Constructing a Prognostic Model for Clear Cell Renal Cell Carcinoma Based on Glycosyltransferase Gene and Verification of Key Gene Identification
Abstract
1. Introduction
2. Results
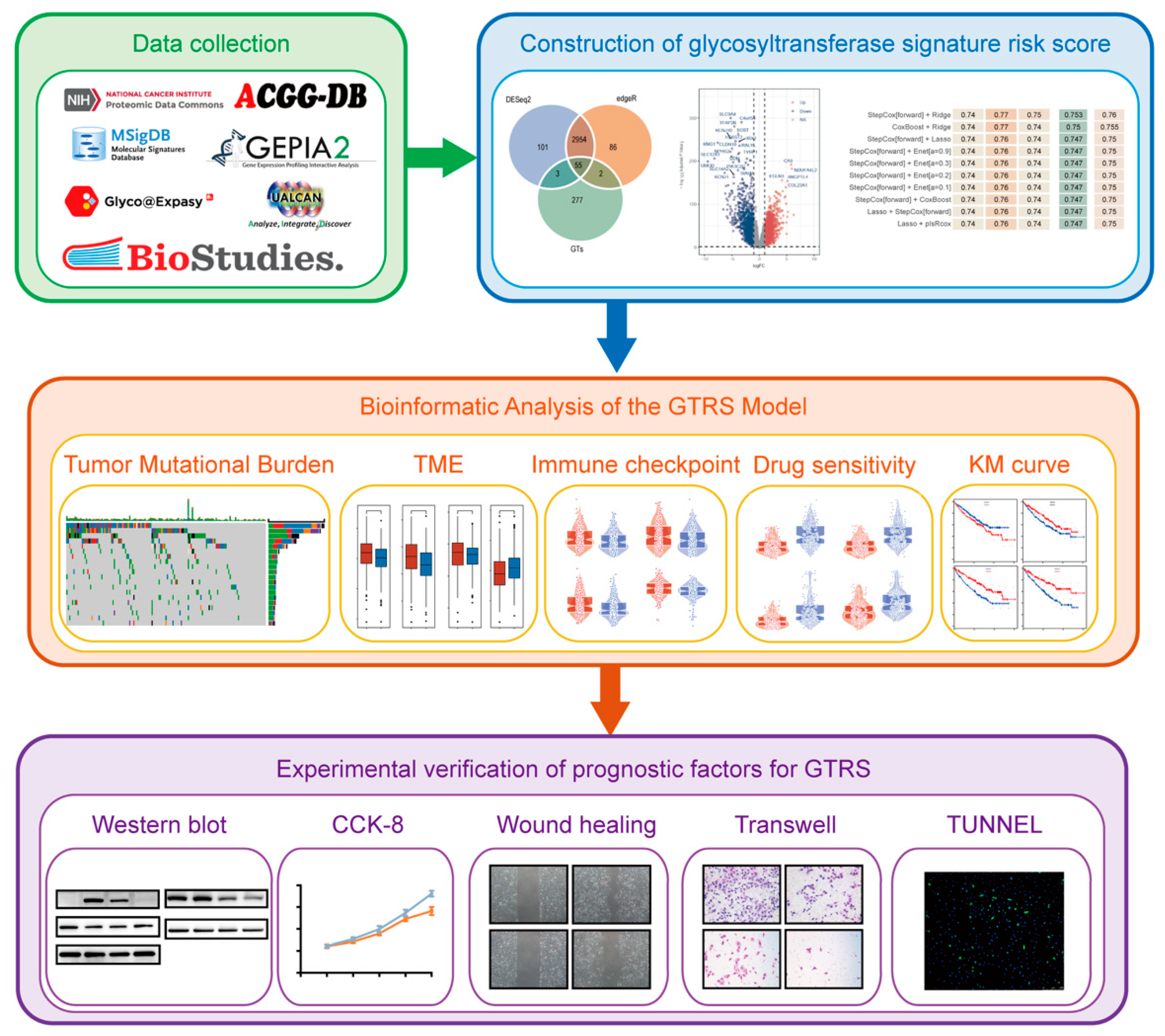
2.1. Identification of Prognosis-Associated Differentially Expressed Glycosyltransferase Genes in ccRCC
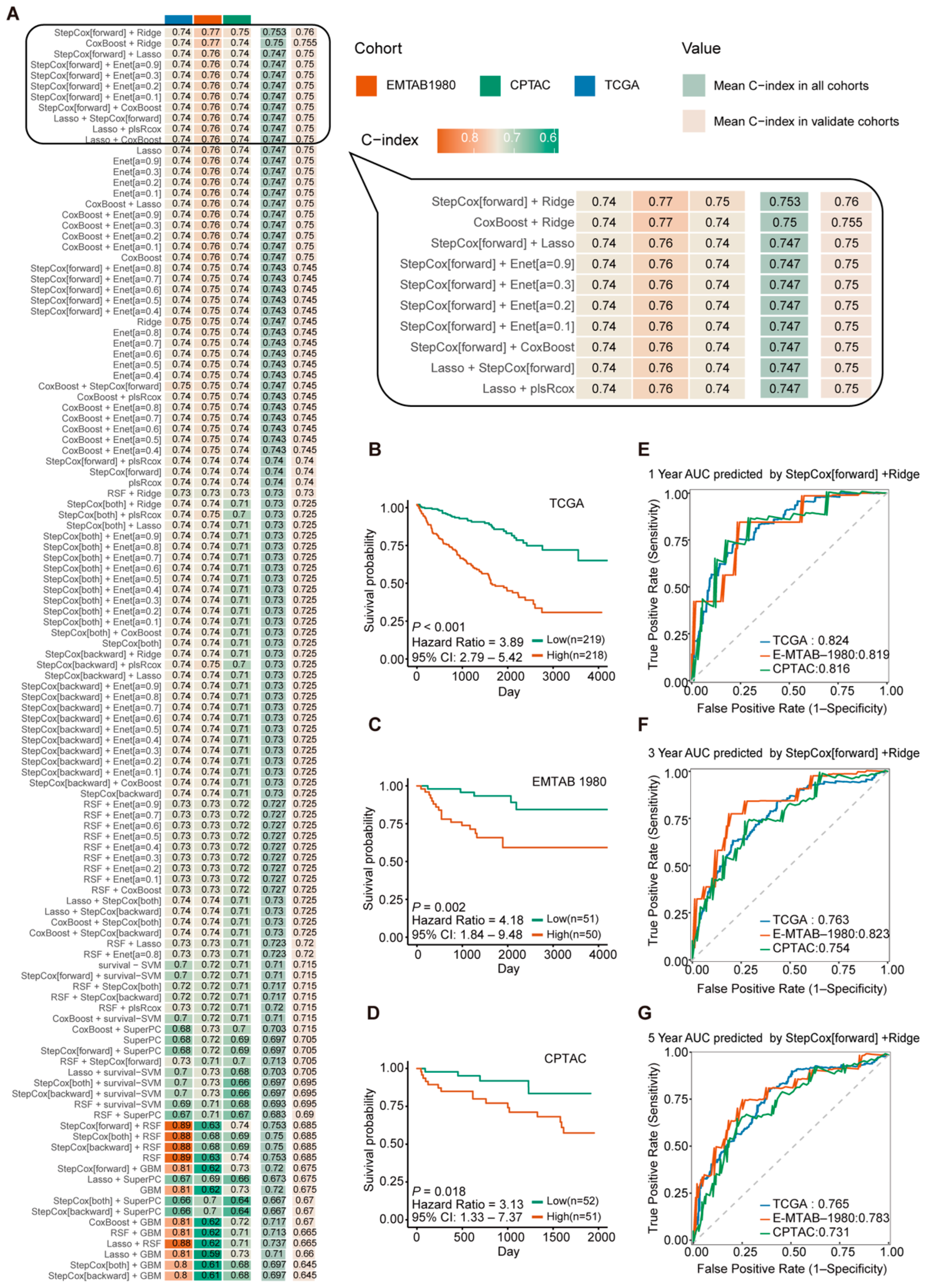
2.2. Construction and Validation of the Glycosyltransferase-Related Signature (GTRS) Prognostic Model for ccRCC
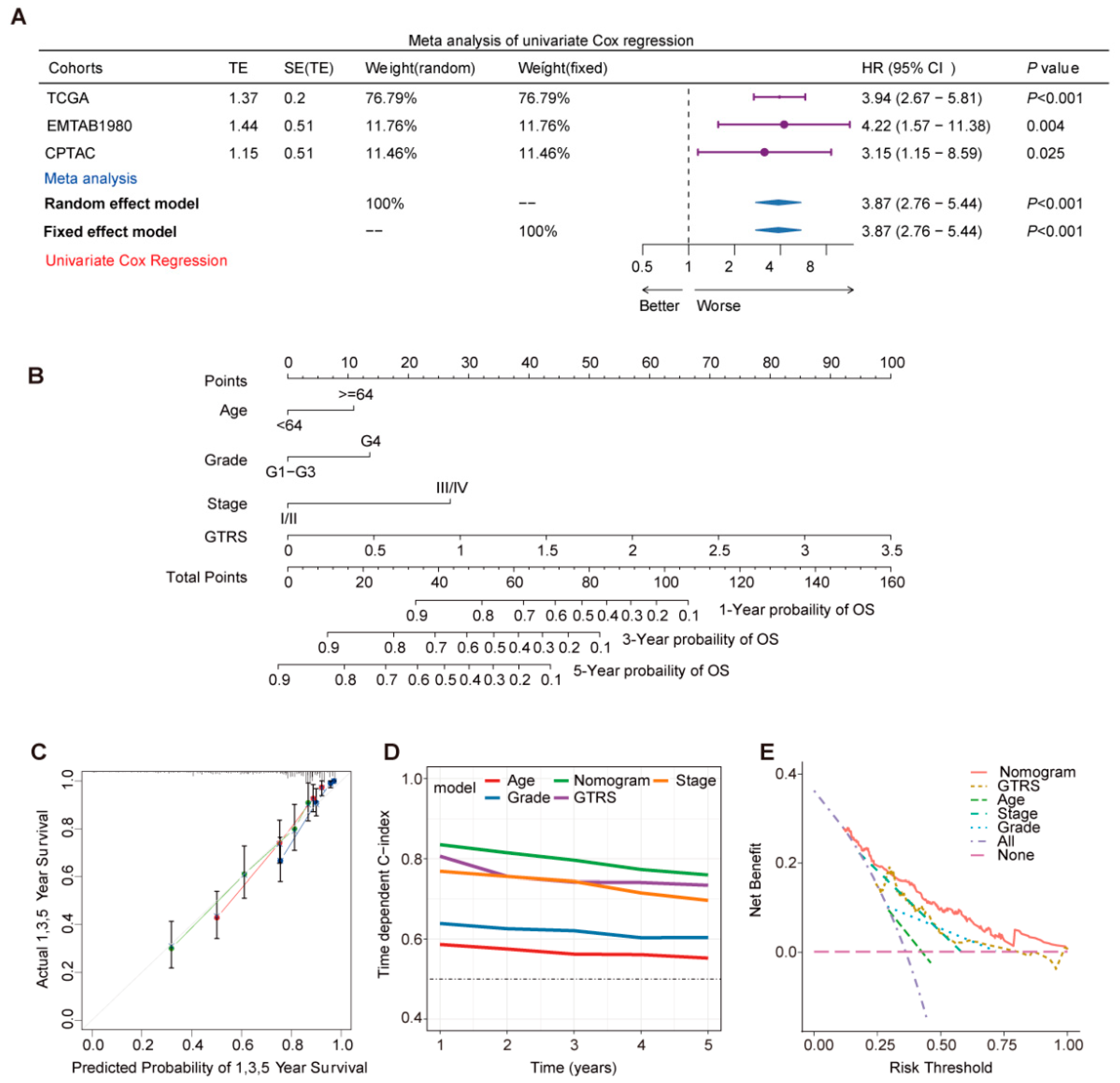
2.3. Prognostic Risk Score GTRS as an Independent Prognostic Factor for ccRCC Patients
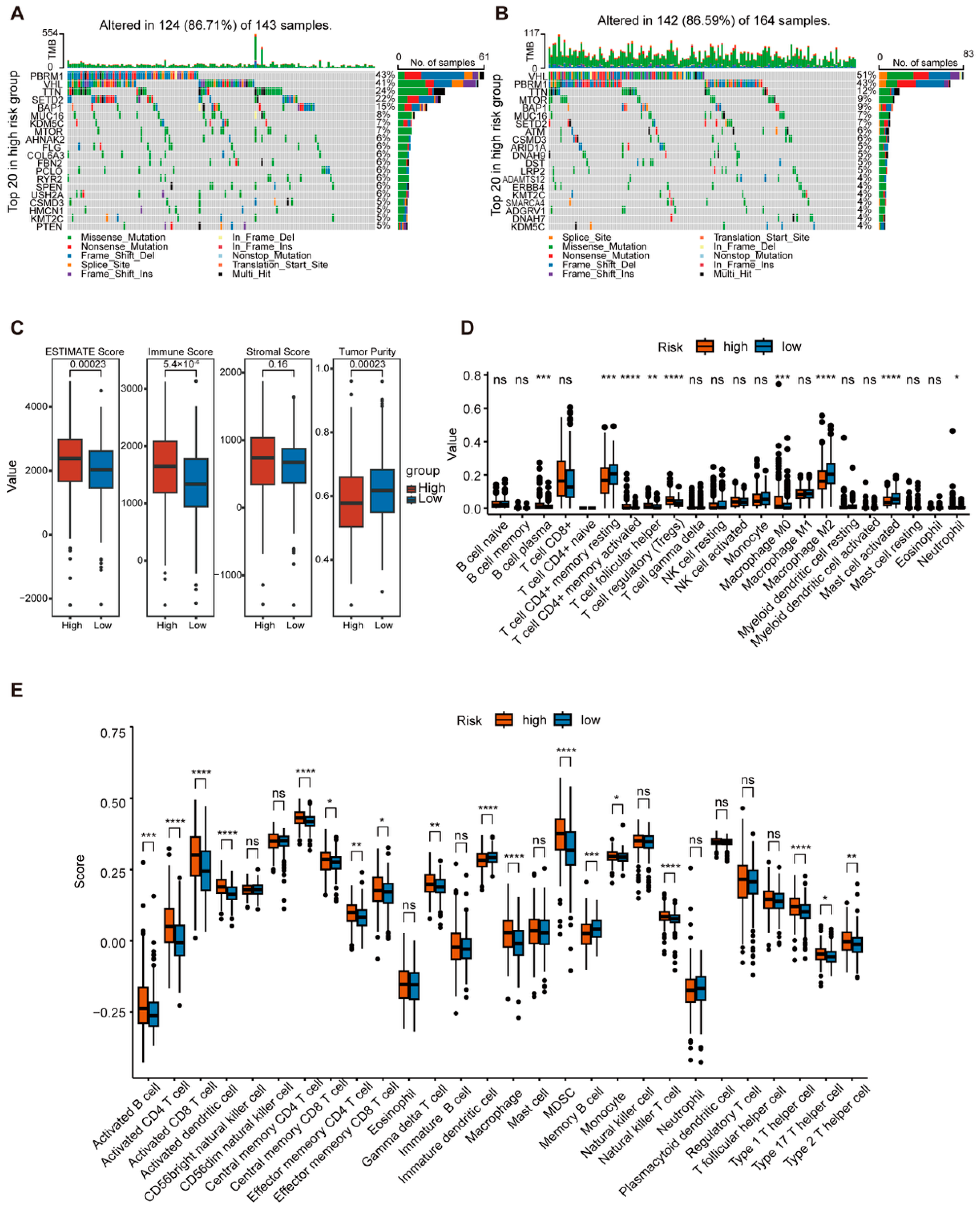
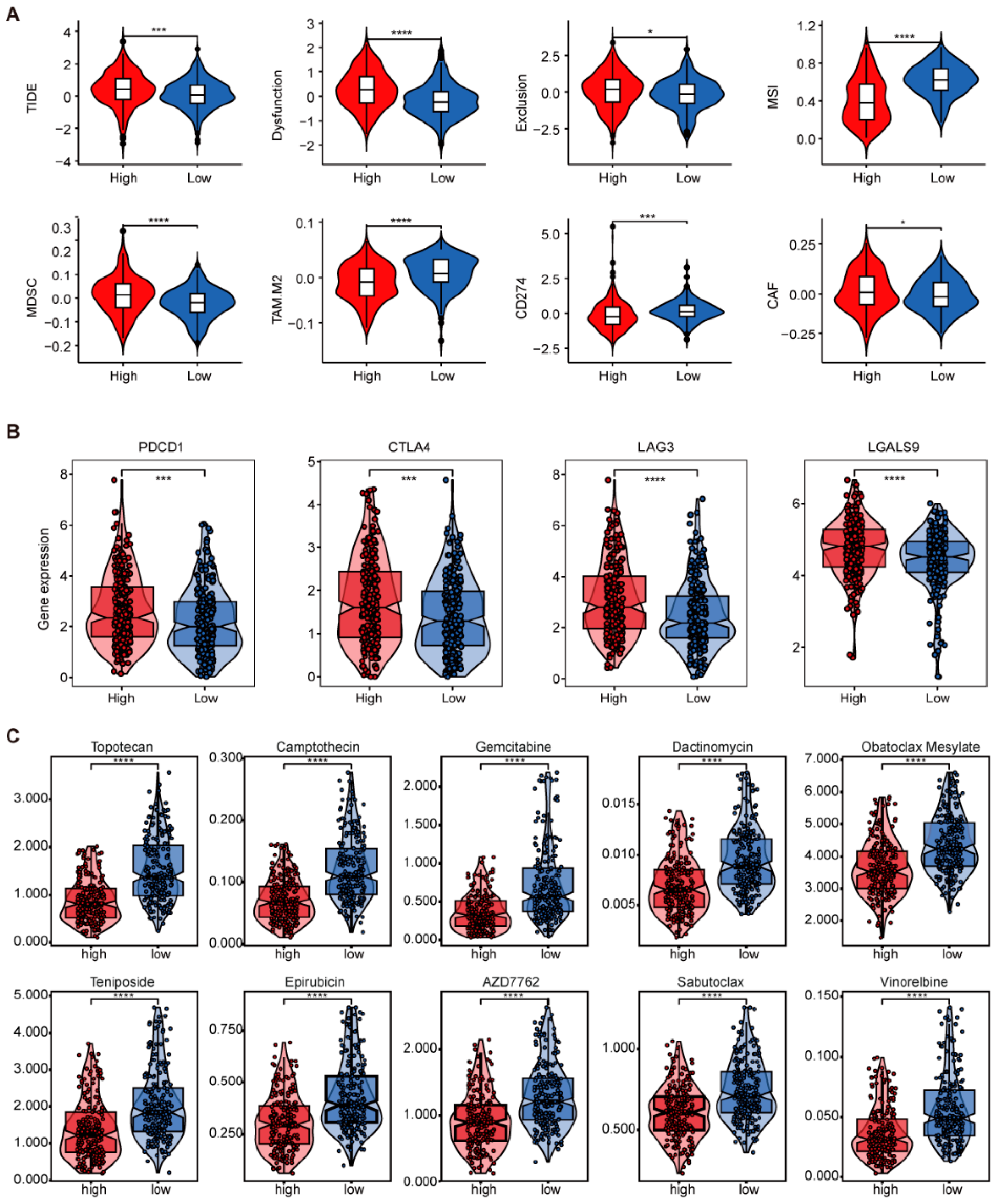
2.4. Comprehensive Analysis of GTRS in Relation to Tumor Mutational Burden and Immune Infilation
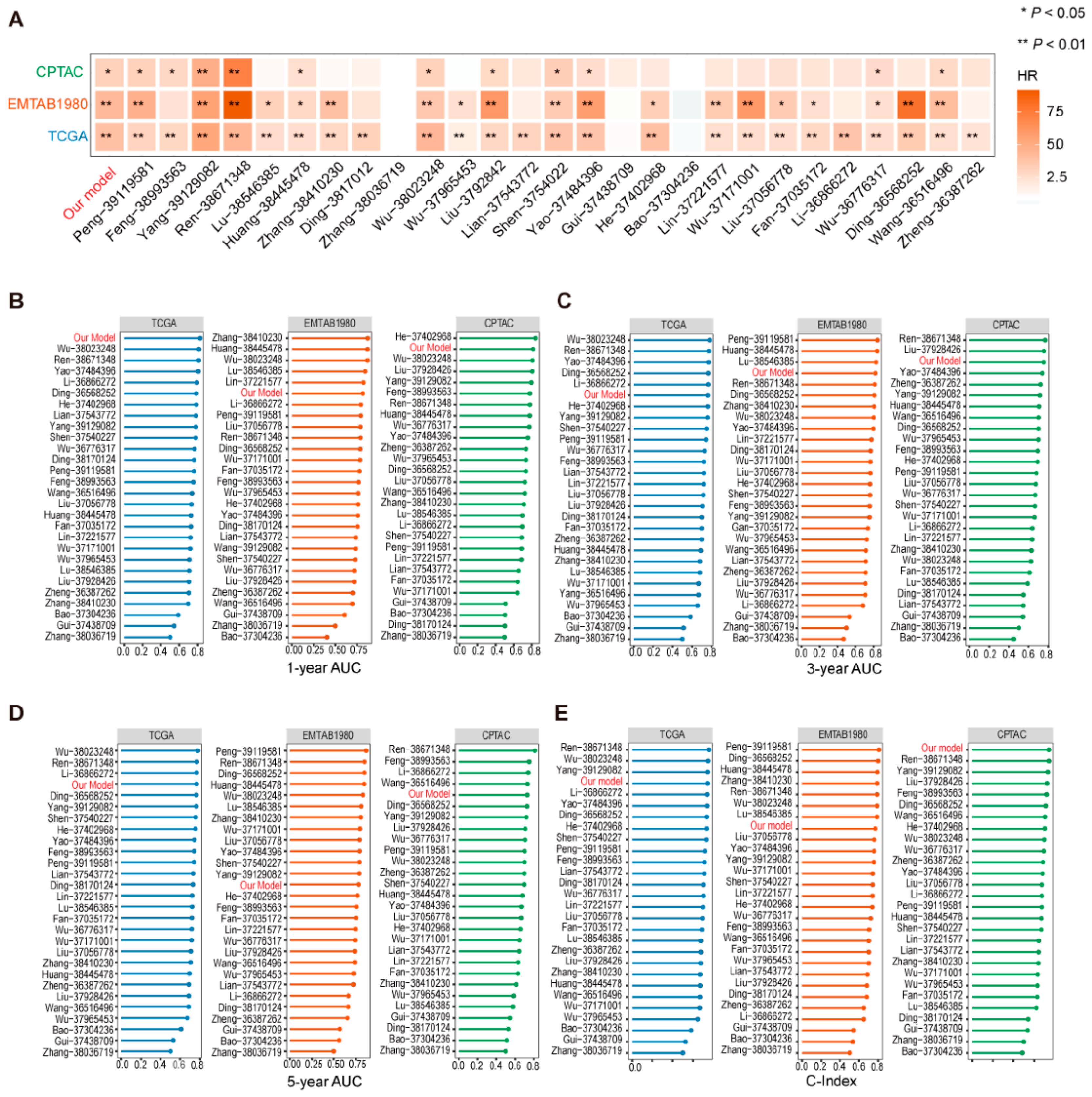
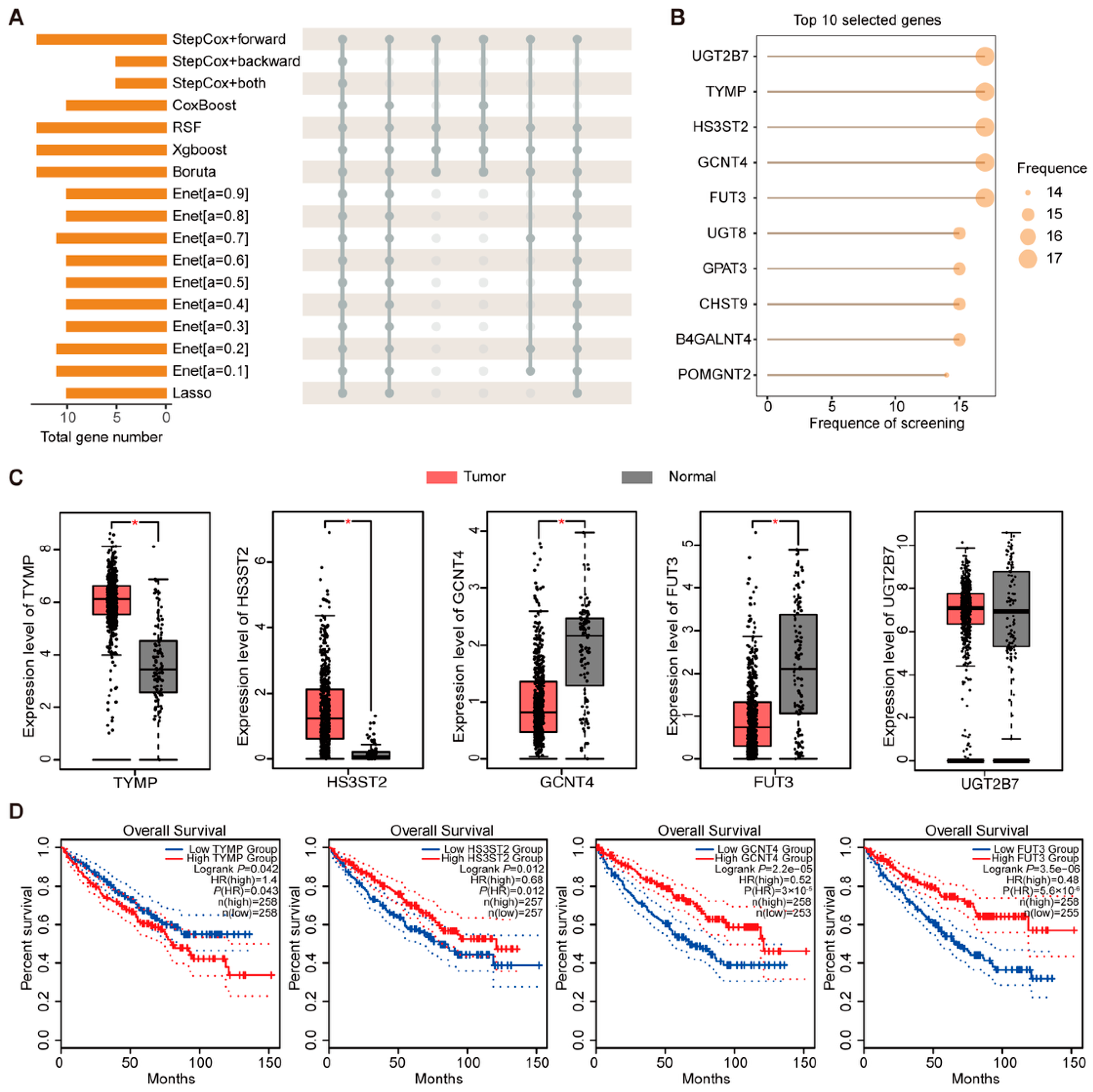
2.5. Screening and Analysis of Key Genes in Prognostic Models
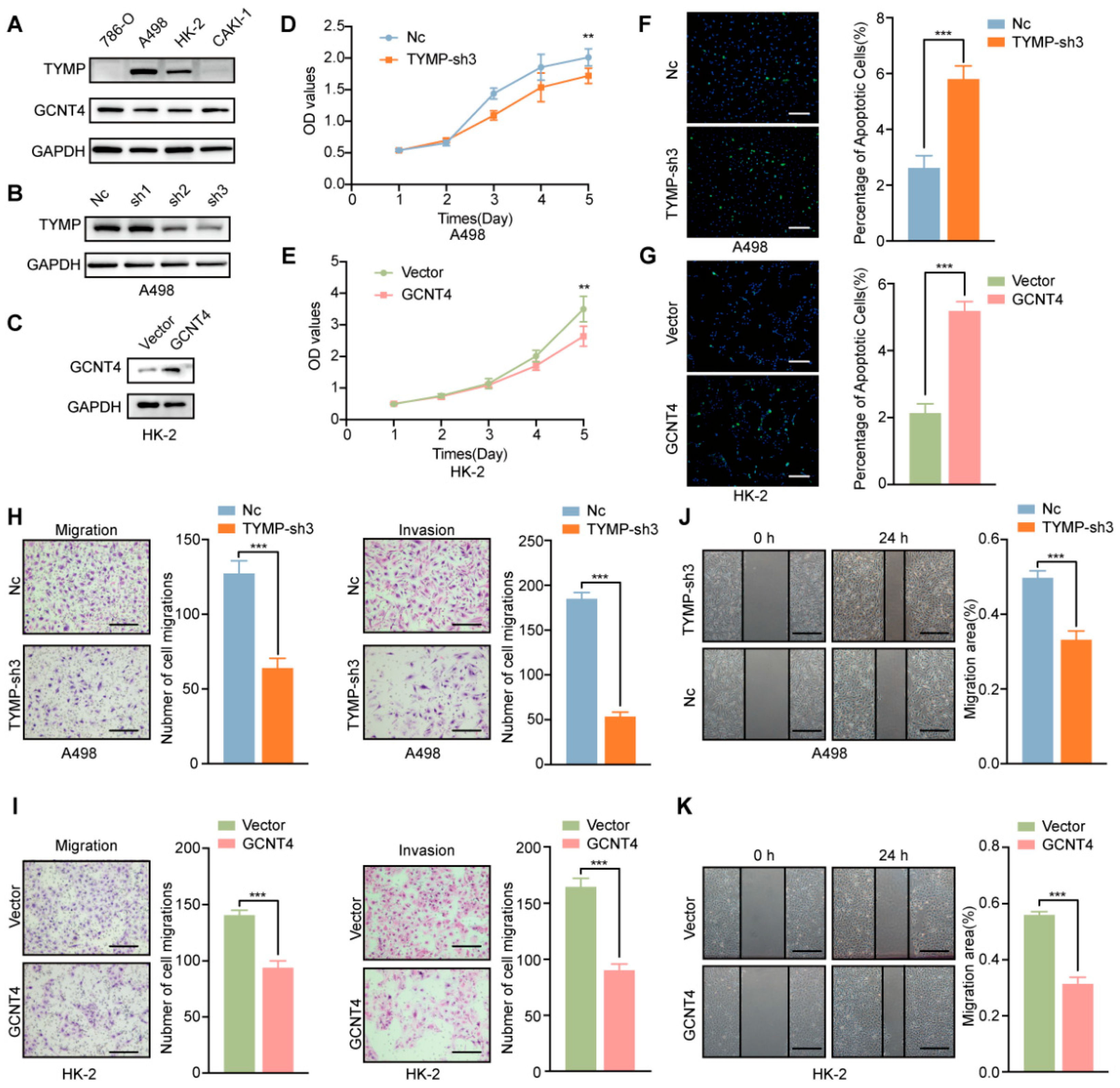
2.6. Functional Validation of TYMP and GCNT4 in ccRCC
3. Discussion
4. Materials and Methods
4.1. Data Acquisition
4.2. Data Normalization
4.3. Integrated Machine Learning Model Construction
4.4. Cox Regression and Nomogram Construction
4.5. TMB Analysis
4.6. Tumor Immune Microenvironment Analysis
4.7. Chemotherapy Drug Sensitivity Analysis
4.8. Functional Enrichment Analysis
4.9. Cell Line Culture
4.10. Cell Counting Kit 8 (CCK-8) Assay and Colony Formation Assay
4.11. Transwell Assay,
4.12. Western Blot
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Quinn, A.E.; Bell, S.D.; Marrah, A.J.; Wakefield, M.R.; Fang, Y. The Current State of the Diagnoses and Treatments for Clear Cell Renal Cell Carcinoma. Cancers 2024, 16, 4034. [Google Scholar] [CrossRef] [PubMed]
- Rose, T.L.; Kim, W.Y. Renal Cell Carcinoma: A Review. Jama 2024, 332, 1001–1010. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Al-Danakh, A.; Zhang, L.; Sun, X.; Jian, Y.; Wu, H.; Feng, D.; Wang, S.; Yang, D. Glycosylation in Renal Cell Carcinoma: Mechanisms and Clinical Implications. Cells 2022, 11, 2598. [Google Scholar] [CrossRef]
- Varadharaj, V.; Petersen, W.; Batra, S.K.; Ponnusamy, M.P. Sugar symphony: Glycosylation in cancer metabolism and stemness. Trends Cell Biol. 2025, 35, 412–425. [Google Scholar] [CrossRef]
- Fisher, P.; Thomas-Oates, J.; Wood, A.J.; Ungar, D. The N-Glycosylation Processing Potential of the Mammalian Golgi Apparatus. Front. Cell Dev. Biol. 2019, 7, 157. [Google Scholar] [CrossRef]
- Ren, X.; Lin, S.; Guan, F.; Kang, H. Glycosylation Targeting: A Paradigm Shift in Cancer Immunotherapy. Int. J. Biol. Sci. 2024, 20, 2607–2621. [Google Scholar] [CrossRef]
- Li, Y.; Chen, H.; Gao, J.; Wu, P.; Hong, S. Glycoengineering in antigen-specific immunotherapies. Curr. Opin. Chem. Biol. 2024, 81, 102503. [Google Scholar] [CrossRef]
- Baro, M.; Lee, H.; Kelley, V.; Lou, R.; Phoomak, C.; Politi, K.; Zeiss, C.J.; Van Zandt, M.; Contessa, J.N. Redundancy of the OST catalytic subunit facilitates therapeutic targeting of N-glycosylation. Cell Chem. Biol. 2025, 32, 839–853.e6. [Google Scholar] [CrossRef]
- Roy, R. Cancer cells and viruses share common glycoepitopes: Exciting opportunities toward combined treatments. Front. Immunol. 2024, 15, 1292588. [Google Scholar] [CrossRef]
- Luo, B.; Liu, X.; Zhang, Q.; Liang, G.; Zhuang, Y. ALG3 predicts poor prognosis and increases resistance to anti-PD-1 therapy through modulating PD-L1 N-link glycosylation in TNBC. Int. Immunopharmacol. 2024, 140, 112875. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cui, K.; Qiang, R.; Wang, L. FUT10 is related to the poor prognosis and immune infiltration in clear cell renal cell carcinoma. Transl. Cancer Res. 2025, 14, 827–842. [Google Scholar] [CrossRef]
- Meng, L.; Xu, L.; Yang, Y.; Zhou, L.; Chang, Y.; Shi, T.; Tan, C.; An, H.; Zhu, Y.; Xu, J. High expression of FUT3 is linked to poor prognosis in clear cell renal cell carcinoma. Oncotarget 2017, 8, 61036–61047. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, H.; Liu, W.; Zhang, W.; An, H.; Xu, J. β1,6-N-acetylglucosaminyltransferase V predicts recurrence and survival of patients with clear-cell renal cell carcinoma after surgical resection. World J. Urol. 2015, 33, 1791–1799. [Google Scholar] [CrossRef]
- Dalal, V.; Carmicheal, J.; Dhaliwal, A.; Jain, M.; Kaur, S.; Batra, S.K. Radiomics in stratification of pancreatic cystic lesions: Machine learning in action. Cancer Lett. 2020, 469, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Du, A.; Li, J.; Yang, Q. An anoikis-related gene signature predicts prognosis in patients with acute myeloid leukemia and immunotherapy. Am. J. Cancer Res. 2024, 14, 5116–5132. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.; Zhong, J.; Yang, Q.; Lin, H.; Chen, B.; Lu, T.; Chen, J.; Luo, N. Single-cell and bulk transcriptomic datasets enable the development of prognostic models based on dynamic changes in the tumor immune microenvironment in patients with hepatocellular carcinoma and portal vein tumor thrombus. Front. Immunol. 2024, 15, 1414121. [Google Scholar] [CrossRef]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef]
- Čaval, T.; Alisson-Silva, F.; Schwarz, F. Roles of glycosylation at the cancer cell surface: Opportunities for large scale glycoproteomics. Theranostics 2023, 13, 2605–2615. [Google Scholar] [CrossRef]
- Gorospe, M.; Egan, J.M.; Zbar, B.; Lerman, M.; Geil, L.; Kuzmin, I.; Holbrook, N.J. Protective function of von Hippel-Lindau protein against impaired protein processing in renal carcinoma cells. Mol. Cell Biol. 1999, 19, 1289–1300. [Google Scholar] [CrossRef]
- Jones, R.B.; Dorsett, K.A.; Hjelmeland, A.B.; Bellis, S.L. The ST6Gal-I sialyltransferase protects tumor cells against hypoxia by enhancing HIF-1α signaling. J. Biol. Chem. 2018, 293, 5659–5667. [Google Scholar] [CrossRef]
- Liu, Y.; Li, M.; Lin, M.; Liu, X.; Guo, H.; Tan, J.; Hu, L.; Li, J.; Zhou, Q. ALKBH1 promotes HIF-1α-mediated glycolysis by inhibiting N-glycosylation of LAMP2A. Cell. Mol. Life Sci. 2024, 81, 130. [Google Scholar] [CrossRef]
- Gao, F.; Ding, J.; Gai, B.; Cai, D.; Hu, C.; Wang, F.A.; He, R.; Liu, J.; Li, Y.; Wu, X.J. Interpretable Multimodal Fusion Model for Bridged Histology and Genomics Survival Prediction in Pan-Cancer. Adv. Sci. 2025, 12, e2407060. [Google Scholar] [CrossRef]
- Audureau, E.; Soubeyran, P.; Martinez-Tapia, C.; Bellera, C.; Bastuji-Garin, S.; Boudou-Rouquette, P.; Chahwakilian, A.; Grellety, T.; Hanon, O.; Mathoulin-Pélissier, S.; et al. Machine Learning to Predict Mortality in Older Patients With Cancer: Development and External Validation of the Geriatric Cancer Scoring System Using Two Large French Cohorts. J. Clin. Oncol. 2025, 43, 1429–1440. [Google Scholar] [CrossRef]
- Wang, H.; Chen, J.; Gao, W.; Wu, Y.; Wang, X.; Lin, F.; Chen, H.; Wang, Y.; Jiang, T.; Pan, Z.; et al. Construction of a nomogram with IrAE and clinic character to predict the survival of advanced G/GEJ adenocarcinoma patients undergoing anti-PD-1 treatment. Front. Immunol. 2024, 15, 1432281. [Google Scholar] [CrossRef]
- Li, Y.; Ge, X.; Peng, F.; Li, W.; Li, J.J. Exaggerated false positives by popular differential expression methods when analyzing human population samples. Genome Biol. 2022, 23, 79. [Google Scholar] [CrossRef]
- Ge, X.; Li, Y.; Li, W.; Li, J.J. Response to “Neglecting normalization impact in semi-synthetic RNA-seq data simulation generates artificial false positives” and “Winsorization greatly reduces false positives by popular differential expression methods when analyzing human population samples”. Genome Biol. 2024, 25, 283. [Google Scholar]
- Ngiam, K.Y.; Khor, I.W. Big data and machine learning algorithms for health-care delivery. Lancet Oncol. 2019, 20, e262–e273, Correctionz in Lancet Oncol. 2019, 20, e262-73. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, W.; Zhang, Y.; Adegboro, A.A.; Fasoranti, D.O.; Dai, L.; Pan, Z.; Liu, H.; Xiong, Y.; Li, W.; et al. Mime: A flexible machine-learning framework to construct and visualize models for clinical characteristics prediction and feature selection. Comput. Struct. Biotechnol. J. 2024, 23, 2798–2810. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Wang, J.; Wang, J.; Wang, G.; Ren, H.; Xu, P.; Yang, M.; Xu, K. Association between conicity index (C-index), relative fat mass (RFM), and osteoarthritis (OA): Evidence from NHANES 2003–2018. Lipids Health Dis. 2025, 24, 140. [Google Scholar] [CrossRef]
- Chen, Q.; Yin, H.; He, J.; Xie, Y.; Wang, W.; Xu, H.; Zhang, L.; Shi, C.; Yu, J.; Wu, W.; et al. Tumor Microenvironment Responsive CD8(+) T Cells and Myeloid-Derived Suppressor Cells to Trigger CD73 Inhibitor AB680-Based Synergistic Therapy for Pancreatic Cancer. Adv. Sci. 2023, 10, e2302498. [Google Scholar] [CrossRef]
- Arpinati, L.; Carradori, G.; Scherz-Shouval, R. CAF-induced physical constraints controlling T cell state and localization in solid tumours. Nat. Rev. Cancer 2024, 24, 676–693. [Google Scholar] [CrossRef]
- Mojtabavi, H.; Fatehi, F.; Shahkarami, S.; Rezaei, N.; Nafissi, S. Novel Mutations of the TYMP Gene in Mitochondrial Neurogastrointestinal Encephalomyopathy: Case Series and Literature Review. J. Mol. Neurosci. 2021, 71, 2526–2533. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yue, H. Thymidine phosphorylase: A potential new target for treating cardiovascular disease. Trends Cardiovasc. Med. 2018, 28, 157–171. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Gigante, A.; Perez-Perez, M.J.; Yue, H.; Hirano, M.; McIntyre, T.M.; Silverstein, R.L. Thymidine phosphorylase participates in platelet signaling and promotes thrombosis. Circ. Res. 2014, 115, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.A.; Zhang, J.P.; Wang, N.; Chen, J. Identifying TYMP as an Immune Prognostic Marker in Clear Cell Renal Cell Carcinoma. Technol. Cancer Res. Treat. 2023, 22, 15330338231194555. [Google Scholar] [CrossRef]
- Li, Y.; Fan, C.; Hu, Y.; Zhang, W.; Li, H.; Wang, Y.; Xu, Z. Multi-cohort validation: A comprehensive exploration of prognostic marker in clear cell renal cell carcinoma. Int. Immunopharmacol. 2024, 135, 112300. [Google Scholar] [CrossRef]
- Gadi, M.R.; Han, J.; Shen, T.; Fan, S.; Xiao, Z.; Li, L. Divergent synthesis of amino acid-linked O-GalNAc glycan core structures. Nat. Protoc. 2025, 20, 480–517. [Google Scholar] [CrossRef]
- Singh, A.; Beaupre, M.; Villegas-Novoa, C.; Shiomitsu, K.; Gaudino, S.J.; Tawch, S.; Damle, R.; Kempen, C.; Choudhury, B.; McAleer, J.P.; et al. IL-22 promotes mucin-type O-glycosylation and MATH1(+) cell-mediated amelioration of intestinal inflammation. Cell Rep. 2024, 43, 114206. [Google Scholar] [CrossRef]
- Yang, L.; Yang, Q.; Lin, L.; Zhang, C.; Dong, L.; Gao, X.; Zhang, Z.; Zeng, C.; Wang, P.G. LectoScape: A Highly Multiplexed Imaging Platform for Glycome Analysis and Biomedical Diagnosis. Anal. Chem. 2024, 96, 6558–6565. [Google Scholar] [CrossRef]
- Tan, Z.; Ning, L.; Cao, L.; Zhou, Y.; Li, J.; Yang, Y.; Lin, S.; Ren, X.; Xue, X.; Kang, H.; et al. Bisecting GlcNAc modification reverses the chemoresistance via attenuating the function of P-gp. Theranostics 2024, 14, 5184–5199. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, A.; Negi, M.; Choi, E.H.; Kaushik, N.K.; Kaushik, N. Upstream-binding protein-1 promotes breast tumorigenesis by inducing NRG2-mediated metastasis, plasticity, and macrophage polarization. Int. J. Biol. Macromol. 2025, 307 Pt 2, 141915. [Google Scholar] [CrossRef]
- Nie, H.; Saini, P.; Miyamoto, T.; Liao, L.; Zielinski, R.J.; Liu, H.; Zhou, W.; Wang, C.; Murphy, B.; Towers, M.; et al. Targeting branched N-glycans and fucosylation sensitizes ovarian tumors to immune checkpoint blockade. Nat. Commun. 2024, 15, 2853. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, T.; Wu, T.; Lei, T.; Li, Y.; Wang, X. GGDB: A Grameneae genome alignment database of homologous genes hierarchically related to evolutionary events. Plant Physiol. 2022, 190, 340–351. [Google Scholar] [CrossRef]
- Seal, R.L.; Braschi, B.; Gray, K.; Jones, T.E.M.; Tweedie, S.; Haim-Vilmovsky, L.; Bruford, E.A. Genenames.org: The HGNC resources in 2023. Nucleic Acids Res. 2023, 51, D1003–D1009. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol. Biol. 2018, 1711, 243–259. [Google Scholar] [PubMed]
- Jia, Q.; Wu, W.; Wang, Y.; Alexander, P.B.; Sun, C.; Gong, Z.; Cheng, J.N.; Sun, H.; Guan, Y.; Xia, X.; et al. Local mutational diversity drives intratumoral immune heterogeneity in non-small cell lung cancer. Nat. Commun. 2018, 9, 5361. [Google Scholar] [CrossRef] [PubMed]
- Maeser, D.; Gruener, R.F.; Huang, R.S. oncoPredict: An R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief. Bioinform. 2021, 22, bbab260. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, C.; Zhou, M.; Luo, Y.; Jiang, R.; Hu, Y.; Zhao, M.; Yan, X.; Xiao, S.; Xue, M.; Wang, M.; et al. Constructing a Prognostic Model for Clear Cell Renal Cell Carcinoma Based on Glycosyltransferase Gene and Verification of Key Gene Identification. Int. J. Mol. Sci. 2025, 26, 10182. https://doi.org/10.3390/ijms262010182
Zhou C, Zhou M, Luo Y, Jiang R, Hu Y, Zhao M, Yan X, Xiao S, Xue M, Wang M, et al. Constructing a Prognostic Model for Clear Cell Renal Cell Carcinoma Based on Glycosyltransferase Gene and Verification of Key Gene Identification. International Journal of Molecular Sciences. 2025; 26(20):10182. https://doi.org/10.3390/ijms262010182
Chicago/Turabian StyleZhou, Chong, Mingzhe Zhou, Yuzhou Luo, Ruohan Jiang, Yushu Hu, Meiqi Zhao, Xu Yan, Shan Xiao, Mengjie Xue, Mengwei Wang, and et al. 2025. "Constructing a Prognostic Model for Clear Cell Renal Cell Carcinoma Based on Glycosyltransferase Gene and Verification of Key Gene Identification" International Journal of Molecular Sciences 26, no. 20: 10182. https://doi.org/10.3390/ijms262010182
APA StyleZhou, C., Zhou, M., Luo, Y., Jiang, R., Hu, Y., Zhao, M., Yan, X., Xiao, S., Xue, M., Wang, M., Jiang, P., Zhou, Y., Huang, X., Sun, D., Zhang, C., Jin, Y., & Wu, N. (2025). Constructing a Prognostic Model for Clear Cell Renal Cell Carcinoma Based on Glycosyltransferase Gene and Verification of Key Gene Identification. International Journal of Molecular Sciences, 26(20), 10182. https://doi.org/10.3390/ijms262010182





