AlphaFold Prediction of Protein–Protein Interactions in the Flaviviridae Proteomes
Abstract
1. Introduction
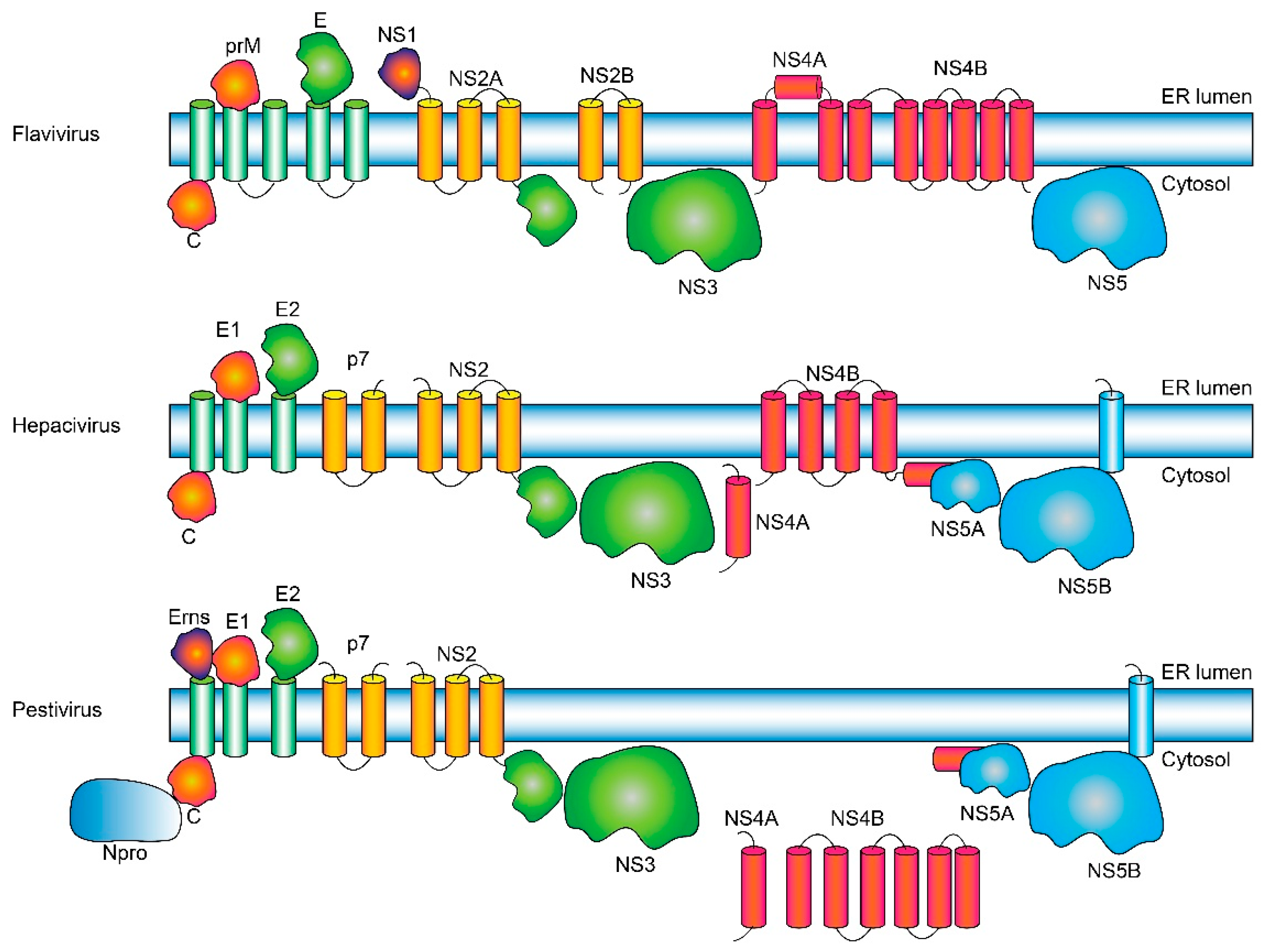
2. Results
2.1. Flavivirus Genus
2.1.1. Polyproteins in Flaviviruses
prM and E
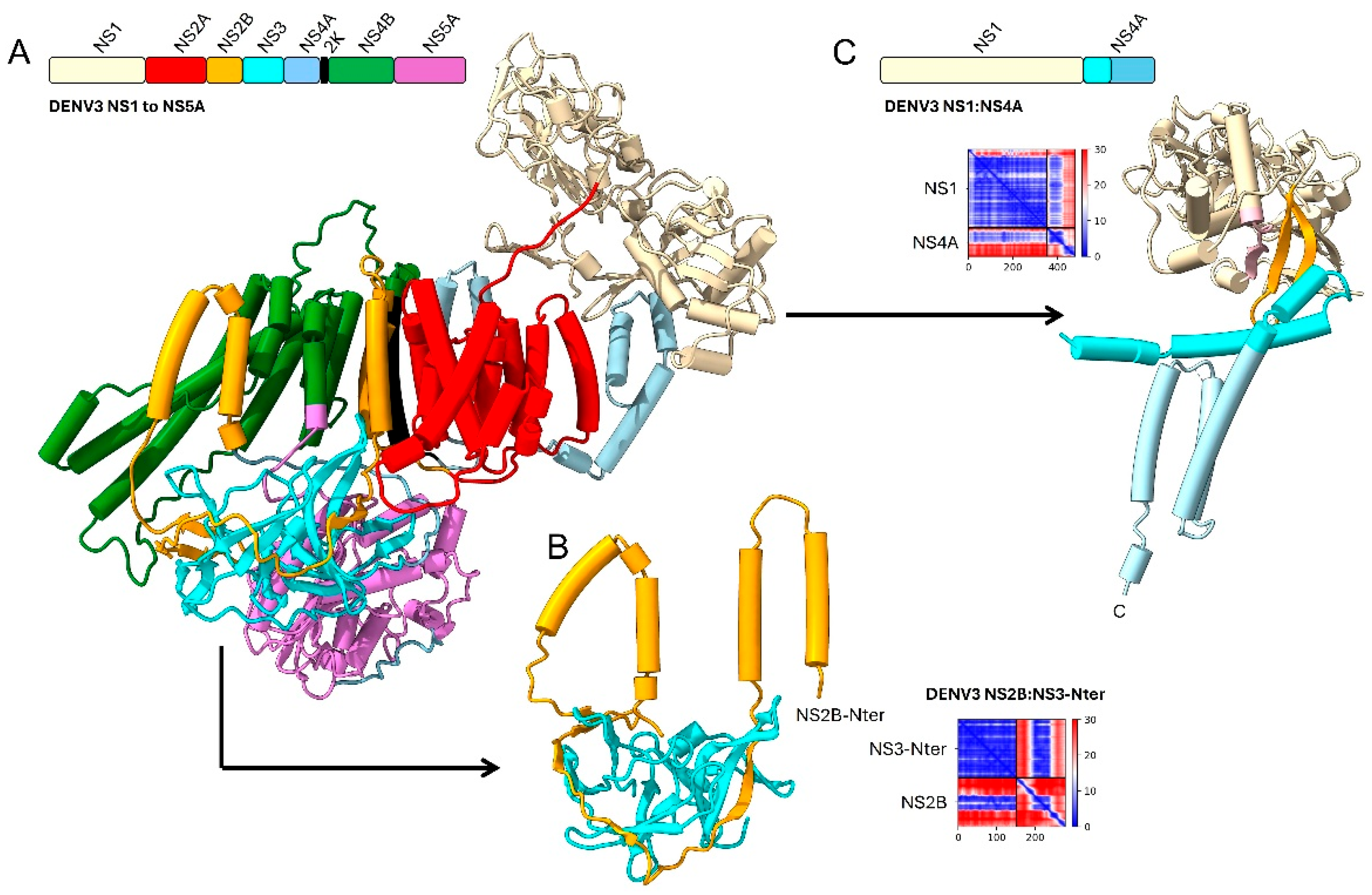
NS1 and NS4A
NS2B and NS4
NS3/NS5 and NS4A
NS4A and NS4B
2.1.2. Separated Proteins in Flaviviruses
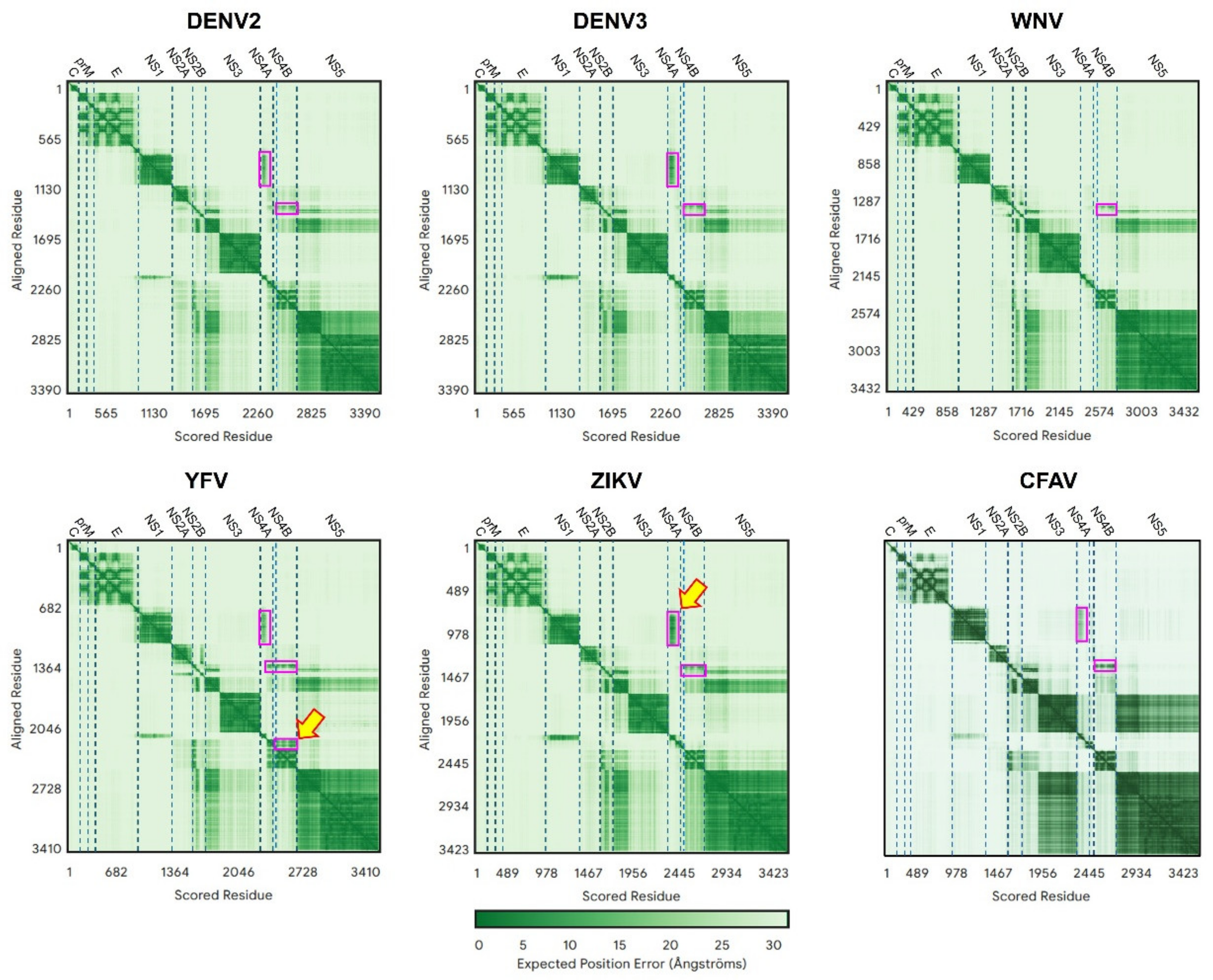
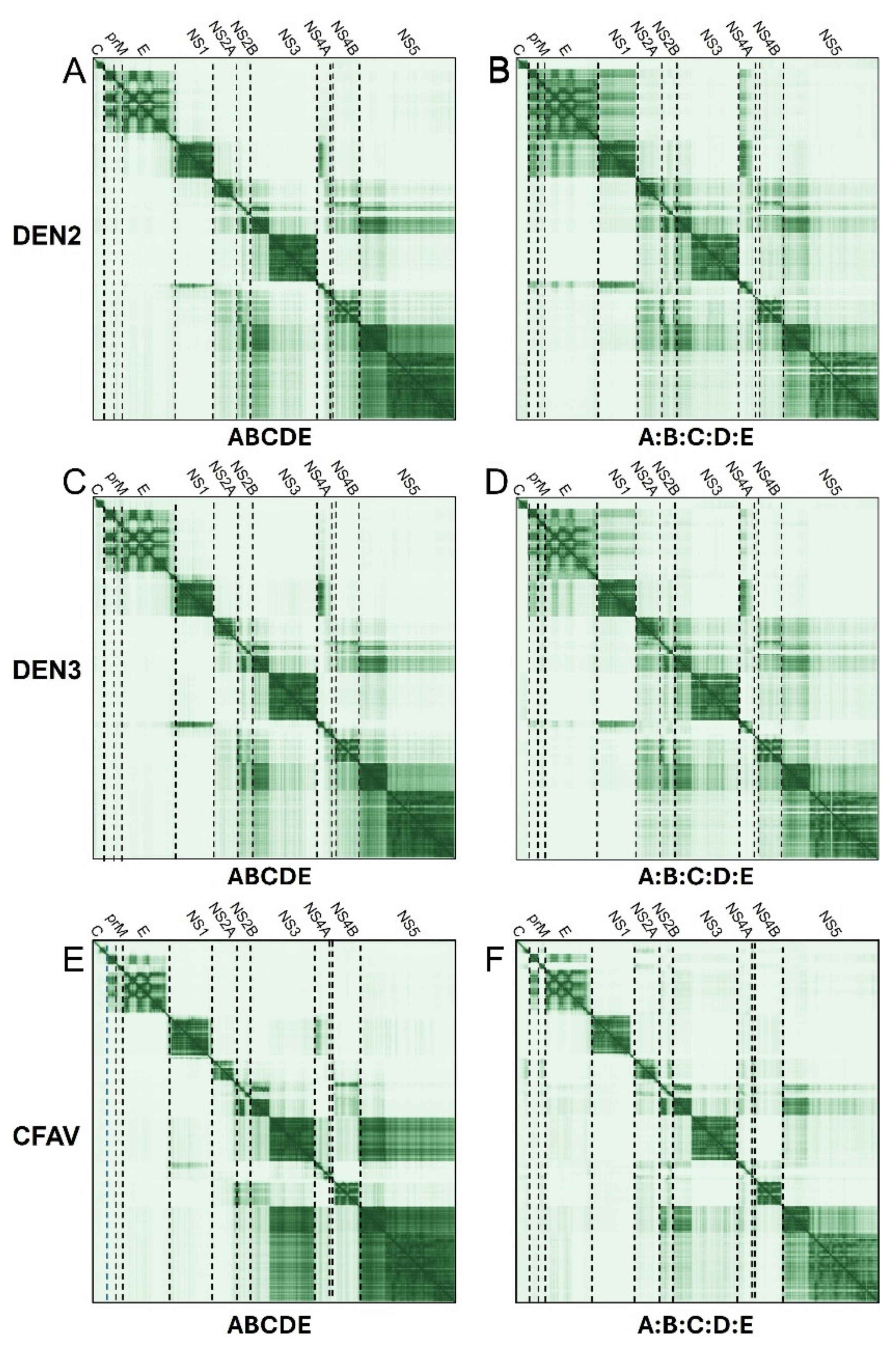
2.1.3. Detailed Interactions
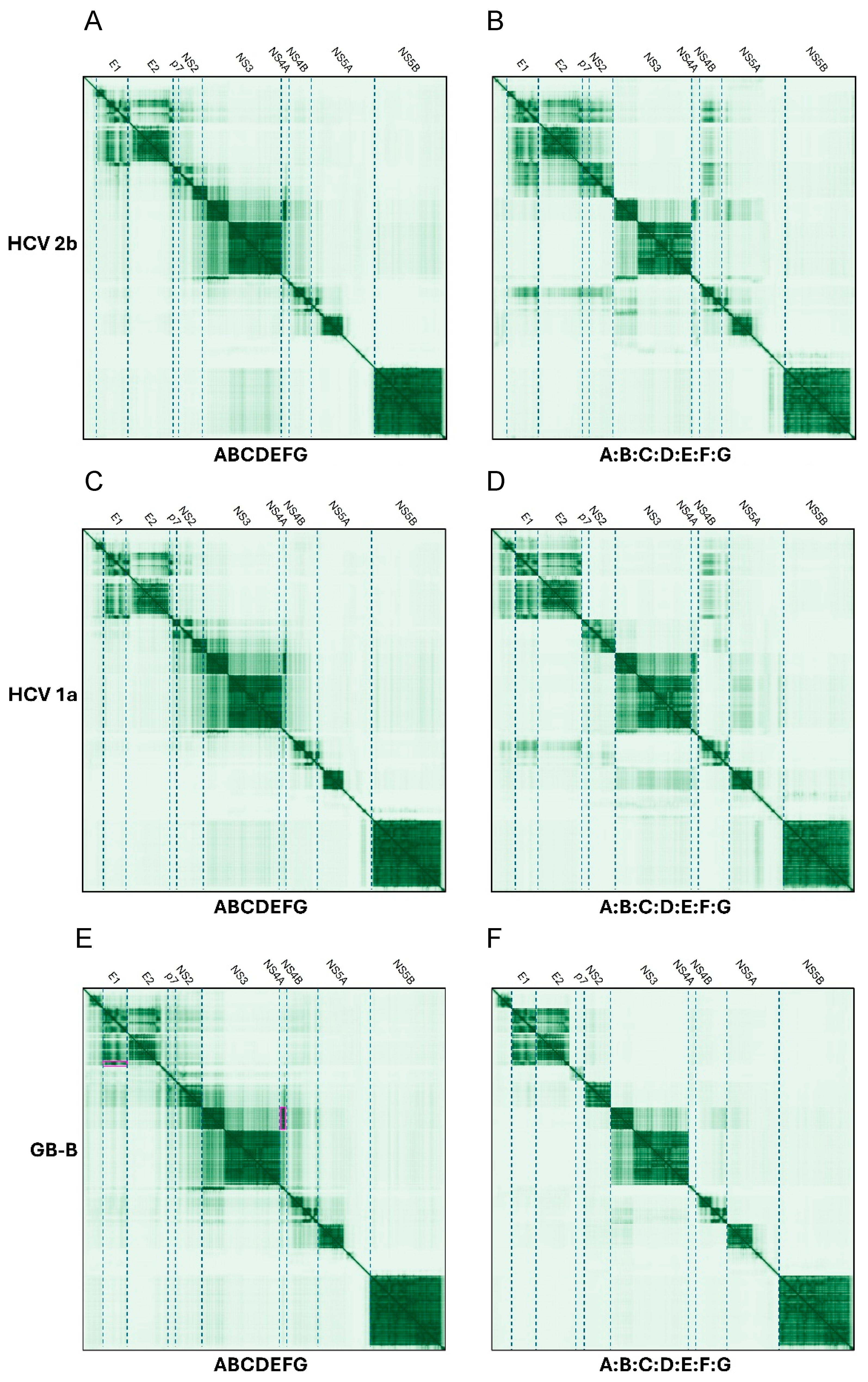
2.2. Hepacivirus Genus
2.2.1. Polyproteins and Separate Proteins in Hepaciviruses
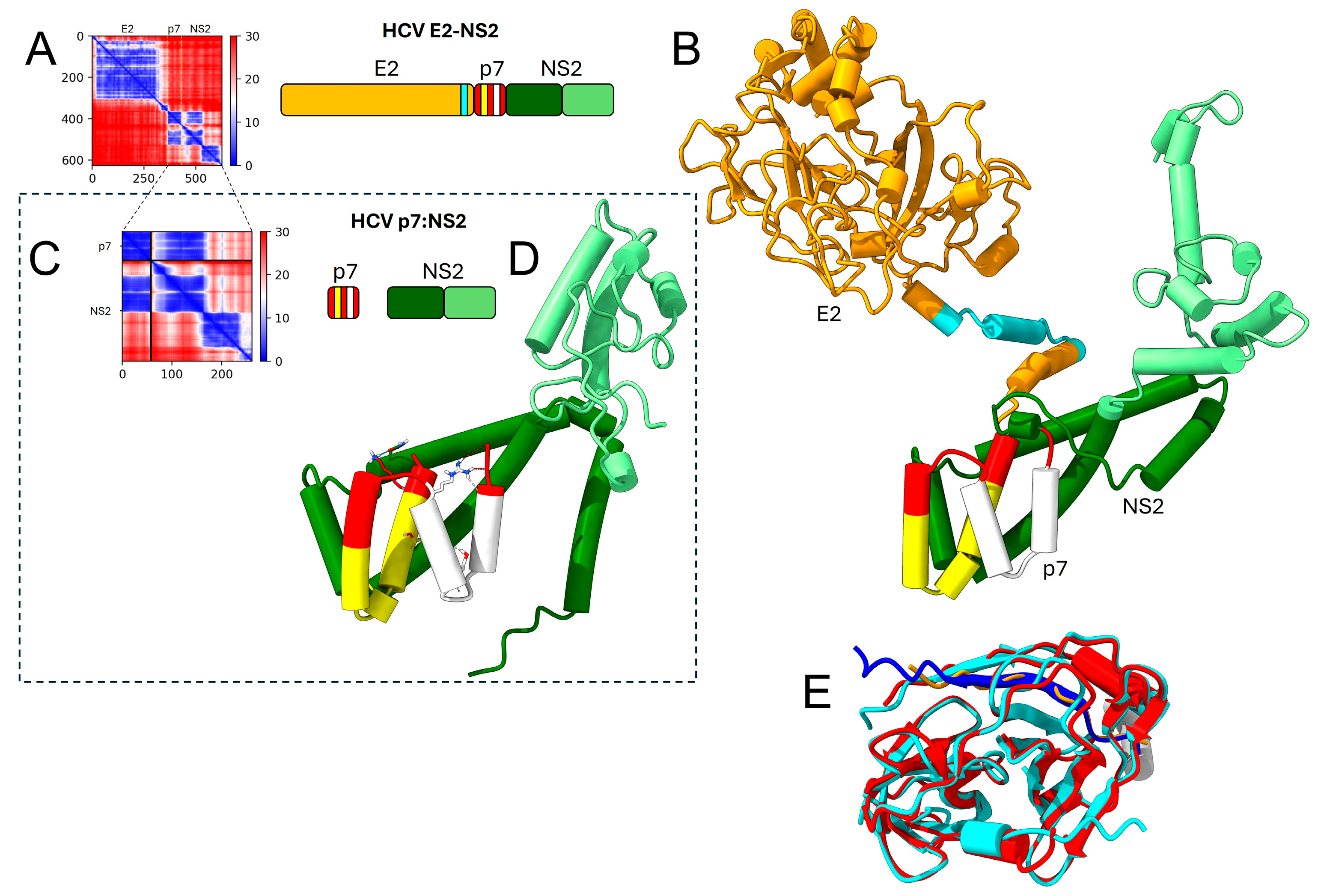
E1 and E2, p7, NS2, NS4A and NS4B
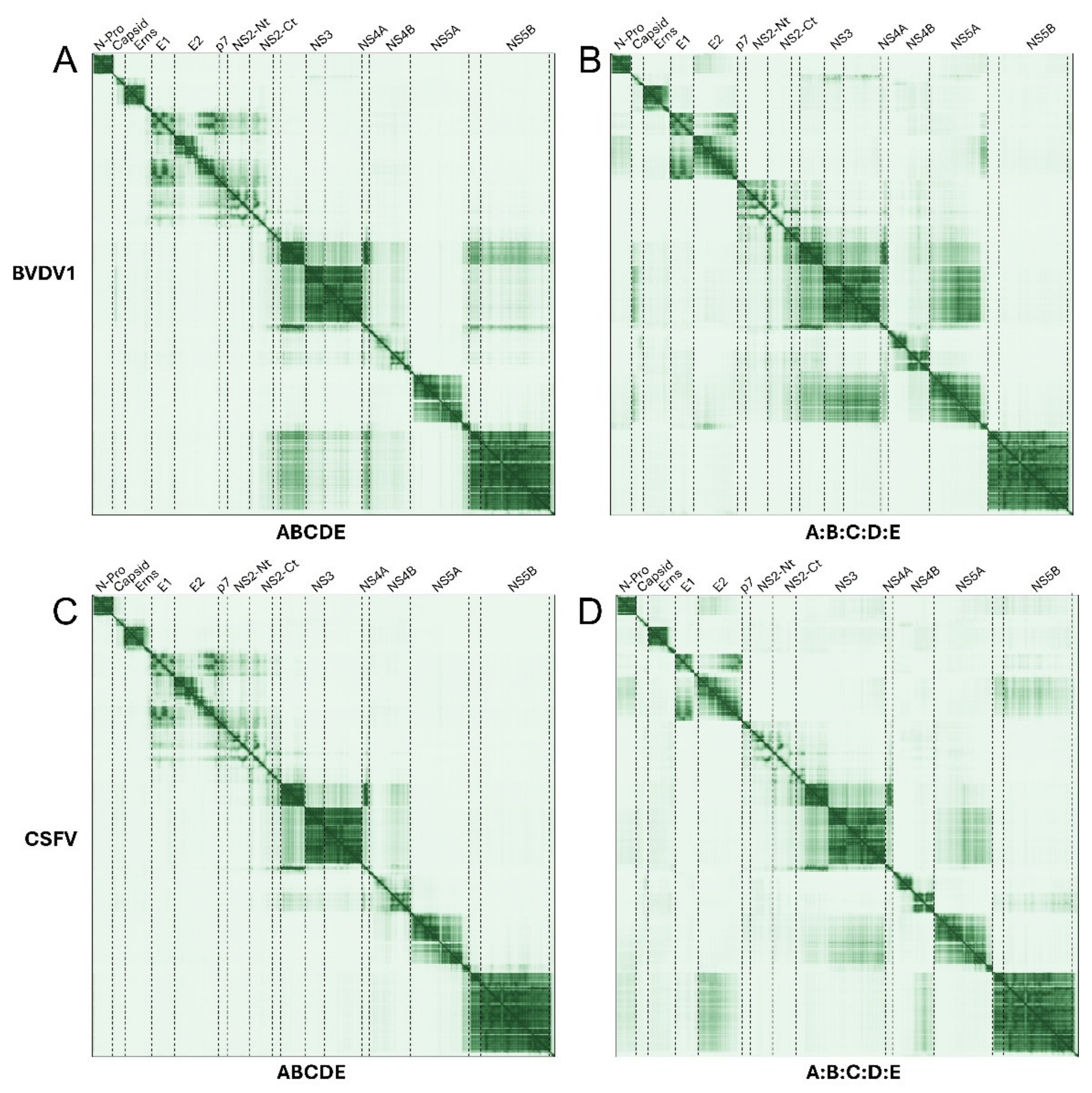
2.3. Pestivirus Genus
2.3.1. Polyprotein and Separate Proteins in Pestiviruses
NS4A and NS3
E1, E2, p7 and NS2
NS5 and NS3
C and NS3
2.3.2. TM Predictions
3. Discussion
4. Materials and Methods
4.1. Sequences Used in AF Structure Prediction
4.2. AF3 Server
4.3. ColabFold Notebook
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miller, S.; Sparacio, S.; Bartenschlager, R. Subcellular localization and membrane topology of the Dengue virus type 2 Non-structural protein 4B. J. Biol. Chem. 2006, 281, 8854–8863. [Google Scholar] [CrossRef]
- Nemesio, H.; Palomares-Jerez, F.; Villalain, J. NS4A and NS4B proteins from dengue virus: Membranotropic regions. Biochim. Biophys. Acta 2012, 1818, 2818–2830. [Google Scholar] [CrossRef]
- Zou, J.; Xie, X.; Lee, L.T.; Chandrasekaran, R.; Reynaud, A.; Yap, L.; Wang, Q.Y.; Dong, H.; Kang, C.; Yuan, Z.; et al. Dimerization of flavivirus NS4B protein. J. Virol. 2014, 88, 3379–3391. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wong, Y.L.; Lee, M.Y.; Li, Q.; Wang, Q.Y.; Lescar, J.; Shi, P.Y.; Kang, C. Secondary structure and membrane topology of the full-length dengue virus NS4B in micelles. Angew. Chem. 2016, 128, 12247–12251. [Google Scholar] [CrossRef]
- Porter, S.S.; Gilchrist, T.M.; Schrodel, S.; Tai, A.W. Dengue and Zika virus NS4B proteins differ in topology and in determinants of ER membrane protein complex dependency. J. Virol. 2025, 99, e0144324. [Google Scholar] [CrossRef]
- Kuhn, R.J.; Zhang, W.; Rossmann, M.G.; Pletnev, S.V.; Corver, J.; Lenches, E.; Jones, C.T.; Mukhopadhyay, S.; Chipman, P.R.; Strauss, E.G.; et al. Structure of dengue virus: Implications for flavivirus organization, maturation, and fusion. Cell 2002, 108, 717–725. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Kim, B.S.; Chipman, P.R.; Rossmann, M.G.; Kuhn, R.J. Structure of West Nile virus. Science 2003, 302, 248. [Google Scholar] [CrossRef]
- Zhang, Y.; Kaufmann, B.; Chipman, P.R.; Kuhn, R.J.; Rossmann, M.G. Structure of immature West Nile virus. J. Virol. 2007, 81, 6141–6145. [Google Scholar] [CrossRef]
- Gastaminza, P.; Kapadia, S.B.; Chisari, F.V. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J. Virol. 2006, 80, 11074–11081. [Google Scholar] [CrossRef]
- Harada, T.; Tautz, N.; Thiel, H.J. E2-p7 region of the bovine viral diarrhea virus polyprotein: Processing and functional studies. J. Virol. 2000, 74, 9498–9506. [Google Scholar] [CrossRef]
- Elbers, K.; Tautz, N.; Becher, P.; Stoll, D.; Rümenapf, T.; Thiel, H.J. Processing in the pestivirus E2-NS2 region: Identification of proteins p7 and E2p7. J. Virol. 1996, 70, 4131–4135. [Google Scholar] [CrossRef] [PubMed]
- Carrère-Kremer, S.; Montpellier-Pala, C.; Cocquerel, L.; Wychowski, C.; Penin, F.; Dubuisson, J. Subcellular localization and topology of the p7 polypeptide of hepatitis C virus. J. Virol. 2002, 76, 3720–3730. [Google Scholar] [CrossRef]
- Murray, C.L.; Jones, C.T.; Rice, C.M. Architects of assembly: Roles of Flaviviridae non-structural proteins in virion morphogenesis. Nat. Rev. Microbiol. 2008, 6, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Griffin, S.D.C.; Beales, L.P.; Clarke, D.S.; Worsfold, O.; Evans, S.D.; Jaeger, J.; Harris, M.P.G.; Rowlands, D.J.; Klenk, H.D. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, Amantadine. FEBS Lett. 2003, 535, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.; Swinscoe, G.; Lefteri, D.A.; Singh, R.; Moran, A.; Thompson, R.F.; Maskell, D.; Beaumont, H.; Bentham, M.J.; Donald, C.; et al. Inhibitors of the small membrane (M) protein viroporin prevent Zika virus infection. eLife 2024, 13, e68404. [Google Scholar] [CrossRef]
- Westaway, E.G.; Mackenzie, J.M.; Kenney, M.T.; Jones, M.K.; Khromykh, A.A. Ultrastructure of Kunjin virus-infected cells: Colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 1997, 71, 6650–6661. [Google Scholar] [CrossRef]
- Mottola, G.; Cardinali, G.; Ceccacci, A.; Trozzi, C.; Bartholomew, L.; Torrisi, M.R.; Pedrazzini, E.; Bonatti, S.; Migliaccio, G. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology 2002, 293, 31–43. [Google Scholar] [CrossRef]
- Xie, X.; Zou, J.; Zhang, X.; Zhou, Y.; Routh, A.L.; Kang, C.; Popov, V.L.; Chen, X.; Wang, Q.Y.; Dong, H.; et al. Dengue NS2A Protein Orchestrates Virus Assembly. Cell Host Microbe 2019, 26, 606–622.e8. [Google Scholar] [CrossRef]
- Klema, V.J.; Padmanabhan, R.; Choi, K.H. Flaviviral replication complex: Coordination between RNA synthesis and 5′-RNA capping. Viruses 2015, 7, 4640–4656. [Google Scholar] [CrossRef]
- Van den Elsen, K.; Chew, B.L.A.; Ho, J.S.; Luo, D. Flavivirus nonstructural proteins and replication complexes as antiviral drug targets. Curr. Opin. Virol. 2023, 59, 101305. [Google Scholar] [CrossRef]
- Welsch, S.; Miller, S.; Romero-Brey, I.; Merz, A.; Bleck, C.K.E.; Walther, P.; Fuller, S.D.; Antony, C.; Krijnse-Locker, J.; Bartenschlager, R. Composition and Three-Dimensional Architecture of the Dengue Virus Replication and Assembly Sites. Cell Host Microbe 2009, 5, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Miorin, L.; Romero-Brey, I.; Maiuri, P.; Hoppe, S.; Krijnse-Locker, J.; Bartenschlager, R.; Marcello, A. Three-dimensional architecture of tick-borne encephalitis virus replication sites and trafficking of the replicated RNA. J. Virol. 2013, 87, 6469–6481. [Google Scholar] [CrossRef] [PubMed]
- Romero-Brey, I.; Bartenschlager, R. Endoplasmic reticulum: The favorite intracellular niche for viral replication and assembly. Viruses 2016, 8, 160. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.; Krijnse-Locker, J. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 2008, 6, 363–374. [Google Scholar] [CrossRef]
- Miller, S.; Kastner, S.; Krijnse-Locker, J.; Bühler, S.; Bartenschlager, R. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 2007, 282, 8873–8882. [Google Scholar] [CrossRef]
- Kuno, G.; Chang, G.J.J. Full-length sequencing and genomic characterization of Bagaza, Kedougou, and Zika viruses. Arch. Virol. 2007, 152, 687–696. [Google Scholar] [CrossRef]
- Lindenbach, B.D.; Thiel, H.J.; Rice, C.M. Flaviviridae: The viruses and their replication. In Fields Virology, 5th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott-Raven Publishers: Philadelphia, PA, USA, 2007; Volume 1, pp. 1101–1152. [Google Scholar]
- Reed, K.E.; Gorbalenya, A.E.; Rice, C.M. The NS5A/NS5 proteins of viruses from three genera of the family Flaviviridae are phosphorylated by associated serine/threonine kinases. J. Virol. 1998, 72, 6199–6206. [Google Scholar] [CrossRef]
- Thai, L.Y.; Xu, T.; Chen, Y.L.; Malet, H.; Egloff, M.P.; Canard, B.; Vasudevan, S.G.; Lescar, J. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J. Virol. 2007, 81, 4753–4765. [Google Scholar] [CrossRef]
- Gould, E.; Solomon, T. Pathogenic flaviviruses. Lancet 2008, 371, 500–509. [Google Scholar] [CrossRef]
- Kuno, G.; Chang, G.J.J.; Tsuchiya, K.R.; Karabatsos, N.; Cropp, C.B. Phylogeny of the genus Flavivirus. J. Virol. 1998, 72, 73–83. [Google Scholar] [CrossRef]
- Pierson, T.C.; Diamond, M.S. The continued threat of emerging flaviviruses. Nat. Microbiol. 2020, 5, 796–812. [Google Scholar] [CrossRef]
- Tautz, N.; Tews, B.A.; Meyers, G. The Molecular Biology of Pestiviruses. Adv. Virus Res. 2015, 93, 47–160. [Google Scholar] [PubMed]
- Becher, P.; Moennig, V.; Tautz, N. Bovine Viral Diarrhea, Border Disease, and Classical Swine Fever Viruses (Flaviviridae). In Encyclopedia of Virology, 4th ed.; Academic Press: New York, NY, USA, 2020; Volume 1–5, pp. 153–164. [Google Scholar]
- Becher, P.; Ramirez, R.A.; Orlich, M.; Rosales, S.C.; König, M.; Schweizer, M.; Stalder, H.; Schirrmeier, H.; Thiel, H.J. Genetic and antigenic characterization of novel pestivirus genotypes: Implications for classification. Virology 2003, 311, 96–104. [Google Scholar] [CrossRef] [PubMed]
- King, A.M.; Lefkowitz, E.J.; Mushegian, A.R.; Adams, M.J.; Dutilh, B.E.; Gorbalenya, A.E.; Harrach, B.; Harrison, R.L.; Junglen, S.; Knowles, N.J. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2018). Arch. Virol. 2018, 163, 2601–2631. [Google Scholar] [CrossRef] [PubMed]
- Lamp, B.; Riedel, C.; Roman-Sosa, G.; Heimann, M.; Jacobi, S.; Becher, P.; Thiel, H.J.; Rümenapf, T. Biosynthesis of classical swine fever virus nonstructural proteins. J. Virol. 2011, 85, 3607–3620. [Google Scholar] [CrossRef]
- Weiland, F.; Weiland, E.; Unger, G.; Saalmüller, A.; Thiel, H.J. Localization of pestiviral envelope proteins E(rns) and E2 at the cell surface and on isolated particles. J. Gen. Virol. 1999, 80, 1157–1165. [Google Scholar] [CrossRef]
- Risager, P.C.; Fahnøe, U.; Gullberg, M.; Rasmussen, T.B.; Belsham, G.J. Analysis of classical swine fever virus RNA replication determinants using replicons. J. Gen. Virol. 2013, 94 Pt 8, 1739–1748. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Op De Beeck, A.; Molenkamp, R.; Caron, M.; Ben Younes, A.; Bredenbeek, P.; Dubuisson, J. Role of the transmembrane domains of prM and E proteins in the formation of yellow fever virus envelope. J. Virol. 2003, 77, 813–820. [Google Scholar] [CrossRef]
- Majowicz, S.A.; Narayanan, A.; Moustafa, I.M.; Bator, C.M.; Hafenstein, S.L.; Jose, J. Zika virus M protein latches and locks the E protein from transitioning to an immature state after prM cleavage. NPJ Viruses 2023, 1, 4. [Google Scholar] [CrossRef]
- Kapoor, M.; Zhang, L.; Ramachandra, M.; Kusukawa, J.; Ebner, K.E.; Padmanabhan, R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J. Biol. Chem. 1995, 270, 19100–19106. [Google Scholar] [CrossRef]
- Le Breton, M.; Meyniel-Schicklin, L.; Deloire, A.; Coutard, B.; Canard, B.; de Lamballerie, X.; Andre, P.; Rabourdin-Combe, C.; Lotteau, V.; Davoust, N. Flavivirus NS3 and NS5 proteins interaction network: A high-throughput yeast two-hybrid screen. BMC Microbiol. 2011, 11, 234. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.L.; Morgenstern, K.A.; Lin, C.; Fox, T.; Dwyer, M.D.; Landro, J.A.; Chambers, S.P.; Markland, W.; Lepre, C.A.; O’Malley, E.T.; et al. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 1996, 87, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Li, Y.; Munshi, S.; Sardana, V.; Cole, J.L.; Sardana, M.; Kuo, L.C.; Chen, Z.; Steinkuehler, C.; Tomei, L.; et al. Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: A 2.2 Å resolution structure in a hexagonal crystal form. Protein Sci. 1998, 7, 837–847. [Google Scholar] [CrossRef] [PubMed]
- Träger, T.K.; Tüting, C.; Kastritis, P.L. The human touch: Utilizing AlphaFold 3 to analyze structures of endogenous metabolons. Structure 2024, 32, 1555–1562. [Google Scholar] [CrossRef]
- Lindenbach, B.D.; Rice, C.M. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J. Virol. 1999, 73, 4611–4621. [Google Scholar] [CrossRef]
- Li, Y.; Lee, M.Y.; Loh, Y.R.; Kang, C. Secondary structure and membrane topology of dengue virus NS4A protein in micelles. Biochim. Biophys. Acta Biomembr. 2018, 1860, 442–450. [Google Scholar] [CrossRef]
- Płaszczyca, A.; Scaturro, P.; Neufeldt, C.J.; Cortese, M.; Cerikan, B.; Ferla, S.; Brancale, A.; Pichlmair, A.; Bartenschlager, R. A novel interaction between dengue virus nonstructural protein 1 and the NS4A-2K-4B precursor is required for viral RNA replication but not for formation of the membranous replication organelle. PLoS Path. 2019, 15, e1007736. [Google Scholar] [CrossRef]
- Lin, C.; Amberg, S.M.; Chambers, T.J.; Rice, C.M. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. J. Virol. 1993, 67, 2327–2335. [Google Scholar] [CrossRef]
- Wicker, J.A.; Whiteman, M.C.; Beasley, D.W.; Davis, C.T.; McGee, C.E.; Lee, J.C.; Higgs, S.; Kinney, R.M.; Huang, C.Y.; Barrett, A.D. Mutational analysis of the West Nile virus NS4B protein. Virology 2012, 426, 22–33. [Google Scholar] [CrossRef]
- Umareddy, I.; Chao, A.; Sampath, A.; Gu, F.; Vasudevan, S.G. Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. J. Gen. Virol. 2006, 87 Pt 9, 2605–2614. [Google Scholar] [CrossRef]
- Xie, X.; Zou, J.; Wang, Q.Y.; Shi, P.Y. Targeting dengue virus NS4B protein for drug discovery. Antivir. Res. 2015, 118, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Roosendaal, J.; Westaway, E.G.; Khromykh, A.; Mackenzie, J.M. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and golgi trafficking of the NS4A protein. J. Virol. 2006, 80, 4623–4632. [Google Scholar] [CrossRef] [PubMed]
- Kiemel, D.; Kroell, A.H.; Denolly, S.; Haselmann, U.; Bonfanti, J.F.; Andres, J.I.; Ghosh, B.; Geluykens, P.; Kaptein, S.J.F.; Wilken, L.; et al. Pan-serotype dengue virus inhibitor JNJ-A07 targets NS4A-2K-NS4B interaction with NS2B/NS3 and blocks replication organelle formation. Nat. Commun. 2024, 15, 6080. [Google Scholar] [CrossRef] [PubMed]
- Youn, S.; Li, T.; McCune, B.T.; Edeling, M.A.; Fremont, D.H.; Cristea, I.M.; Diamond, M.S. Evidence for a genetic and physical interaction between nonstructural proteins NS1 and NS4B that modulates replication of west nile virus. J. Virol. 2012, 86, 7360–7371. [Google Scholar] [CrossRef]
- Yu, L.; Takeda, K.; Markoff, L. Protein-protein interactions among West Nile non-structural proteins and transmembrane complex formation in mammalian cells. Virology 2013, 446, 365–377. [Google Scholar] [CrossRef]
- Li, X.D.; Deng, C.L.; Ye, H.Q.; Zhang, H.L.; Zhang, Q.Y.; Chen, D.D.; Zhang, P.T.; Shi, P.Y.; Yuan, Z.M.; Zhang, B. Transmembrane domains of NS2B contribute to both viral RNA replication and particle formation in Japanese encephalitis virus. J. Virol. 2016, 90, 5735–5749. [Google Scholar] [CrossRef]
- Wang, Y.; Xie, X.; Shi, P.Y. Flavivirus NS4B protein: Structure, function, and antiviral discovery. Antivir. Res. 2022, 207, 105423. [Google Scholar] [CrossRef]
- Li, Q.; Kang, C. Dengue virus NS4B protein as a target for developing antivirals. Front. Cell. Infect. Microbiol. 2022, 12, 959727. [Google Scholar] [CrossRef]
- Muñoz-Jordán, J.L.; Sánchez-Burgos, G.G.; Laurent-Rolle, M.; García-Sastre, A. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 2003, 100 (Suppl. 2), 14333–14338. [Google Scholar] [CrossRef]
- Muñoz-Jordán, J.L.; Laurent-Rolle, M.; Ashour, J.; Martínez-Sobrido, L.; Ashok, M.; Lipkin, W.I.; García-Sastre, A. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 2005, 79, 8004–8013. [Google Scholar] [CrossRef] [PubMed]
- Naik, N.G.; Wu, H.N. Mutation of putative N-glycosylation sites on dengue virus NS4B decreases RNA replication. J. Virol. 2015, 89, 6746–6760. [Google Scholar] [CrossRef] [PubMed]
- Gopala Reddy, S.B.; Chin, W.X.; Shivananju, N.S. Dengue virus NS2 and NS4: Minor proteins, mammoth roles. Biochem. Pharmacol. 2018, 154, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Liu, Q.; Zhou, J.; Xie, W.; Chen, C.; Wang, Z.; Yang, H.; Cui, J. Zika virus evades interferon-mediated antiviral response through the co-operation of multiple nonstructural proteins in vitro. Cell Discov. 2017, 3, 17006, Erratum in Cell Discov. 2020, 11, 17014. [Google Scholar] [CrossRef]
- Zou, J.; Lee, L.T.; Wang, Q.Y.; Xie, X.; Lu, S.; Yau, Y.H.; Yuan, Z.; Shochat, S.G.; Kang, C.; Lescar, J.; et al. Mapping the interactions between the NS4B and NS3 proteins of dengue virus. J. Virol. 2015, 89, 3471–3483. [Google Scholar] [CrossRef]
- Lu, H.; Zhan, Y.; Li, X.; Bai, X.; Yuan, F.; Ma, L.; Wang, X.; Xie, M.; Wu, W.; Chen, Z. Novel insights into the function of an N-terminal region of DENV2 NS4B for the optimal helicase activity of NS3. Virus Res. 2021, 295, 198318. [Google Scholar] [CrossRef]
- Chatel-Chaix, L.; Fischl, W.; Scaturro, P.; Cortese, M.; Kallis, S.; Bartenschlager, M.; Fischer, B.; Bartenschlager, R. A Combined genetic-proteomic approach identifies residues within dengue virus NS4B critical for interaction with NS3 and viral replication. J. Virol. 2015, 89, 7170–7186. [Google Scholar] [CrossRef]
- Zou, J.; Xie, X.; Wang, Q.Y.; Dong, H.; Lee, M.Y.; Kang, C.; Yuan, Z.; Shi, P.Y. Characterization of dengue virus NS4A and NS4B protein interaction. J. Virol. 2015, 89, 3455–3470. [Google Scholar] [CrossRef]
- Tajima, S.; Takasaki, T.; Kurane, I. Restoration of replication-defective dengue type 1 virus bearing mutations in the N-terminal cytoplasmic portion of NS4A by additional mutations in NS4B. Arch. Virol. 2011, 156, 63–69. [Google Scholar] [CrossRef]
- Li, X.D.; Ye, H.Q.; Deng, C.L.; Liu, S.Q.; Zhang, H.L.; Shang, B.D.; Shi, P.Y.; Yuan, Z.M.; Zhang, B. Genetic interaction between NS4A and NS4B for replication of Japanese encephalitis virus. J. Gen. Virol. 2015, 96, 1264–1275. [Google Scholar] [CrossRef]
- Surya, W.; Honey, S.S.; Torres, J. Flavivirus Zika NS4A protein forms large oligomers in liposomes and in mild detergent. Sci. Rep. 2024, 14, 12533. [Google Scholar] [CrossRef] [PubMed]
- Amberg, S.M.; Nestorowicz, A.; McCourt, D.W.; Rice, C.M. NS2B-3 proteinase-mediated processing in the yellow fever virus structural region: In vitro and in vivo studies. J. Virol. 1994, 68, 3794–3802. [Google Scholar] [CrossRef] [PubMed]
- Khromykh, A.A.; Sedlak, P.L.; Westaway, E.G. cis- and trans-acting elements in flavivirus RNA replication. J. Virol. 2000, 74, 3253–3263. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, J.M.; Khromykh, A.A.; Jones, M.K.; Westaway, E.G. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 1998, 245, 203–215. [Google Scholar] [CrossRef]
- Kümmerer, B.M.; Rice, C.M. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 2002, 76, 4773–4784. [Google Scholar] [CrossRef]
- Agapov, E.V.; Murray, C.L.; Frolov, I.; Qu, L.; Myers, T.M.; Rice, C.M. Uncleaved NS2-3 Is Required for Production of Infectious Bovine Viral Diarrhea Virus. J. Virol. 2004, 78, 2414–2425. [Google Scholar] [CrossRef]
- Moulin, H.R.; Seuberlich, T.; Bauhofer, O.; Bennett, L.C.; Tratschin, J.D.; Hofmann, M.A.; Ruggli, N. Nonstructural proteins NS2-3 and NS4A of classical swine fever virus: Essential features for infectious particle formation. Virology 2007, 365, 376–389. [Google Scholar] [CrossRef]
- Yu, G.Y.; Lee, K.J.; Gao, L.; Lai, M.M. Palmitoylation and polymerization of hepatitis C virus NS4B protein. J. Virol. 2006, 80, 6013–6023. [Google Scholar] [CrossRef]
- Grakoui, A.; McCourt, D.W.; Wychowski, C.; Feinstone, S.M.; Rice, C.M. A second hepatitis C virus-encoded proteinase. Proc. Natl. Acad. Sci. USA 1993, 90, 10583–10587. [Google Scholar] [CrossRef]
- Jones, C.T.; Murray, C.L.; Eastman, D.K.; Tassello, J.; Rice, C.M. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 2007, 81, 8374–8383. [Google Scholar] [CrossRef]
- Yi, M.; Ma, Y.; Yates, J.; Lemon, S.M. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chirneric hepatitis C virus. J. Virol. 2007, 81, 629–638. [Google Scholar] [CrossRef]
- Ma, Y.; Yates, J.; Liang, Y.; Lemon, S.M.; Yi, M. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J. Virol. 2008, 82, 7624–7639. [Google Scholar] [CrossRef]
- Pietschmann, T.; Kaul, A.; Koutsoudakis, G.; Shavinskaya, A.; Kallis, S.; Steinmann, E.; Abid, K.; Negro, F.; Dreux, M.; Cosset, F.L.; et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA 2006, 103, 7408–7413. [Google Scholar] [CrossRef]
- Kiiver, K.; Merits, A.; Ustav, M.; Žusinaite, E. Complex formation between hepatitis C virus NS2 and NS3 proteins. Virus Res. 2006, 117, 264–272. [Google Scholar] [CrossRef]
- Jirasko, V.; Montserret, R.; Appel, N.; Janvier, A.; Eustachi, L.; Brohm, C.; Steinmann, E.; Pietschmann, T.; Penin, F.; Bartenschlagerd, R. Structural and functional characterization of nonstructural protein 2 for its role in hepatitis C virus assembly. J. Biol. Chem. 2008, 283, 28546–28562. [Google Scholar] [CrossRef] [PubMed]
- Jirasko, V.; Montserret, R.; Lee, J.Y.; Gouttenoire, J.; Moradpour, D.; Penin, F.; Bartenschlager, R. Structural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of virion assembly. PLoS Path. 2010, 6, e1001233. [Google Scholar] [CrossRef] [PubMed]
- Hagen, N.; Bayers, K.; Rösch, K.; Schindler, M. The intraviral protein interaction network of Hepatitis C virus. Mol. Cell. Proteom. 2014, 13, 1676–1689. [Google Scholar] [CrossRef] [PubMed]
- Boson, B.; Granio, O.; Bartenschlager, R.; CossetFranç, F.L. A concerted action of hepatitis C virus P7 and nonstructural protein 2 regulates core localization at the endoplasmic reticulum and virus assembly. PLoS Path. 2011, 7, e1002144. [Google Scholar] [CrossRef]
- Guo, H.C.; Sun, S.Q.; Sun, D.H.; Wei, Y.Q.; Xu, J.; Huang, M.; Liu, X.T.; Liu, Z.X.; Luo, J.X.; Yin, H.; et al. Viroporin activity and membrane topology of classic swine fever virus p7 protein. Int. J. Biochem. Cell Biol. 2013, 45, 1186–1194. [Google Scholar] [CrossRef]
- Gladue, D.P.; Holinka, L.G.; Largo, E.; Sainz, I.F.; Carrillo, C.; O’Donnell, V.; Baker-Branstetter, R.; Lu, Z.; Ambroggio, X.; Risatti, G.R.; et al. Classical swine fever virus p7 protein is a viroporin involved in virulence in swine. J. Virol. 2012, 86, 6778–6791. [Google Scholar] [CrossRef]
- Steinmann, E.; Penin, F.; Kallis, S.; Patel, A.H.; Bartenschlager, R.; Pietschmann, T. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Path. 2007, 3, e103. [Google Scholar] [CrossRef] [PubMed]
- Atoom, A.M.; Jones, D.M.; Russell, R.S. Evidence suggesting that HCV p7 protects E2 glycoprotein from premature degradation during virus production. Virus Res. 2013, 176, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Atoom, A.M.; Taylor, N.G.A.; Russell, R.S. The elusive function of the hepatitis C virus p7 protein. Virology 2014, 462–463, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Bentham, M.J.; Foster, T.L.; McCormick, C.; Griffin, S. Mutations in hepatitis C virus p7 reduce both the egress and infectivity of assembled particles via impaired proton channel function. J. Gen. Virol. 2013, 94 Pt 10, 2236–2248. [Google Scholar] [CrossRef]
- Vieyres, G.; Brohm, C.; Friesland, M.; Gentzsch, J.; Wölk, B.; Roingeard, P.; Steinmann, E.; Pietschmann, T. Subcellular localization and function of an epitope-tagged p7 viroporin in hepatitis C virus-producing cells. J. Virol. 2013, 87, 1664–1678. [Google Scholar] [CrossRef]
- Lin, C.; Lindenbach, B.D.; Prágai, B.M.; McCourt, D.W.; Rice, C.M. Processing in the hepatitis C virus E2-NS2 region: Identification of p7 and two distinct E2-specific products with different C termini. J. Virol. 1994, 68, 5063–5073. [Google Scholar] [CrossRef]
- Mizushima, H.; Hijikata, M.; Asabe, S.I.; Hirota, M.; Kimura, K.; Shimotohno, K. Two hepatitis C virus glycoprotein E2 products with different C termini. J. Virol. 1994, 68, 6215–6222. [Google Scholar] [CrossRef]
- Shanmugam, S.; Yi, M. Efficiency of E2-p7 processing modulates production of infectious hepatitis C virus. J. Virol. 2013, 87, 11255–11266. [Google Scholar] [CrossRef]
- Carrère-Kremer, S.; Montpellier, C.; Lorenzo, L.; Brulin, B.; Cocquerel, L.; Belouzard, S.; Penin, F.; Dubuisson, J. Regulation of hepatitis C virus polyprotein processing by signal peptidase involves structural determinants at the p7 sequence junctions. J. Biol. Chem. 2004, 279, 41384–41392. [Google Scholar] [CrossRef]
- Zhao, C.; Shen, X.; Wu, R.; Li, L.; Pan, Z. Classical swine fever virus nonstructural protein p7 modulates infectious virus production. Sci. Rep. 2017, 7, 12995. [Google Scholar] [CrossRef]
- Ma, Y.; Anantpadma, M.; Timpe, J.M.; Shanmugam, S.; Singh, S.M.; Lemon, S.M.; Yi, M. Hepatitis C virus NS2 protein serves as a scaffold for virus assembly by interacting with both structural and nonstructural proteins. J. Virol. 2011, 85, 86–97. [Google Scholar] [CrossRef]
- Popescu, C.I.; Callens, N.; Trinel, D.; Roingeard, P.; Moradpour, D.; Descamps, V.; Duverlie, G.; Penin, F.; Héliot, L.; Rouillé, Y.; et al. NS2 protein of hepatitis C virus interacts with structural and non-structural proteins towards virus assembly. PLoS Path. 2011, 7, e1001278. [Google Scholar] [CrossRef] [PubMed]
- Stapleford, K.A.; Lindenbach, B.D. Hepatitis C virus NS2 coordinates virus particle assembly through physical interactions with the E1–E2 glycoprotein and NS3-NS4A enzyme complexes. J. Virol. 2011, 85, 1706–1717. [Google Scholar] [CrossRef] [PubMed]
- Dubrau, D.; Tortorici, M.A.; Rey, F.A.; Tautz, N. A positive-strand RNA virus uses alternative protein-protein interactions within a viral protease/cofactor complex to switch between RNA replication and virion morphogenesis. PLoS Path. 2017, 13, e1006134. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; McShan, A.C. The power and pitfalls of AlphaFold2 for structure prediction beyond rigid globular proteins. Nat. Chem. Biol. 2024, 20, 950–959. [Google Scholar] [CrossRef]
- Elfmann, C.; Stülke, J. PAE viewer: A webserver for the interactive visualization of the predicted aligned error for multimer structure predictions and crosslinks. Nucleic Acids Res. 2023, 51, W404–W410. [Google Scholar] [CrossRef]
- Mirdita, M.; Schutze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Torres, J.; Pervushin, K.; Surya, W. Prediction of conformational states in a coronavirus channel using Alphafold-2 and DeepMSA2: Strengths and limitations. Comput. Struct. Biotechnol. J. 2024, 23, 3730–3740. [Google Scholar] [CrossRef]
- Mariani, V.; Biasini, M.; Barbato, A.; Schwede, T. lDDT: A local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics 2013, 29, 2722–2728. [Google Scholar] [CrossRef]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Zidek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Surya, W.; Yong, C.P.Y.; Tyagi, A.; Bhushan, S.; Torres, J. Anomalous Oligomerization Behavior of E. coli Aquaporin Z in Detergent and in Nanodiscs. Int. J. Mol. Sci. 2023, 24, 8098. [Google Scholar] [CrossRef]
- Sonnhammer, E.L.; von Heijne, G.; Krogh, A. A hidden Markov model for predicting transmembrane helices in protein sequences. Int. Conf. Intell. Syst. Mol. Biol. 1998, 6, 175–182. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Surya, W.; Goh, J.; Ponniah, C.; Torres, J. AlphaFold Prediction of Protein–Protein Interactions in the Flaviviridae Proteomes. Int. J. Mol. Sci. 2025, 26, 10159. https://doi.org/10.3390/ijms262010159
Surya W, Goh J, Ponniah C, Torres J. AlphaFold Prediction of Protein–Protein Interactions in the Flaviviridae Proteomes. International Journal of Molecular Sciences. 2025; 26(20):10159. https://doi.org/10.3390/ijms262010159
Chicago/Turabian StyleSurya, Wahyu, Justin Goh, Caleb Ponniah, and Jaume Torres. 2025. "AlphaFold Prediction of Protein–Protein Interactions in the Flaviviridae Proteomes" International Journal of Molecular Sciences 26, no. 20: 10159. https://doi.org/10.3390/ijms262010159
APA StyleSurya, W., Goh, J., Ponniah, C., & Torres, J. (2025). AlphaFold Prediction of Protein–Protein Interactions in the Flaviviridae Proteomes. International Journal of Molecular Sciences, 26(20), 10159. https://doi.org/10.3390/ijms262010159





