micropatterning fluid lipid bilayers on solid supports. Science 1997, 275, 651–653. [Google Scholar] [CrossRef]
- Mossman, K.D.; Campi, G.; Groves, J.T.; Dustin, M.L. Altered TCR signaling from geometrically repatterned immunological synapses. Science 2005, 310, 1191–1193. [Google Scholar] [CrossRef]
- Biswas, K.H.; Groves, J.T. Hybrid Live Cell-Supported Membrane Interfaces for Signaling Studies. Annu. Rev. Biophys. 2019, 48, 537–562. [Google Scholar] [CrossRef]
- Chen, Z.; Oh, D.; Biswas, K.H.; Yu, C.H.; Zaidel-Bar, R.; Groves, J.T. Spatially modulated ephrinA1:EphA2 signaling increases local contractility and global focal adhesion dynamics to promote cell motility. Proc. Natl. Acad. Sci. USA 2018, 115, E5696–E5705. [Google Scholar] [CrossRef]
- Yu, C.H.; Rafiq, N.B.; Cao, F.; Zhou, Y.; Krishnasamy, A.; Biswas, K.H.; Ravasio, A.; Chen, Z.; Wang, Y.H.; Kawauchi, K.; et al. Integrin-beta3 clusters recruit clathrin-mediated endocytic machinery in the absence of traction force. Nat. Commun. 2015, 6, 8672. [Google Scholar] [CrossRef]
- Biswas, K.H.; Zaidel-Bar, R. Early events in the assembly of E-cadherin adhesions. Exp. Cell Res. 2017, 358, 14–19. [Google Scholar] [CrossRef]
- Biswas, K.H.; Zhongwen, C.; Dubey, A.K.; Oh, D.; Groves, J.T. Multicomponent Supported Membrane Microarray for Monitoring Spatially Resolved Cellular Signaling Reactions. Adv. Biosyst. 2018, 2, 1800015. [Google Scholar] [CrossRef]
- Biswas, K.H.; Hartman, K.L.; Zaidel-Bar, R.; Groves, J.T. Sustained alpha-catenin Activation at E-cadherin Junctions in the Absence of Mechanical Force. Biophys. J. 2016, 111, 1044–1052. [Google Scholar] [CrossRef] [PubMed]
- Biswas, K.H.; Cho, N.J.; Groves, J.T. Fabrication of multicomponent, spatially segregated DNA and protein functionalized supported membrane microarray. Langmuir 2018, 34, 9781–9788. [Google Scholar] [CrossRef]
- Chen, Z.; Biswas, K.H.; Groves, J.T. Patterned Substrate of Mobile and Immobile Ligands to Probe EphA2 Receptor Clustering. Bio-Protocol 2022, 12, e4434. [Google Scholar] [CrossRef] [PubMed]
- Biswas, K.H.; Hartman, K.L.; Yu, C.H.; Harrison, O.J.; Song, H.; Smith, A.W.; Huang, W.Y.; Lin, W.C.; Guo, Z.; Padmanabhan, A.; et al. E-cadherin junction formation involves an active kinetic nucleation process. Proc. Natl. Acad. Sci. USA 2015, 112, 10932–10937, Erratum in Proc. Natl. Acad. Sci. USA 2016, 113, E7640. [Google Scholar] [CrossRef]
- Salaita, K.; Nair, P.M.; Petit, R.S.; Neve, R.M.; Das, D.; Gray, J.W.; Groves, J.T. Restriction of receptor movement alters cellular response: Physical force sensing by EphA2. Science 2010, 327, 1380–1385. [Google Scholar] [CrossRef]
- DeMond, A.L.; Mossman, K.D.; Starr, T.; Dustin, M.L.; Groves, J.T. T cell receptor microcluster transport through molecular mazes reveals mechanism of translocation. Biophys. J. 2008, 94, 3286–3292. [Google Scholar] [CrossRef]
- Yamazaki, V.; Sirenko, O.; Schafer, R.J.; Nguyen, L.; Gutsmann, T.; Brade, L.; Groves, J.T. Cell membrane array fabrication and assay technology. BMC Biotechnol. 2005, 5, 18. [Google Scholar] [CrossRef]
- Pautot, S.; Lee, H.; Isacoff, E.Y.; Groves, J.T. Neuronal synapse interaction reconstituted between live cells and supported lipid bilayers. Nat. Chem. Biol. 2005, 1, 283–289. [Google Scholar] [CrossRef]
- Biswas, K.H. Regulation of α-catenin conformation at cadherin adhesions. J. Biomech. Sci. Eng. 2018, 13, 17–00699. [Google Scholar] [CrossRef]
- Chen, Z.; Oh, D.; Biswas, K.H.; Zaidel-Bar, R.; Groves, J.T. Probing the effect of clustering on EphA2 receptor signaling efficiency by subcellular control of ligand-receptor mobility. eLife 2021, 10, e67379. [Google Scholar] [CrossRef] [PubMed]
- Knyazev, E.; Nersisyan, S.; Tonevitsky, A. Endocytosis and Transcytosis of SARS-CoV-2 Across the Intestinal Epithelium and Other Tissue Barriers. Front. Immunol. 2021, 12, 636966. [Google Scholar] [CrossRef]
- Prichard, K.; Chau, N.; Xue, J.; Krauss, M.; Sakoff, J.A.; Gilbert, J.; Bahnik, C.; Muehlbauer, M.; Radetzki, S.; Robinson, P.J.; et al. Inhibition Clathrin Mediated Endocytosis: Pitstop 1 and Pitstop 2 Chimeras. ChemMedChem 2024, 19, e202400253. [Google Scholar] [CrossRef] [PubMed]
- Hollopeter, G. Stepwise assembly of the AP2 endocytic clathrin adaptor complex. Proc. Natl. Acad. Sci. USA 2024, 121, e2415313121. [Google Scholar] [CrossRef] [PubMed]
- Kovács, M.; Tóth, J.; Hetényi, C.; Málnási-Csizmadia, A.; Sellers, J.R. Mechanism of Blebbistatin Inhibition of Myosin II *. J. Biol. Chem. 2004, 279, 35557–35563. [Google Scholar] [CrossRef]
- Hornung, F.; Köse-Vogel, N.; Le Saux, C.J.; Häder, A.; Herrmann, L.; Schulz, L.; Radosa, L.; Lauf, T.; Sandhaus, T.; Samson, P.; et al. Uncovering a unique pathogenic mechanism of SARS-CoV-2 omicron variant: Selective induction of cellular senescence. Aging 2023, 15, 13593–13607. [Google Scholar] [CrossRef]
- Fujimoto, A.; Kawai, H.; Kawamura, R.; Kitamura, A. Interaction of Receptor-Binding Domain of the SARS-CoV-2 Omicron Variant with hACE2 and Actin. Cells 2024, 13, 1318. [Google Scholar] [CrossRef]
- Planas, D.; Staropoli, I.; Michel, V.; Lemoine, F.; Donati, F.; Prot, M.; Porrot, F.; Guivel-Benhassine, F.; Jeyarajah, B.; Brisebarre, A.; et al. Distinct evolution of SARS-CoV-2 Omicron XBB and BA.2.86/JN.1 lineages combining increased fitness and antibody evasion. Nat. Commun. 2024, 15, 2254. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shi, K.; Gu, Y.; Xu, Z.; Shu, C.; Li, D.; Sun, J.; Cong, M.; Li, X.; Zhao, X.; et al. Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants. Structure 2024, 32, 1055–1067.e6. [Google Scholar] [CrossRef]
- Kim, S.; Liu, Y.; Ziarnik, M.; Seo, S.; Cao, Y.; Zhang, X.F.; Im, W. Binding of human ACE2 and RBD of Omicron enhanced by unique interaction patterns among SARS-CoV-2 variants of concern. J. Comput. Chem. 2023, 44, 594–601. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Huo, J.; Zhou, D.; Zahradník, J.; Supasa, P.; Liu, C.; Duyvesteyn, H.M.E.; Ginn, H.M.; Mentzer, A.J.; Tuekprakhon, A.; et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 2022, 185, 467–484.e15. [Google Scholar] [CrossRef] [PubMed]
- Cameroni, E.; Bowen, J.E.; Rosen, L.E.; Saliba, C.; Zepeda, S.K.; Culap, K.; Pinto, D.; VanBlargan, L.A.; Marco, A.D.; Iulio, J.D.; et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 2022, 602, 664–670. [Google Scholar] [CrossRef]
- Parsons, R.J.; Acharya, P. Evolution of the SARS-CoV-2 Omicron spike. Cell Rep. 2023, 42, 113444. [Google Scholar] [CrossRef]
- Zhao, H.; Lu, L.; Peng, Z.; Chen, L.L.; Meng, X.; Zhang, C.; Ip, J.D.; Chan, W.M.; Chu, A.W.; Chan, K.H.; et al. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg. Microbes Infect. 2022, 11, 277–283. [Google Scholar] [CrossRef]
- Saito, A.; Irie, T.; Suzuki, R.; Maemura, T.; Nasser, H.; Uriu, K.; Kosugi, Y.; Shirakawa, K.; Sadamasu, K.; Kimura, I.; et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 2022, 602, 300–306. [Google Scholar] [CrossRef]
- Ray, A.; Minh Tran, T.T.; Santos Natividade, R.d.; Moreira, R.A.; Simpson, J.D.; Mohammed, D.; Koehler, M.; Petitjean, S.J.L.; Zhang, Q.; Bureau, F.; et al. Single-Molecule Investigation of the Binding Interface Stability of SARS-CoV-2 Variants with ACE2. ACS Nanosci. Au 2024, 4, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guo, Y.; Iketani, S.; Nair, M.S.; Li, Z.; Mohri, H.; Wang, M.; Yu, J.; Bowen, A.D.; Chang, J.Y.; et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature 2022, 608, 603–608. [Google Scholar] [CrossRef]
- Pastorio, C.; Noettger, S.; Nchioua, R.; Zech, F.; Sparrer, K.M.J.; Kirchhoff, F. Impact of mutations defining SARS-CoV-2 Omicron subvariants BA.2.12.1 and BA.4/5 on Spike function and neutralization. iScience 2023, 26, 108299. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Iketani, S.; Li, Z.; Guo, Y.; Yeh, A.Y.; Liu, M.; Yu, J.; Sheng, Z.; Huang, Y.; Liu, L.; et al. Antigenic characterization of the SARS-CoV-2 Omicron subvariant BA.2.75. Cell Host Microbe 2022, 30, 1512–1517.e4. [Google Scholar] [CrossRef] [PubMed]
- Campbell, B.C.; Nabel, E.M.; Murdock, M.H.; Lao-Peregrin, C.; Tsoulfas, P.; Blackmore, M.G.; Lee, F.S.; Liston, C.; Morishita, H.; Petsko, G.A. mGreenLantern: A bright monomeric fluorescent protein with rapid expression and cell filling properties for neuronal imaging. Proc. Natl. Acad. Sci. USA 2020, 117, 30710–30721. [Google Scholar] [CrossRef] [PubMed]
- Campbell, B.C.; Paez-Segala, M.G.; Looger, L.L.; Petsko, G.A.; Liu, C.F. Chemically stable fluorescent proteins for advanced microscopy. Nat. Methods 2022, 19, 1612–1621. [Google Scholar] [CrossRef]
- Oh, D.; Chen, Z.; Biswas, K.H.; Bai, F.; Ong, H.T.; Sheetz, M.P.; Groves, J.T. Competition for shared downstream signaling molecules establishes indirect negative feedback between EGFR and EphA2. Biophys. J. 2022, 121, 1897–1908. [Google Scholar] [CrossRef]
- Huang, W.Y.C.; Alvarez, S.; Kondo, Y.; Lee, Y.K.; Chung, J.K.; Lam, H.Y.M.; Biswas, K.H.; Kuriyan, J.; Groves, J.T. A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science 2019, 363, 1098–1103. [Google Scholar] [CrossRef]
- Biswas, K.H.; Groves, J.T. A Microbead Supported Membrane-Based Fluorescence Imaging Assay Reveals Intermembrane Receptor-Ligand Complex Dimension with Nanometer Precision. Langmuir 2016, 32, 6775–6780. [Google Scholar] [CrossRef]
- Azioune, A.; Carpi, N.; Tseng, Q.; Thery, M.; Piel, M. Protein micropatterns: A direct printing protocol using deep UVs. Methods Cell Biol. 2010, 97, 133–146. [Google Scholar] [CrossRef]
- Wang, L.; Biswas, K.H.; Yoon, B.K.; Kawakami, L.M.; Park, S.; Groves, J.T.; Li, L.; Huang, W.; Cho, N.J. Membrane Reconstitution of Monoamine Oxidase Enzymes on Supported Lipid Bilayers. Langmuir 2018, 34, 10764–10773. [Google Scholar] [CrossRef]
- Taylor, M.J.; Perrais, D.; Merrifield, C.J. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011, 9, e1000604. [Google Scholar] [CrossRef]
- Hall, T.E.; Martel, N.; Ariotti, N.; Xiong, Z.; Lo, H.P.; Ferguson, C.; Rae, J.; Lim, Y.W.; Parton, R.G. In vivo cell biological screening identifies an endocytic capture mechanism for T-tubule formation. Nat. Commun. 2020, 11, 3711. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Naranjo, J.C.; Ugaz, V.M. A nanometre-scale resolution interference-based probe of interfacial phenomena between microscopic objects and surfaces. Nat. Commun. 2013, 4, 1919. [Google Scholar] [CrossRef]
- Mani, N.; Marchan, M.F.; Subramanian, R. Simultaneous Visualization of the Dynamics of Crosslinked and Single Microtubules In Vitro by TIRF Microscopy. J. Vis. Exp. 2022, 180, e63377. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Philip, A.M.; Uddin, S.M.N.; Islam, Z.; Kolatkar, P.R.; Biswas, K.H. Selective Endocytosis-Mediated Omicron S1-RBD Internalization Revealed by Reconstitution of ACE2-S1-RBD Interaction on Micropatterned Membrane Substrates. Int. J. Mol. Sci. 2025, 26, 10216. https://doi.org/10.3390/ijms262010216
Philip AM, Uddin SMN, Islam Z, Kolatkar PR, Biswas KH. Selective Endocytosis-Mediated Omicron S1-RBD Internalization Revealed by Reconstitution of ACE2-S1-RBD Interaction on Micropatterned Membrane Substrates. International Journal of Molecular Sciences. 2025; 26(20):10216. https://doi.org/10.3390/ijms262010216
Chicago/Turabian StylePhilip, Angelin M., S. M. Nasir Uddin, Zeyaul Islam, Prasanna R. Kolatkar, and Kabir H. Biswas. 2025. "Selective Endocytosis-Mediated Omicron S1-RBD Internalization Revealed by Reconstitution of ACE2-S1-RBD Interaction on Micropatterned Membrane Substrates" International Journal of Molecular Sciences 26, no. 20: 10216. https://doi.org/10.3390/ijms262010216
APA StylePhilip, A. M., Uddin, S. M. N., Islam, Z., Kolatkar, P. R., & Biswas, K. H. (2025). Selective Endocytosis-Mediated Omicron S1-RBD Internalization Revealed by Reconstitution of ACE2-S1-RBD Interaction on Micropatterned Membrane Substrates. International Journal of Molecular Sciences, 26(20), 10216. https://doi.org/10.3390/ijms262010216






