2.1. Monte Carlo Simulation 1: DepMap Dataset
A Monte Carlo simulation was conducted to assess the performance of DD-CC-II based on generated synthetic datasets with the ground truth known. Statistical evaluation results were provided through repeated sampling, ensuring that performance metrics were not artifacts of a specific dataset.
We used a publicly available CCLE expression dataset from the DepMap database (
https://depmap.org/portal/ (accessed on 1 February 2020)), comprising 19,221 genes and 1406 cells from more than 20 types of cancer (i.e., lineages). The lineage subtypes of each cancer were defined as distinct cell groups, selecting only cancer types with diverse sub-lineages and a sufficient number of cells. Specifically, sub-lineages comprising more than ten cells were considered individual cell groups; only cancer types containing more than two sub-lineages were included in the analysis. Consequently, CCI inferences were made for cells from lung, blood, lymphoid tissue, breast, soft tissue, and bone cancers. The cell groups for each cancer type are presented in
Table 1. Previous studies demonstrated enhanced ligand–receptor communication within the same lineage subpopulations in various cancer types, including glioma, lung adenocarcinoma, and colorectal cancer [
12,
13,
14]. Tang et al. [
12] revealed numerous significant ligand–receptor interactions among neoplastic cells, including those associated with autocrine and paracrine signaling within the same tumor lineage. Meanwhile, Yang et al. [
13] demonstrated that tumor cell sub-lineages within the same lung adenocarcinoma lineage engage in direct communication via shared ligand–receptor expression. Similarly, Lin et al. [
14] defined active ligand–receptor interactions between different subpopulations of malignant epithelial cells, indicating robust communication among sub-lineages within the same colorectal cancer lineage.
In the current study, the interactions between cell groups (i.e., sub-lineages) in the same lineage were considered true positives of the CCI inference. For instance, interactions between groups of cells in non-small cell lung cancer (NSCLC), small-cell lung cancer (SCLC), and mesothelioma were considered true positives of CCI inference for lung cancer. The eigen-cells of a specific sub-lineage were estimated using the expression levels of cells in the sub-lineage and a randomly selected 5% of cells for other lineages. The eigen-cells that did not belong to a sub-lineage were estimated by using SVD based on the expression levels of randomly selected cells for other lineages. That is, eigen-cells for NSCLC, SCLC, and mesothelioma (EGs-NSCLC, EGs-SCL, and EGs-mes) were estimated based on 141 cells (135 NSCLC cells + 5% of 135 non-lung cells), 52 (50 SCLC cells + 5% of 50 non-lung cells) and 21 (20 mesothelioma cells + 5% of 20 non-lung cells), respectively. For false positives, eigen-cells not involved in lung cancer (EGs-nLC1, EGs-nLC2, and EGs-nLC3) were estimated by randomly selecting 141, 52, and 21 cells of lineages other than lung cancer. The interactions among EGs-nLC1, EGs-nLC2, and EGs-nLC3 were considered CCI false positives.
The selection of singular values was generally guided by the criterion that their cumulative variance contribution exceeds a threshold within the 70–90% range [
15,
16]. In line with previous research, we selected the number of eigen-cells (
) based on the number of singular values required to capture 75% of the cumulative variance in the expression profiles of each sub-lineage.
Table 2 presents the numbers of eigen-cells (i.e.,
) capturing 75% variance of cell groups, where EGs-
x and EGs-n
x indicate the cell groups for cancer sub-lineages (e.g., EGs-1: NSCLC, EGs-2: SCLC, EGs-3: mesothelioma in lung cancer) and randomly generated groups of cells, respectively.
Eigen-cell correlation networks between EGs-NSCLC, EGs-SCL, EGs-mes, EGs-nLC1, EGs-nLC2, and EGs-nLC3 constructed using significant correlation coefficients with a significant level
(i.e.,
p-value
). The significance of associations between groups was determined using the hypergeometric test. Similar procedures were applied to infer the CCI of other cancers (blood, lymphocyte, breast, soft tissue, and bone). The proposed strategy was evaluated by comparing it with existing strategies for CCI analysis: CellCall, CellChat, iTALK, and REMI. Using existing methods, the strength of the association (i.e., edge weight) was computed between cell groups. In CellCall, CCI strength is measured as the ligand–receptor score between groups based on intercellular signaling (ligand and receptor expression levels) and intracellular signaling (activity of downstream transcription factors). CellChat computes the edge weight based on total communication probability of ligand–receptor pairs between groups. The edge weights of CCIs in iTALK represent the average expression levels of the ligand gene in a given cell type (i.e., value of “
cell_from_mean_exprs ” in the
iTALK package version 0.1.0 of
R). In REMI, the strength of the association between cell groups is expressed as the number of ligand–receptor interactions detected between cell types. Detailed descriptions of the existing methods for CCI inference have been provided elsewhere by [
2,
3,
4,
5]. We measured the significance of the association based on FDR-q. values and described the edge weights as −log(FDR-q.value). The simulation was performed over 50 iterations.
Figure 1 shows the strength of the association between groups of cells computed by CellCall, CellChat, iTALK, REMI, and DD-CC-II in the 50 simulations, where the green boxes indicate the true positives of CCIs. DD-CC-II appropriately achieved CCI inference, as evidenced by the designation of relatively larger edge weights for true positive interactions between cell groups (i.e., P
x*P
x) than for false positive interactions (i.e., N
x*P
x, N
x*N
x). In contrast, existing methods (i.e., CellCall, CellChat, iTALK, and REMI) failed to effectively perform CCI inference; the strength of the association was not described accurately.
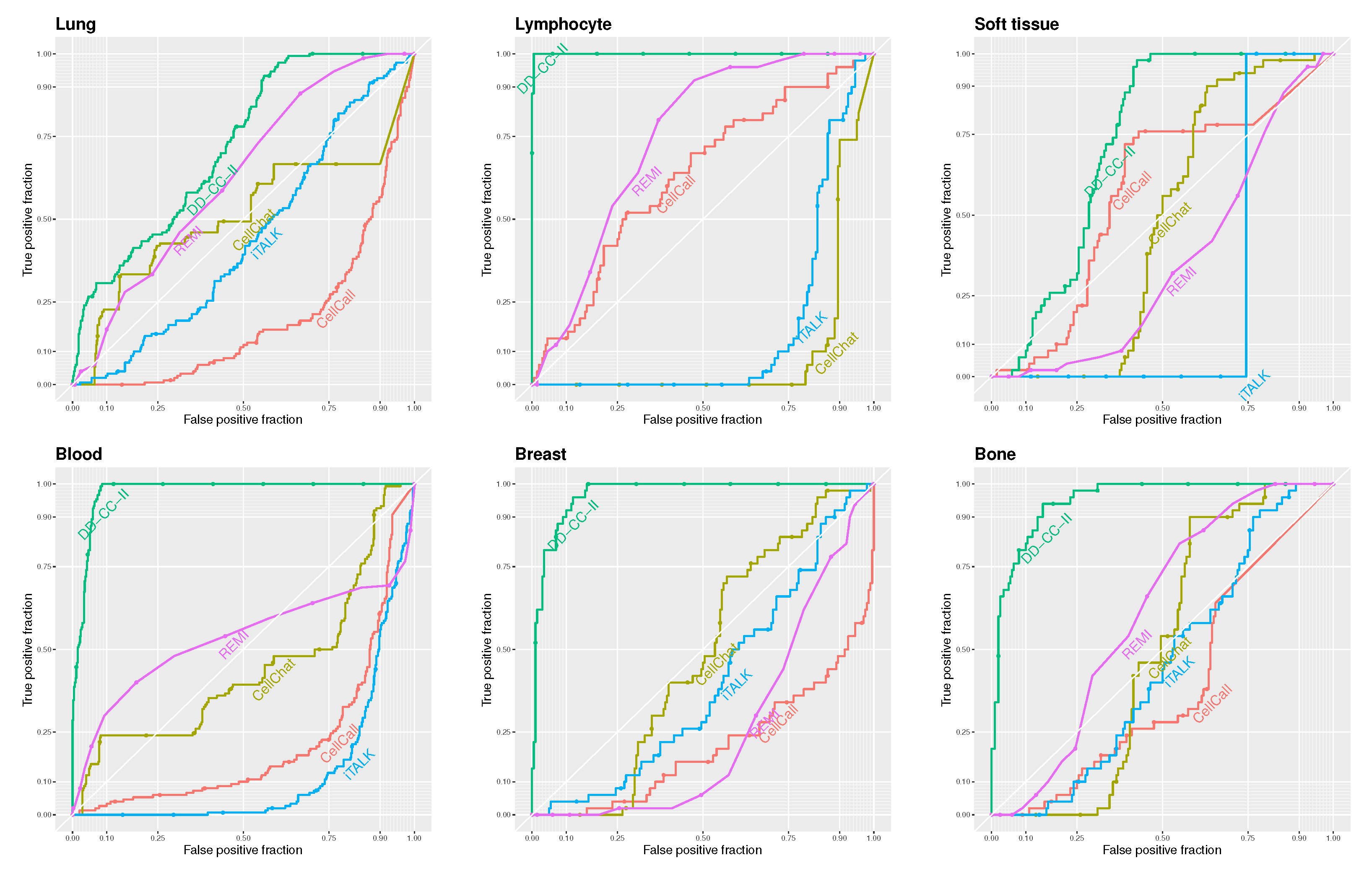
Figure 2 shows the receiver operator characteristic (ROC) curves for DD-CC-II, CellCall, CellChat, iTALK, and REMI based on the threshold of the edge weights described in
Figure 2. Our strategy outperformed other methods in CCI inference. In particular, DD-CC-II exhibited an outstanding performance for CCI inference of lung, lymphocyte, blood, breast, and bone cancers. In contrast, the other methods, particularly iTALK, performed poorly.
We evaluated the methods for inferring CCIs based on the area under the curve (AUC) of the ROC curves. Given that our scenarios involve a small number of true CCIs relative to all possible pairings (i.e., a class imbalance), we additionally assessed performance using the AUC of the precision–recall (PR) curves, which are more appropriate for imbalanced datasets.
Table 3 reports the AUC values of both ROC and PR curves; values in parentheses indicate the standard deviations of AUC scores across 50 simulation runs. To assess the sensitivity of the results of DD-CC-II to significant level
for correlation coefficient network, we also described the results (AUC values) based on
in
Table 3, where columns
and
indicate AUC values of DD-CC-II based on the correlation coefficient network with
and
, respectively.
The results also show that our strategy will be a useful tool for CCI inference and evaluating cell signaling. Furthermore,
Table 3 also demonstrates that using a threshold of
in the construction of the correlation coefficient network yields better results than using
. This result suggests that the performance of DD-CC-II is sensitive to the threshold used in constructing the correlation coefficient network. Therefore, selecting an appropriate threshold is a critical issue that warrants careful consideration, and the application of multiple testing correction should be taken into account. However, given that DD-CC-II demonstrated more effective results with a relatively higher threshold (i.e.,
) compared to
, multiple testing correction was not applied in this analysis.
2.2. Monte Carlo Simulation 2: GDSC Database
DD-CC-II was also applied to the “Sanger Genomics of Drug Sensitivity in Cancer (GDSC) dataset from the Cancer Genome Project”. The gene expression data (Cell_line_RMA_proc_basalExp.txt) comprised 9764 genes in 968 cells from more than 30 cancer types. Cell groups were generated based on primary tissue type classification (i.e., GDSC Tissue descriptor 1) related to more than two cancer types with more than ten cells, i.e., aero_dig_tract (AERO), leukaemia (LEUL), digestive_system (DIG), nervous_system (NERV).
Table 4 lists the cell groups for each cancer type.
Similar to the analysis of the DeepMap dataset, the interactions between cell groups (i.e., cancer types) in the same lineage/primary tissue type classification were considered true positives of the CCI inference. That is, the eigen-cells of a specific cancer type were estimated using the expression levels of cells in the cancer type and 5% randomly selected cells for other cancer types. The eigen-cells that did not belong to a cancer type were also estimated based on the expression levels of randomly selected cells for other categories of primary tissue type classification (i.e., true negative scenario). The number of eigen-cells (e.g.,
) was set to the number of singular values capturing 75% variance in the expression levels of each sub-lineage, as shown in
Table 5. The eigen-cell correlation networks were constructed with a significant level
. The CCI inference based on correlation coefficient networks were performed similar to the CCI inference of DeepMap data.
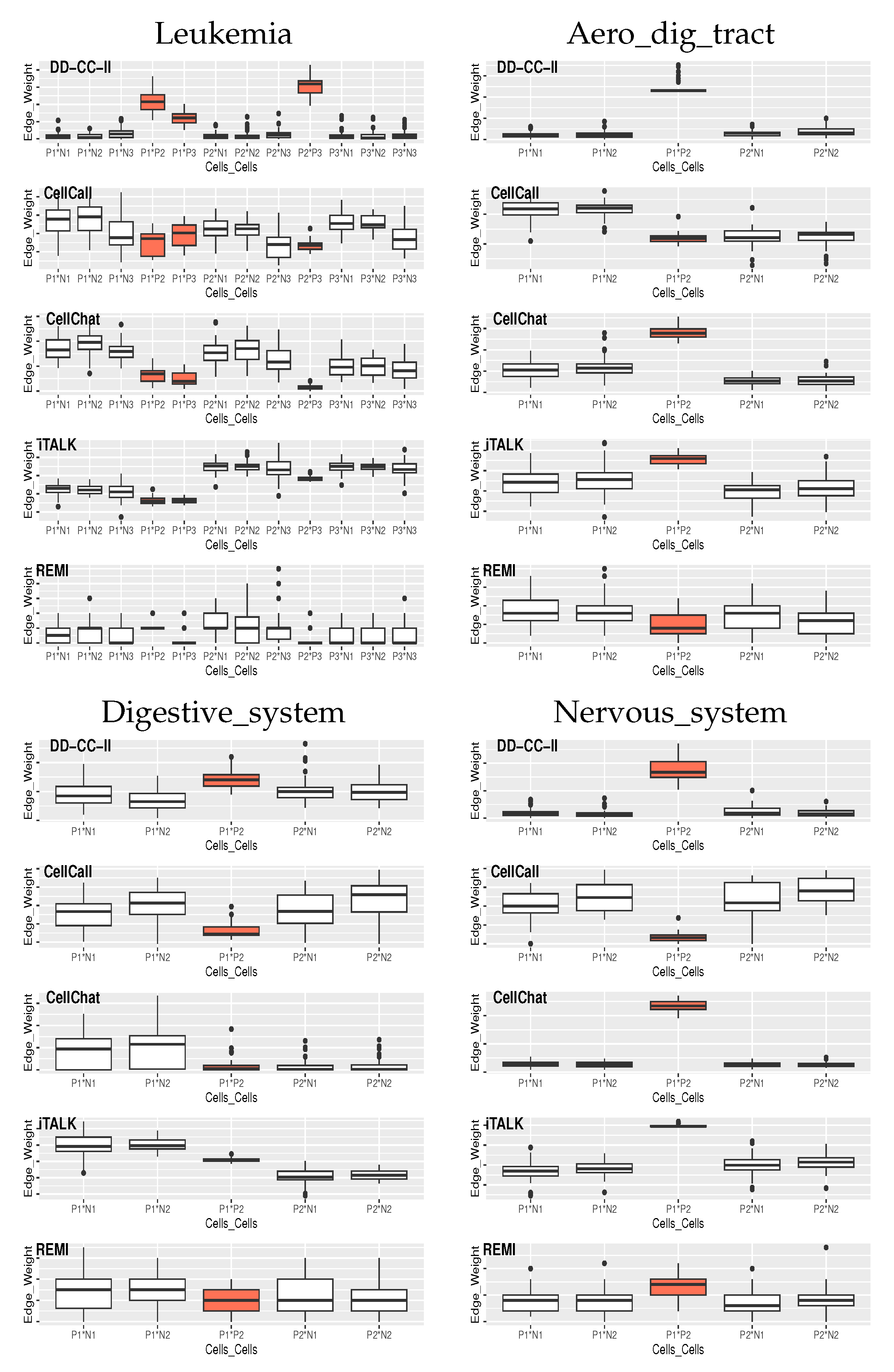
Figure 3 shows the edge weights between the groups of cells estimated by the CellCall, CellChat, iTALK, REMI, and DD-CC-II in the 50 simulations, where the orange boxes indicate the true positives of CCIs.
DD-CC-II also exhibited an outstanding performance for CCI inference, i.e., edge weight estimation in CCIs (
Figure 3). Although the CellChat and iTALK also appropriately achieved CCI inference for Aero_dig_tract and Nervous_system, they did not generate effective CCI results for Leukemia and Digestive_system.
Table 6 shows the AUC values of the ROC and PR curves, where the numbers in parentheses correspond to the standard deviation of the AUC values obtained from 50 simulations. Consistent with the results of the DeepMap dataset, the proposed DD-CC-II shows outstanding performances for the CCI inference of cancer types.
Finally, CCI inference in terms of computational complexity was evaluated, where the CCI execution times were assessed based on DD-CC-II, CellCall, CellChat, iTALK, and REMI for CCIs for DepMap and GDSC datasets using the R package (see
Table 7). The proposed DD-CC-II demonstrated competitive performance in terms of computational complexity compared to the existing methods. In contrast, CellCall demonstrated a considerable computational burden.
2.3. Uncovering Disease Trajectory Correlations Between COVID-19 Severity Stages
COVID-19 is a severe infectious disease, particularly for those with critical illnesses who are at high risk of rapid deterioration. Dinsay et al. [
17] reported in-hospital mortality rates of 5.4%, 8.1%, 27.0%, and 80.3%, for mild, moderate, severe, and critical COVID-19 cases in the Philippines, respectively. Meanwhile, in Turkey, mortality rates were 4.7% for mild-to-moderate cases, 23.9% for severe cases, and 100% for critical cases [
18]. Preventing COVID-19 progression is crucial for better clinical outcomes. Accordingly, we sought to characterize correlations between COVID-19 severity stages and key markers involved in disease progression. We considered samples from each severity stage as a cell group and measured the strength of the association between the COVID-19 severity stages based on the eigen-cell links for each stage. DD-CC-II was applied to the whole blood RNA-seq data of 1102 genotyped samples provided by the Japan COVID-19 Task Force; the COVID severity stages were defined as “critical (Level 4: patients in intensive care unit or requiring intubation and ventilation),” “severe (Level 3: others requiring oxygen support),” “mild (Level 2: other symptomatic patients),” and “asymptomatic (Level 1: without COVID-19 related symptoms)” [
11]. The RNA-seq expression data of COVID-19 samples are available at the National Bioscience Database Center (NBDC) Human Database (accession code: hum0343;
https://humandbs.biosciencedbc.jp/en/hum0343, (accessed on 1 April 2022)).
The RNA-seq data for 71 asymptomatic, 241 mild, 404 severe, and 303 critical samples were considered as four groups of cells and applied to construct eigen-cell correlation networks. Particular focus was placed on genes involved in the “
Coronavirus disease-COVID-19” pathway, i.e., COVID-19 genes in the KEGG pathway database. Subsequently, disease-trajectory correlations between COVID-19 stages were inferred based on the eigen-cells of each stage computed by the expression levels of the COVID-19 genes. To elucidate the mechanisms associated with immune damage in COVID-19, DD-CC-II was also applied to disease-trajectory correlations between COVID-19 severity stages based on the genes involved in “
immune disease” pathways, i.e., immune disease-related genes.
Table 8 presents the KEGG database “
Coronavirus disease-COVID-19” and “
immune disease” pathways. For the genes involved in each pathway, eigen-cells were estimated for samples corresponding to each COVID-19 stage and used to construct eigen-cell correlation coefficient networks. Finally, DD-CC-II was applied to identify disease-trajectory correlations for COVID-19 severity stages. For each pathway, we examined whether severe COVID-19 groups showed significant associations. To control the false positive rate from multiple comparisons, Bonferroni correction was applied, and associations with an FDR-q.value less than 0.05 were considered significant. All genes were included in the eigen-cell construction. Lowly expressed genes were not excluded; their contributions to the eigenvectors are naturally weighted according to their expression levels, allowing all genes to influence the representation while reducing the dominance of highly variable genes.
Table 9 lists the FDR-q.value of the disease-trajectory correlations analysis. As shown in
Table 9, the severity stages of COVID-19 computed by the COVID-19 genes show a relatively strong association with those computed by immune disease-related genes.
Figure 4 (upper right) presents the disease-trajectory correlations between COVID-19 severity stages based on COVID-19 genes. In COVID-19 severity stage interactions, asymptomatic samples (Level 1) were strongly associated with mild (Level 2), severe (Level 3), and critical (Level 4) samples. This implies that mild, severe, and critical stages of COVID-19 may have similar gene transcription patterns as asymptomatic samples. Hence, genes with key roles in the initial stages of COVID-19 may also be critical for later-stage disease progression.
Table 10 presents the crucial genes in eigen-cell estimation for each COVID-19 severity stage, where rank indicates the ranking of the absolute loading values for the first eigen-cell estimation. The highly ranked genes can be considered crucial markers for understanding COVID-19 mechanism. The crucial genes for the eigen-cell estimation of the initial stages (asymptomatic samples; Level 1), that is, HLAB, HLAC, NFKBIA, RPS11, RPS27, and RPL41, were also identified in the eigen-cell estimation for higher stages (mild: Level 2, severe: Level 3, critical: Level 4 samples). These common genes have been suggested previously as COVID-19 markers (
Table 10).
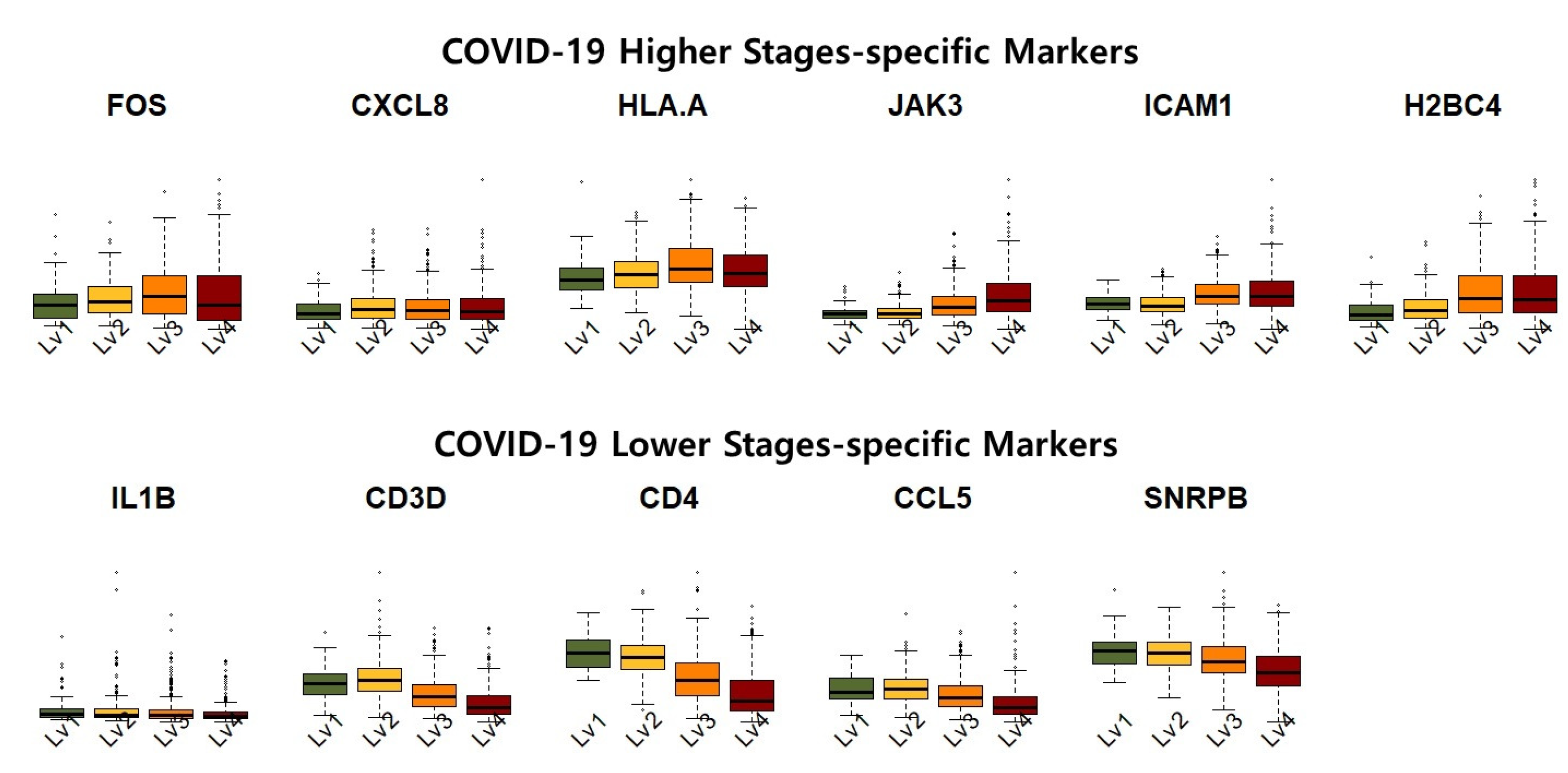
In contrast, FOS, CXCL8, and HLA-A were revealed as high-severity-specific markers that were identified not in asymptomatic patients but in mild, severe, and critical patients.
FOS
High FOS expression is a key feature of COVID-19 patients [
27], making it a potentially promising target for managing SARS-CoV-2 infection [
28,
29]. Similarly, Lu et al. [
30] observed a strong association between FOS and nonalcoholic steatohepatitis and COVID-19.
CXCL8
Elevated CXCL8 levels have also been reported in early COVID-19 patients’ blood and alveolar spaces [
31], with higher levels in severe cases but no significant increase in mild cases compared to healthy controls [
32]. Hence, downregulated inflammatory marker genes, particularly CXCL8, may serve as powerful biomarkers for managing COVID-19 infection [
33]. According to Park and Lee [
32], HLAA also significantly influences COVID-19 severity across ethnicities.
Collectively, these results suggest that suppressing high severity-specific markers (i.e., FOS, CXCL8, and HLA-A) may help prevent COVID-19 progression.
The disease-trajectory correlations between COVID-19 stages computed by immune-related genes are also presented in
Figure 4. The COVID-19 stages show relatively weak associations with the immune-related genes compared with the COVID-19 genes. Moreover, “
inflammatory bowel disease”, “
primary immunodeficiency”, “
Rheumatoid arthritis”, and “
systemic lupus erythematosus” were identified as immune damage pathways underlying COVID-19, with significant COVID-19 stage interactions estimated based on genes involved in these four pathways. The association between mild and critical samples were common for immune-related pathways. Additionally, the COVID-19 severity stage cells for genes involved in the “
Systemic lupus erythematosus” pathway exhibited relatively strong association and active interplay (i.e., numerous edges). This implies that the “
Systemic lupus erythematosus” pathway is crucial in defining the mechanism and progression of COVID-19 stages. The “
Systemic lupus erythematosus” pathway was highlighted due to its relevance to immune dysregulation in COVID-19, including aberrant type I interferon signaling [
34]. Shared molecular mechanisms and severity-specific enrichment suggest its role in modulating immune responses during disease progression [
35]. The “
Immune disease” genes that are crucial for eigen-cell estimation are listed in
Table 11. Similarly to eigen-cell estimation based on the COVID-19 genes, many common genes were identified as crucial markers. IL1B, CD3D, CD4, CCL5, and SNRPB were identified as low-severity-specific markers.
IL1
The elevated levels of intestinal IL-1
have been linked to the longer survival and lower levels of intestinal SARS-CoV-2 [
36]. Moreover, patients with severe COVID-19 and poor prognosis have lower levels of IL1B, IL2, and IL8 compared to those with favorable outcomes [
37]. Hence, IL-1
could serve as a key marker for targeted treatment in patients with COVID-19 [
38,
39].
CD3D
CD3D was also identified as a core gene linked to immune infiltration, with potential diagnostic utility in COVID-19 patients with sepsis. Zhang et al. [
40] proposed that a risk score based on CD3D, CD3E, LCK, and EVL could serve as a predictive model for severe COVID-19.
CD4
CD4+ T cells are significantly diminished in severe COVID-19 cases [
40]. Meanwhile, CD4-mediated SARS-CoV-2 infection of T helper cells can contribute to a weakened immune response in patients with COVID-19 [
41]. However, SARS-CoV-2-specific, TNF-
-producing CD4+ T cells are crucial in maintaining antibody titres after COVID-19 infection [
42].
CCL5
CCL5 has been described as the optimal indicator of COVID-19 severity [
43]. CCL5 levels negatively correlate with mortality in COVID-19, suggesting that it may protect against severe disease progression [
44]. In particular, CCL5 is significantly upregulated from the early stages of infection in those with mild disease, but not in severe cases [
45]. Thus, enhancing CCL5 expression early in COVID-19 may reduce the risk of severe illness. Therefore, monitoring CCL5 levels could predict infection severity [
46] and be applied to inform treatment strategies [
45].
JAK3, ICAM1, and H2BC4 were identified as markers of higher COVID-19 severity. These results are strongly supported by those of existing research, implying that our strategy provides biologically reliable results for the disease-trajectory correlations of COVID-19 stages and related marker identification. The role of SNRPB in COVID-19 has not yet been explored, indicating that it could be considered a novel potential biomarker for the disease.
JAK3
Sbruzzi et al. [
47] identified a novel homozygous JAK3 variant in a patient with severe COVID-19, suggesting that JAK3 may represent a key marker for persistent infection. In patients with cirrhosis, elevated plasma ICAM1 acts as an independent predictor of severe COVID-19 [
48].
ICAM1
ICAM1 serves as a prognostic marker for long-term complications or sequelae due to COVID-19 infection [
49] and an effective biomarker for predicting COVID-19 severity [
50].
Figure 5 shows the expression levels of the identified COVID-19 high-severity and low-severity-specific markers. The COVID-19 high-stage-specific markers (i.e., FOS, CXCL8, HLAA, JAK3, ICAM1, and H2BC4) were overexpressed in higher-stage samples, that is, increased expression levels of the markers were observed in asymptomatic to critical samples. In contrast, the low-stage-specific markers (i.e., IL1B, CD3D, CD4, CCL5, SNRPB) were relatively upregulated in the lower-stage samples. Furthermore, the expression of high (low)-stage-specific markers exhibited considerable variance in severe (non-severe) samples. This result implies that high (low)-stage-specific markers exhibited high transcriptional activity in samples of COVID-19 high(low) stages.
Based on our results, we suggest that controlling high-severity-specific markers (FOS, CXCL8, HLA-A, JAK3, ICAM1, and H2BC4) and low-severity-specific markers (IL1B, CD3D, CD4, CCL5, and SNRPB) may prevent COVID-19 progression. We also suggest that the “Systemic lupus erythematosus” pathway is crucial to understanding the mechanisms underlying COVID-19 stage progression.