Plasma Protein Biomarkers to Detect Early Gastric Preneoplasia and Cancer: A Prospective Study
Abstract
1. Introduction
2. Results
2.1. Characteristics of the Studied Cohort and Patient Diagnosis
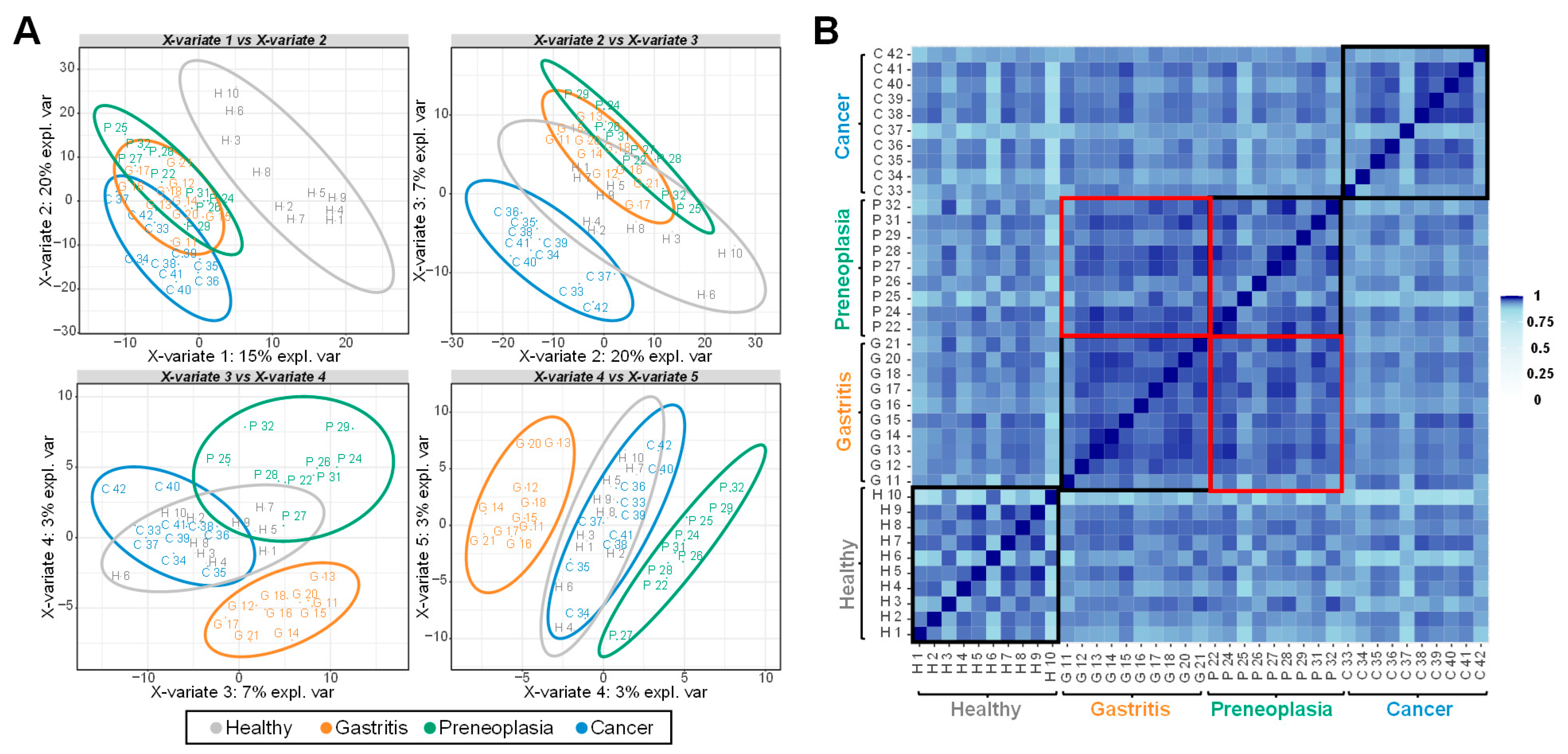
2.2. Multivariate Analysis of the LC-MS/MS-Based Proteomics Data
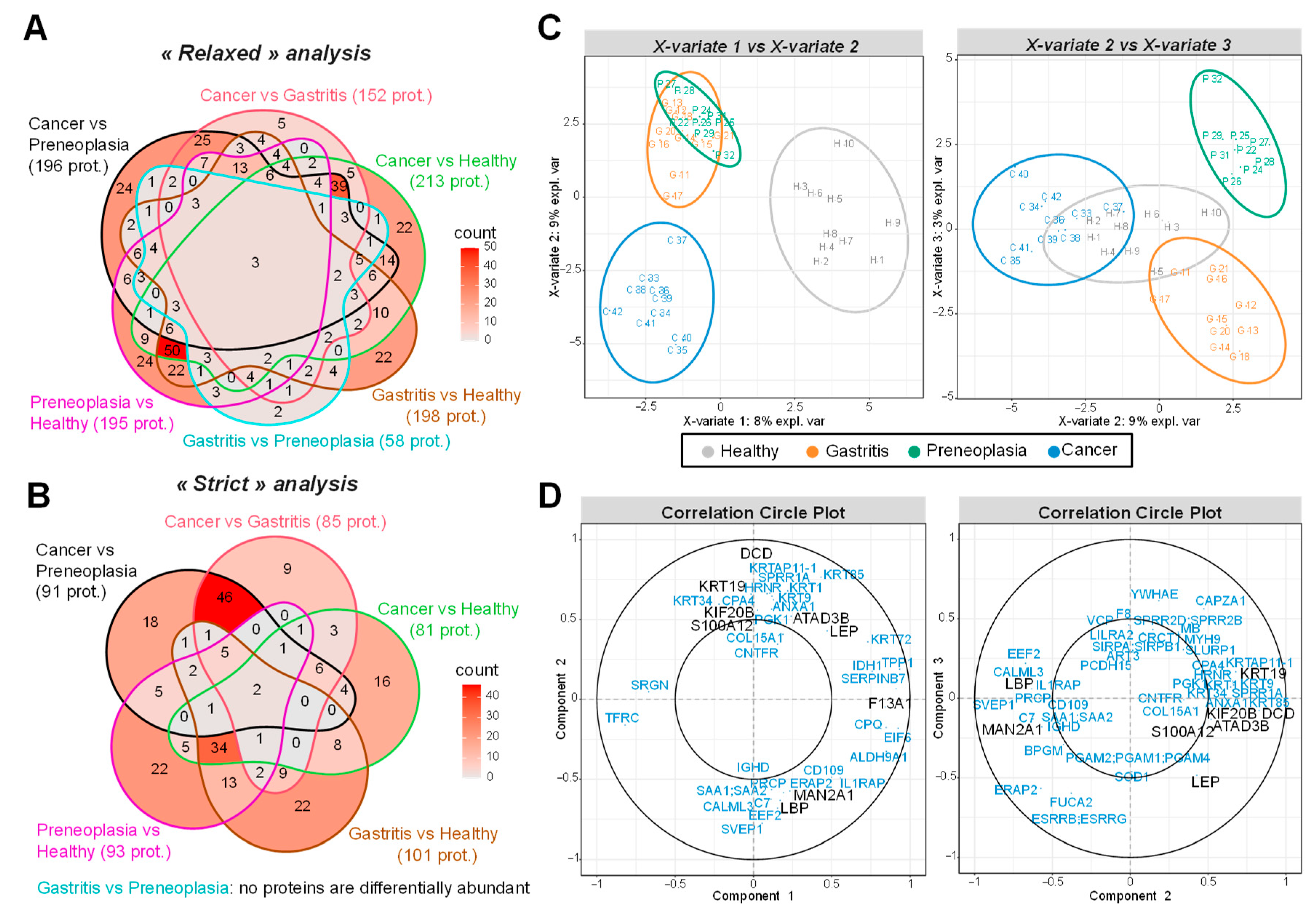
2.3. Biomarker Candidates Identified by LC-MS/MS-Based Data
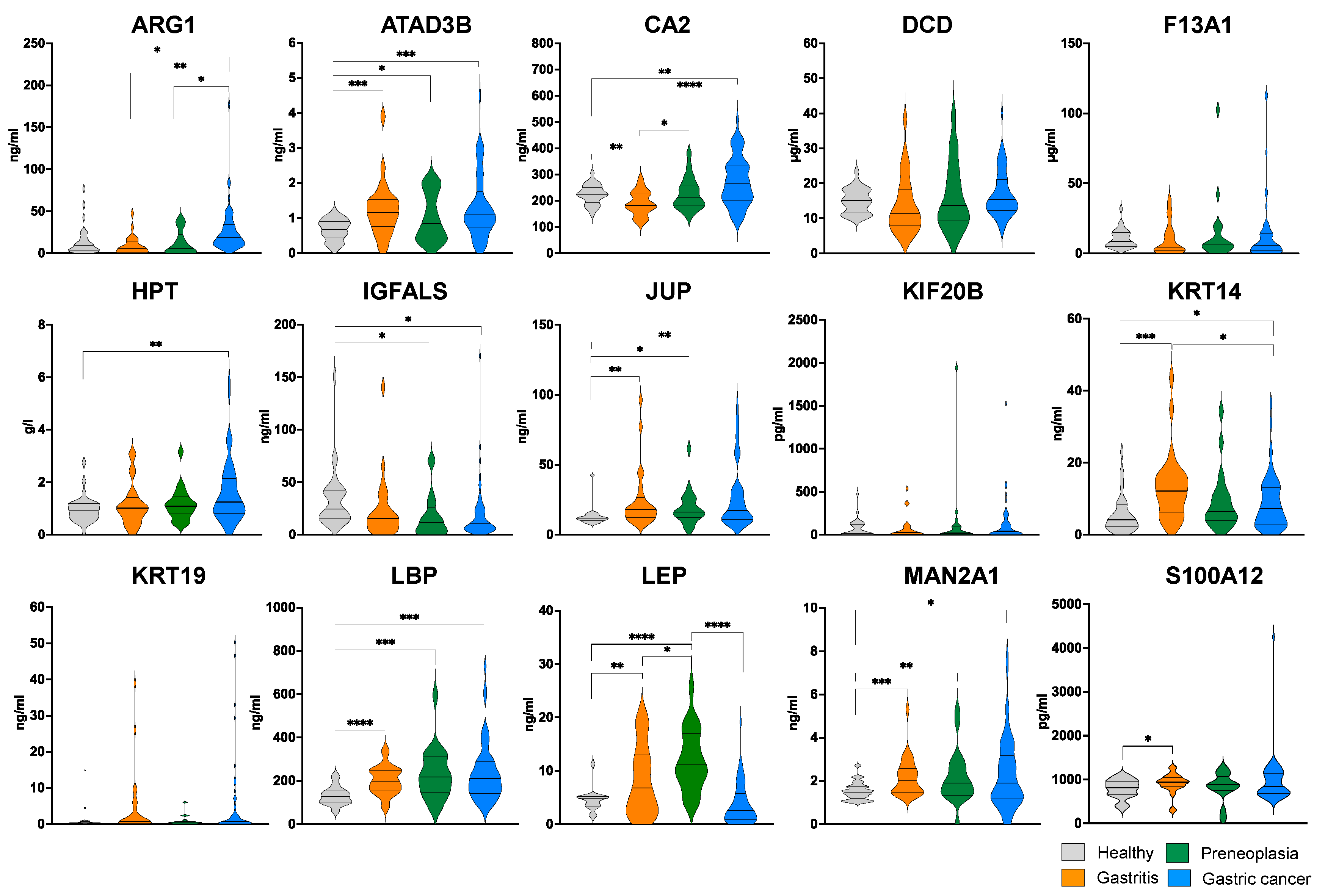
2.4. ELISA-Based Validation of 15 Biomarker Candidates Highlighted by LC-MS/MS
2.5. Correlations Between Plasma Concentration Levels of the Biomarkers
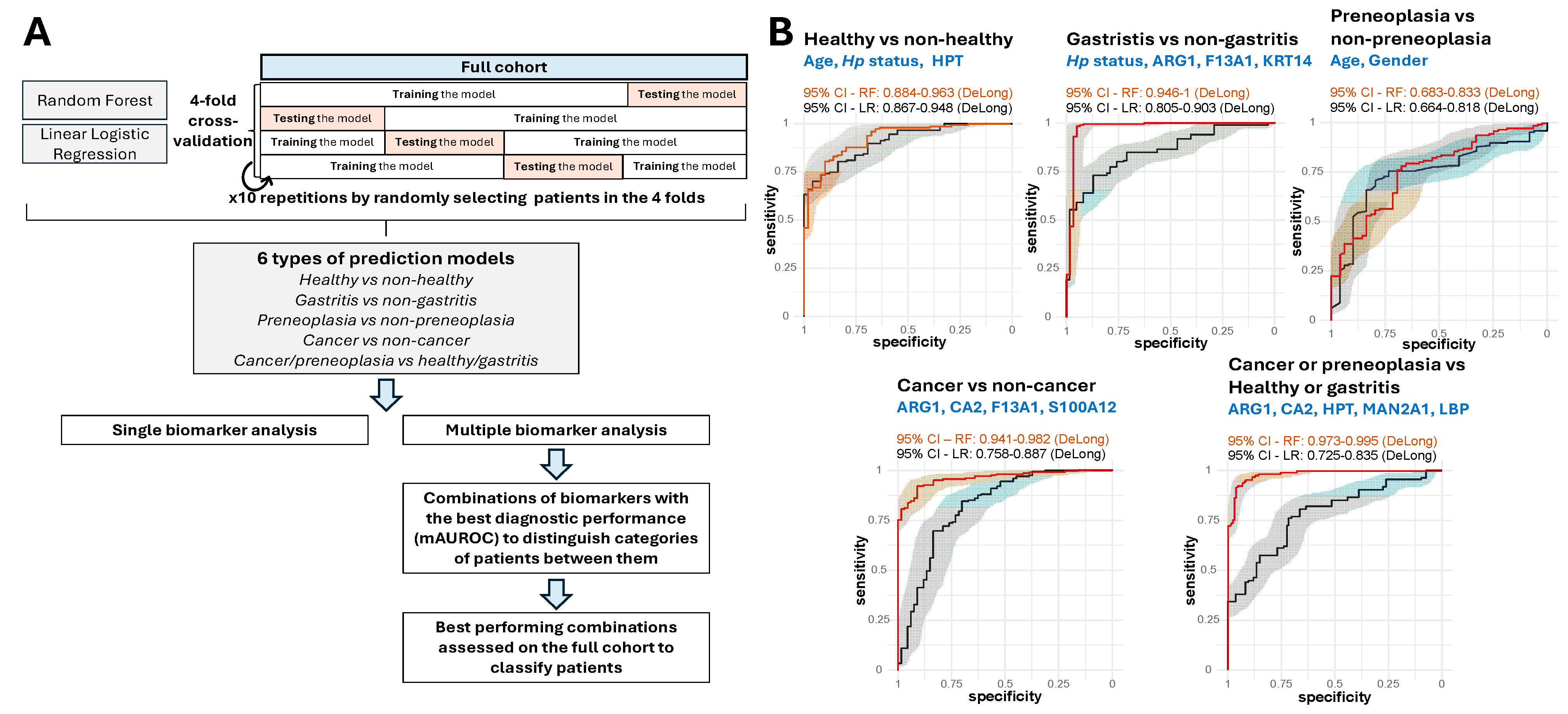
2.6. Performance of Prediction Models with Single and Multiple Biomarkers
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Identification and Quantification of Plasma Proteomes Using LC-MS/MS
4.3. Statistical Analysis of Large-Scale LC-MS/MS Proteomic Data for Biomarker Identification
4.4. Validation of LC-MS/MS-Identified Proteins Using Enzyme-Linked Immunosorbent Assays (ELISA), Statistical Analysis and Prediction Models
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AG | Atrophic Gastritis |
| AKT | Protein kinase B |
| AUROC | Area Under the Receiver Operating Characteristics |
| ATAD3B | ATPase family AAA domain containing 3B |
| ARG1 | Arginase-1 |
| CA2 | Carbonic anhydrase 2 |
| CA12-5 | Carbohydrate Antigen 12-5 |
| CA19-9 | Carbohydrate Antigen 19-9 |
| CAPZA1 | Capping Actin Protein of Muscle Z-Line Subunit alpha |
| CEA | Carcinoembryonic Antigen |
| DCD | Dermcidin |
| DDA | Data-Dependent Acquisition |
| DIA | Data-Independent Acquisition |
| DYS | Dysplasia |
| ELISA | Enzyme-Linked Immunosorbent Assays |
| ERAP2 | Endoplasmic Reticulum Aminopeptidase 2 |
| F13A1 | Coagulation factor XIII A chain |
| FC | Fold-Change |
| FDR | False Discovery Rate |
| FUCA2 | Alpha-L-Fucosidase 1 |
| GC | Gastric Cancer |
| GI | Gastrointestinal |
| GPLD1 | Glycosylphosphatidylinositol phospholipase D |
| HPT | Haptoglobin |
| IGFALS | Insulin-like growth factor-binding protein complex acid labile subunit |
| IGHG1 | Immunoglobulin heavy constant gamma 1 |
| IL1RAP | Interleukin-1 receptor accessory protein |
| IM | Intestinal Metaplasia |
| JUP | Junction Plakoglobin |
| KIF20B | Kinesin-like protein KIF20B |
| KRT14 | Keratin, type I cytoskeletal 14 |
| KRT19 | Keratin, type I cytoskeletal 19 |
| LBP | Lipopolysaccharide-binding protein |
| LC-MS/MS | Liquid Chromatography coupled to tandem Mass Spectrometry |
| LEP | Leptin |
| MAN2A1 | Alpha-mannosidase 2A1 |
| MAPK | Mitogen-activated protein kinase |
| mTOR | Mammalian target of rapamycin |
| PG | Pepsinogen |
| PI3K | phosphoinositide 3-kinase |
| PLS-DA | Partiel Least Square-Discriminated Analysis |
| PRCP | Lysosomal Pro-X carboxypeptidase |
| PRSS3 | Serine protease 3 |
| S100A12 | S100-A12 protein |
| SAA1/2 | Serum amyloid A-1 protein/Serum amyloid A-2 protein |
| STAT3 | Signal transducer and activator of transcription 3 |
| SVEP1 | Sushi Von Willebrand Factor Type A, EGF &Pentraxin Domain containing 1 |
| TFRC | Transferrin Receptor Protein 1 |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Mukaisho, K.-I.; Nakayama, T.; Hagiwara, T.; Hattori, T.; Sugihara, H. Two Distinct Etiologies of Gastric Cardia Adenocarcinoma: Interactions among pH, Helicobacter Pylori, and Bile Acids. Front. Microbiol. 2015, 6, 412. [Google Scholar] [CrossRef]
- Moss, S.F. The Clinical Evidence Linking Helicobacter Pylori to Gastric Cancer. Cell Mol. Gastroenterol. Hepatol. 2017, 3, 183–191. [Google Scholar] [CrossRef]
- Arnold, M.; Park, J.Y.; Camargo, M.C.; Lunet, N.; Forman, D.; Soerjomataram, I. Is Gastric Cancer Becoming a Rare Disease? A Global Assessment of Predicted Incidence Trends to 2035. Gut 2020, 69, 823–829. [Google Scholar] [CrossRef] [PubMed]
- Anderson, W.F.; Rabkin, C.S.; Turner, N.; Fraumeni, J.F.J.; Rosenberg, P.S.; Camargo, M.C. The Changing Face of Noncardia Gastric Cancer Incidence Among US Non-Hispanic Whites. J. Natl. Cancer Inst. 2018, 110, 608–615. [Google Scholar] [CrossRef]
- Sun, D.; Mülder, D.T.; Li, Y.; Nieboer, D.; Park, J.Y.; Suh, M.; Hamashima, C.; Han, W.; O’Mahony, J.F.; Lansdorp-Vogelaar, I. The Effect of Nationwide Organized Cancer Screening Programs on Gastric Cancer Mortality: A Synthetic Control Study. Gastroenterology 2024, 166, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Shan, Y.-S.; Hu, H.-M.; Price, T.J.; Sirohi, B.; Yeh, K.-H.; Yang, Y.-H.; Sano, T.; Yang, H.-K.; Zhang, X.; et al. Management of Gastric Cancer in Asia: Resource-Stratified Guidelines. Lancet Oncol. 2013, 14, e535–e547. [Google Scholar] [CrossRef] [PubMed]
- Correa, P. Human Gastric Carcinogenesis: A Multistep and Multifactorial Process--First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992, 52, 6735–6740. [Google Scholar]
- Rugge, M.; Genta, R.M.; Di Mario, F.; El-Omar, E.M.; El-Serag, H.B.; Fassan, M.; Hunt, R.H.; Kuipers, E.J.; Malfertheiner, P.; Sugano, K.; et al. Gastric Cancer as Preventable Disease. Clin. Gastroenterol. Hepatol. 2017, 15, 1833–1843. [Google Scholar] [CrossRef]
- Graham, D.Y.; Lee, Y.-C. Commentary on: The Management of Patients with Gastric Intestinal Metaplasia. Gut 2024, 74, 699–700. [Google Scholar] [CrossRef]
- Liou, J.-M.; Malfertheiner, P.; Smith, S.I.; El-Omar, E.M.; Wu, M.-S. 40 Years after the Discovery of Helicobacter Pylori: Towards Elimination of H Pylori for Gastric Cancer Prevention. Lancet 2024, 403, 2570–2572. [Google Scholar] [CrossRef]
- Ford, A.C.; Yuan, Y.; Moayyedi, P. Long-Term Impact of Helicobacter Pylori Eradication Therapy on Gastric Cancer Incidence and Mortality in Healthy Infected Individuals: A Meta-Analysis Beyond 10 Years of Follow-Up. Gastroenterology 2022, 163, 754–756.e1. [Google Scholar] [CrossRef] [PubMed]
- Pimentel-Nunes, P.; Libânio, D.; Marcos-Pinto, R.; Areia, M.; Leja, M.; Esposito, G.; Garrido, M.; Kikuste, I.; Megraud, F.; Matysiak-Budnik, T.; et al. Management of Epithelial Precancerous Conditions and Lesions in the Stomach (MAPS II): European Society of Gastrointestinal Endoscopy (ESGE), European Helicobacter and Microbiota Study Group (EHMSG), European Society of Pathology (ESP), and Sociedade Portuguesa de Endoscopia Digestiva (SPED) Guideline Update 2019. Endoscopy 2019, 51, 365–388. [Google Scholar] [CrossRef] [PubMed]
- Dinis-Ribeiro, M.; Shah, S.; El-Serag, H.; Banks, M.; Uedo, N.; Tajiri, H.; Coelho, L.G.; Libanio, D.; Lahner, E.; Rollan, A.; et al. The Road to a World-Unified Approach to the Management of Patients with Gastric Intestinal Metaplasia: A Review of Current Guidelines. Gut 2024, 73, 1607–1617, Correction in Gut 2024, 73, e1. [Google Scholar] [CrossRef]
- Beck, M.; Bringeland, E.A.; Qvigstad, G.; Fossmark, R. Gastric Cancers Missed at Upper Endoscopy in Central Norway 2007 to 2016-A Population-Based Study. Cancers 2021, 13, 5628. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and Localization of Surgically Resectable Cancers with a Multi-Analyte Blood Test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef]
- Desai, S.; Guddati, A.K. Carcinoembryonic Antigen, Carbohydrate Antigen 19-9, Cancer Antigen 125, Prostate-Specific Antigen and Other Cancer Markers: A Primer on Commonly Used Cancer Markers. World J. Oncol. 2023, 14, 4–14. [Google Scholar] [CrossRef]
- Feng, F.; Tian, Y.; Xu, G.; Liu, Z.; Liu, S.; Zheng, G.; Guo, M.; Lian, X.; Fan, D.; Zhang, H. Diagnostic and Prognostic Value of CEA, CA19-9, AFP and CA125 for Early Gastric Cancer. BMC Cancer 2017, 17, 737. [Google Scholar] [CrossRef] [PubMed]
- Agréus, L.; Kuipers, E.J.; Kupcinskas, L.; Malfertheiner, P.; Di Mario, F.; Leja, M.; Mahachai, V.; Yaron, N.; van Oijen, M.; Perez Perez, G.; et al. Rationale in Diagnosis and Screening of Atrophic Gastritis with Stomach-Specific Plasma Biomarkers. Scand. J. Gastroenterol. 2012, 47, 136–147. [Google Scholar] [CrossRef]
- Loong, T.H.; Soon, N.C.; Nik Mahmud, N.R.K.; Naidu, J.; Rani, R.A.; Abdul Hamid, N.; Elias, M.H.; Mohamed Rose, I.; Tamil, A.; Mokhtar, N.M.; et al. Serum Pepsinogen and Gastrin-17 as Potential Biomarkers for Pre-Malignant Lesions in the Gastric Corpus. Biomed. Rep. 2017, 7, 460–468. [Google Scholar] [CrossRef]
- Shen, Q.; Polom, K.; Williams, C.; de Oliveira, F.M.S.; Guergova-Kuras, M.; Lisacek, F.; Karlsson, N.G.; Roviello, F.; Kamali-Moghaddam, M. A Targeted Proteomics Approach Reveals a Serum Protein Signature as Diagnostic Biomarker for Resectable Gastric Cancer. EBioMedicine 2019, 44, 322–333. [Google Scholar] [CrossRef]
- Bazin, T.; Nozeret, K.; Julié, C.; Lamarque, D.; Touati, E. Protein Biomarkers of Gastric Preneoplasia and Cancer Lesions in Blood: A Comprehensive Review. Cancers 2024, 16, 3019. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Capelle, L.G.; de Vries, A.C.; Haringsma, J.; Steyerberg, E.W.; Looman, C.W.N.; Nagtzaam, N.M.A.; van Dekken, H.; ter Borg, F.; de Vries, R.A.; Kuipers, E.J. Serum Levels of Leptin as Marker for Patients at High Risk of Gastric Cancer. Helicobacter 2009, 14, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Tas, F.; Karabulut, S.; Erturk, K.; Duranyildiz, D. Clinical Significance of Serum Leptin Level in Patients with Gastric Cancer. Eur. Cytokine Netw. 2018, 29, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Young, E.; Philpott, H.; Singh, R. Endoscopic Diagnosis and Treatment of Gastric Dysplasia and Early Cancer: Current Evidence and What the Future May Hold. World J. Gastroenterol. 2021, 27, 5126–5151. [Google Scholar] [CrossRef]
- Kim, Y.-I.; Park, J.Y.; Kim, B.J.; Hwang, H.W.; Hong, S.A.; Kim, J.G. Risk of Metachronous Gastric Neoplasm Occurrence during Intermediate-Term Follow-up Period after Endoscopic Submucosal Dissection for Gastric Dysplasia. Sci. Rep. 2020, 10, 6747. [Google Scholar] [CrossRef]
- Ramesh, P.; Nisar, M.; Neha, M.; Ammankallu, S.; Babu, S.; Nandakumar, R.; Abhinand, C.S.; Prasad, T.S.K.; Codi, J.A.K.; Raju, R. Delineating Protein Biomarkers for Gastric Cancers: A Catalogue of Mass Spectrometry-Based Markers and Assessment of Their Suitability for Targeted Proteomics Applications. J. Proteom. 2024, 306, 105262. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-H.; Kim, H. Role of Leptin in the Digestive System. Front. Pharmacol. 2021, 12, 660040. [Google Scholar] [CrossRef]
- Niu, F.; Yu, Y.; Li, Z.; Ren, Y.; Li, Z.; Ye, Q.; Liu, P.; Ji, C.; Qian, L.; Xiong, Y. Arginase: An Emerging and Promising Therapeutic Target for Cancer Treatment. Biomed. Pharmacother. 2022, 149, 112840. [Google Scholar] [CrossRef]
- Wang, X.; Xu, Y.; Wang, R.; Dai, N.; Zhang, W.; Li, F. The Significance of Arginase-1 Expression in the Diagnosis of Liver Cancer: A Protocol for a Systematic Review. Medicine 2020, 99, e19159. [Google Scholar] [CrossRef]
- Li, S.; Rousseau, D. ATAD3, a Vital Membrane Bound Mitochondrial ATPase Involved in Tumor Progression. J. Bioenerg. Biomembr. 2012, 44, 189–197. [Google Scholar] [CrossRef]
- Liu, X.; Li, G.; Ai, L.; Ye, Q.; Yu, T.; Yang, B. Prognostic Value of ATAD3 Gene Cluster Expression in Hepatocellular Carcinoma. Oncol. Lett. 2019, 18, 1304–1310. [Google Scholar] [CrossRef]
- Kivelä, A.-J.; Kivelä, J.; Saarnio, J.; Parkkila, S. Carbonic Anhydrases in Normal Gastrointestinal Tract and Gastrointestinal Tumours. World J. Gastroenterol. 2005, 11, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Huang, Z.; Liao, Z.; He, C.; Fang, X. Low CA II Expression Is Associated with Tumor Aggressiveness and Poor Prognosis in Gastric Cancer Patients. Int. J. Clin. Exp. Pathol. 2014, 7, 6716–6724. [Google Scholar] [PubMed]
- Jeong, S.; Oh, M.J.; Kim, U.; Lee, J.; Kim, J.-H.; An, H.J. Glycosylation of Serum Haptoglobin as a Marker of Gastric Cancer: An Overview for Clinicians. Expert. Rev. Proteom. 2020, 17, 109–117. [Google Scholar] [CrossRef]
- Ghuman, S.; Van Hemelrijck, M.; Garmo, H.; Holmberg, L.; Malmström, H.; Lambe, M.; Hammar, N.; Walldius, G.; Jungner, I.; Wulaningsih, W. Serum Inflammatory Markers and Colorectal Cancer Risk and Survival. Br. J. Cancer 2017, 116, 1358–1365. [Google Scholar] [CrossRef]
- Xu, L.; Xiong, L.; Chen, Y.; Chen, J.; Liu, X.; Xu, Y.; Shen, Y.; Wang, S.; Yu, S.; Xu, X. IGFALS Suppresses Hepatocellular Carcinoma Progression by Stabilizing PPAR-γ. Int. Immunopharmacol. 2024, 143 Pt 2, 113414. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, L.; Qin, Y.; Liu, S.; Qiao, Y.; Wan, X.; Zeng, H.; Tang, X.; Liu, M.; Hou, Y. Effects of Differential Distributed-JUP on the Malignancy of Gastric Cancer. J. Adv. Res. 2021, 28, 195–208. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhao, G.; Xu, B.; Liu, C.; Li, C.; Zhang, X.; Chang, X. PADI4 Has Genetic Susceptibility to Gastric Carcinoma and Upregulates CXCR2, KRT14 and TNF-α Expression Levels. Oncotarget 2016, 7, 62159–62176. [Google Scholar] [CrossRef]
- Lv, H.; Lv, M.; Guo, X.; Zhu, X.; Chao, Y.; Li, D. Lipopolysaccharide-Binding Protein (LBP): A Prognostic Biomarker for Gastric Cancer Linked to Immune Infiltration. BMC Gastroenterol. 2025, 25, 205. [Google Scholar] [CrossRef]
- Xie, L.; Qiu, S.; Lu, C.; Gu, C.; Wang, J.; Lv, J.; Fang, L.; Chen, Z.; Li, Y.; Jiang, T.; et al. Gastric Cancer-Derived LBP Promotes Liver Metastasis by Driving Intrahepatic Fibrotic Pre-Metastatic Niche Formation. J. Exp. Clin. Cancer Res. 2023, 42, 258. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, J.; Fu, G.; Sheng, C.; Zhu, J.; Zhong, T.; Yang, F.; Jiang, Z. MAN2A1 Predicts Prognosis and Progression through Cancer-Related Pathways in Colorectal Cancer. Transl. Cancer Res. 2022, 11, 3686–3697. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zeng, Z.; Yu, T.; Qin, J.; Wu, J.; Song, J.-C.; Zhou, Z.-Y.; Yuan, J.-P. Expression and Clinical Implication of S100A12 in Gastric Carcinoma. Tumour Biol. 2016, 37, 6551–6559. [Google Scholar] [CrossRef]
- Ercan, H.; Mauracher, L.-M.; Grilz, E.; Hell, L.; Hellinger, R.; Schmid, J.A.; Moik, F.; Ay, C.; Pabinger, I.; Zellner, M. Alterations of the Platelet Proteome in Lung Cancer: Accelerated F13A1 and ER Processing as New Actors in Hypercoagulability. Cancers 2021, 13, 2260. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Suh, I.B.; Lee, E.J.; Hur, G.Y.; Lee, S.Y.; Lee, S.Y.; Shin, C.; Shim, J.J.; In, K.H.; Kang, K.H.; et al. Relationships of Coagulation Factor XIII Activity with Cell-Type and Stage of Non-Small Cell Lung Cancer. Yonsei Med. J. 2013, 54, 1394–1399. [Google Scholar] [CrossRef]
- Raval, J.S.; Berg, A.N.; Djokic, M.; Roth, C.G.; Rollins-Raval, M.A. Factor XIII Subunit A Immunohistochemical Expression Is Associated With Inferior Outcomes in Acute Promyelocytic Leukemia. Appl. Immunohistochem. Mol. Morphol. 2018, 26, 202–205. [Google Scholar] [CrossRef]
- Liu, Y.; Deng, H.; Song, P.; Zhang, M. Constructing a Glioblastoma Prognostic Model Related to Fatty Acid Metabolism Using Machine Learning and Identifying F13A1 as a Potential Target. Biomedicines 2025, 13, 256. [Google Scholar] [CrossRef]
- Wang, X.; Luo, Y.; Zhou, Q.; Ma, J. The Roles of S100A8/A9 and S100A12 in Autoimmune Diseases: Mechanisms, Biomarkers, and Therapeutic Potential. Autoimmun. Rev. 2025, 24, 103920. [Google Scholar] [CrossRef] [PubMed]
- Muta, T.; Takeshige, K. Essential Roles of CD14 and Lipopolysaccharide-Binding Protein for Activation of Toll-like Receptor (TLR)2 as Well as TLR4 Reconstitution of TLR2- and TLR4-Activation by Distinguishable Ligands in LPS Preparations. Eur. J. Biochem. 2001, 268, 4580–4589. [Google Scholar] [CrossRef]
- Parkkila, S.; Lasota, J.; Fletcher, J.A.; Ou, W.-B.; Kivelä, A.J.; Nuorva, K.; Parkkila, A.-K.; Ollikainen, J.; Sly, W.S.; Waheed, A.; et al. Carbonic Anhydrase II. A Novel Biomarker for Gastrointestinal Stromal Tumors. Mod. Pathol. 2010, 23, 743–750. [Google Scholar] [CrossRef]
- Shi, S.; Gu, S.; Han, T.; Zhang, W.; Huang, L.; Li, Z.; Pan, D.; Fu, J.; Ge, J.; Brown, M.; et al. Inhibition of MAN2A1 Enhances the Immune Response to Anti-PD-L1 in Human Tumors. Clin. Cancer Res. 2020, 26, 5990–6002. [Google Scholar] [CrossRef] [PubMed]
- Grohmann, U.; Bronte, V. Control of Immune Response by Amino Acid Metabolism. Immunol. Rev. 2010, 236, 243–264. [Google Scholar] [CrossRef]
- Di, R.; Murray, A.F.; Xiong, J.; Esposito, D.; Komarnytsky, S.; Gianfagna, T.J.; Munafo, J.P. Lily Steroidal Glycoalkaloid Promotes Early Inflammatory Resolution in Wounded Human Fibroblasts. J. Ethnopharmacol. 2020, 258, 112766. [Google Scholar] [CrossRef]
- Monteiro, L.d.B.; Prodonoff, J.S.; Favero de Aguiar, C.; Correa-da-Silva, F.; Castoldi, A.; Bakker, N.v.T.; Davanzo, G.G.; Castelucci, B.; Pereira, J.A.d.S.; Curtis, J.; et al. Leptin Signaling Suppression in Macrophages Improves Immunometabolic Outcomes in Obesity. Diabetes 2022, 71, 1546–1561. [Google Scholar] [CrossRef]
- Kamikubo, Y.; Dellas, C.; Loskutoff, D.J.; Quigley, J.P.; Ruggeri, Z.M. Contribution of Leptin Receptor N-Linked Glycans to Leptin Binding. Biochem. J. 2008, 410, 595–604. [Google Scholar] [CrossRef]
- Dinis-Ribeiro, M.; Libânio, D.; Uchima, H.; Spaander, M.C.W.; Bornschein, J.; Matysiak-Budnik, T.; Tziatzios, G.; Santos-Antunes, J.; Areia, M.; Chapelle, N.; et al. Management of Epithelial Precancerous Conditions and Early Neoplasia of the Stomach (MAPS III): European Society of Gastrointestinal Endoscopy (ESGE), European Helicobacter and Microbiota Study Group (EHMSG) and European Society of Pathology (ESP) Guideline Update 2025. Endoscopy 2025, 57, 504–554. [Google Scholar] [CrossRef]
- Suzuki, R.; Shimodaira, H. Pvclust: An R Package for Assessing the Uncertainty in Hierarchical Clustering. Bioinformatics 2006, 22, 1540–1542. [Google Scholar] [CrossRef] [PubMed]
- Gianetto, Q.G.; Wieczorek, S.; Couté, Y.; Burger, T. A Peptide-Level Multiple Imputation Strategy Accounting for the Different Natures of Missing Values in Proteomics Data. bioRxiv 2020. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.-A. mixOmics: An R Package for ‘omics Feature Selection and Multiple Data Integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef]
- Giai Gianetto, Q. Statistical Analysis of Post-Translational Modifications Quantified by Label-Free Proteomics Across Multiple Biological Conditions with R: Illustration from SARS-CoV-2 Infected Cells. In Statistical Analysis of Proteomic Data; Burger, T., Ed.; Methods in Molecular Biology; Springer US: New York, NY, USA, 2023; Volume 2426, pp. 267–302. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Giai Gianetto, Q.; Combes, F.; Ramus, C.; Bruley, C.; Couté, Y.; Burger, T. Calibration Plot for Proteomics: A Graphical Tool to Visually Check the Assumptions Underlying FDR Control in Quantitative Experiments. Proteomics 2016, 16, 29–32. [Google Scholar] [CrossRef] [PubMed]
- Pounds, S.; Cheng, C. Robust Estimation of the False Discovery Rate. Bioinformatics 2006, 22, 1979–1987. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: An Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the Areas under Two or More Correlated Receiver Operating Characteristic Curves: A Nonparametric Approach. Biometrics 1988, 44, 837. [Google Scholar] [CrossRef]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for Micro-Purification, Enrichment, Pre-Fractionation and Storage of Peptides for Proteomics Using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef]
- Kulak, N.A.; Pichler, G.; Paron, I.; Nagaraj, N.; Mann, M. Minimal, Encapsulated Proteomic-Sample Processing Applied to Copy-Number Estimation in Eukaryotic Cells. Nat. Methods 2014, 11, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef]


| Group | n | Mean Age (Range) | Sex Ratio (M/F) | H. pylori Positive (%) |
|---|---|---|---|---|
| Healthy | 32 10 | 42 (21–70) 38 (22–69) | 0.4 0.4 | 0 0 |
| Gastritis | 26 | 59 (27–88) | 0.7 | 58% |
| p = 0.0003 | ||||
| 10 | 38 (22–69) | 0.6 | 58% | |
| p = 0.0015 | ||||
| Preneoplasia | 20 | 70 (50–83) | 0.4 | 61% |
| p < 0.0001 | ||||
| 9 | 70 (54–83) | 0.5 | 50% | |
| p = 0.0006 | ||||
| Gastric cancer | 60 | 62 (29–84) | 2 | 28% |
| p < 0.0001 | ||||
| 10 | 62 (51–74) | 4 | 40% | |
| p = 0.0044 | ||||
| Total | 138 | |||
| 39 |
| Protein Name | Uniprot Code | Protein Description | KEGG Pathways | Human Protein Atlas (TCGA) |
|---|---|---|---|---|
| ARG-1 | P05089 | Arginase-1 | hsa01100 Metabolic pathways hsa00220 Arginine biosynthesis | Cancer enriched Liver Hepatocellular Carcinoma. No prognostic found. |
| ATAD3B | Q5T9A4 | ATPase family AAA domain containing 3B | hsa03029 Mitochondrial Biogenesis | Cancer enhanced Testicular Germ Cell Tumor. Potential prognostic * Liver Hepatocellular Carcinoma, Kidney Renal Clear Cell Carcinoma. |
| CA2 | P00918 | Carbonic anhydrase 2 | hsa00910 Nitrogen metabolism hsa01100 Metabolic pathways hsa04971 Gastric acid secretion | Cancer enriched Kidney Chromophobe Renal Cell Carcinoma. Potential prognostic * Lung Squamous Cell Carcinoma. |
| DCD | P81605 | Dermcidin | hsa09193 Unclassified: signaling and cellular processes | Cancer enriched Breast Invasive Carcinoma. No prognostic found. |
| F13A1 | P00488 | Coagulation Factor XIII A Chain | hsa09151 Immune system hsa04610 Complement and coagulation cascades | Low cancer specificity. Potential prognostic * Lung Squamous Cell Carcinoma. |
| HPT | P00738 | Haptoglobin | hsa09181 Protein families: metabolism hsa01002 Peptidases and inhibitors >Serine peptidases hsa04147 Exosomal proteins of other cancer cells | Cancer enriched Liver Hepatocellular Carcinoma. Validated prognostic * Kidney Renal Clear Cell Carcinoma, Stomach Adenocarcinoma Lung Adenocarcinoma. Potential prognostic ** Hepatocellular Carcinoma. |
| IGFALS | P35858 | Insulin-like growth factor-binding protein complex acid labile subunit | hsa04935 Growth hormone synthesis, secretion and action hsa04147 Exosomal proteins of haemopoietic cells | Cancer enhanced Hepatocellular Carcinoma. Potential prognostic ** Hepatocellular Carcinoma, Lung Adenocarcinoma |
| JUP | P14923 | Junction plakoglobin | hsa04820 Cytoskeleton in muscle cells hsa05200 Pathways in cancer hsa05202 Transcriptional misregulation in cancer hsa05221 Acute myeloid leukemia hsa05226 Gastric cancer hsa05412 Arrhythmogenic right ventricular cardiomyopathy | Low cancer specificity. Potential prognostic *, Hepatocellular Carcinoma Potential prognostic ** Kidney Renal Clear Cell Carcinoma. |
| KIF20B | Q96Q89 | Kinesin-like protein KIF20B | hsa04814 Motor proteins hsa04131 Membrane trafficking hsa04812 Cytoskeleton proteins | Low cancer specificity. Potential prognostic * Lung Adenocarcinoma, Potential prognostic ** Kidney Renal Clear Cell Carcinoma Validated prognostic * Pancreatic Adenocarcinoma. |
| KRT14 | P02533 | Keratin, type I cytoskeletal 14 | hsa04915 Estrogen signaling pathway hsa05150 Staphylococcus aureus infection hsa04147 Exosomal proteins of epithelial, colorectal cancer, melanoma cells | Cancer enriched Head and Neck Squamous Cell Carcinoma. Potential prognostic ** Breast Invasive Carcinoma Potential prognostic * Lung Adenocarcinoma. |
| KRT19 | P08727 | Keratin, type I cytoskeletal 19 | hsa04915 Estrogen signaling pathway hsa05150 Staphylococcus aureus infection hsa04147 Exosomal proteins of epithelial, colorectal cancer, melanoma cells | Low cancer specificity. Potential prognostic * Pancreatic Adenocarcinoma. Validated prognostic * Kidney Renal Clear Cell Carcinoma. |
| LBP | P18428 | Lipopolysaccharide- binding protein | hsa04064 NF-kB signaling pathway hsa04620 Toll-like receptor signaling pathway hsa04936 Alcoholic liver disease hsa05152 Tuberculosis hsa05417 Lipid and atherosclerosis hsa04147 Exosomal proteins of other cancer cells | Cancer enriched Liver Hepatocellular Carcinoma. Validated prognostic * Kidney Renal Clear Cell Carcinoma Potential prognostic * Kidney Renal Papillary Cell Carcinoma. |
| LEP | P41159 | Leptin | hsa04060 Cytokine–cytokine receptor interaction hsa04080 Neuroactive ligand–receptor interaction hsa04081 Hormone signaling hsa04152 AMPK signaling pathway hsa04630 JAK-STAT signaling pathway hsa04920 Adipocytokine signaling pathway hsa04932 Non-alcoholic fatty liver disease | Cancer enhanced Breast Invasive Carcinoma. No prognostic found. |
| MAN2A1 | Q16706 | Alpha-mannosidase 2 | hsa00510 N-Glycan biosynthesis hsa00513 Various types of N-glycan biosynthesis hsa01100 Metabolic pathways | Low cancer specificity. Potential prognostic ** Kidney Renal Clear Cell Carcinoma Potential prognostic * Thyroid Carcinoma |
| S100A12 | P80511 | Protein S100-A12 | hsa04990 EF-hand domain-containing proteins >S100 proteins | Cancer enriched Head and Neck Squamous Cell Carcinoma. Potential prognostic * Stomach Adenocarcinoma. |
| Healthy vs. Non-Healthy | Gastritis vs. Non-Gastritis | Preneoplasia vs. Non-Preneoplasia | Cancer vs. Non-Cancer | Cancer or Preneoplasia vs. Healthy or Gastritis | All Stages | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mAUROC | sd | mAUROC | sd | mAUROC | sd | mAUROC | sd | mAUROC | sd | mAUROC | sd | |
| Age | 80.1% | 6.9% | 60.2% | 8.1% | 71.8% | 8.8% | 62.2% | 10.0% | 75.1% | 8.1% | 63.2% | 4.8% |
| Gender | 57.0% | 10.9% | 57.7% | 7.7% | 49.6% | 2.4% | 66.0% | 10.4% | 55.4% | 9.5% | 56.6% | 4.0% |
| Hpstatus | 48.2% | 4.9% | 67.2% | 15.3% | 50.9% | 3.3% | 56.8% | 6.1% | 54.5% | 4.8% | 59.6% | 5.0% |
| DCD | 57.4% | 7.0% | 60.5% | 8.2% | 63.7% | 8.7% | 63.6% | 10.2% | 55.7% | 5.9% | 57.1% | 5.0% |
| IGFALS | 63.4% | 8.7% | 59.6% | 8.0% | 62.5% | 9.3% | 58.3% | 8.0% | 61.2% | 7.8% | 54.1% | 4.9% |
| LEP | 77.0% | 8.5% | 65.1% | 11.4% | 62.4% | 8.9% | 68.4% | 8.8% | 63.2% | 9.1% | 62.7% | 6.5% |
| KRT14 | 63.3% | 7.9% | 72.3% | 8.4% | 61.3% | 8.0% | 58.1% | 6.6% | 55.8% | 5.0% | 63.3% | 4.4% |
| MAN2A1 | 66.0% | 8.0% | 60.3% | 7.8% | 60.7% | 8.3% | 58.8% | 8.2% | 59.5% | 7.4% | 57.8% | 4.8% |
| KIF20B | 57.7% | 8.2% | 57.4% | 7.7% | 60.4% | 7.6% | 62.1% | 8.8% | 57.4% | 7.9% | 49.9% | 5.7% |
| ARG1 | 60.2% | 5.9% | 68.0% | 10.9% | 60.1% | 8.6% | 75.5% | 6.3% | 66.1% | 7.0% | 59.7% | 5.9% |
| F13A1 | 65.6% | 7.8% | 60.5% | 7.1% | 60.0% | 7.4% | 59.1% | 6.6% | 55.3% | 6.0% | 50.4% | 5.1% |
| S100A12 | 57.1% | 8.4% | 59.5% | 8.3% | 59.9% | 8.3% | 61.6% | 10.9% | 58.3% | 6.5% | 53.8% | 5.7% |
| LBP | 70.7% | 6.9% | 60.6% | 9.3% | 59.5% | 7.1% | 68.4% | 9.3% | 67.6% | 6.2% | 61.1% | 4.4% |
| ATAD3B | 61.3% | 8.4% | 61.0% | 7.9% | 59.5% | 8.0% | 59.7% | 8.2% | 57.3% | 8.4% | 50.6% | 7.4% |
| JUP | 71.4% | 8.1% | 58.6% | 7.7% | 59.3% | 6.8% | 63.2% | 9.9% | 65.2% | 6.6% | 58.8% | 5.4% |
| CA2 | 64.9% | 8.6% | 63.9% | 10.8% | 58.8% | 7.8% | 78.1% | 8.7% | 69.4% | 7.7% | 62.0% | 6.1% |
| HPT | 62.0% | 7.9% | 61.1% | 8.9% | 58.5% | 6.2% | 64.1% | 10.0% | 64.8% | 8.1% | 55.6% | 4.9% |
| KRT19 | 63.3% | 9.5% | 61.0% | 9.3% | 57.7% | 8.5% | 60.4% | 7.7% | 56.0% | 7.2% | 53.3% | 3.8% |
| Max mAUROC | 80.1% | 6.9% | 72.3% | 8.4% | 71.8% | 8.8% | 78.1% | 8.7% | 75.1% | 8.1% | 63.3% | 4.4% |
| Min mAUROC | 48.2% | 4.9% | 57.4% | 7.7% | 49.6% | 2.4% | 56.8% | 6.1% | 54.5% | 4.8% | 49.9% | 5.7% |
| Marker with best mAUROC | Age | KRT14 | Age | CA2 | Age | KRT14 | ||||||
| Prediction Task | Number of Biomarkers in the Prediction Model | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |||||||
| mAUROC | sd | mAUROC | sd | mAUROC | sd | mAUROC | sd | mAUROC | sd | mAUROC | sd | |
| Healthy vs. non-healthy | 80.1% | 6.9% | 87.7% | 5.0% | 88.0% | 5.3% | 87.4% | 6.9% | 86.2% | 7.9% | 85.2% | 8.2% |
| Age | Age, Hp status | Age, Hp status, HPT | Age, Hp status, HPT, KRT19 | Age, Gender, Hp status, HPT, KRT19 | Age, Gender, Hp status, ARG1, F13A1 | |||||||
| Gastritis vs. non-gastritis | 72.3% | 8.4% | 81.4% | 7.3% | 80.6% | 7.5% | 82.2% | 9.2% | 79.6% | 9.2% | 78.0% | 8.0% |
| KRT14 | Hp status, KRT14 | Hp status, F13A1, KRT14 | Hp status, ARG1, KRT14, F13A1 | Hp status, ARG1, KRT14, F13A1, KIF20B | Hp status, HPT, KIF20B, ARG1, F13A1, KRT14 | |||||||
| Preneoplasia vs. non-preneoplasia | 71.8% | 8.8% | 75.7% | 8.9% | 74.4% | 10.6% | 73.4% | 10.8% | 72.7% | 7.9% | 70.5% | 11.3% |
| Age | Age, Gender | Age, Gender, KRT14 | Age, Gender, KRT14, ARG1 | Age, Gender, KRT14, KRT19, F13A1 | HPT, Leptin, S100A12, KIF20B, ATAD3B, CA2 | |||||||
| Cancer vs. non-cancer | 78.1% | 8.7% | 83.0% | 9.4% | 85.2% | 7.0% | 85.3% | 6.4% | 85.0% | 5.7% | 84.9% | 7.4% |
| CA2 | CA2, Leptin | ARG1, CA2, S100A12 | ARG1, CA2, F13A1, S100A12 | Gender, ARG1, CA2, MAN2A1, IGFALS | Age, Gender, ARG1, CA2, MAN2A1, IGFALS | |||||||
| Cancer/preneoplasia vs. healthy/gastritis | 75.1% | 8.1% | 76.7% | 7.7% | 78.8% | 6.8% | 81.2% | 6.8% | 83.9% | 5.1% | 81.4% | 6.3% |
| Age | Age, Leptin | Age, HPT, ARG1 | ARG1, CA2, HPT, LBP | ARG1, CA2, HPT, MAN2A1, LBP | ARG1, CA2, HPT, MAN2A1, LBP, DCD | |||||||
| All stages | 63.3% | 4.4% | 71.6% | 5.4% | 74.8% | 4.2% | 76.2% | 6.9% | 77.9% | 7.7% | 75.6% | 6.3% |
| KRT14 | Age, Hp status | Age, Gender, Hp status | Age, Hp status, ARG1, KRT14 | Age, Gender, Hp Status, ARG1, KRT14 | Age, Gender, Hp Status, ARG1, KRT14, F13A1 | |||||||
| Number of tested combinations | 18 | 153 | 816 | 3060 | 8568 | 18,564 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giai Gianetto, Q.; Michel, V.; Douché, T.; Nozeret, K.; Zaanan, A.; Colussi, O.; Trouilloud, I.; Pernot, S.; Ungeheuer, M.-N.; Julié, C.; et al. Plasma Protein Biomarkers to Detect Early Gastric Preneoplasia and Cancer: A Prospective Study. Int. J. Mol. Sci. 2025, 26, 10114. https://doi.org/10.3390/ijms262010114
Giai Gianetto Q, Michel V, Douché T, Nozeret K, Zaanan A, Colussi O, Trouilloud I, Pernot S, Ungeheuer M-N, Julié C, et al. Plasma Protein Biomarkers to Detect Early Gastric Preneoplasia and Cancer: A Prospective Study. International Journal of Molecular Sciences. 2025; 26(20):10114. https://doi.org/10.3390/ijms262010114
Chicago/Turabian StyleGiai Gianetto, Quentin, Valérie Michel, Thibaut Douché, Karine Nozeret, Aziz Zaanan, Oriane Colussi, Isabelle Trouilloud, Simon Pernot, Marie-Noelle Ungeheuer, Catherine Julié, and et al. 2025. "Plasma Protein Biomarkers to Detect Early Gastric Preneoplasia and Cancer: A Prospective Study" International Journal of Molecular Sciences 26, no. 20: 10114. https://doi.org/10.3390/ijms262010114
APA StyleGiai Gianetto, Q., Michel, V., Douché, T., Nozeret, K., Zaanan, A., Colussi, O., Trouilloud, I., Pernot, S., Ungeheuer, M.-N., Julié, C., Jolly, N., Taïeb, J., Lamarque, D., Matondo, M., & Touati, E. (2025). Plasma Protein Biomarkers to Detect Early Gastric Preneoplasia and Cancer: A Prospective Study. International Journal of Molecular Sciences, 26(20), 10114. https://doi.org/10.3390/ijms262010114








