High-Resolution Assembly of the Human Y Chromosome Identifies a Vast Landscape of Inverted Repeats Associated with Structural and Functional Genomic Features
Abstract
1. Introduction
2. Results
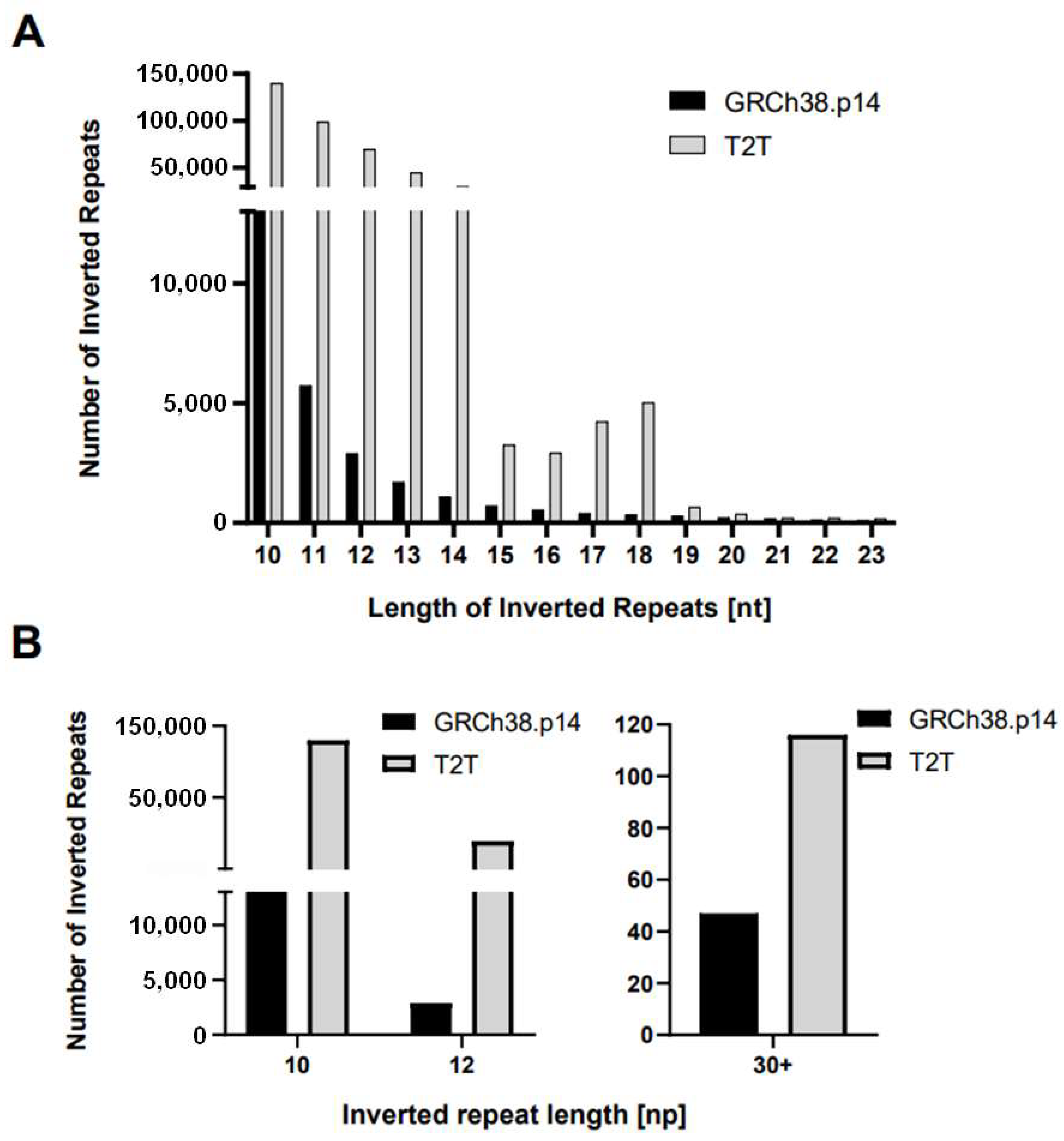
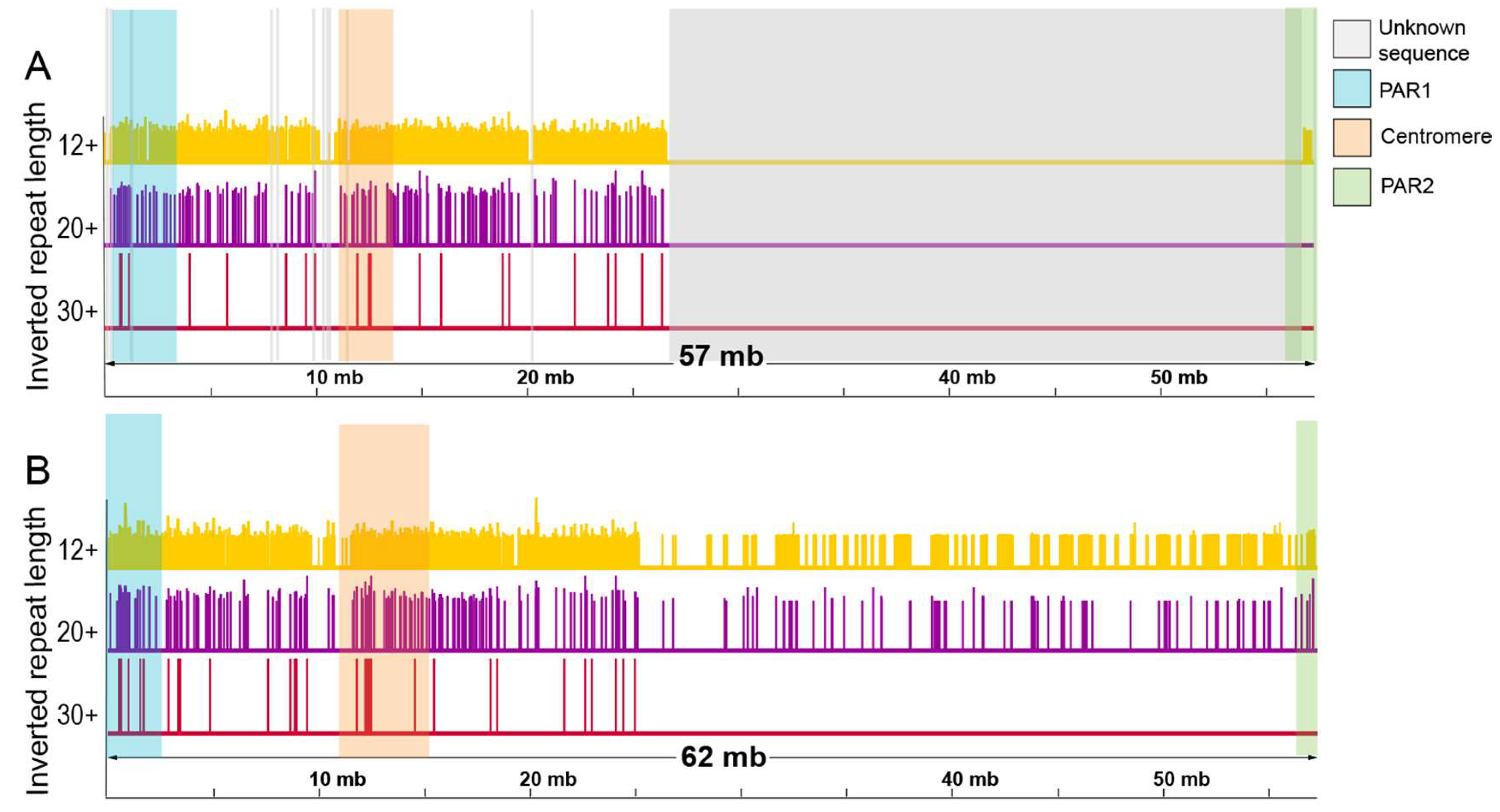
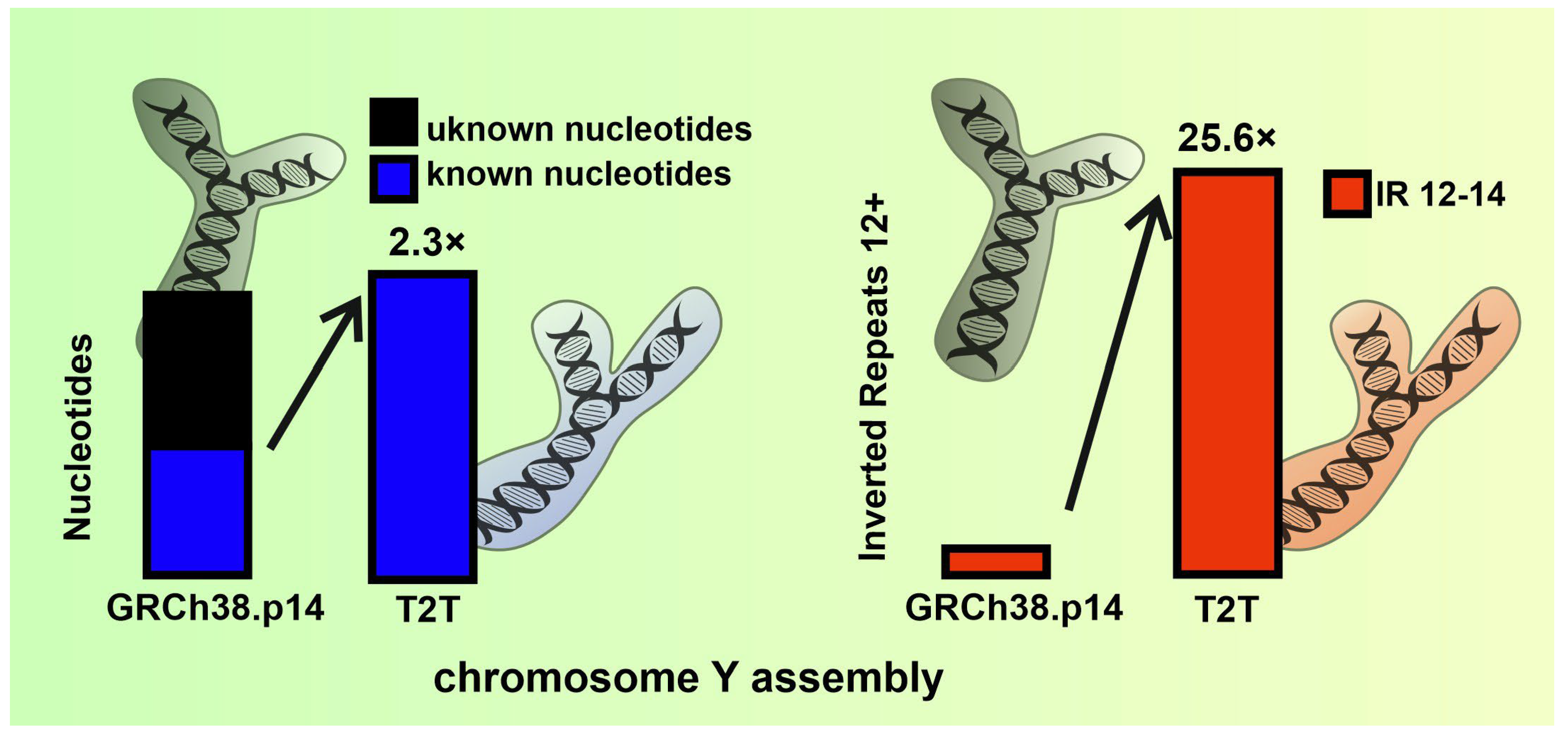
2.1. Variation in IR Occurrence and Frequency in Chromosome Y Assemblies
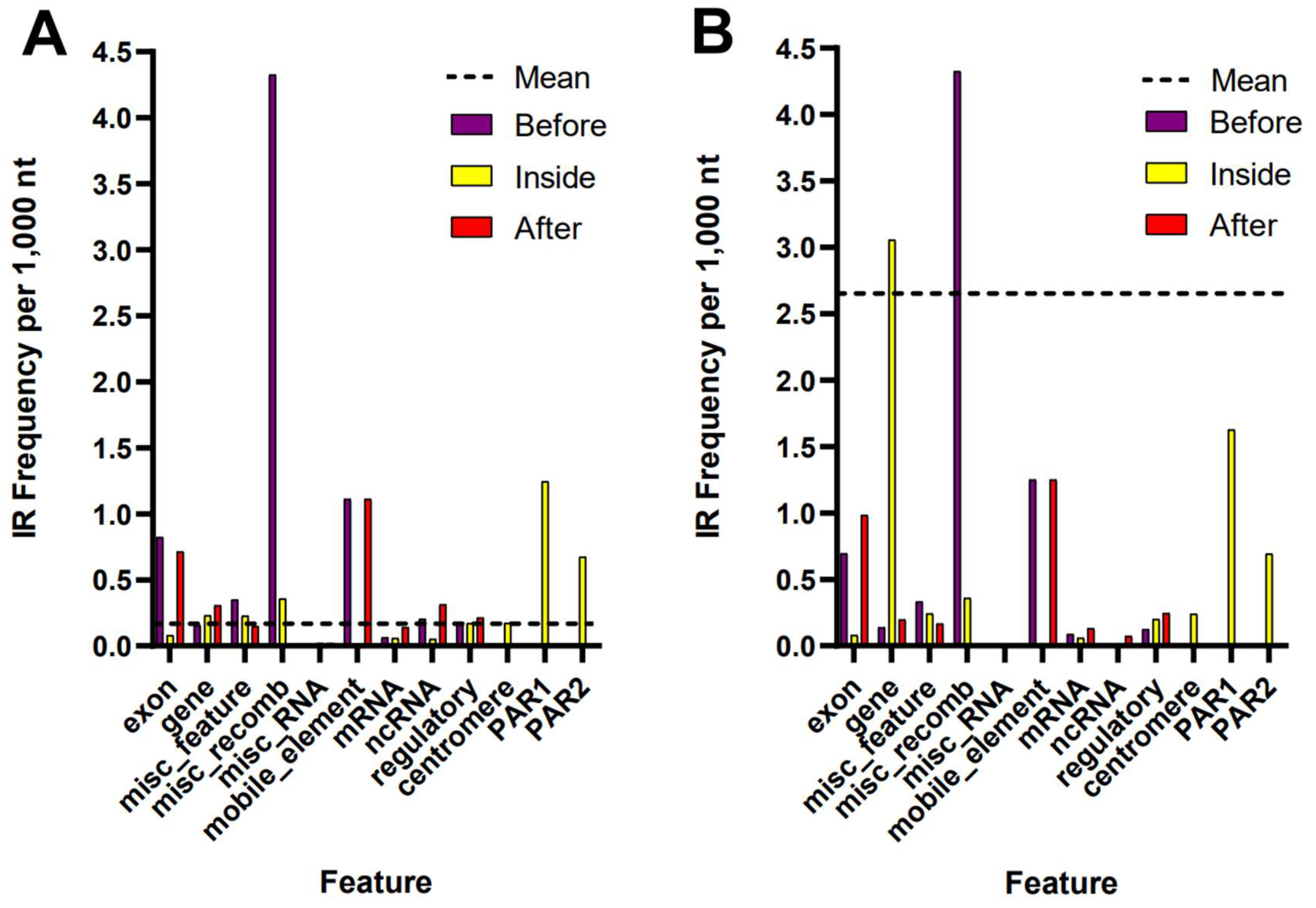
2.2. Comparison of IR Occurrence Around Annotated Features of the Genomes
3. Discussion
4. Materials and Methods
4.1. Genomes
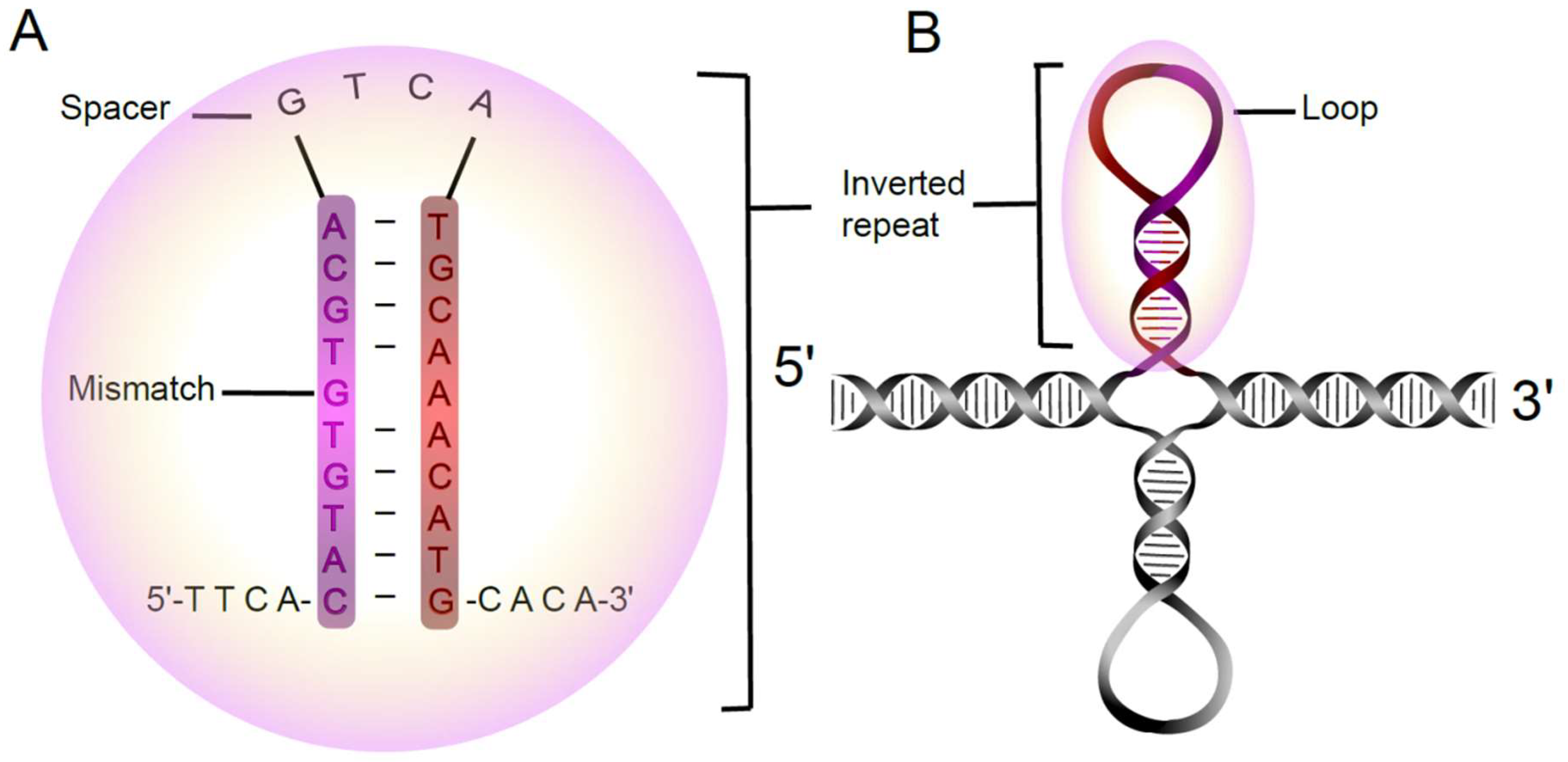
4.2. Analyses of Short Inverted Repeats
4.3. Analyses of Genomic Features Overlap with Inverted Repeats
4.4. Sequence Identity Analyses and Transcription Factor-Binding Sites Prediction
4.5. Statistical Evaluation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| bp | Base Pair |
| Chr | Chromosome |
| Crx | Cone-Rod Homeobox |
| DAZ1 | Deleted in Azoospermia 1 |
| DOID | Disease Ontology Identifier |
| DNA | Deoxyribonucleic acid |
| ENCODE | Encyclopedia of DNA Elements |
| ENSEMBL | European Bioinformatics Institute Genome Database |
| GC | Guanine–Cytosine (content) |
| GR-β | Glucocorticoid Receptor Beta |
| GRCh38 | Genome Reference Consortium Human Build 38 |
| HOXD8 | Homeobox D8 |
| IGV | Integrative Genomics Viewer |
| IR(s) | Inverted Repeat(s) |
| kbp | Kilobase Pair |
| KW | UniProt Keyword |
| misc_RNA | Miscellaneous Small RNA |
| mRNA | Messenger RNA |
| ncRNA | Non-coding RNA |
| NCBI | National Center for Biotechnology Information |
| nt | Nucleotide |
| PAR(s) | Pseudoautosomal Region(s) |
| POU1F1a/POU1F1s | POU Class 1 Homeobox 1 (isoforms a/s) |
| p-value | Probability Value |
| PROMO | Transcription Factor Binding Site Prediction Tool |
| RNA | Ribonucleic Acid |
| RPS4Y1 | Ribosomal Protein S4, Y-linked 1 |
| SRY | Sex-determining Region Y |
| STRING | Search Tool for the Retrieval of Interacting Genes/Proteins |
| T2T | Telomere-to-Telomere |
| UTR | Untranslated Region |
| USP9Y | Ubiquitin-Specific Peptidase 9, Y-linked |
References
- Quintana-Murci, L.; Fellous, M. The Human Y Chromosome: The Biological Role of a “Functional Wasteland”. J. Biomed. Biotechnol. 2001, 1, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Koren, S.; Rhie, A.; Rautiainen, M.; Bzikadze, A.V.; Mikheenko, A.; Vollger, M.R.; Altemose, N.; Uralsky, L.; Gershman, A.; et al. The Complete Sequence of a Human Genome. Science 2022, 376, 44–53. [Google Scholar] [CrossRef]
- Schneider, V.A.; Graves-Lindsay, T.; Howe, K.; Bouk, N.; Chen, H.-C.; Kitts, P.A.; Murphy, T.D.; Pruitt, K.D.; Thibaud-Nissen, F.; Albracht, D.; et al. Evaluation of GRCh38 and de Novo Haploid Genome Assemblies Demonstrates the Enduring Quality of the Reference Assembly. Genome Res. 2017, 27, 849–864. [Google Scholar] [CrossRef]
- Rhie, A.; Nurk, S.; Cechova, M.; Hoyt, S.J.; Taylor, D.J.; Altemose, N.; Hook, P.W.; Koren, S.; Rautiainen, M.; Alexandrov, I.A.; et al. The Complete Sequence of a Human Y Chromosome. Nature 2023, 621, 344–354. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.H.J.; Seidl, M.F.; Shi-Kunne, X.; Cook, D.E.; Bolton, M.D.; van Kan, J.A.L.; Faino, L. Mind the Gap; Seven Reasons to Close Fragmented Genome Assemblies. Fungal Genet. Biol. 2016, 90, 24–30. [Google Scholar] [CrossRef]
- Hallast, P.; Ebert, P.; Loftus, M.; Yilmaz, F.; Audano, P.A.; Logsdon, G.A.; Bonder, M.J.; Zhou, W.; Höps, W.; Kim, K.; et al. Assembly of 43 Human Y Chromosomes Reveals Extensive Complexity and Variation. Nature 2023, 621, 355–364. [Google Scholar] [CrossRef]
- Weissensteiner, M.H.; Cremona, M.A.; Guiblet, W.M.; Stoler, N.; Harris, R.S.; Cechova, M.; Eckert, K.A.; Chiaromonte, F.; Huang, Y.-F.; Makova, K.D. Accurate Sequencing of DNA Motifs Able to Form Alternative (Non-B) Structures. Genome Res. 2023, 33, 907–922. [Google Scholar] [CrossRef]
- Cer, R.Z.; Donohue, D.E.; Mudunuri, U.S.; Temiz, N.A.; Loss, M.A.; Starner, N.J.; Halusa, G.N.; Volfovsky, N.; Yi, M.; Luke, B.T.; et al. Non-B DB v2.0: A Database of Predicted Non-B DNA-Forming Motifs and Its Associated Tools. Nucleic Acids Res. 2013, 41, D94–D100. [Google Scholar] [CrossRef]
- Alamro, H.; Alzamel, M.; Iliopoulos, C.S.; Pissis, S.P.; Watts, S. IUPACpal: Efficient Identification of Inverted Repeats in IUPAC-Encoded DNA Sequences. BMC Bioinform. 2021, 22, 51. [Google Scholar] [CrossRef]
- Pyne, A.L.B.; Noy, A.; Main, K.H.S.; Velasco-Berrelleza, V.; Piperakis, M.M.; Mitchenall, L.A.; Cugliandolo, F.M.; Beton, J.G.; Stevenson, C.E.M.; Hoogenboom, B.W.; et al. Base-Pair Resolution Analysis of the Effect of Supercoiling on DNA Flexibility and Major Groove Recognition by Triplex-Forming Oligonucleotides. Nat. Commun. 2021, 12, 1053. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, C.; Hastie, C.; Metera, K.; Scott, S.; Zhang, Z.; Chen, S.; Gu, G.; Weber, L.; Munsky, B.; Kouzine, F.; et al. Non-Equilibrium Structural Dynamics of Supercoiled DNA Plasmids Exhibits Asymmetrical Relaxation. Nucleic Acids Res. 2022, 50, 2754–2764. [Google Scholar] [CrossRef]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Brázda, V.; Kolomazník, J.; Lýsek, J.; Hároníková, L.; Coufal, J.; Št’astný, J. Palindrome Analyser—A New Web-Based Server for Predicting and Evaluating Inverted Repeats in Nucleotide Sequences. Biochem. Biophys. Res. Commun. 2016, 478, 1739–1745. [Google Scholar] [CrossRef]
- Brauburger, K.; Boehmann, Y.; Krähling, V.; Mühlberger, E. Transcriptional Regulation in Ebola Virus: Effects of Gene Border Structure and Regulatory Elements on Gene Expression and Polymerase Scanning Behavior. J. Virol. 2016, 90, 1898–1909. [Google Scholar] [CrossRef]
- Ladoukakis, E.D.; Eyre-Walker, A. The Excess of Small Inverted Repeats in Prokaryotes. J. Mol. Evol. 2008, 67, 291–300. [Google Scholar] [CrossRef]
- Shamanskiy, V.; Mikhailova, A.A.; Tretiakov, E.O.; Ushakova, K.; Mikhailova, A.G.; Oreshkov, S.; Knorre, D.A.; Ree, N.; Overdevest, J.B.; Lukowski, S.W.; et al. Secondary Structure of the Human Mitochondrial Genome Affects Formation of Deletions. BMC Biol. 2023, 21, 103. [Google Scholar] [CrossRef]
- Xu, R.; Pan, Z.; Nakagawa, T. Gross Chromosomal Rearrangement at Centromeres. Biomolecules 2023, 14, 28. [Google Scholar] [CrossRef] [PubMed]
- Bastos, C.A.C.; Afreixo, V.; Rodrigues, J.M.O.S.; Pinho, A.J. Concentration of Inverted Repeats along Human DNA. J. Integr. Bioinform. 2023, 20, 20220052. [Google Scholar] [CrossRef] [PubMed]
- Brewer, B.J.; Dunham, M.J.; Raghuraman, M.K. A Unifying Model That Explains the Origins of Human Inverted Copy Number Variants. PLoS Genet. 2024, 20, e1011091. [Google Scholar] [CrossRef] [PubMed]
- Brázda, V.; Laister, R.C.; Jagelská, E.B.; Arrowsmith, C. Cruciform Structures Are a Common DNA Feature Important for Regulating Biological Processes. BMC Mol. Biol. 2011, 12, 33. [Google Scholar] [CrossRef]
- Soto, D.C.; Uribe-Salazar, J.M.; Shew, C.J.; Sekar, A.; McGinty, S.P.; Dennis, M.Y. Genomic Structural Variation: A Complex but Important Driver of Human Evolution. Am. J. Biol. Anthropol. 2023, 181 (Suppl. S76), 118–144. [Google Scholar] [CrossRef]
- Kolb, J.; Chuzhanova, N.A.; Högel, J.; Vasquez, K.M.; Cooper, D.N.; Bacolla, A.; Kehrer-Sawatzki, H. Cruciform-Forming Inverted Repeats Appear to Have Mediated Many of the Microinversions That Distinguish the Human and Chimpanzee Genomes. Chromosome Res. 2009, 17, 469–483. [Google Scholar] [CrossRef]
- Yu, X.-W.; Wei, Z.-T.; Jiang, Y.-T.; Zhang, S.-L. Y Chromosome Azoospermia Factor Region Microdeletions and Transmission Characteristics in Azoospermic and Severe Oligozoospermic Patients. Int. J. Clin. Exp. Med. 2015, 8, 14634–14646. [Google Scholar]
- Ait Saada, A.; Guo, W.; Costa, A.B.; Yang, J.; Wang, J.; Lobachev, K.S. Widely Spaced and Divergent Inverted Repeats Become a Potent Source of Chromosomal Rearrangements in Long Single-Stranded DNA Regions. Nucleic Acids Res. 2023, 51, 3722–3734. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Helena Mangs, A.; Morris, B.J. The Human Pseudoautosomal Region (PAR): Origin, Function and Future. Curr. Genom. 2007, 8, 129–136. [Google Scholar] [CrossRef]
- Mosig, A.; Guofeng, M.; Stadler, B.M.R.; Stadler, P.F. Evolution of the Vertebrate Y RNA Cluster. Theory Biosci. 2007, 126, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, M.P.; Krude, T. Functional Roles of Non-Coding Y RNAs. Int. J. Biochem. Cell Biol. 2015, 66, 20. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.W.; Amode, M.R.; Austine-Orimoloye, O.; Azov, A.G.; Barba, M.; Barnes, I.; Becker, A.; Bennett, R.; Berry, A.; Bhai, J.; et al. Ensembl 2024. Nucleic Acids Res. 2024, 52, D891–D899. [Google Scholar] [CrossRef]
- Fu, X.-F.; Cheng, S.-F.; Wang, L.-Q.; Yin, S.; De Felici, M.; Shen, W. DAZ Family Proteins, Key Players for Germ Cell Development. Int. J. Biol. Sci. 2015, 11, 1226–1235. [Google Scholar] [CrossRef]
- Hughes, J.F.; Skaletsky, H.; Koutseva, N.; Pyntikova, T.; Page, D.C. Sex Chromosome-to-Autosome Transposition Events Counter Y-Chromosome Gene Loss in Mammals. Genome Biol. 2015, 16, 104. [Google Scholar] [CrossRef]
- Vollger, M.R.; Guitart, X.; Dishuck, P.C.; Mercuri, L.; Harvey, W.T.; Gershman, A.; Diekhans, M.; Sulovari, A.; Munson, K.M.; Lewis, A.P.; et al. Segmental Duplications and Their Variation in a Complete Human Genome. Science 2022, 376, eabj6965. [Google Scholar] [CrossRef]
- Skaletsky, H.; Kuroda-Kawaguchi, T.; Minx, P.J.; Cordum, H.S.; Hillier, L.; Brown, L.G.; Repping, S.; Pyntikova, T.; Ali, J.; Bieri, T.; et al. The Male-Specific Region of the Human Y Chromosome Is a Mosaic of Discrete Sequence Classes. Nature 2003, 423, 825–837. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H.; Marszalek, J.D.; Minx, P.J.; Cordum, H.S.; Waterston, R.H.; Wilson, R.K.; Page, D.C. Abundant Gene Conversion between Arms of Palindromes in Human and Ape Y Chromosomes. Nature 2003, 423, 873–876. [Google Scholar] [CrossRef] [PubMed]
- Bonito, M.; Ravasini, F.; Novelletto, A.; D’Atanasio, E.; Cruciani, F.; Trombetta, B. Disclosing Complex Mutational Dynamics at a Y Chromosome Palindrome Evolving through Intra- and Inter-Chromosomal Gene Conversion. Hum. Mol. Genet. 2023, 32, 65–78. [Google Scholar] [CrossRef]
- Carvalho, C.M.B.; Zhang, F.; Lupski, J.R. Structural Variation of the Human Genome: Mechanisms, Assays, and Role in Male Infertility. Syst. Biol. Reprod. Med. 2011, 57, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Dobrovolná, M.; Mergny, J.-L.; Brázda, V. Complete Analysis of G-Quadruplex Forming Sequences in the Gapless Assembly of Human Chromosome Y. Biochimie 2024, 229, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Brázda, V.; Bohálová, N.; Bowater, R.P. New Telomere to Telomere Assembly of Human Chromosome 8 Reveals a Previous Underestimation of G-Quadruplex Forming Sequences and Inverted Repeats. Gene 2022, 810, 146058. [Google Scholar] [CrossRef]
- Bohálová, N.; Mergny, J.-L.; Brázda, V. Novel G-Quadruplex Prone Sequences Emerge in the Complete Assembly of the Human X Chromosome. Biochimie 2021, 191, 87–90. [Google Scholar] [CrossRef]
- Nguyen, T.; Li, S.; Chang, J.T.-H.; Watters, J.W.; Ng, H.; Osunsade, A.; David, Y.; Liu, S. Chromatin Sequesters Pioneer Transcription Factor Sox2 from Exerting Force on DNA. Nat. Commun. 2022, 13, 3988. [Google Scholar] [CrossRef]
- Lu, S.; Wang, G.; Bacolla, A.; Zhao, J.; Spitser, S.; Vasquez, K.M. Short Inverted Repeats Are Hotspots for Genetic Instability: Relevance to Cancer Genomes. Cell Rep. 2015, 10, 1674–1680. [Google Scholar] [CrossRef]
- Smeds, L.; Kamali, K.; Kejnovská, I.; Kejnovský, E.; Chiaromonte, F.; Makova, K.D. Non-Canonical DNA in Human and Other Ape Telomere-to-Telomere Genomes. Nucleic Acids Res. 2025, 53, gkaf298. [Google Scholar] [CrossRef]
- Swiel, Y.; Kelso, J.; Peyrégne, S. Resolving the Source of Branch Length Variation in the Y Chromosome Phylogeny. Genome Biol. 2025, 26, 4. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genomics Viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Farré, D.; Roset, R.; Huerta, M.; Adsuara, J.E.; Roselló, L.; Albà, M.M.; Messeguer, X. Identification of Patterns in Biological Sequences at the ALGGEN Server: PROMO and MALGEN. Nucleic Acids Res. 2003, 31, 3651–3653. [Google Scholar] [CrossRef]
- Messeguer, X.; Escudero, R.; Farré, D.; Nuñez, O.; Martínez, J.; Albà, M.M. PROMO: Detection of Known Transcription Regulatory Elements Using Species-Tailored Searches. Bioinformatics 2002, 18, 333–334. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S. The STRING Database in 2023: Protein–Protein Association Networks and Functional Enrichment Analyses for Any Sequenced Genome of Interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]

| GRCh38.p14 | T2T | Δ | Δ% | |
|---|---|---|---|---|
| Length | 57,227,415 | 62,460,029 | 5,232,614 | 9.1 |
| A | 7,886,192 | 21,954,563 | 14,068,371 | 178.4 |
| T | 7,956,168 | 17,929,049 | 9,972,881 | 125.4 |
| G | 5,286,894 | 13,373,414 | 8,086,520 | 152.9 |
| C | 5,285,789 | 9,203,003 | 3,917,214 | 74.1 |
| N | 30,812,372 | 0 | −3.1 × 107 | −100 |
| GC | 10,572,683 | 22,576,417 | 12,003,734 | 113.5 |
| GC [%] | 40.03 * | 36.15 | −3.88 | −9.7 |
| A | ||||
| IR length | GRCh38.p14 | T2T | Δ | Δ% |
| all | 28,003 | 399,394 | 371,391 | 1426.3 |
| 12+ | 8962 | 160,726 | 151,764 | 1793.4 |
| 20+ | 982 | 1423 | 441 | 144.9 |
| B | ||||
| IR length | GRCh38.p14 | T2T | Δ | Δ% |
| 10 | 13,313 | 139,977 | 126,664 | 1051.4 |
| 11 | 5728 | 98,691 | 92,963 | 1723 |
| 12 | 2898 | 69,430 | 66,532 | 2395.8 |
| 13 | 1700 | 44,452 | 42,752 | 2614.8 |
| 14 | 1096 | 29,347 | 28,251 | 2677.6 |
| 15 | 713 | 3247 | 2534 | 455.4 |
| 16 | 542 | 2924 | 2382 | 539.5 |
| 17 | 396 | 4227 | 3831 | 1067.4 |
| 18 | 341 | 5023 | 4682 | 1473 |
| 19 | 294 | 653 | 359 | 222.1 |
| 20 | 213 | 373 | 160 | 175.1 |
| 21 | 176 | 195 | 19 | 110.8 |
| 22 | 136 | 194 | 58 | 142.6 |
| 23 | 100 | 160 | 60 | 160 |
| 24 | 104 | 126 | 22 | 121.2 |
| 25 | 66 | 80 | 14 | 121.2 |
| 26 | 48 | 56 | 8 | 116.7 |
| 27 | 41 | 43 | 2 | 104.9 |
| 28 | 30 | 45 | 15 | 150 |
| 29 | 21 | 35 | 14 | 166.7 |
| 30+ | 47 | 116 | 69 | 246.8 |
| C | ||||
| IR length | GRCh38.p14 | T2T | Δ | Δ% |
| all | 0.489 | 6.394 | 5.915 | 1306.8 |
| 12+ | 0.157 | 2.573 | 2.417 | 1643.2 |
| 20+ | 0.017 | 0.023 | 0.006 | 132.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dobrovolná, M.; Bowater, R.P.; Pečinka, P.; Brázda, V.; Bartas, M. High-Resolution Assembly of the Human Y Chromosome Identifies a Vast Landscape of Inverted Repeats Associated with Structural and Functional Genomic Features. Int. J. Mol. Sci. 2025, 26, 10180. https://doi.org/10.3390/ijms262010180
Dobrovolná M, Bowater RP, Pečinka P, Brázda V, Bartas M. High-Resolution Assembly of the Human Y Chromosome Identifies a Vast Landscape of Inverted Repeats Associated with Structural and Functional Genomic Features. International Journal of Molecular Sciences. 2025; 26(20):10180. https://doi.org/10.3390/ijms262010180
Chicago/Turabian StyleDobrovolná, Michaela, Richard P. Bowater, Petr Pečinka, Václav Brázda, and Martin Bartas. 2025. "High-Resolution Assembly of the Human Y Chromosome Identifies a Vast Landscape of Inverted Repeats Associated with Structural and Functional Genomic Features" International Journal of Molecular Sciences 26, no. 20: 10180. https://doi.org/10.3390/ijms262010180
APA StyleDobrovolná, M., Bowater, R. P., Pečinka, P., Brázda, V., & Bartas, M. (2025). High-Resolution Assembly of the Human Y Chromosome Identifies a Vast Landscape of Inverted Repeats Associated with Structural and Functional Genomic Features. International Journal of Molecular Sciences, 26(20), 10180. https://doi.org/10.3390/ijms262010180










