DNA Damage Response and Redox Status in the Resistance of Multiple Myeloma Cells to Genotoxic Treatment
Abstract
1. Introduction
2. Results
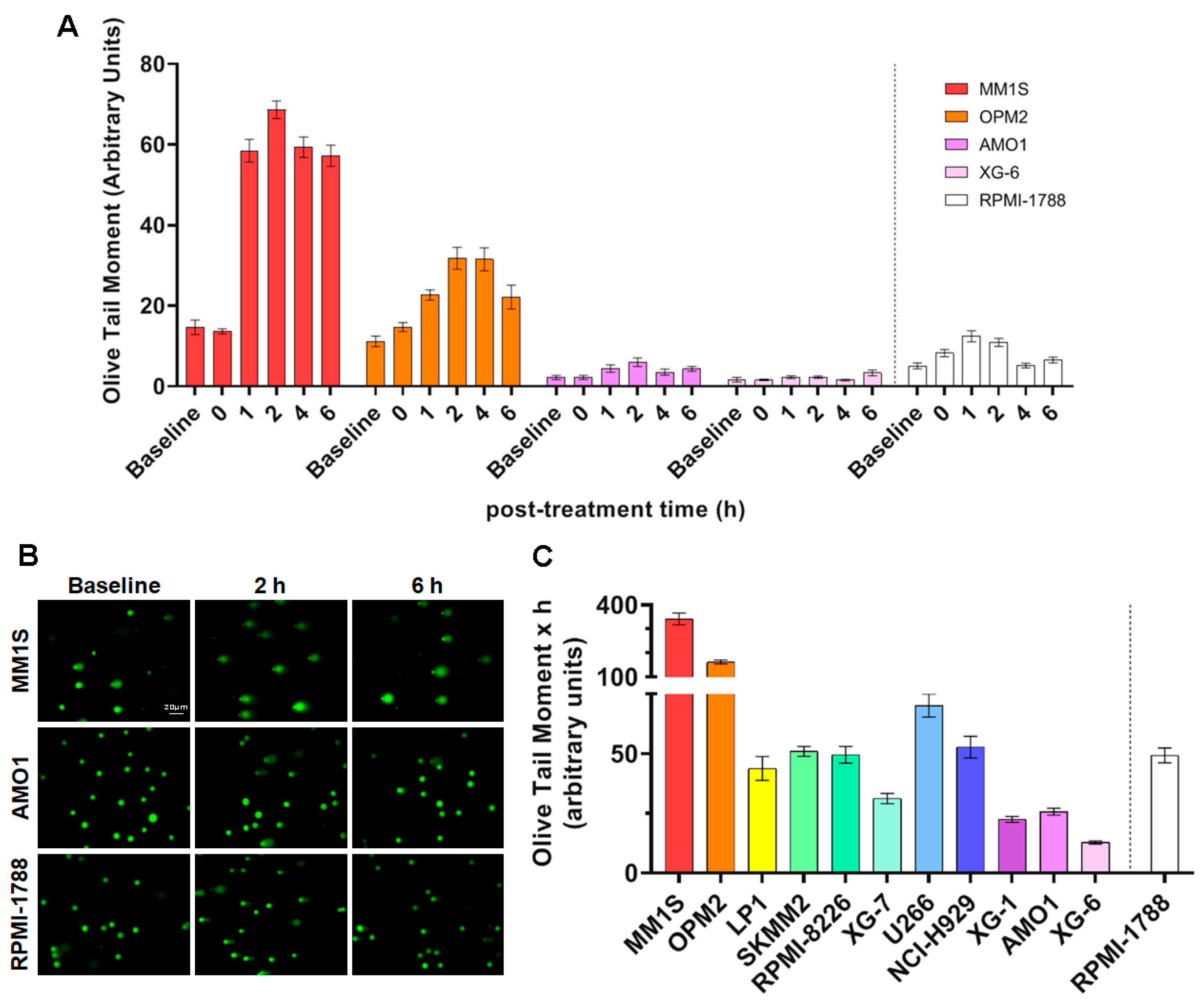
2.1. MM Cell Lines Showed Differential Sensitivity to Genotoxic Insults
2.2. DDR-Related Parameters in MM Cell Lines at Baseline
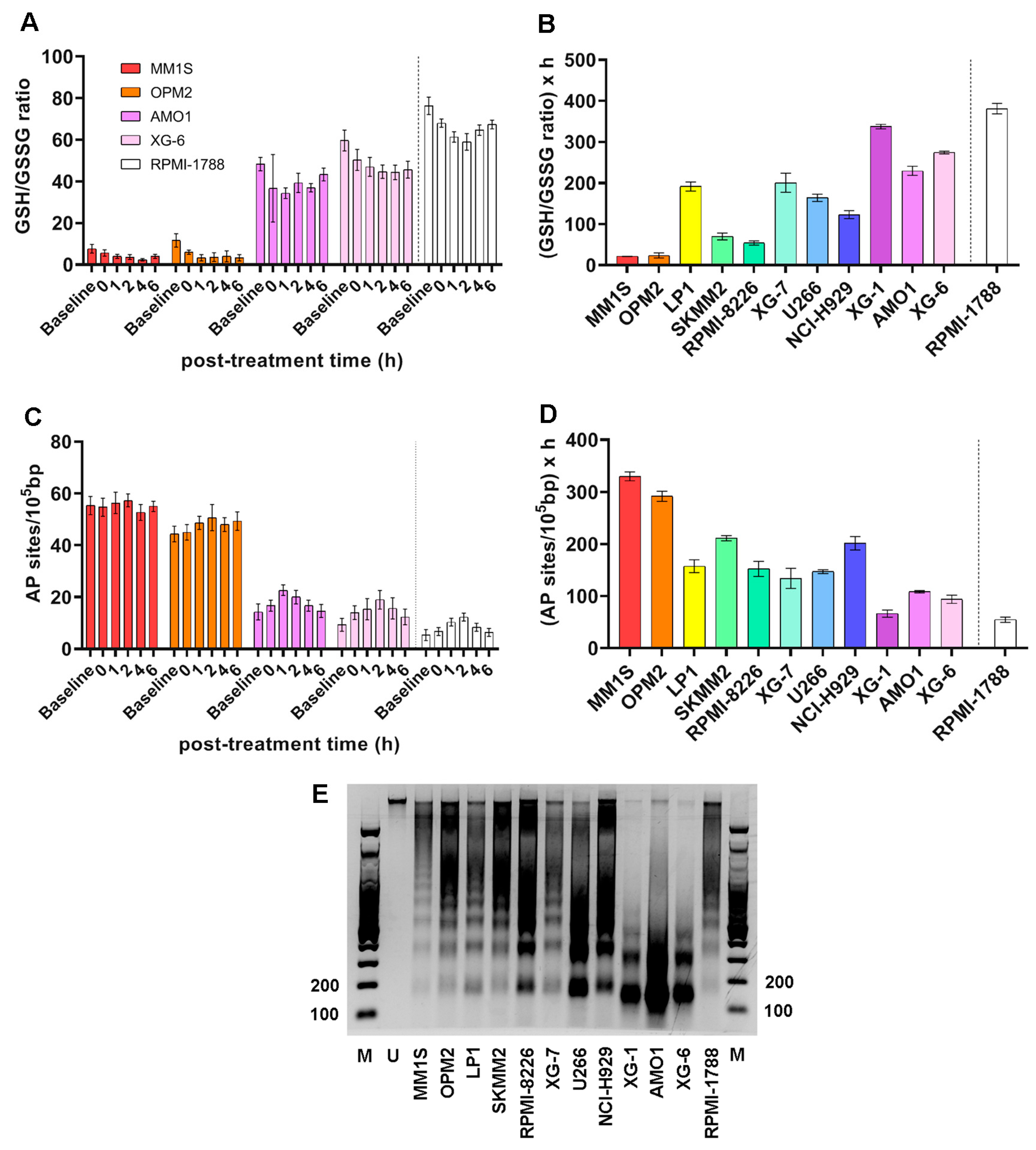
2.3. NER Capacity of MM Cell Lines
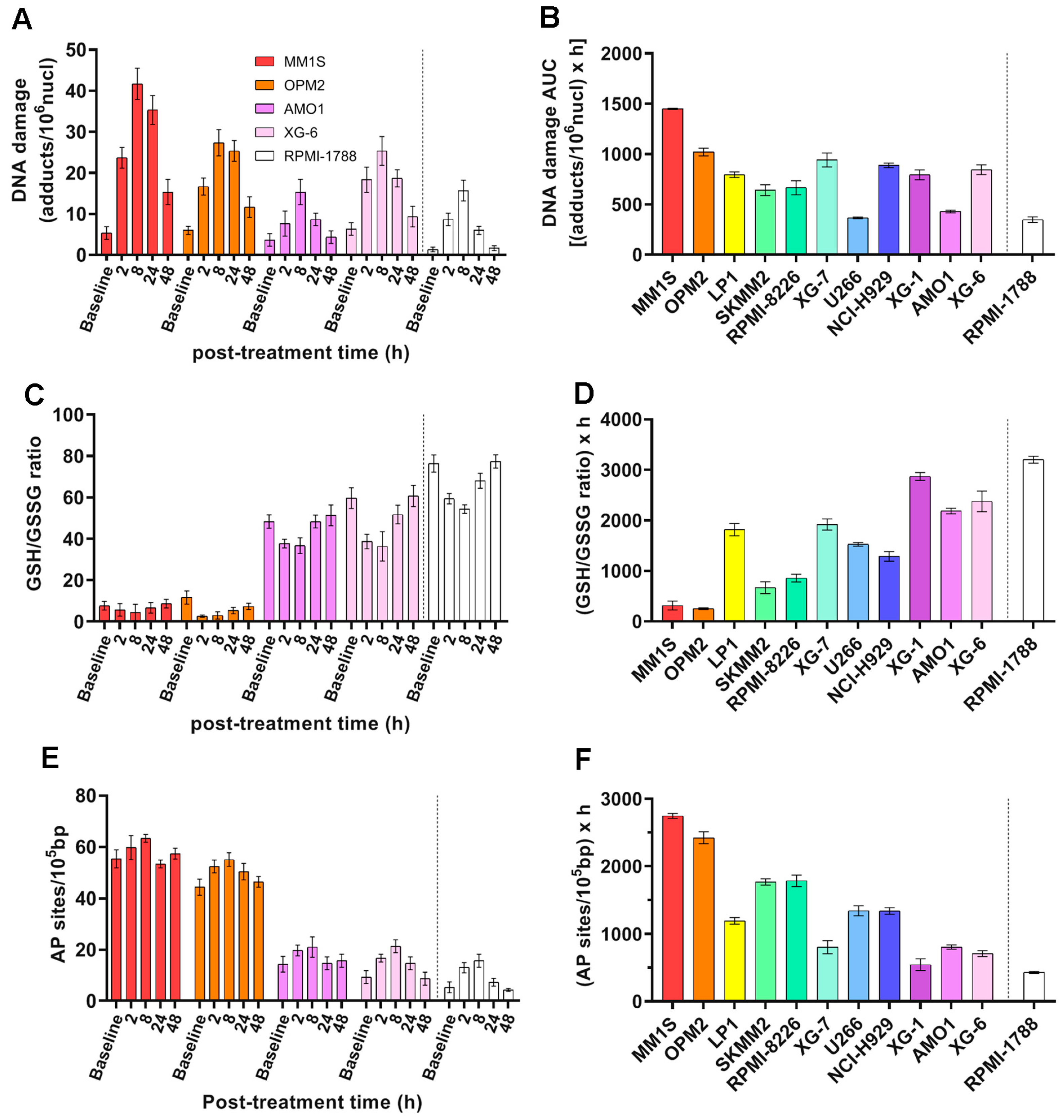
2.4. ICL Formation and Repair Capabilities in MM Cell Lines
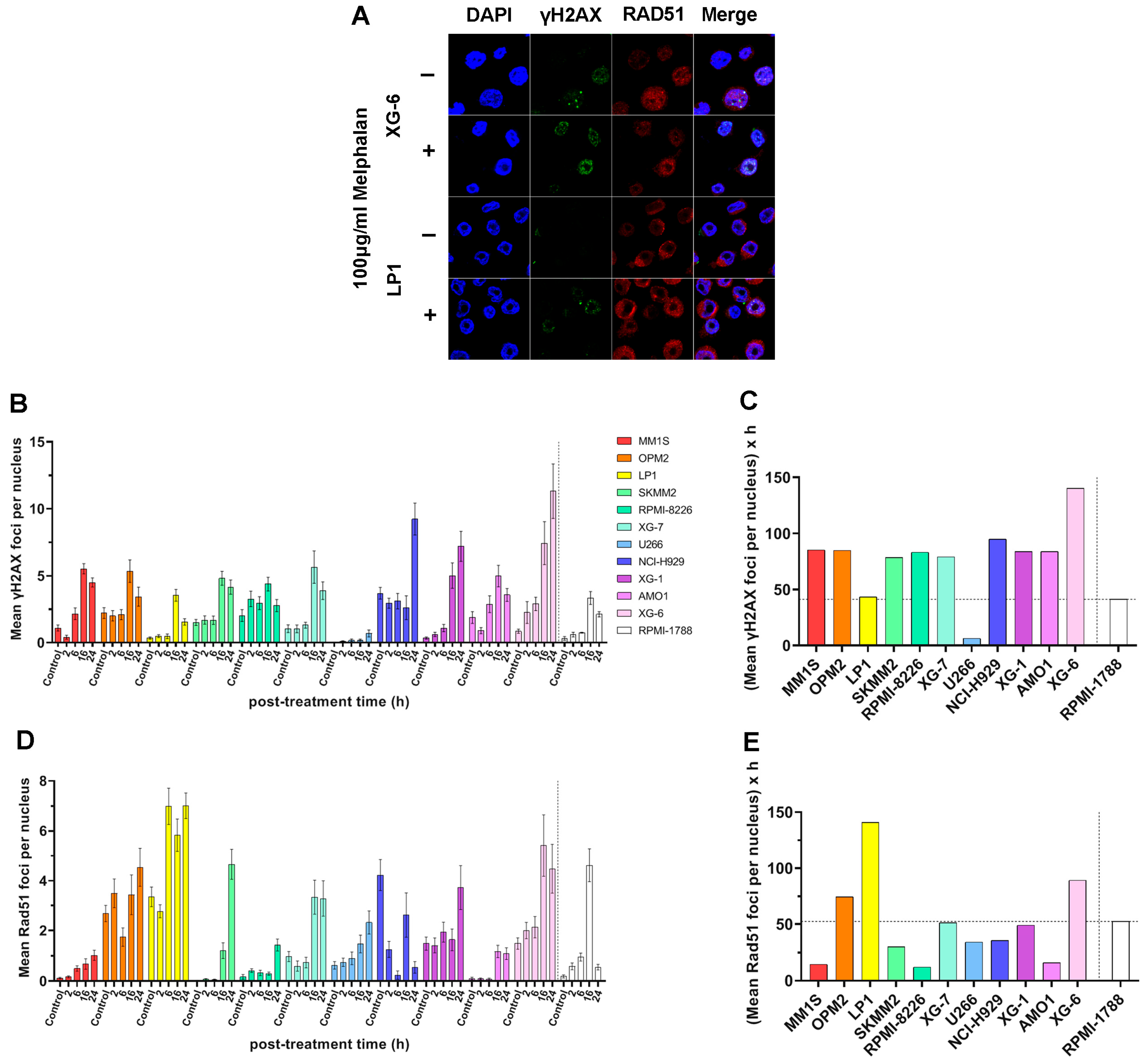
2.5. DSB Repair Capacity of MM Cell Lines
2.6. Statistical Analysis of DDR Parameters Across MM Cell Lines
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. MTT Viability Assay
4.3. Measurement of NER
4.4. Measurement of ICL Repair
4.5. Measurement of DSB Repair
4.6. Oxidative Stress and Apurinic/Apyrimidinic Sites Assessment
4.7. Apoptosis Rates
4.8. Measurement of Chromatin Condensation
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Saade, C.; Ghobrial, I.M. Updates on Mechanisms of Disease Progression in Precursor Myeloma: Monoclonal Gammopathy of Undermined Significance and Smoldering Myeloma. Presse Méd. 2025, 54, 104268. [Google Scholar] [CrossRef]
- Kyle, R.A.; Larson, D.R.; Therneau, T.M.; Dispenzieri, A.; Kumar, S.; Cerhan, J.R.; Rajkumar, S.V. Long-Term Follow-up of Monoclonal Gammopathy of Undetermined Significance. N. Engl. J. Med. 2018, 378, 241–249. [Google Scholar] [CrossRef]
- Rajkumar, S.V. Multiple Myeloma: 2022 Update on Diagnosis, Risk Stratification, and Management. Am. J. Hematol. 2022, 97, 1086–1107. [Google Scholar] [CrossRef] [PubMed]
- Kazandjian, D. Multiple Myeloma Epidemiology and Survival: A Unique Malignancy. Semin. Oncol. 2016, 43, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, H.; Durie, S.N.; Meckl, A.; Hinke, A.; Durie, B. Multiple Myeloma Incidence and Mortality Around the Globe; Interrelations Between Health Access and Quality, Economic Resources, and Patient Empowerment. Oncologist 2020, 25, e1406–e1413. [Google Scholar] [CrossRef]
- Fonseca, R.; Abouzaid, S.; Bonafede, M.; Cai, Q.; Parikh, K.; Cosler, L.; Richardson, P. Trends in Overall Survival and Costs of Multiple Myeloma, 2000–2014. Leukemia 2017, 31, 1915–1921. [Google Scholar] [CrossRef]
- Morgan, G.J.; Walker, B.A.; Davies, F.E. The Genetic Architecture of Multiple Myeloma. Nat. Rev. Cancer 2012, 12, 335–348. [Google Scholar] [CrossRef]
- Malamos, P.; Papanikolaou, C.; Gavriatopoulou, M.; Dimopoulos, M.A.; Terpos, E.; Souliotis, V.L. The Interplay between the DNA Damage Response (DDR) Network and the Mitogen-Activated Protein Kinase (MAPK) Signaling Pathway in Multiple Myeloma. Int. J. Mol. Sci. 2024, 25, 6991. [Google Scholar] [CrossRef]
- Elbezanti, W.O.; Challagundla, K.B.; Jonnalagadda, S.C.; Budak-Alpdogan, T.; Pandey, M.K. Past, Present, and a Glance into the Future of Multiple Myeloma Treatment. Pharmaceuticals 2023, 16, 415. [Google Scholar] [CrossRef]
- Sousa, M.M.L.; Zub, K.A.; Aas, P.A.; Hanssen-Bauer, A.; Demirovic, A.; Sarno, A.; Tian, E.; Liabakk, N.B.; Slupphaug, G. An Inverse Switch in DNA Base Excision and Strand Break Repair Contributes to Melphalan Resistance in Multiple Myeloma Cells. PLoS ONE 2013, 8, e55493. [Google Scholar] [CrossRef] [PubMed]
- Zub, K.A.; Sousa, M.M.L.d.; Sarno, A.; Sharma, A.; Demirovic, A.; Rao, S.; Young, C.; Aas, P.A.; Ericsson, I.; Sundan, A.; et al. Modulation of Cell Metabolic Pathways and Oxidative Stress Signaling Contribute to Acquired Melphalan Resistance in Multiple Myeloma Cells. PLoS ONE 2015, 10, e0119857. [Google Scholar] [CrossRef]
- Petrilla, C.; Galloway, J.; Kudalkar, R.; Ismael, A.; Cottini, F. Understanding DNA Damage Response and DNA Repair in Multiple Myeloma. Cancers 2023, 15, 4155. [Google Scholar] [CrossRef]
- Besse, A.; Besse, L.; Kraus, M.; Mendez-Lopez, M.; Bader, J.; Xin, B.-T.; Bruin, G.d.; Maurits, E.; Overkleeft, H.S.; Driessen, C. Proteasome Inhibition in Multiple Myeloma: Head-to-Head Comparison of Currently Available Proteasome Inhibitors. Cell Chem. Biol. 2019, 26, 340–351.e3. [Google Scholar] [CrossRef]
- Holstein, S.A.; McCarthy, P.L. Immunomodulatory Drugs in Multiple Myeloma: Mechanisms of Action and Clinical Experience. Drugs 2017, 77, 505–520. [Google Scholar] [CrossRef]
- Sammartano, V.; Franceschini, M.; Fredducci, S.; Caroni, F.; Ciofini, S.; Pacelli, P.; Bocchia, M.; Gozzetti, A. Anti-BCMA Novel Therapies for Multiple Myeloma. Cancer Drug Resist. 2023, 6, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Parikh, R.H.; Lonial, S. Chimeric Antigen Receptor T-Cell Therapy in Multiple Myeloma: A Comprehensive Review of Current Data and Implications for Clinical Practice. CA Cancer J. Clin. 2023, 73, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Devarakonda, S.; Efebera, Y.; Sharma, N. Role of Stem Cell Transplantation in Multiple Myeloma. Cancers 2021, 13, 863. [Google Scholar] [CrossRef] [PubMed]
- Lipchick, B.C.; Fink, E.E.; Nikiforov, M.A. Oxidative Stress and Proteasome Inhibitors in Multiple Myeloma. Pharmacol. Res. 2016, 105, 210–215. [Google Scholar] [CrossRef]
- Athanasopoulou, S.; Kapetanou, M.; Magouritsas, M.G.; Mougkolia, N.; Taouxidou, P.; Papacharalambous, M.; Sakellaridis, F.; Gonos, E. Antioxidant and Antiaging Properties of a Novel Synergistic Nutraceutical Complex: Readouts from an In Cellulo Study and an In Vivo Prospective, Randomized Trial. Antioxidants 2022, 11, 468. [Google Scholar] [CrossRef]
- Caillot, M.; Dakik, H.; Mazurier, F.; Sola, B. Targeting Reactive Oxygen Species Metabolism to Induce Myeloma Cell Death. Cancers 2021, 13, 2411. [Google Scholar] [CrossRef]
- Xiong, S.; Chng, W.-J.; Zhou, J. Crosstalk between Endoplasmic Reticulum Stress and Oxidative Stress: A Dynamic Duo in Multiple Myeloma. Cell. Mol. Life Sci. 2021, 78, 3883–3906. [Google Scholar] [CrossRef]
- Kul, A.N.; Kurt, B.O. Multiple Myeloma from the Perspective of Pro- and Anti-Oxidative Parameters: Potential for Diagnostic and/or Follow-Up Purposes? J. Pers. Med. 2024, 14, 221. [Google Scholar] [CrossRef]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA Damage, Repair, and Mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef]
- O’Connor, M.J. Targeting the DNA Damage Response in Cancer. Mol. Cell 2015, 60, 547–560. [Google Scholar] [CrossRef]
- Saitoh, T.; Oda, T. DNA Damage Response in Multiple Myeloma: The Role of the Tumor Microenvironment. Cancers 2021, 13, 504. [Google Scholar] [CrossRef]
- Alagpulinsa, D.A.; Szalat, R.E.; Poznansky, M.C.; Reis, R.J.S. Genomic Instability in Multiple Myeloma. Trends Cancer 2020, 6, 858–873. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Heuck, C.J.; Fazzari, M.J.; Mehta, J.; Singhal, S.; Greally, J.M.; Verma, A. DNA Methylation Alterations in Multiple Myeloma as a Model for Epigenetic Changes in Cancer. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010, 2, 654–669. [Google Scholar] [CrossRef]
- Feng, Z.; Hu, W.; Chasin, L.A.; Tang, M. Effects of Genomic Context and Chromatin Structure on Transcription-coupled and Global Genomic Repair in Mammalian Cells. Nucleic Acids Res. 2003, 31, 5897–5906. [Google Scholar] [CrossRef] [PubMed]
- Papanikolaou, C.; Economopoulou, P.; Spathis, A.; Kotsantis, I.; Gavrielatou, N.; Anastasiou, M.; Moutafi, M.; Kyriazoglou, A.; Foukas, G.-R.P.; Lelegiannis, I.M.; et al. Association of DNA Damage Response Signals and Oxidative Stress Status with Nivolumab Efficacy in Patients with Head and Neck Squamous Cell Carcinoma. Br. J. Cancer 2025, 133, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Gkotzamanidou, M.; Terpos, E.; Bamia, C.; Kyrtopoulos, S.A.; Sfikakis, P.P.; Dimopoulos, M.A.; Souliotis, V.L. Progressive Changes in Chromatin Structure and DNA Damage Response Signals in Bone Marrow and Peripheral Blood during Myelomagenesis. Leukemia 2014, 28, 1113–1121. [Google Scholar] [CrossRef]
- Xu, X.; Lai, Y.; Hua, Z.-C. Apoptosis and Apoptotic Body: Disease Message and Therapeutic Target Potentials. Biosci. Rep. 2019, 39, BSR20180992. [Google Scholar] [CrossRef]
- Enoiu, M.; Jiricny, J.; Schärer, O.D. Repair of Cisplatin-Induced DNA Interstrand Crosslinks by a Replication-Independent Pathway Involving Transcription-Coupled Repair and Translesion Synthesis. Nucleic Acids Res. 2012, 40, 8953–8964. [Google Scholar] [CrossRef] [PubMed]
- Souliotis, V.L.; Dimopoulos, M.A.; Sfikakis, P.P. Gene-Specific Formation and Repair of DNA Monoadducts and Interstrand Cross-Links after Therapeutic Exposure to Nitrogen Mustards. Clin. Cancer Res. 2003, 9, 4465–4474. [Google Scholar]
- Herrero, A.B.; Gutiérrez, N.C. Targeting Ongoing DNA Damage in Multiple Myeloma: Effects of DNA Damage Response Inhibitors on Plasma Cell Survival. Front. Oncol. 2017, 7, 98. [Google Scholar] [CrossRef]
- Kumar, S.; Gkotzamanidou, M.; Talluri, S.; Day, M.; Neptune, M.; Potluri, L.B.; Chakraborty, C.; Mills, K.; Shammas, M.A.; Munshi, N.C. Ongoing Spontaneous DNA Damage Creates Synthetic Lethality Targeted By Novel RAD51 Inhibitors in Multiple Myeloma. Blood 2019, 134, 4378. [Google Scholar] [CrossRef]
- Walters, D.K.; Wu, X.; Tschumper, R.C.; Arendt, B.K.; Huddleston, P.M.; Henderson, K.J.; Dispenzieri, A.; Jelinek, D.F. Evidence for Ongoing DNA Damage in Multiple Myeloma Cells as Revealed by Constitutive Phosphorylation of H2AX. Leukemia 2011, 25, 1344–1353. [Google Scholar] [CrossRef]
- Nakamura, H.; Takada, K. Reactive Oxygen Species in Cancer: Current Findings and Future Directions. Cancer Sci. 2021, 112, 3945–3952. [Google Scholar] [CrossRef]
- Attique, I.; Haider, Z.; Khan, M.; Hassan, S.; Soliman, M.M.; Ibrahim, W.N.; Anjum, S. Reactive Oxygen Species: From Tumorigenesis to Therapeutic Strategies in Cancer. Cancer Med. 2025, 14, e70947. [Google Scholar] [CrossRef]
- Wang, J.; Lin, D.; Peng, H.; Huang, Y.; Huang, J.; Gu, J. Cancer-Derived Immunoglobulin G Promotes Tumor Cell Growth and Proliferation through Inducing Production of Reactive Oxygen Species. Cell Death Dis. 2013, 4, e945. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.-H.; Yin, H.; Marincas, M.; Xie, L.-L.; Bu, L.-L.; Guo, M.-H.; Zheng, X.-L. From DNA Repair to Redox Signaling: The Multifaceted Role of APEX1 (Apurinic/Apyrimidinic Endonuclease 1) in Cardiovascular Health and Disease. Int. J. Mol. Sci. 2025, 26, 3034. [Google Scholar] [CrossRef] [PubMed]
- Thompson, P.S.; Cortez, D. New Insights into Abasic Site Repair and Tolerance. DNA Repair 2020, 90, 102866. [Google Scholar] [CrossRef] [PubMed]
- Mavroeidi, D.; Georganta, A.; Stefanou, D.T.; Papanikolaou, C.; Syrigos, K.N.; Souliotis, V.L. DNA Damage Response Network and Intracellular Redox Status in the Clinical Outcome of Patients with Lung Cancer. Cancers 2024, 16, 4218. [Google Scholar] [CrossRef]
- Papanikolaou, C.; Economopoulou, P.; Gavrielatou, N.; Mavroeidi, D.; Psyrri, A.; Souliotis, V.L. UVC-Induced Oxidative Stress and DNA Damage Repair Status in Head and Neck Squamous Cell Carcinoma Patients with Different Responses to Nivolumab Therapy. Biology 2025, 14, 195. [Google Scholar] [CrossRef]
- Souliotis, V.L.; Vlachogiannis, N.I.; Pappa, M.; Argyriou, A.; Ntouros, P.A.; Sfikakis, P.P. DNA Damage Response and Oxidative Stress in Systemic Autoimmunity. Int. J. Mol. Sci. 2020, 21, 55. [Google Scholar] [CrossRef]
- Vlachogiannis, N.I.; Pappa, M.; Ntouros, P.A.; Nezos, A.; Mavragani, C.P.; Souliotis, V.L.; Sfikakis, P.P. Association Between DNA Damage Response, Fibrosis and Type I Interferon Signature in Systemic Sclerosis. Front. Immunol. 2020, 11, 582401. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Richa; Kumar, A.; Tyagi, M.B.; Sinha, R.P. Molecular Mechanisms of Ultraviolet Radiation-Induced DNA Damage and Repair. J. Nucleic Acids 2010, 2010, 592980. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.; Møller, P.; Gajski, G.; Vodenková, S.; Abdulwahed, A.; Anderson, D.; Bankoglu, E.E.; Bonassi, S.; Boutet-Robinet, E.; Brunborg, G.; et al. Measuring DNA Modifications with the Comet Assay: A Compendium of Protocols. Nat. Protoc. 2023, 18, 929–989. [Google Scholar] [CrossRef]
- Zheng, W.; He, J.-L.; Jin, L.-F.; Lou, J.-L.; Wang, B.-H. Assessment of Human DNA Repair (NER) Capacity with DNA Repair Rate (DRR) by Comet Assay. Biomed. Environ. Sci. 2005, 18, 117–123. [Google Scholar]
- Souliotis, V.L.; Vlachogiannis, N.I.; Pappa, M.; Argyriou, A.; Sfikakis, P.P. DNA Damage Accumulation, Defective Chromatin Organization and Deficient DNA Repair Capacity in Patients with Rheumatoid Arthritis. Clin. Immunol. 2019, 203, 28–36. [Google Scholar] [CrossRef]
- Stefanou, D.T.; Kouvela, M.; Stellas, D.; Voutetakis, K.; Papadodima, O.; Syrigos, K.; Souliotis, V.L. Oxidative Stress and Deregulated DNA Damage Response Network in Lung Cancer Patients. Biomedicines 2022, 10, 1248. [Google Scholar] [CrossRef] [PubMed]
- Pappa, M.; Ntouros, P.A.; Papanikolaou, C.; Sfikakis, P.P.; Souliotis, V.L.; Tektonidou, M.G. Augmented Oxidative Stress, Accumulation of DNA Damage and Impaired DNA Repair Mechanisms in Thrombotic Primary Antiphospholipid Syndrome. Clin. Immunol. 2023, 254, 109693. [Google Scholar] [CrossRef]
- Dimopoulos, M.A.; Souliotis, V.L.; Anagnostopoulos, A.; Papadimitriou, C.; Sfikakis, P.P. Extent of Damage and Repair in the P53 Tumor-Suppressor Gene After Treatment of Myeloma Patients with High-Dose Melphalan and Autologous Blood Stem-Cell Transplantation Is Individualized and May Predict Clinical Outcome. J. Clin. Oncol. 2005, 23, 4381–4389. [Google Scholar] [CrossRef]
- Gkotzamanidou, M.; Terpos, E.; Bamia, C.; Munshi, N.C.; Dimopoulos, M.A.; Souliotis, V.L. DNA Repair of Myeloma Plasma Cells Correlates with Clinical Outcome: The Effect of the Nonhomologous End-Joining Inhibitor SCR7. Blood 2016, 128, 1214–1225. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, K.S.; Kuttikrishnan, S.; Ahmad, N.; Habeeba, U.; Mariyam, Z.; Suleman, M.; Bhat, A.A.; Uddin, S. H2AX: A Key Player in DNA Damage Response and a Promising Target for Cancer Therapy. Biomed. Pharmacother. 2024, 175, 116663. [Google Scholar] [CrossRef]
- Cruz, C.; Castroviejo-Bermejo, M.; Gutiérrez-Enríquez, S.; Llop-Guevara, A.; Ibrahim, Y.H.; Gris-Oliver, A.; Bonache, S.; Morancho, B.; Bruna, A.; Rueda, O.M.; et al. RAD51 Foci as a Functional Biomarker of Homologous Recombination Repair and PARP Inhibitor Resistance in Germline BRCA-Mutated Breast Cancer. Ann. Oncol. 2018, 29, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Szalat, R.; Samur, M.K.; Fulciniti, M.; Lopez, M.; Nanjappa, P.; Cleynen, A.; Wen, K.; Kumar, S.; Perini, T.; Calkins, A.S.; et al. Nucleotide Excision Repair Is a Potential Therapeutic Target in Multiple Myeloma. Leukemia 2018, 32, 111–119. [Google Scholar] [CrossRef]
- Deans, A.J.; West, S.C. DNA Interstrand Crosslink Repair and Cancer. Nat. Rev. Cancer 2011, 11, 467–480. [Google Scholar] [CrossRef] [PubMed]
- Shukla, P.; Solanki, A.; Ghosh, K.; Vundinti, B.R. DNA Interstrand Cross-Link Repair: Understanding Role of Fanconi Anemia Pathway and Therapeutic Implications. Eur. J. Haematol. 2013, 91, 381–393. [Google Scholar] [CrossRef]
- Herrero, A.B.; Miguel, J.S.; Gutierrez, N.C. Deregulation of DNA Double-Strand Break Repair in Multiple Myeloma: Implications for Genome Stability. PLoS ONE 2015, 10, e0121581. [Google Scholar] [CrossRef]
- Gourzones-Dmitriev, C.; Kassambara, A.; Sahota, S.; Rème, T.; Moreaux, J.; Bourquard, P.; Hose, D.; Pasero, P.; Constantinou, A.; Klein, B. DNA Repair Pathways in Human Multiple Myeloma. Cell Cycle 2013, 12, 2760–2773. [Google Scholar] [CrossRef]
- Gillyard, T.; Davis, J. Chapter Two—DNA Double-Strand Break Repair in Cancer: A Path to Achieving Precision Medicine. In International Review of Cell and Molecular Biology; Weyemi, U., Galluzzi, L., Eds.; Chromatin and Genomic Instability in Cancer; Academic Press: Cambridge, MA, USA, 2021; Volume 364, pp. 111–137. [Google Scholar]
- Chakraborty, C.; Mukherjee, S. Molecular Crosstalk between Chromatin Remodeling and Tumor Microenvironment in Multiple Myeloma. Curr. Oncol. 2022, 29, 9535–9549. [Google Scholar] [CrossRef]
- Ohguchi, H.; Hideshima, T.; Anderson, K.C. The Biological Significance of Histone Modifiers in Multiple Myeloma: Clinical Applications. Blood Cancer J. 2018, 8, 83. [Google Scholar] [CrossRef] [PubMed]
- Tigu, A.B.; Ivancuta, A.; Uhl, A.; Sabo, A.C.; Nistor, M.; Mureșan, X.-M.; Cenariu, D.; Timis, T.; Diculescu, D.; Gulei, D. Epigenetic Therapies in Melanoma—Targeting DNA Methylation and Histone Modification. Biomedicines 2025, 13, 1188. [Google Scholar] [CrossRef] [PubMed]
- De Mel, S.; Lee, A.R.; Tan, J.H.I.; Tan, R.Z.Y.; Poon, L.M.; Chan, E.; Lee, J.; Chee, Y.L.; Lakshminarasappa, S.R.; Jaynes, P.W.; et al. Targeting the DNA Damage Response in Hematological Malignancies. Front. Oncol. 2024, 14, 1307839. [Google Scholar] [CrossRef]
- Kwok, M.; Agathanggelou, A.; Stankovic, T. DNA Damage Response Defects in Hematologic Malignancies: Mechanistic Insights and Therapeutic Strategies. Blood 2024, 143, 2123–2144. [Google Scholar] [CrossRef]
- Shim, Y.J. Therapeutic Targeting of DNA Damage Response Pathways in TP53- and ATM-Mutated Tumors. Brain Tumor Res. Treat. 2025, 13, 73–80. [Google Scholar] [CrossRef]
- Bandini, C.; Mereu, E.; Paradzik, T.; Labrador, M.; Maccagno, M.; Cumerlato, M.; Oreglia, F.; Prever, L.; Manicardi, V.; Taiana, E.; et al. Lysin (K)-Specific Demethylase 1 Inhibition Enhances Proteasome Inhibitor Response and Overcomes Drug Resistance in Multiple Myeloma. Exp. Hematol. Oncol. 2023, 12, 71. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.; Wang, Y.; Yang, X.; Chen, Z.; Wu, F.; Ren, R.; Sun, Y.; Lai, Y.; Peng, L.; Yu, L.; et al. Discovery of a First-in-Class PROTAC Degrader of Histone Lysine Demethylase KDM4. Eur. J. Med. Chem. 2025, 288, 117410. [Google Scholar] [CrossRef]
- Nastaranpour, M.; Damara, A.; Grabbe, S.; Shahneh, F. Lysine Demethylase 6 (KDM6): A Promising Therapeutic Target in Autoimmune Disorders and Cancer. Biomed. Pharmacother. 2025, 189, 118254. [Google Scholar] [CrossRef]
- Avramis, I.A.; Christodoulopoulos, G.; Suzuki, A.; Laug, W.E.; Gonzalez-Gomez, I.; McNamara, G.; Sausville, E.A.; Avramis, V.I. In Vitro and in Vivo Evaluations of the Tyrosine Kinase Inhibitor NSC 680410 against Human Leukemia and Glioblastoma Cell Lines. Cancer Chemother. Pharmacol. 2002, 50, 479–489. [Google Scholar] [CrossRef]
- Lapytsko, A.; Kollarovic, G.; Ivanova, L.; Studencka, M.; Schaber, J. FoCo: A Simple and Robust Quantification Algorithm of Nuclear Foci. BMC Bioinform. 2015, 16, 392. [Google Scholar] [CrossRef] [PubMed]
- Souliotis, V.L.; Dimopoulos, M.A.; Episkopou, H.G.; Kyrtopoulos, S.A.; Sfikakis, P.P. Preferential In Vivo DNA Repair of Melphalan-Induced Damage in Human Genes Is Greatly Affected by the Local Chromatin Structure. DNA Repair 2006, 5, 972–985. [Google Scholar] [CrossRef] [PubMed]

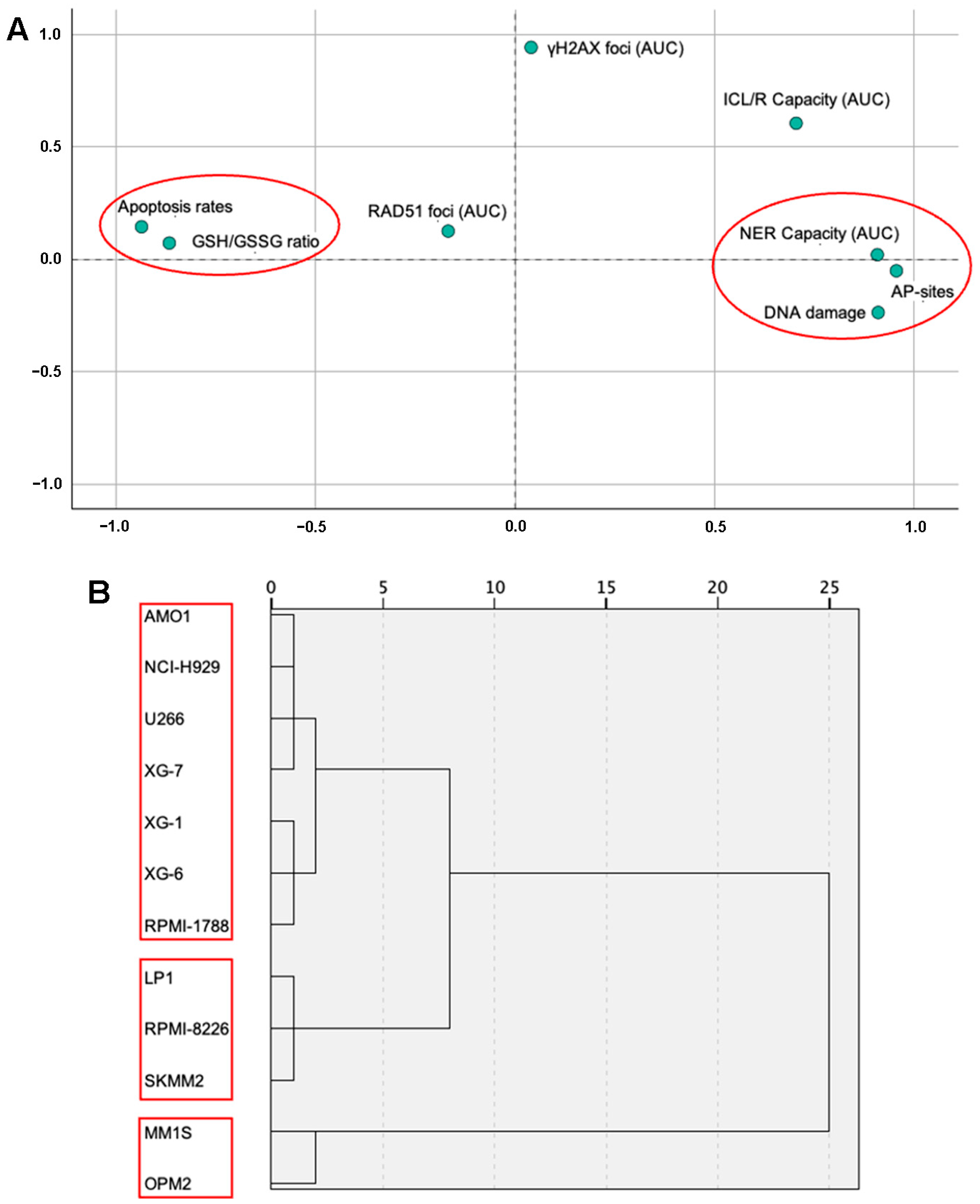
| Apoptosis Rates | DNA Damage | GSH/GSSG Ratio | AP-Sites | NER Capacity (AUC) | ICL/R Capacity (AUC) | γH2AX Foci (AUC) | |
|---|---|---|---|---|---|---|---|
| DNA damage | −0.918 ** | ||||||
| GSH/GSSG ratio | 0.792 ** | −0.804 ** | |||||
| AP-sites | −0.864 ** | 0.864 ** | −0.814 ** | ||||
| NER Capacity (AUC) # | −0.792 ** | 0.762 ** | −0.648 * | 0.892 ** | |||
| ICL/R Capacity (AUC) | −0.617 * | 0.456 | −0.485 | 0.592 * | 0.707 * | ||
| γH2AX foci (AUC) | 0.135 | −0.145 | −0.053 | 0.021 | −0.003 | 0.483 | |
| RAD51 foci (AUC) | −0.032 | −0.033 | 0.286 | −0.215 | −0.244 | 0.073 | −0.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malamos, P.; Papanikolaou, C.; Deligianni, E.; Mavroeidi, D.; Koutoulogenis, K.; Gavriatopoulou, M.; Terpos, E.; Souliotis, V.L. DNA Damage Response and Redox Status in the Resistance of Multiple Myeloma Cells to Genotoxic Treatment. Int. J. Mol. Sci. 2025, 26, 10171. https://doi.org/10.3390/ijms262010171
Malamos P, Papanikolaou C, Deligianni E, Mavroeidi D, Koutoulogenis K, Gavriatopoulou M, Terpos E, Souliotis VL. DNA Damage Response and Redox Status in the Resistance of Multiple Myeloma Cells to Genotoxic Treatment. International Journal of Molecular Sciences. 2025; 26(20):10171. https://doi.org/10.3390/ijms262010171
Chicago/Turabian StyleMalamos, Panagiotis, Christina Papanikolaou, Elisavet Deligianni, Dimitra Mavroeidi, Konstantinos Koutoulogenis, Maria Gavriatopoulou, Evangelos Terpos, and Vassilis L. Souliotis. 2025. "DNA Damage Response and Redox Status in the Resistance of Multiple Myeloma Cells to Genotoxic Treatment" International Journal of Molecular Sciences 26, no. 20: 10171. https://doi.org/10.3390/ijms262010171
APA StyleMalamos, P., Papanikolaou, C., Deligianni, E., Mavroeidi, D., Koutoulogenis, K., Gavriatopoulou, M., Terpos, E., & Souliotis, V. L. (2025). DNA Damage Response and Redox Status in the Resistance of Multiple Myeloma Cells to Genotoxic Treatment. International Journal of Molecular Sciences, 26(20), 10171. https://doi.org/10.3390/ijms262010171






