Measuring the Invisible: Microbial Diagnostics for Periodontitis—A Narrative Review
Abstract
1. Introduction
2. Detection Methods for Periodontopathic Bacteria
2.1. Sample Collection Techniques
2.2. Enzymatic Activity Assays
2.3. Immunological Methods
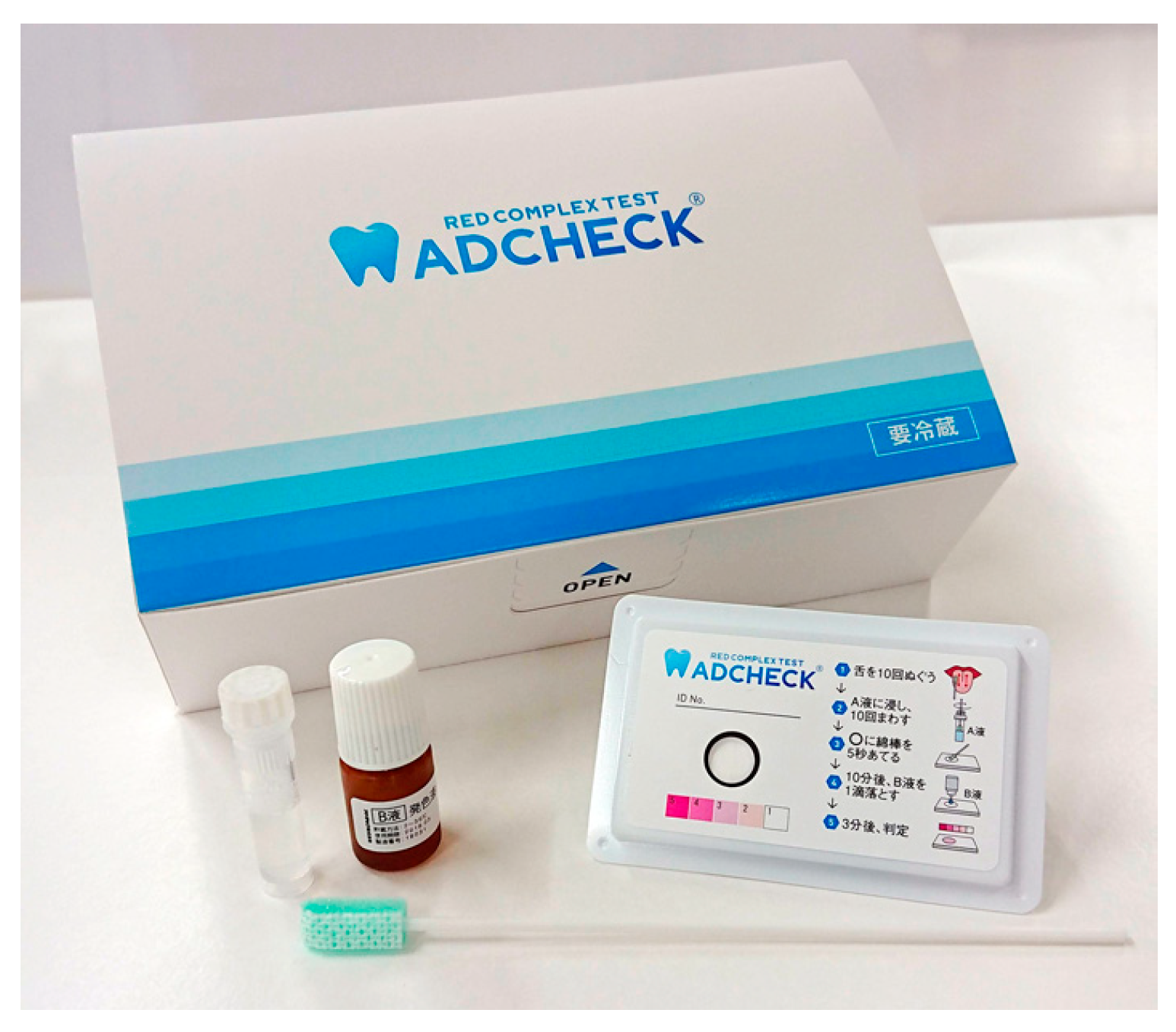
2.3.1. Immunochromatography
2.3.2. Enzyme-Linked Immunosorbent Assay
2.4. Real-Time PCR (Quantitative PCR)
2.5. Checkerboard DNA–DNA Hybridization
| Reference | Sample Size | Relationship with PPD | Association with Disease Status |
|---|---|---|---|
| Socransky et al., 1998 (ref. [69]) | 185 subjects, 13,261 subgingival samples | Distinct microbial complexes (red, orange, etc.) strongly associated with increasing pocket depth | Red complex species linked to periodontitis; distribution patterns distinguished health vs. disease |
| Haffajee et al., 1997 (ref. [70]) | 57 periodontitis patients | Scaling and root planing reduced mean pocket depth; reductions correlated with decreases in red/orange complex species | Microbial shifts after therapy paralleled clinical improvements |
| do Nascimento et al., 2009 (ref. [74]) | 30 implant patients | Not directly assessed at periodontal pockets; focus on internal implant contamination | Checkerboard detected peri-implant microbial leakage; compared cast vs. pre-machined abutments |
| Haffajee et al., 2009 (ref. [76]) | 187 subjects, 9182 subgingival samples | Checkerboard and PCR showed consistent detection trends; bacterial levels increased with pocket depth | Both methods identified higher prevalence of pathogens in periodontitis compared with health |
2.6. Loop-Mediated Isothermal Amplification
2.7. Microarray-Based Assays
2.8. Fluorescence In Situ Hybridization
2.9. Next-Generation Sequencing
2.10. Fourier Transform Infrared Spectroscopy
3. Discussion
3.1. Sampling Bias and Site-Level Heterogeneity
3.2. Discordance Between Presence and Activity
3.3. Semi-Quantitation and Dynamic Range Constraints
3.4. Molecular Biases and Normalization Challenges
3.5. Limited Strain-Level and Functional Resolution
3.6. Confounding by Host and Behavior
3.7. Low-Biomass Artifacts and Contamination
3.8. Lack of Validated Clinical Cutoffs
3.9. Cost, Turnaround, and Workflow Fit
3.10. Practical Implications
3.11. Future Directions
4. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PPD | Probing Pocket Depth |
| BspA | Bacteroides surface protein A |
| PCR | Polymerase chain reaction |
| LAMP | Loop mediated isothermal amplification |
| FISH | Fluorescence in situ hybridization |
| BANA | N-benzoyl-DL-arginine-2-naphthylamide |
| CAL | Chemical attainment level |
| ELISA | Enzyme-Linked Immunosorbent Assay |
| Ig | Immunoglobulin |
| qPCR | Quantitative polymerase chain reaction |
| GCF | Gingival crevicular fluid |
| CKB | Checkerboard DNA–DNA hybridization |
| MDA | Multiple-displacement amplification |
| HOMIM | Human oral microbe identification microassay |
| NGS | Next-generation sequencing |
| FTIR | Fourier transform infrared |
| QA/QC | Quality assurance/quality control |
| SOPs | Standard operating procedures |
| PMA | Propidium monoazide |
| MIQE | Minimum Information for Publication of Quantitative Real-Time PCR Experiments |
| DIG | Digoxigenin |
References
- Usui, M.; Onizuka, S.; Sato, T.; Kokabu, S.; Ariyoshi, W.; Nakashima, K. Mechanism of alveolar bone destruction in periodontitis—Periodontal bacteria and inflammation. Jpn. Dent. Sci. Rev. 2021, 57, 201–208. [Google Scholar] [CrossRef]
- Gerritsen, A.E.; Allen, P.F.; Witter, D.J.; Bronkhorst, E.M.; Creugers, N.H. Tooth loss and oral health-related quality of life: A systematic review and meta-analysis. Health Qual. Life Outcomes 2010, 8, 126. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.D.; Papapanou, P.N.; Philips, K.H.; Offenbacher, S. Periodontal medicine: 100 years of progress. J. Dent. Res. 2019, 98, 1053–1062. [Google Scholar] [CrossRef]
- Kalhan, A.C.; Wong, M.L.; Allen, F.; Gao, X. Periodontal disease and systemic health: An update for medical practitioners. Ann. Acad. Med. Singap. 2022, 51, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.; Hassan, N.; Khatoon, K.; Mirza, M.A.; Naseef, P.P.; Kuruniyan, M.S.; Iqbal, Z. Periodontitis and systemic disorder—An overview of relation and novel treatment modalities. Pharmaceutics 2021, 13, 1175. [Google Scholar] [CrossRef]
- Shinjo, T.; Nishimura, F. The bidirectional association between diabetes and periodontitis, from basic to clinical. Jpn. Dent. Sci. Rev. 2024, 60, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Marruganti, C.; Suvan, J.E.; D’Aiuto, F. Periodontitis and metabolic diseases (diabetes and obesity): Tackling multimorbidity. Periodontology 2000 2023. [Google Scholar] [CrossRef]
- Chapple, I.L.C.; Hirschfeld, J.; Cockwell, P.; Dietrich, T.; Sharma, P. Interplay between periodontitis and chronic kidney disease. Nat. Rev. Nephrol. 2025, 21, 226–240. [Google Scholar] [CrossRef]
- Serni, L.; Caroti, L.; Barbato, L.; Nieri, M.; Serni, S.; Cirami, C.L.; Cairo, F. Association between chronic kidney disease and periodontitis: A systematic review and metanalysis. Oral Dis. 2023, 29, 40–50. [Google Scholar] [CrossRef]
- Hasegawa, K.; Furuichi, Y.; Shimotsu, A.; Nakamura, M.; Yoshinaga, M.; Kamitomo, M.; Hatae, M.; Maruyama, I.; Izumi, Y. Associations between systemic status, periodontal status, serum cytokine levels, and delivery outcomes in pregnant women with a diagnosis of threatened premature labor. J. Periodontol. 2003, 74, 1764–1770. [Google Scholar] [CrossRef]
- Jajoo, N.S.; Shelke, A.U.; Bajaj, R.S.; Patil, P.P.; Patil, M.A. Association of periodontitis with pre term low birth weight—A review. Placenta 2020, 95, 62–68. [Google Scholar] [CrossRef]
- Hasan, F.; Tandon, A.; AlQallaf, H.; John, V.; Sinha, M.; Gibson, M.P. Inflammatory association between periodontal disease and systemic health. Inflammation 2025. [Google Scholar] [CrossRef]
- Bosshardt, D.D. The periodontal pocket: Pathogenesis, histopathology and consequences. Periodontology 2000 2018, 76, 43–50. [Google Scholar] [CrossRef]
- Bosshardt, D.D.; Lang, N.P. The junctional epithelium: From health to disease. J. Dent. Res. 2005, 84, 9–20. [Google Scholar] [CrossRef]
- Darveau, R.P. Periodontitis: A polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 2010, 8, 481–490. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D. Dental biofilms: Difficult therapeutic targets. Periodontology 2000 2002, 28, 12–55. [Google Scholar] [CrossRef] [PubMed]
- Goetting-Minesky, M.P.; Godovikova, V.; Fenno, J.C. Approaches to understanding mechanisms of dentilisin protease complex expression in Treponema denticola. Front. Cell. Infect. Microbiol. 2021, 11, 668287. [Google Scholar] [CrossRef] [PubMed]
- Mysak, J.; Podzimek, S.; Sommerova, P.; Lyuya-Mi, Y.; Bartova, J.; Janatova, T.; Prochazkova, J.; Duskova, J. Porphyromonas gingivalis: Major periodontopathic pathogen overview. J. Immunol. Res. 2014, 2014, 476068. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhou, W.; Wang, H.; Liang, S. Roles of Porphyromonas gingivalis and its virulence factors in periodontitis. Adv. Protein Chem. Struct. Biol. 2020, 120, 45–84. [Google Scholar] [CrossRef]
- Rosen, G.; Genzler, T.; Sela, M.N. Coaggregation of Treponema denticola with Porphyromonas gingivalis and Fusobacterium nucleatum is mediated by the major outer sheath protein of Treponema denticola. FEMS Microbiol. Lett. 2008, 289, 59–66. [Google Scholar] [CrossRef]
- Inagaki, S.; Onishi, S.; Kuramitsu, H.K.; Sharma, A. Porphyromonas gingivalis vesicles enhance attachment, and the leucine-rich repeat BspA protein is required for invasion of epithelial cells by “Tannerella forsythia”. Infect. Immun. 2006, 74, 5023–5028. [Google Scholar] [CrossRef] [PubMed]
- Scannapieco, F.A.; Dongari-Bagtzoglou, A. Dysbiosis revisited: Understanding the role of the oral microbiome in the pathogenesis of gingivitis and periodontitis: A critical assessment. J. Periodontol. 2021, 92, 1071–1078. [Google Scholar] [CrossRef]
- Kumar, P.S. Microbial dysbiosis: The root cause of periodontal disease. J. Periodontol. 2021, 92, 1079–1087. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Darveau, R.P.; Curtis, M.A. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 2012, 10, 717–725. [Google Scholar] [CrossRef]
- Hajishengallis, G. Immunomicrobial pathogenesis of periodontitis: Keystones, pathobionts, and host response. Trends Immunol. 2014, 35, 3–11. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Lamont, R.J. Polymicrobial communities in periodontal disease: Their quasi-organismal nature and dialogue with the host. Periodontology 2000 2021, 86, 210–230. [Google Scholar] [CrossRef] [PubMed]
- Loesche, W.J.; Bretz, W.A.; Kerschensteiner, D.; Stoll, J.; Socransky, S.S.; Hujoel, P.; Lopatin, D.E. Development of a diagnostic test for anaerobic periodontal infections based on plaque hydrolysis of benzoyl-DL-arginine-naphthylamide. J. Clin. Microbiol. 1990, 28, 1551–1559. [Google Scholar] [CrossRef]
- Loesche, W.J.; Lopatin, D.E.; Giordano, J.; Alcoforado, G.; Hujoel, P.P. Comparison of the benzoyl-DL-arginine-naphthylamide (BANA) test, DNA probes, and immunological reagents for ability to detect anaerobic periodontal infections due to Porphyromonas gingivalis, Treponema denticola, and Bacteroides forsythus. J. Clin. Microbiol. 1992, 30, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Lamster, I.B.; Harper, D.S.; Fiorello, L.A.; Oshrain, R.L.; Gordon, J.M. Evaluation and comparison of salivary and crevicular fluid enzyme activities in periodontitis patients. J. Clin. Periodontol. 1988, 15, 347–352. [Google Scholar] [CrossRef]
- Loesche, W.J.; Giordano, J.; Hujoel, P.P. The utility of the BANA test for monitoring anaerobic infections due to spirochetes (Treponema denticola) in periodontal disease. J. Dent. Res. 1990, 69, 1696–1702. [Google Scholar] [CrossRef]
- Grisi, M.F.d.M.; Novaes, A.B., Jr.; Ito, I.Y.; Salvador, S.L. Relationship between clinical probing depth and reactivity to the BANA test of samples of subgingival microbiota from patients with periodontitis. Braz. Dent. J. 1998, 9, 77–84. [Google Scholar]
- Usui, M.; Iwasaki, M.; Ariyoshi, W.; Kobayashi, K.; Kasai, S.; Yamanaka, R.; Nakashima, K.; Nishihara, T. The ability of a novel trypsin-like peptidase activity assay kit to detect red-complex species. Diagnostics 2022, 12, 2172. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, M.; Usui, M.; Ariyoshi, W.; Nakashima, K.; Nagai-Yoshioka, Y.; Inoue, M.; Kobayashi, K.; Nishihara, T. Evaluation of the ability of the trypsin-like peptidase activity assay to detect severe periodontitis. PLoS ONE 2021, 16, e0256538. [Google Scholar] [CrossRef]
- Iwasaki, M.; Usui, M.; Ariyoshi, W.; Nakashima, K.; Nagai-Yoshioka, Y.; Inoue, M.; Nishihara, T. A preliminary study on the ability of the trypsin-like peptidase activity assay kit to detect periodontitis. Dent. J. 2020, 8, 98. [Google Scholar] [CrossRef]
- Iwasaki, M.; Inoue, M.; Usui, M.; Ariyoshi, W.; Nakashima, K.; Nagai-Yoshioka, Y.; Nishihara, T. The association between trypsin-like protease activity in the oral cavity and kidney function in Japanese workers. J. Clin. Periodontol. 2024, 51, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Mikami, R.; Takeuchi, Y.; Ishihara, K.; Tsuchiya, Y.; Nagai, T.; Kobayashi, R.; Shiba, T.; Iwata, T. Evaluation of trypsin-like enzyme activity using a novel device for monitoring Porphyromonas gingivalis in periodontal pockets: A pilot study. Cureus 2025, 17, e83832. [Google Scholar] [CrossRef]
- Yamazaki, T.; Miyamoto, M.; Yamada, S.; Okuda, K.; Ishihara, K. Surface protease of Treponema denticola hydrolyzes C3 and influences function of polymorphonuclear leukocytes. Microbes Infect. 2006, 8, 1758–1763. [Google Scholar] [CrossRef] [PubMed]
- Ravishankar, P.L.; Chaitanya, N.C.; Anitha, A.; Devanna, R.; Dwarakanath, C.D. Chairside diagnostics in periodontics. SRM J. Res. Dent. Sci. 2017, 8, 102–106. [Google Scholar] [CrossRef]
- Mani, A. Diagnostic kits: An aid to periodontal diagnosis. J. Int. Clin. Dent. Res. Organ. 2016, 8, 140–146. [Google Scholar] [CrossRef]
- Singh, V.; Kumar, V.; Singh, N.; Tiwari, P.; Pandey, A. Chairside diagnostic kits in periodontal practice. Int. J. Sci. Res. 2020, 9, 44–47. [Google Scholar] [CrossRef]
- Imamura, K.; Takayama, S.; Saito, A.; Inoue, E.; Nakayama, Y.; Ogata, Y.; Shirakawa, S.; Nagano, T.; Gomi, K.; Morozumi, T.; et al. Evaluation of a novel immunochromatographic device for rapid and accurate clinical detection of Porphyromonas gingivalis in subgingival plaque. J. Microbiol. Methods 2015, 117, 4–10. [Google Scholar] [CrossRef]
- O’Brien-Simpson, N.M.; Burgess, K.; Lenzo, J.C.; Brammar, G.C.; Darby, I.B.; Reynolds, E.C. Rapid chair-side test for detection of Porphyromonas gingivalis. J. Dent. Res. 2017, 96, 618–625. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, R.; Usui, M.; Kobayashi, K.; Onizuka, S.; Kasai, S.; Sano, K.; Hironaka, S.; Yamasaki, R.; Yoshii, S.; Sato, T.; et al. Evaluation of a novel immunochromatographic device for detecting Porphyromonas gingivalis in patients with periodontal disease. Int. J. Mol. Sci. 2024, 25, 8187. [Google Scholar] [CrossRef] [PubMed]
- Griffith, A.; Chande, C.; Kulkarni, S.; Morel, J.; Cheng, Y.H.; Shimizu, E.; Cugini, C.; Basuray, S.; Kumar, V. Point-of-care diagnostic devices for periodontitis—Current trends and urgent need. Sens. Diagn. 2024, 3, 1119–1134. [Google Scholar] [CrossRef] [PubMed]
- He, W.; You, M.; Wan, W.; Xu, F.; Li, F.; Li, A. Point-of-care periodontitis testing: Biomarkers, current technologies, and perspectives. Trends Biotechnol. 2018, 36, 1127–1144. [Google Scholar] [CrossRef]
- Gadekar, N.B.; Hosmani, J.V.; Bhat, K.G.; Kotrashetti, V.S.; Nayak, R.S.; Babji, D.V.; Pattanshetty, S.M.; Joshi, V.M.; Bansode, R.A. Detection of antibodies against Aggregatibacter actinomycetemcomitans in serum and saliva through ELISA in periodon tally healthy individuals and individuals with chronic periodontitis. Microb. Pathog. 2018, 125, 438–442. [Google Scholar] [CrossRef]
- Lakio, L.; Antinheimo, J.; Paju, S.; Buhlin, K.; Pussinen, P.J.; Alfthan, G. Tracking of plasma antibodies against Aggregatibacter actinomycetemcomitans and Porphyromonas gingivalis during 15 years. J. Oral Microbiol. 2009, 1, 1979. [Google Scholar] [CrossRef]
- Hall, L.M.; Dunford, R.G.; Genco, R.J.; Sharma, A. Levels of serum immunoglobulin G specific to bacterial surface protein A of Tannerella forsythia are related to periodontal status. J. Periodontol. 2012, 83, 228–234. [Google Scholar] [CrossRef]
- Kakuta, E.; Nomura, Y.; Morozumi, T.; Nakagawa, T.; Nakamura, T.; Noguchi, K.; Yoshimura, A.; Hara, Y.; Fujise, O.; Nishimura, F.; et al. Assessing the progression of chronic periodontitis using subgingival pathogen levels: A 24-month prospective multicenter cohort study. BMC Oral Health 2017, 17, 46. [Google Scholar] [CrossRef]
- Fatima, T.; Khurshid, Z.; Rehman, A.; Imran, E.; Srivastava, K.C.; Shrivastava, D. Gingival crevicular fluid (GCF): A diagnostic tool for the detection of periodontal health and diseases. Molecules 2021, 26, 1208. [Google Scholar] [CrossRef]
- Kinney, J.S.; Morelli, T.; Oh, M.; Braun, T.M.; Ramseier, C.A.; Sugai, J.V.; Giannobile, W.V. Crevicular fluid biomarkers and periodontal disease progression. J. Clin. Periodontol. 2014, 41, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Lyons, S.R.; Griffen, A.L.; Leys, E.J. Quantitative real-time PCR for Porphyromonas gingivalis and total bacteria. J. Clin. Microbiol. 2000, 38, 2362–2365. [Google Scholar] [CrossRef]
- Sanz, M.; Lau, L.; Herrera, D.; Morillo, J.M.; Silva, A. Methods of detection of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis and Tannerella forsythensis in periodontal microbiology, with special emphasis on advanced molecular techniques: A review. J. Clin. Periodontol. 2004, 31, 1034–1047. [Google Scholar] [CrossRef]
- Boutaga, K.; van Winkelhoff, A.J.; Vandenbroucke-Grauls, C.M.; Savelkoul, P.H.M. Comparison of real-time PCR and culture for detection of Porphyromonas gingivalis in subgingival plaque samples. J. Clin. Microbiol. 2003, 41, 4950–4954. [Google Scholar] [CrossRef]
- Lau, L.; Sanz, M.; Herrera, D.; Morillo, J.M.; Martín, C.; Silva, A. Quantitative real-time polymerase chain reaction versus culture: A comparison between two methods for the detection and quantification of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis and Tannerella forsythensis in subgingival plaque samples. J. Clin. Periodontol. 2004, 31, 1061–1069. [Google Scholar] [CrossRef]
- Jervøe-Storm, P.M.; Koltzscher, M.; Falk, W.; Dörfler, A.; Jepsen, S. Comparison of culture and real-time PCR for detection and quantification of five putative periodontopathogenic bacteria in subgingival plaque samples. J. Clin. Periodontol. 2005, 32, 778–783. [Google Scholar] [CrossRef]
- Nonnenmacher, C.; Dalpke, A.; Mutters, R.; Heeg, K. Quantitative detection of periodontopathogens by real-time PCR. J. Microbiol. Methods 2004, 59, 117–125. [Google Scholar] [CrossRef]
- Kuboniwa, M.; Amano, A.; Kimura, K.R.; Sekine, S.; Kato, S.; Yamamoto, Y.; Okahashi, N.; Iida, T.; Shizukuishi, S. Quantitative detection of periodontal pathogens using real-time polymerase chain reaction with TaqMan probes. Oral Microbiol. Immunol. 2004, 19, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Holland, P.M.; Abramson, R.D.; Watson, R.; Gelfand, D.H. Detection of specific polymerase chain reaction product by utilizing the 5′→3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 1991, 88, 7276–7280. [Google Scholar] [CrossRef]
- Heid, C.A.; Stevens, J.; Livak, K.J.; Williams, P.M. Real time quantitative PCR. Genome Res. 1996, 6, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Ririe, K.M.; Rasmussen, R.P.; Wittwer, C.T. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 1997, 245, 154–160. [Google Scholar] [CrossRef]
- Morrison, T.B.; Weis, J.J.; Wittwer, C.T. Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques 1998, 24, 954–958. [Google Scholar]
- Maeda, H.; Fujimoto, C.; Haruki, Y.; Maeda, T.; Kokeguchi, S.; Petelin, M.; Arai, H.; Tanimoto, I.; Nishimura, F.; Takashiba, S. Quantitative real-time PCR using TaqMan and SYBR Green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol. Med. Microbiol. 2003, 39, 81–86. [Google Scholar] [CrossRef]
- Sánchez, M.C.; Marín, M.J.; Figuero, E.; Llama-Palacios, A.; Herrera, D.; Sanz, M. Analysis of viable vs. dead Aggregatibacter actinomycetemcomitans and Porphyromonas gingivalis using selective quantitative real-time PCR with propidium monoazide. J. Periodontal Res. 2013, 48, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, M.C.; Marín, M.J.; Figuero, E.; Llama-Palacios, A.; León, R.; Blanc, V.; Herrera, D.; Sanz, M. Quantitative real-time PCR combined with propidium monoazide for the selective quantification of viable periodontal pathogens in an in vitro subgingival biofilm model. J. Periodontal Res. 2014, 49, 20–28. [Google Scholar] [CrossRef]
- Socransky, S.S.; Smith, C.; Martin, L.; Paster, B.J.; Dewhirst, F.E.; Levin, A.E. “Checkerboard” DNA–DNA hybridization. Biotechniques 1994, 17, 788–792. [Google Scholar]
- Gellen, L.S.; Wall-Manning, G.M.; Sissons, C.H. Checkerboard DNA–DNA hybridization technology using digoxigenin detection. Methods Mol. Biol. 2007, 353, 39–67. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D.; Cugini, M.A.; Smith, C.; Kent, R.L., Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998, 25, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Haffajee, A.D.; Cugini, M.A.; Dibart, S.; Smith, C.; Kent, R.L., Jr.; Socransky, S.S. The effect of scaling and root planing on the clinical and microbiological parameters of periodontal diseases. J. Clin. Periodontol. 1997, 24, 324–334. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D.; Smith, C.; Martin, L.; Haffajee, J.A.; Uzel, N.G.; Goodson, J.M. Use of checkerboard DNA–DNA hybridization to study complex microbial ecosystems. Oral Microbiol. Immunol. 2004, 19, 352–362. [Google Scholar] [CrossRef]
- do Nascimento, C.; Santos Barbosa, R.E.; Mardegan Issa, J.P.; Watanabe, E.; Yoko Ito, I.; Monesi, N.; de Albuquerque Júnior, R.F. The use of fluorescein for labeling genomic probes in the checkerboard DNA–DNA hybridization method. Microbiol. Res. 2008, 163, 403–407. [Google Scholar] [CrossRef]
- Teles, F.; Haffajee, A.D.; Socransky, S.S. Multiple displacement amplification as an aid in checkerboard DNA–DNA hybridization. Oral Microbiol. Immunol. 2007, 22, 118–125. [Google Scholar] [CrossRef]
- do Nascimento, C.; Santos Barbosa, R.E.; Mardegan Issa, J.P.; Watanabe, E.; Ito, I.Y.; de Albuquerque Júnior, R.F. Use of checkerboard DNA–DNA hybridization to evaluate the internal contamination of dental implants and comparison of bacterial leakage with cast or pre-machined abutments. Clin. Oral Implant. Res. 2009, 20, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Paster, B.J.; Dewhirst, F.E. Molecular microbial diagnosis. Periodontology 2000 2009, 51, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Haffajee, A.D.; Yaskell, T.; Torresyap, G.; Teles, R.; Socransky, S.S. Comparison between polymerase chain reaction-based and checkerboard DNA hybridization techniques for microbial assessment of subgingival plaque samples. J. Clin. Periodontol. 2009, 36, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef]
- Notomi, T.; Mori, Y.; Tomita, N.; Kanda, H. Loop-mediated isothermal amplification (LAMP): Principle, features, and future prospects. J. Microbiol. 2015, 53, 1–5. [Google Scholar] [CrossRef]
- Yoshida, A.; Nagashima, S.; Ansai, T.; Tachibana, M.; Kato, H.; Watari, H.; Notomi, T.; Takehara, T. Loop-mediated isothermal amplification method for rapid detection of the periodontopathic bacteria Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola. J. Clin. Microbiol. 2005, 43, 2418–2424. [Google Scholar] [CrossRef]
- Osawa, R.; Yoshida, A.; Masakiyo, Y.; Nagashima, S.; Ansai, T.; Watari, H.; Notomi, T.; Takehara, T. Rapid detection of Actinobacillus actinomycetemcomitans using a loop-mediated isothermal amplification method. Oral Microbiol. Immunol. 2007, 22, 252–259. [Google Scholar] [CrossRef]
- Lenkowski, M.; Nijakowski, K.; Kaczmarek, M.; Surdacka, A. The loop-mediated isothermal amplification technique in periodontal diagnostics: A systematic review. J. Clin. Med. 2021, 10, 1189. [Google Scholar] [CrossRef]
- Do, T.; Devine, D.; Marsh, P.D. Oral biofilms: Molecular analysis, challenges, and future prospects in dental diagnostics. Clin. Cosmet. Investig. Dent. 2013, 5, 11–19. [Google Scholar] [CrossRef]
- Colombo, A.P.; Boches, S.K.; Cotton, S.L.; Goodson, J.M.; Kent, R.; Haffajee, A.D.; Socransky, S.S.; Hasturk, H.; Van Dyke, T.E.; Dewhirst, F.; et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J. Periodontol. 2009, 80, 1421–1432. [Google Scholar] [CrossRef] [PubMed]
- Colombo, A.P.; Bennet, S.; Cotton, S.L.; Goodson, J.M.; Kent, R.; Haffajee, A.D.; Socransky, S.S.; Hasturk, H.; Van Dyke, T.E.; Dewhirst, F.E.; et al. Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: Comparison between good responders and individuals with refractory periodontitis using the human oral microbe identification microarray. J. Periodontol. 2012, 83, 1279–1287. [Google Scholar] [CrossRef] [PubMed]
- Udoh, S.; Adukwu, E.; Varadi, A.; Saad, S. Effectiveness of the human oral microbe identification microarray in identifying periodontal pathogens: A systematic review. Appl. Microbiol. 2022, 2, 614–625. [Google Scholar] [CrossRef]
- Huyghe, A.; Francois, P.; Charbonnier, Y.; Tangomo-Bento, M.; Bonetti, E.J.; Paster, B.J.; Bolivar, I.; Baratti-Mayer, D.; Pittet, D.; Schrenzel, J.; et al. Novel microarray design strategy to study complex bacterial communities. Appl. Environ. Microbiol. 2008, 74, 1876–1885. [Google Scholar] [CrossRef]
- Parolin, C.; Giordani, B.; Ñahui Palomino, R.A.; Biagi, E.; Severgnini, M.; Consolandi, C.; Caredda, G.; Storelli, S.; Strohmenger, L.; Vitali, B. Design and validation of a DNA-microarray for phylogenetic analysis of bacterial communities in different oral samples and dental implants. Sci. Rep. 2017, 7, 6280. [Google Scholar] [CrossRef]
- Starke, E.M.; Smoot, J.C.; Smoot, L.M.; Liu, W.T.; Chandler, D.P.; Lee, H.H.; Stahl, D.A. Technology development to explore the relationship between oral health and the oral microbial community. BMC Oral Health 2006, 6, S10. [Google Scholar] [CrossRef]
- Topcuoglu, N.; Kulekci, G. 16S rRNA based microarray analysis of ten periodontal bacteria in patients with different forms of periodontitis. Anaerobe 2015, 35, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, A.; Oshima, H.; Togawa, N.; Nozaki, T.; Murakami, S. Development of Oral Care Chip, a novel device for quantitative detection of the oral microbiota associated with periodontal disease. PLoS ONE 2020, 15, e0229485. [Google Scholar] [CrossRef]
- Amann, R.; Fuchs, B.M. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 2008, 6, 339–348. [Google Scholar] [CrossRef]
- Mark Welch, J.L.; Rossetti, B.J.; Rieken, C.W.; Dewhirst, F.E.; Borisy, G.G. Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. USA 2016, 113, E791–E800. [Google Scholar] [CrossRef] [PubMed]
- Zijnge, V.; van Leeuwen, M.B.M.; Degener, J.E.; Abbas, F.; Thurnheer, T.; Gmür, R.; Harmsen, H.J.M. Oral biofilm architecture on natural teeth. PLoS ONE 2010, 5, e9321. [Google Scholar] [CrossRef] [PubMed]
- Griffen, A.L.; Beall, C.J.; Campbell, J.H.; Firestone, N.D.; Kumar, P.S.; Yang, Z.K.; Podar, M.; Leys, E.J. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012, 6, 1176–1185. [Google Scholar] [CrossRef]
- Liu, B.; Faller, L.L.; Klitgord, N.; Mazumdar, V.; Ghodsi, M.; Sommer, D.D.; Gibbons, T.R.; Treangen, T.J.; Chang, Y.-C.; Li, S.; et al. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS ONE 2012, 7, e37919. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Shi, L.; Zhao, C. Using next-generation sequencing to detect oral microbiome changes following periodontal interventions: A systematic review. Oral Dis. 2021, 27, 1971–1986. [Google Scholar] [CrossRef]
- Nagai, T.; Horigome, T.; Seto, K.; Yamada, M. Optimal 16S rRNA gene amplicon sequencing analysis for oral microbiome studies. Microbiol. Spectr. 2024, 12, e03512-23. [Google Scholar] [CrossRef]
- Fujii, S.; Sato, S.; Fukuda, K.; Okinaga, T.; Ariyoshi, W.; Usui, M.; Nakashima, K.; Nishihara, T.; Takenaka, S. Diagnosis of periodontal disease from saliva samples using Fourier transform infrared microscopy coupled with partial least squares discriminant analysis. Anal. Sci. 2016, 32, 225–231. [Google Scholar] [CrossRef]
- Nogueira, M.S.; Barreto, A.L.; Furukawa, M.; Rovai, E.S.; Bastos, A.; Bertoncello, G.; Carvalho, L.F. FTIR spectroscopy as a point-of-care diagnostic tool for diabetes and periodontitis: A saliva analysis approach. Photodiagn. Photodyn. Ther. 2022, 40, 103036. [Google Scholar] [CrossRef]
- Kaneda, T.; Watanabe, M.; Honda, H.; Yamamoto, M.; Inagaki, T.; Hironaka, S. Fourier transform infrared spectroscopy and machine learning for Porphyromonas gingivalis detection in oral bacteria. Anal. Sci. 2024, 40, 691–699. [Google Scholar] [CrossRef] [PubMed]

| Method | Principle | Advantages | Limitations | Main Clinical Applications |
|---|---|---|---|---|
| Culture method (aerobic/anaerobic) | Culturing specimens on selective or enriched media; identification based on colony morphology and biochemical characteristics | Well-established standard; enables antimicrobial susceptibility testing | Cannot detect fastidious or uncultivable species; labor-intensive and time-consuming | Identification of major periodontal pathogens; guidance for antibiotic selection |
| Enzymatic Activity Assays | Determination of enzyme activity | No specialized equipment required; cost-effective | Lower sensitivity and specificity compared with molecular methods | Identification of red complex pathogens |
| Immunological methods (Enzyme-Linked Immunosorbent Assay, Immunochromatography) | Detection of specific antigens using antigen–antibody reactions | Rapid turnaround; applicable to chairside testing | Possible cross-reactivity leading to false positives; semi-quantitative at best | Rapid screening for Porphyromonas gingivalis, Treponema denticola |
| Real Time Polymerase Chain Reaction (PCR) | Amplification of species-specific genetic sequences | High sensitivity and specificity; culture-independent | Requires laboratory infrastructure; risk of contamination | Quantitative monitoring of target pathogens; assessment of treatment efficacy |
| DNA–DNA hybridization (checkerboard method) | Hybridization between immobilized DNA probes and target DNA sequences | High-throughput analysis of multiple samples and species | Requires specialized equipment and expertise | Longitudinal epidemiological studies; microbiota shift analysis |
| LAMP (loop-mediated isothermal amplification) | Isothermal amplification of target genes | Simple, rapid, portable | Complex primer design; limited multiplexing | Point-of-care screening for specific pathogens |
| Microarray | Hybridization of sample DNA (16S ribosomal RNA targets) to hundreds–thousands of arrayed probes | High throughput (many species × many samples) | Fixed probe set—misses novel/strain-level variation; Requires good DNA quality; lab-based | Profiling established oral panels (health vs. periodontitis); monitoring post-therapy shifts |
| Next-Generation Sequencing (metagenomic analysis) | High-throughput sequencing of total microbial DNA | Broad-spectrum, culture-independent; detects rare and novel species | Expensive; requires bioinformatics expertise | Basic microbiome research; discovery of novel periodontal pathogens |
| FISH (fluorescence in situ hybridization) | Fluorescent probe hybridization to visualize target bacteria in situ | Provides spatial distribution information | Lower sensitivity compared with PCR | Visualization of plaque biofilm architecture |
| Reference | Sample Size | Enzymatic Activity Assessed | Relationship with PPD | Association with Disease Status |
|---|---|---|---|---|
| Lamster et al., 1988 (ref. [29]) | 36 patients | GCF lysosomal enzymes (β-glucuronidase, etc.) | Increased activity at sites with higher PPD | Whole-mouth β-glucuronidase distinguished progressive disease |
| Loesche et al., 1990 (ref. [30]) | Not specified | BANA (subgingival plaque) | BANA-positive teeth showed greater PPD reduction after SRP + MTZ | Associated with spirochetal infection and responsive periodontitis |
| Grisi et al., 1998 (ref. [31]) | 28 patients, 513 sites | BANA (subgingival plaque) | Deeper pockets showed higher BANA positivity (100% at ≥8 mm) | Positive sites associated with periodontitis |
| Usui et al., 2021 (ref. [32]) | 347 adults | TLP (tongue swab) | Not site-level; disease-level only | High values associated with severe periodontitis (AUC 0.83, sens. 83%, spec. 77%) |
| Iwasaki et al., 2020 (ref. [33]) | 105 adults | TLP (tongue swab) | Not site-level; disease-level only | Excellent diagnostic ability for severe periodontitis (AUC 0.93) |
| Mikami et al., 2025 (ref. [36]) | 30 patients | TLP (subgingival plaque) | Strong correlation with PPD (ρ = 0.80, p < 0.001) | Also correlated with P. gingivalis counts |
| Reference | Sample Size | Relationship with PPD | Association with Disease Status |
|---|---|---|---|
| Imamura et al., 2015 (ref. [41]) | 63 chronic periodontitis + 28 healthy | Device score positively correlated with PPD (r = 0.317, p < 0.01) | Sensitivity 96.2%, specificity 91.8% vs. PCR; no positives among healthy controls at cut-off ≥0.25 |
| O’Brien-Simpson et al., 2017 (ref. [42]) | 50 periodontitis patients + 50 healthy | Device result correlated with probing depth (r = 0.695, p < 0.01) | Positive test strongly associated with high salivary P. gingivalis (>105 cells/mL); overall accuracy 94.0% (Se 95.0%, Sp 93.3%) |
| Yamanaka et al., 2024 (ref. [43]) | 72 chronic periodontitis + 53 healthy | Device score significantly higher at deeper pockets; positive correlation with PPD | Periodontitis classification: AUC 0.73; IC score correlated with qPCR counts (r = 0.73) |
| Reference | Sample Size | Relationship with PPD | Association with Disease Status |
|---|---|---|---|
| Lyons et al., 2000 (ref. [53]) | 20 periodontitis patients, 20 healthy controls | Higher P. gingivalis counts by qPCR in deep pockets; qPCR more sensitive than culture | qPCR discriminated periodontitis patients from controls based on P. gingivalis load |
| Boutaga et al., 2003 (ref. [55]) | 59 periodontitis patients (181 subgingival samples) | qPCR more frequently detected P. gingivalis in deeper sites than culture | Higher detection in periodontitis compared with health |
| Lau et al., 2004 (ref. [56]) | 40 periodontitis patients | Bacterial loads by qPCR increased with pocket depth for A. actinomycetemcomitans, P. gingivalis, T. forsythia | qPCR distinguished disease from health more effectively than culture |
| Jervøe-Storm et al., 2005 (ref. [57]) | 33 periodontitis patients | qPCR detected higher levels of pathogens in deeper pockets compared with culture | Higher prevalence of red complex bacteria in periodontitis |
| Nonnenmacher et al., 2004 (ref. [58]) | 21 periodontitis patients, 20 healthy controls | qPCR showed higher pathogen counts in diseased sites with deeper PPD | Pathogen levels significantly associated with periodontitis vs. health |
| Kuboniwa et al., 2004 (ref. [59]) | 18 periodontitis patients, 18 healthy controls | TaqMan qPCR showed correlation between pathogen counts and probing depth | Strong association between bacterial load and periodontal status (disease vs. healthy) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Usui, M.; Miyagi, S.; Yamanaka, R.; Oka, Y.; Kobayashi, K.; Sato, T.; Sano, K.; Onizuka, S.; Inoue, M.; Fujii, W.; et al. Measuring the Invisible: Microbial Diagnostics for Periodontitis—A Narrative Review. Int. J. Mol. Sci. 2025, 26, 10172. https://doi.org/10.3390/ijms262010172
Usui M, Miyagi S, Yamanaka R, Oka Y, Kobayashi K, Sato T, Sano K, Onizuka S, Inoue M, Fujii W, et al. Measuring the Invisible: Microbial Diagnostics for Periodontitis—A Narrative Review. International Journal of Molecular Sciences. 2025; 26(20):10172. https://doi.org/10.3390/ijms262010172
Chicago/Turabian StyleUsui, Michihiko, Suzuka Miyagi, Rieko Yamanaka, Yuichiro Oka, Kaoru Kobayashi, Tsuyoshi Sato, Kotaro Sano, Satoru Onizuka, Maki Inoue, Wataru Fujii, and et al. 2025. "Measuring the Invisible: Microbial Diagnostics for Periodontitis—A Narrative Review" International Journal of Molecular Sciences 26, no. 20: 10172. https://doi.org/10.3390/ijms262010172
APA StyleUsui, M., Miyagi, S., Yamanaka, R., Oka, Y., Kobayashi, K., Sato, T., Sano, K., Onizuka, S., Inoue, M., Fujii, W., Iwasaki, M., Ariyoshi, W., Nakashima, K., & Nishihara, T. (2025). Measuring the Invisible: Microbial Diagnostics for Periodontitis—A Narrative Review. International Journal of Molecular Sciences, 26(20), 10172. https://doi.org/10.3390/ijms262010172







