Clinical Variability and Genotype–Phenotype Correlation in Spanish Patients with Type 1 Gaucher Disease: A Focus on Non-c.[1226A>G]; [1448T>C] Genotypes
Abstract
1. Introduction
2. Results
2.1. Patients Genotype and Characteristics
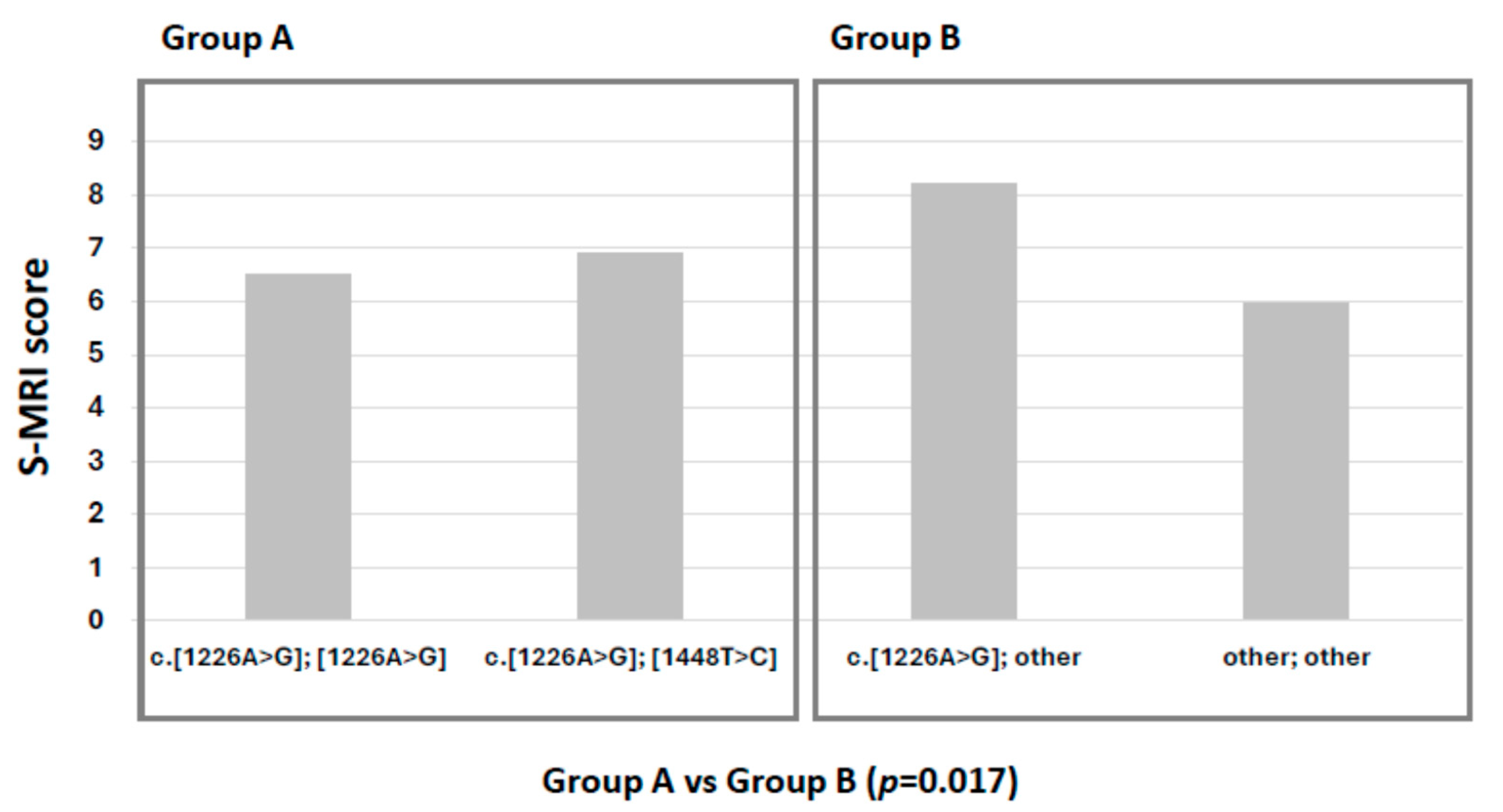
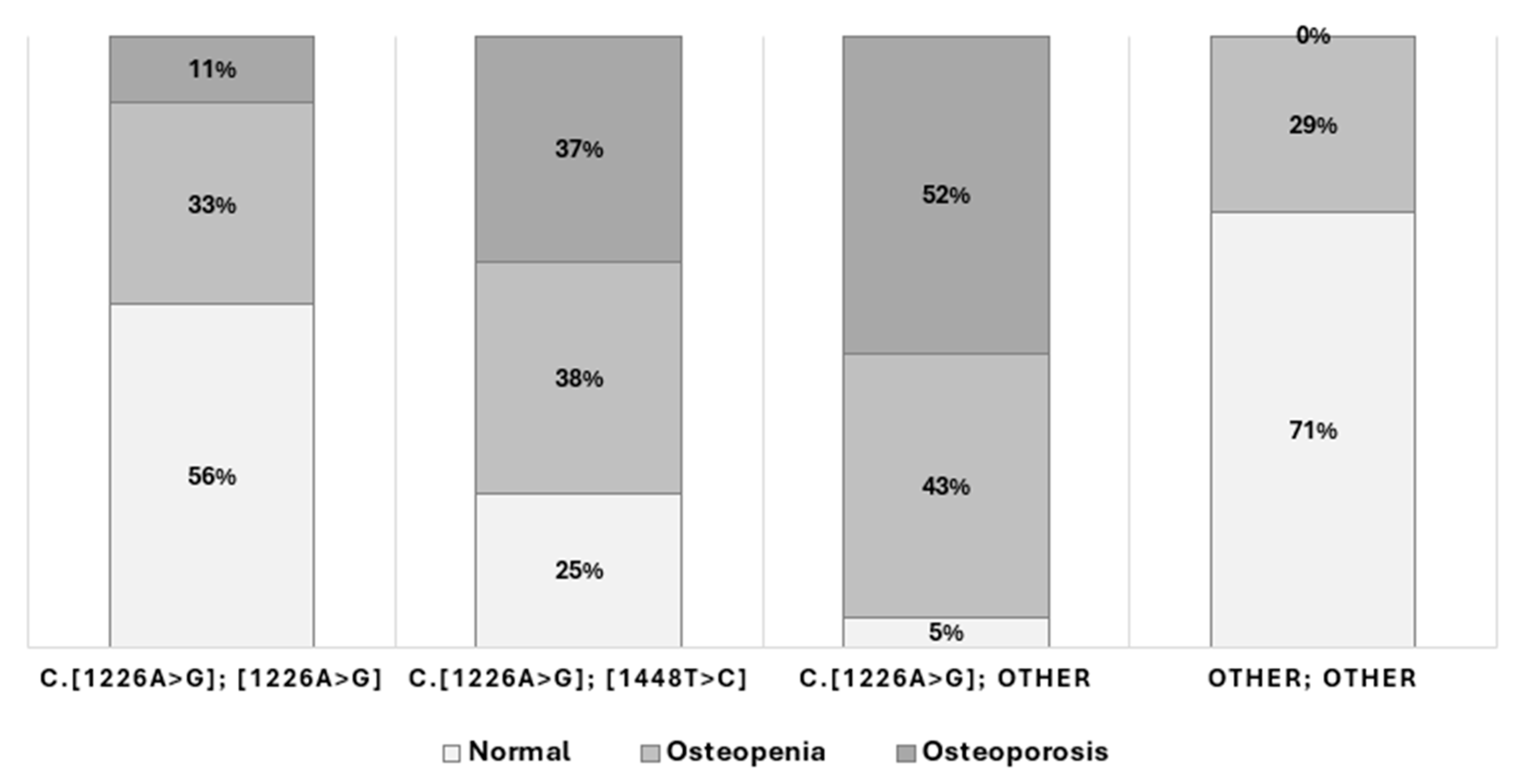
2.2. Bone Involvement
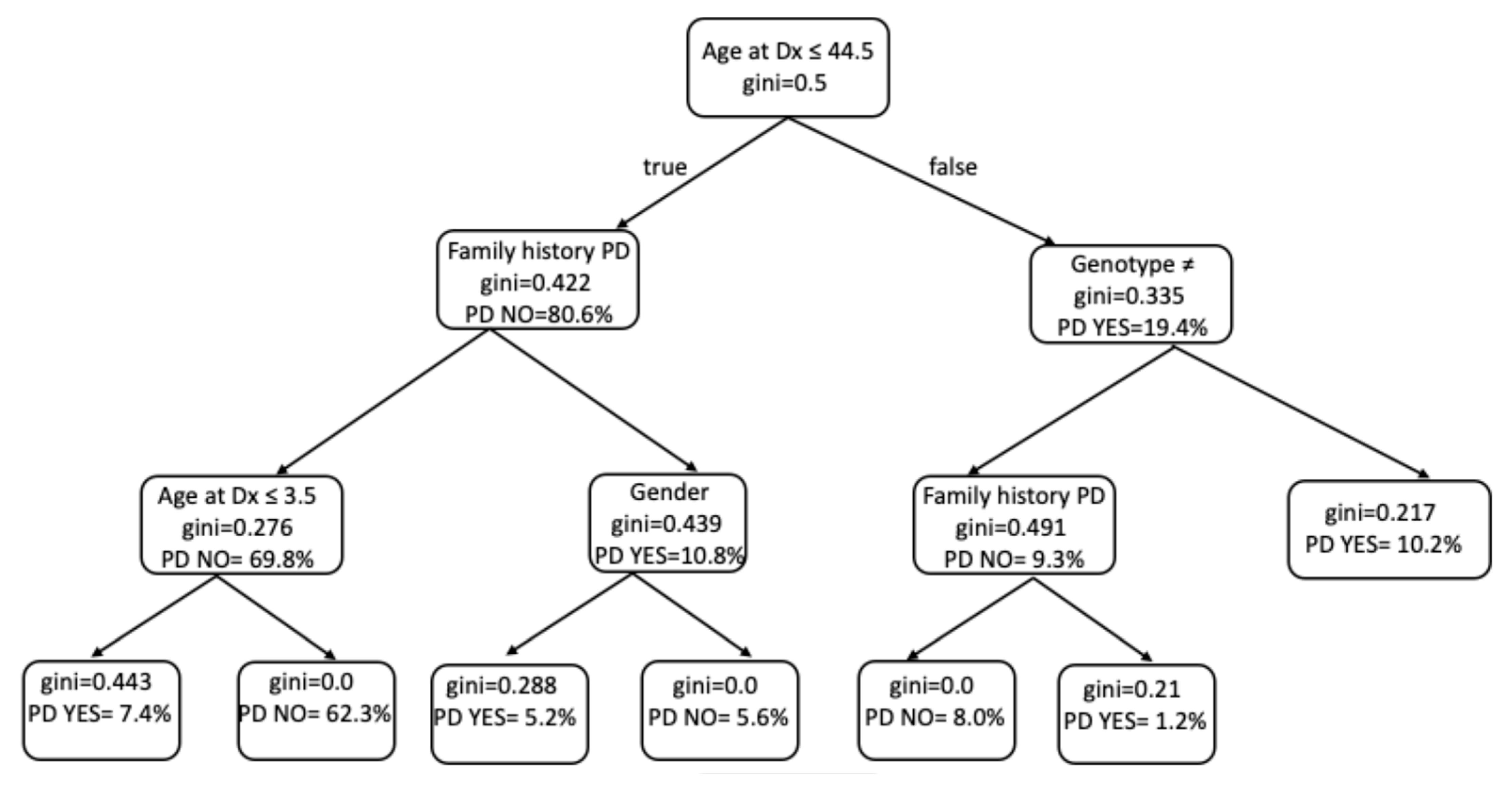
2.3. Development of Parkinson’s Disease
3. Discussion
4. Patients and Methods
4.1. Study Design and Population
4.2. Genetic Analysis
4.3. Biomarker Analysis
4.4. Bone Involvement Assessment
4.5. Statistical Analyses
4.6. Predictive Modeling
5. Conclusions
6. Highlights
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koprivica, V.; Stone, D.L.; Park, J.K.; Callahan, M.; Frisch, A.; Cohen, I.J.; Tayebi, N.; Sidransky, E. Analysis and Classification of 304 Mutant Alleles in Patients with Type 1 and Type 3 Gaucher Disease. Am. J. Hum. Genet. 2000, 66, 1777–1786. [Google Scholar] [CrossRef]
- Nalysnyk, L.; Rotella, P.; Simeone, J.C.; Hamed, A.; Weinreb, N. Gaucher disease epidemiology and natural history: A comprehensive review of the literature. Hematology 2017, 22, 65–73. [Google Scholar] [CrossRef]
- Mistry, P.K.; Weinthal, J.A.; Weinreb, N.J. Disease state awareness in Gaucher disease: A Q&A expert roundtable discussion. Clin. Adv. Hematol. Oncol. H&O 2012, 10 (Suppl. 8), S1–S6. [Google Scholar]
- Dimitriou, E.; Moraitou, M.; Cozar, M.; Serra-Vinardell, J.; Vilageliu, L.; Grinberg, D.; Mavridou, I.; Michelakakis, H. Gaucher disease: Biochemical and molecular findings in 141 patients diagnosed in Greece. Mol. Genet. Metab. Rep. 2020, 24, 100614. [Google Scholar] [CrossRef] [PubMed]
- Fairley, C.; Zimran, A.; Phillips, M.; Cizmarik, M.; Yee, J.; Weinreb, N.; Packman, S. Phenotypic heterogeneity of N370S homozygotes with type I Gaucher disease: An analysis of 798 patients from the ICGG Gaucher Registry. J. Inherit. Metab. Dis. 2008, 31, 738–744. [Google Scholar] [CrossRef]
- Grabowski, G.A. Gaucher disease: Gene frequencies and genotype/phenotype correlations. Genet. Test. 1997, 1, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, P.; Alfonso, P.; Irún, P.; Gort, L.; Chabás, A.; Vilageliu, L.; Grinberg, D.; Sá Miranda, C.M.; Pocovi, M. Mapping the genetic and clinical characteristics of Gaucher disease in the Iberian Peninsula. Orphanet. J. Rare Dis. 2012, 7, 17. [Google Scholar] [CrossRef]
- Hruska, K.S.; LaMarca, M.E.; Scott, C.R.; Sidransky, E. Gaucher disease: Mutation and polymorphism spectrum in the glucocerebrosidase gene (GBA). Hum. Mutat. 2008, 29, 567–583. [Google Scholar] [CrossRef]
- Davidson, B.A.; Hassan, S.; Garcia, E.J.; Tayebi, N.; Sidransky, E. Exploring genetic modifiers of Gaucher disease: The next horizon. Hum. Mutat. 2018, 39, 1739–1751. [Google Scholar] [CrossRef]
- Menkovic, I.; Boutin, M.; Lavoie, P.; Auray-Blais, C. Multiplex Quantification of Plasma Biomarkers for Patients with Gaucher Disease Type 1. Curr. Protoc. 2023, 3, e696. [Google Scholar] [CrossRef]
- Bettioui, T.; Chipeaux, C.; Ben Arfa, K.; Héron, S.; Belmatoug, N.; Franco, M.; de Person, M.; Moussa, F. Development of a new online SPE-HPLC-MS/MS method for the profiling and quantification of sphingolipids and phospholipids in red blood cells—Application to the study of Gaucher’s disease. Anal. Chim. Acta 2023, 1278, 341719. [Google Scholar] [CrossRef]
- Li, W.; Cologna, S.M. Mass spectrometry-based proteomics in neurodegenerative lysosomal storage disorders. Mol. Omics 2022, 18, 256–278. [Google Scholar] [CrossRef]
- Woo, E.G.; Tayebi, N.; Sidransky, E. Next-Generation Sequencing Analysis of GBA1: The Challenge of Detecting Complex Recombinant Alleles. Front. Genet. 2021, 12, 684067. [Google Scholar] [CrossRef]
- Cebolla, J.J.; Giraldo, P.; Gómez, J.; Montoto, C.; Gervas-Arruga, J. Machine Learning-Driven Biomarker Discovery for Skeletal Complications in Type 1 Gaucher Disease Patients. Int. J. Mol. Sci. 2024, 25, 8586. [Google Scholar] [CrossRef]
- Andrade-Campos, M.M.; de Frutos, L.L.; Cebolla, J.J.; Serrano-Gonzalo, I.; Medrano-Engay, B.; Roca-Espiau, M.; Gomez-Barrera, B.; Pérez-Heredia, J.; Iniguez, D.; Giraldo, P. Identification of risk features for complication in Gaucher’s disease patients: A machine learning analysis of the Spanish registry of Gaucher disease. Orphanet J. Rare Dis. 2020, 15, 256. [Google Scholar] [CrossRef]
- Hurvitz, N.; Dinur, T.; Revel-Vilk, S.; Agus, S.; Berg, M.; Zimran, A.; Ilan, Y. A Feasibility Open-Labeled Clinical Trial Using a Second-Generation Artificial-Intelligence-Based Therapeutic Regimen in Patients with Gaucher Disease Treated with Enzyme Replacement Therapy. J. Clin. Med. 2024, 13, 3325. [Google Scholar] [CrossRef]
- Valero-Tena, E.; Roca-Espiau, M.; Verdú-Díaz, J.; Diaz-Manera, J.; Andrade-Campos, M.; Giraldo, P. Advantages of digital technology in the assessment of bone marrow involvement in Gaucher’s disease. Front. Med. 2023, 10, 1098472. [Google Scholar] [CrossRef] [PubMed]
- Basiri, M.; Ghaffari, M.E.; Ruan, J.; Murugesan, V.; Kleytman, N.; Belinsky, G.; Akhavan, A.; Lischuk, A.; Guo, L.; Klinger, K.; et al. Osteonecrosis in Gaucher disease in the era of multiple therapies: Biomarker set for risk stratification from a tertiary referral center. Elife 2023, 12, e87537. [Google Scholar] [CrossRef] [PubMed]
- Charrow, J.; Andersson, H.C.; Kaplan, P.; Kolodny, E.H.; Mistry, P.; Pastores, G.; Rosenbloom, B.E.; Scott, C.R.; Wappner, R.S.; Weinreb, N.J. The Gaucher registry: Demographics and disease characteristics of 1698 patients with Gaucher disease. Arch. Intern. Med. 2000, 160, 2835–2843. [Google Scholar] [CrossRef]
- Elstein, D.; Belmatoug, N.; Bembi, B.; Deegan, P.; Fernandez-Sasso, D.; Giraldo, P.; Göker-Alpan, Ö.; Hughes, D.; Lau, H.; Lukina, E. Twelve Years of the Gaucher Outcomes Survey (GOS): Insights, Achievements, and Lessons Learned from a Global Patient Registry. J. Clin. Med. 2024, 13, 3588. [Google Scholar] [CrossRef] [PubMed]
- Dinur, T.; Zimran, A.; Becker-Cohen, M.; Arkadir, D.; Cozma, C.; Hovakimyan, M.; Oppermann, S.; Demuth, L.; Rolfs, A.; Revel-Vilk, S.; et al. Long Term Follow-Up of 103 Untreated Adult Patients with Type 1 Gaucher Disease. J. Clin. Med. 2019, 8, 1662. [Google Scholar] [CrossRef]
- Choon, Y.W.; Choon, Y.F.; Nasarudin, N.A.; Al Jasmi, F.; Remli, M.A.; Alkayali, M.H.; Mohamad, M.S. Artificial intelligence and database for NGS-based diagnosis in rare disease. Front. Genet. 2024, 14, 1258083. [Google Scholar] [CrossRef]
- Maitra, C.; Seal, D.B.; Das, V.; De, R.K. Unsupervised neural network for single cell Multi-omics INTegration (UMINT): An application to health and disease. Front. Mol. Biosci. 2023, 10, 1184748. [Google Scholar] [CrossRef] [PubMed]
- Tenenbaum, A.; Revel-Vilk, S.; Gazit, S.; Roimi, M.; Gill, A.; Gilboa, D.; Paltiel, O.; Manor, O.; Shalev, V.; Chodick, G.; et al. A machine learning model for early diagnosis of type 1 Gaucher disease using real-life data. J. Clin. Epidemiol. 2024, 175, 111517. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.; Chiorean, A.; Aguiar, M.; Sekulic, D.; Pavlick, P.; Shah, N.; Sniderman King, L.; Génin, M.; Rollot, M.; Blanchon, M.; et al. Development of a rare disease algorithm to identify persons at risk of Gaucher disease using electronic health records in the United States. Orphanet J. Rare Dis. 2023, 18, 280. [Google Scholar] [CrossRef] [PubMed]
- Sidransky, E. Gaucher disease: Complexity in a "simple" disorder. Mol. Genet. Metab. 2004, 83, 6–15. [Google Scholar] [CrossRef]
- Pimenta, J.; Lopes, A.M.; Carracedo, A.; Arenas, M.; Amorim, A.; Comas, D. Spatially explicit analysis reveals complex human genetic gradients in the Iberian Peninsula. Sci. Rep. 2019, 9, 7825. [Google Scholar] [CrossRef]
- Gustavsson, E.K.; Sethi, S.; Gao, Y.; Brenton, J.W.; García-Ruiz, S.; Zhang, D.; Garza, R.; Reynolds, R.H.; Evans, J.R.; Chen, Z.; et al. The annotation of GBA1 has been concealed by its protein-coding pseudogene GBAP1. Sci. Adv. 2024, 10, eadk1296. [Google Scholar] [CrossRef]
- Grabowski, G.A.; Kishnani, P.S.; Alcalay, R.N.; Prakalapakorn, S.G.; Rosenbloom, B.E.; Tuason, D.A.; Weinreb, N.J. Challenges in Gaucher disease: Perspectives from an expert panel. Mol. Genet. Metab. 2025, 145, 109074. [Google Scholar] [CrossRef]
- Londoño, M.A.; Vallejo, J.M.; Manzano, A.C. Normal Development and Maturation of Bone Marrow. Assessment by Magnetic Resonance Imaging. Rev. Colomb. Radiol. 2015, 26, 4206–4212. [Google Scholar]
- Hughes, D.; Mikosch, P.; Belmatoug, N.; Carubbi, F.; Cox, T.; Goker-Alpan, O.; Kindmark, A.; Mistry, P.; Poll, L.; Weinreb, N.; et al. Gaucher Disease in Bone: From Pathophysiology to Practice. J. Bone Miner. Res. 2019, 34, 996–1013. [Google Scholar] [CrossRef]
- Arends, M.; van Dussen, L.; Biegstraaten, M.; Hollak, C.E. Malignancies and monoclonal gammopathy in Gaucher disease; a systematic review of the literature. Br. J. Haematol. 2013, 161, 832–842. [Google Scholar] [CrossRef]
- Huh, Y.E.; Usnich, T.; Scherzer, C.R.; Klein, C.; Chung, S.J. GBA1 Variants and Parkinson’s Disease: Paving the Way for Targeted Therapy. J. Mov. Disord. 2023, 16, 261–278. [Google Scholar] [CrossRef]
- Barrett, M.J.; Giraldo, P.; Capablo, J.L.; Alfonso, P.; Irun, P.; Garcia-Rodriguez, B.; Pocovi, M.; Pastores, G.M. Greater risk of parkinsonism associated with non-N370S GBA1 mutations. J. Inherit. Metab. Dis. 2013, 36, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Schaake, S.; Usnich, T.; Boehm, J.; Steffen, N.; Schell, N.; Krüger, C.; Gül-Demirkale, T.; Bahr, N.; Kleinz, T.; et al. Classification and Genotype-Phenotype Relationships of GBA1 Variants: MDSGene Systematic Review. Mov. Disord. 2025, 40, 605–618. [Google Scholar] [CrossRef]
- Alfonso, P.; Aznarez, S.; Giralt, M.; Pocovi, M.; Giraldo, P.; Spanish Gaucher’s Disease Registry. Mutation analysis and genotype/phenotype relationships of Gaucher disease patients in Spain. J. Hum. Genet. 2007, 52, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Hollak, C.E.; van Weely, S.; van Oers, M.H.; Aerts, J.M. Marked elevation of plasma chitotriosidase activity. A novel hallmark of Gaucher disease. J. Clin. Investig. 1994, 93, 1288–1292. [Google Scholar] [CrossRef]
- Boot, R.G.; Verhoek, M.; de Fost, M.; Hollak, C.E.; Maas, M.; Bleijlevens, B.; van Breemen, M.J.; van Meurs, M.; Boven, L.A.; Laman, J.D.; et al. Marked elevation of the chemokine CCL18/PARC in Gaucher disease: A novel surrogate marker for assessing therapeutic intervention. Blood 2004, 103, 33–39. [Google Scholar] [CrossRef]
- Nilsson, O.; Svennerholm, L. Accumulation of glucosylceramide and glucosylsphingosine (psychosine) in cerebrum and cerebellum in infantile and juvenile Gaucher disease. J. Neurochem. 1982, 39, 709–718. [Google Scholar] [CrossRef] [PubMed]
- Irún, P.; Cebolla, J.J.; López de Frutos, L.; De Castro-Orós, I.; Roca-Espiau, M.; Giraldo, P. LC-MS/MS analysis of plasma glucosylsphingosine as a biomarker for diagnosis and follow-up monitoring in Gaucher disease in the Spanish population. Clin. Chem. Lab. Med. 2020, 58, 798–809. [Google Scholar] [CrossRef]
- Roca, M.; Mota, J.; Alfonso, P.; Pocoví, M.; Giraldo, P. S-MRI score: A simple method for assessing bone marrow involvement in Gaucher disease. Eur. J. Radiol. 2007, 62, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Roca-Espiau, M.; Valero-Tena, E.; Ereño-Ealo, M.J.; Giraldo, P. Structured bone marrow report as an assessment tool in patients with hematopoietic disorders. Quant. Imaging Med. Surg. 2022, 12, 3717–3724. [Google Scholar] [CrossRef] [PubMed]
- Porter, J.L.; Varacallo, M. Osteoporosis. 12 February 2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK441901/ (accessed on 7 April 2022).
- Ganz, M.L.; Stern, S.; Ward, A.; Nalysnyk, L.; Selzer, M.; Hamed, A.; Weinreb, N. A new framework for evaluating the health impacts of treatment for Gaucher disease type 1. Orphanet J. Rare Dis. 2017, 12, 38. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Therneau, T.; Atkinson, B. R Package, Version 4.1–15. rpart: Recursive Partitioning and Regression Trees. R Foundation for Statistical Computing: Vienna, Austria, 2019.
- Bishop, C.M. Pattern Recognition and Machine Learning; Springer: New York, NY, USA, 2006. [Google Scholar]
- Kohavi, R. A study of cross-validation and bootstrap for accuracy estimation and model selection. IJCAI 1995, 14, 1137–1145. [Google Scholar]

| Genotype c.[1226A>G]; [1448T>C] (N = 113) | Genotype c.[1226A>G]; [Other] (N = 195) | ||
|---|---|---|---|
| DEMOGRAPHY | |||
| Gender | ♀/♂ | 55/58 | 92/103 |
| Age at diagnosis (years) | ♀ Mean (min–max) ♂ Mean (min–max) | 28.2 (3–70) 27.4 (3–78) | 26.8 (1–77) 22.7 (1–76) |
| Family history of Parkinson’s disease | Yes/No | 16/97 | 27/168 |
| Dead | Yes/No | 15/107 | 14/180 |
| Survival (years) | ♀ Median (Q1–Q3) Mean (min–max) ♂ Median (Q1–Q3) Mean (min–max) | 64.5 (50–68) 68 (54–82) 64.0 (35.5–76.5) 63 (32–78) | 66.5 (58–70.5) 63.7 (56–73 47.0 (44–48) 55.9 (41.5–76.2) |
| CLINICAL CHARACTERISTICS | |||
| Classification according to GD-DS3 score | Mild (%) Moderate (%) Severe (%) | ♀; ♂ 41.8; 36.2 30.9; 39.6 27.3; 24.1 | ♀; ♂ 34.4; 40.0 30.0; 24.7 35.5; 34.3 |
| Spleen removal | N (%) | 20 (17.7) | 38 (19.5) |
| Liver Volume | MN (multiples of normal) | 1–2 | 1–3 |
| Spleen Volume | MN multiples of normal) | 5–13 | 5–15 |
| Previous Bone Crisis | N (%) | 26 (23.0) | 59 (30.2) |
| IMAGE STUDIES | |||
| S-MRI score | ♀ Mean (min–max) ♂ Mean (min–max) | 7.0 (0–17) 7.0 (0–24) | 8.0 (0–24) 8.0 (0–25) |
| Bone Mineral Density (DXA) * | T score > −1 N (%) T score (−1 to −2.5) N (%) T score < −2.5 N (%) | 64 (59.8) 28 (26.2) 15 (14.0) | 60 (49.2) 33 (27.0) 29 (23.8) |
| ANALYTICAL DATA Mean (min–max) | |||
| Hemoglobin (g/dL) | 12.2 (4.5–17.0) | 12.3 (6.5–18.5) | |
| Leucocytes × 109 (/L) | 6.5 (1.3–23.0 | 6.2 (1.4–18.1) | |
| Platelets × 109 (/L) | 106.1 (4.0–363.0) | 93.1 (7.0–410) | |
| Ferritin (mcg/L) | 620.6 (9.5–1850.0 | 534.9 (14.0–2811.0) | |
| Iron (mg/dL) | 126.3 (13.0–230.2) | 88.2 (24.0–1553.0) | |
| Cholesterol (mg/dL) | 145.7 (33.0–285.0) | 147.6 (64–348) | |
| Triglycerides (mg/dL) | 141.0 (32.0–362.0) | 146.8 (14.0–583.0) | |
| Cholesterol HDL (mg/dL) | 33.7 (14.0–234.0) | 38.6 (11.0–297.0) | |
| Cholesterol LDL (mg/dL) | 86.0 (5.0–148.0) | 87.4 (12.0–300.0) | |
| AST (UI) | 35.9 (9.2–89.0) | 34.1 (12.0–100.0) | |
| ALT (UI) | 28.6 (4.0–82.0) | 29.5 (6.0–142.0) | |
| GGT (UI) | 41.2 (2.4–297.0) | 35.7 (7.0–174.0) | |
| Bilirubin (mg/dL) | 0.98 (0.11–4.0) | 1.06 (0.17–4.70) | |
| IgG (mg/dL) | 1332.8 (523.0–2453.0)) | 1302.0 (565.0–2520.0) | |
| IgA (mg/dL) | 254.0 (22.0–765.0) | 273.0 (90.0–2108.0) | |
| IgM (mg/dL) | 245.8 (49.0–949.0) | 209.0 (25.0–532.0) | |
| Monoclonal Gammopathy | Yes/No | 3/103 | 8/167 |
| Polyclonal Gammopathy | Yes/No | 10/93 | 12/55 |
| DIAGNOSIS | |||
| GCase activity (nmol/mL/h) | Mean (min–max) | 0.9 (0.1–2.0) | 0.76 (0.1–2.2) |
| BIOMARKERS | |||
| Chitotriosidase ChT (nmol/mL/h) | Mean (min–max) | 11,322.0 (190.0–65,497.0) | 10,290.0 (526.0–57,466.0) |
| CCL18/PARC (ng/mL) | Mean (min–max) | 568.3 (52.0–3763.0) | 716.6 (51.0–3895.0) |
| Glucosylsphingosine (ng/mL) | Mean (min–max) | 100.5 (5.1–320.0) | 126.7 (0.88–836.0) |
| FOLLOW-UP (5–26 YEARS) | |||
| Age at start therapy (years) | Mean (min–max) | 29.0 (2–69) | 27.9 (1–74) |
| Cumulated time on therapy (years) | Mean (min–max) | 19.3 (2–30) | 20.7 (1–31) |
| New bone crisis | N (%) | 8 (7.1) | 17 (9.5) |
| Joint replacement | N (%) | 6 (5.3) | 14 (7.2) |
| Neoplasia | N (%) | 9 (7.9) | 10 (5.1) |
| Parkinson’s disease | N (%) | 2 (1.7) | 12 (6.1) |
| Other comorbidities ** | N (%) | 34 (30.0) | 31(31.0) |
| Type of therapy | ERT N (%) SRT N (%) None N (%) | 64 (56.6) 34 (30) 15 (13.4) | 123 (63.1) 63 (32.3) 9 (4.6) |
| cDNA NM_000157.4 | Protein NP_000148 | Protein (-39 aa) | N Alleles | % | Functional Severity |
|---|---|---|---|---|---|
| c.[1226A>G] | p.Asn409Ser | N370S | 195 | ||
| Deletions | 21 | 10.8 | SEVERE 74 (37.9%) | ||
| Recombinations | 16 | 8.2 | |||
| Insertions | 16 | 8.2 | |||
| c.[108G>A] | p.TrpW36Ter | W(-4)X | 4 | 2.1 | |
| c.[256C>T] | p.Arg86Ter | R47X | 4 | 2.1 | |
| IVS4-2A>G +c.(-203)A>G | 3 | 1.5 | |||
| c.[604C>T] | p.Arg202Ter | R163X | 3 | 1.5 | |
| c.[886C>T] | p.Arg296Ter | R257X | 3 | 1.5 | |
| c.[662C>T] | p.Pro221Leu | P182L | 2 | 1.0 | |
| c.[622C>T] | p.Gln208Ter | Q169X | 1 | 0.5 | |
| c.[1992C>T] | p.Arg398Ter | R359X | 1 | 0.5 | |
| c.[721G>A] | p.Gly241Arg | G202R | 14 | 7.2 | MODERATE 82 (42.1%) |
| c.[475C>T] | p.Arg159Trp | R120W | 12 | 6.2 | |
| c.[700G>T] | p.Gly234Trp | G195W | 8 | 4.1 | |
| c.[1054T>C] | p.Tyr352His | Y313H | 7 | 3.6 | |
| c.[1246G>A] | p.Gly416Ser | G377S | 7 | 3.6 | |
| c.[680A>G] | p.Asn227Ser | N188S | 7 | 3.6 | |
| c.[1289C>T] | p.Pro430Leu | P391L | 6 | 3.1 | |
| c.[517A>C] | p.Thr173Pro | T134P | 4 | 2.1 | |
| c.[701G>A] | p.Gly234Glu | G195E | 3 | 1.5 | |
| c.[887G>A] | p.Arg296Gln | R257Q | 3 | 1.5 | |
| c.[1124T>C] | p.Leu375Pro | L336P | 3 | 1.5 | |
| c.[1090G>T] | p.Gly364Trp | G325W | 2 | 1.0 | |
| c.[1051T>A] | p.Trp351Arg | W312R | 1 | 0.5 | |
| c.[1300C>T] | p.Arg434Cys | R395C | 1 | 0.5 | |
| c.[1304A>C] | p.Asn435Thr | N396T | 1 | 0.5 | |
| c.[1309G>A] | p.Val437Ile | V398I | 1 | 0.5 | |
| c.[1583T>C] | p.Ile528Thr | I489T | 1 | 0.5 | |
| c.[1604G>A] | p.Arg535His | R496H | 1 | 0.5 | |
| c.[1505G>A] | p.Arg502His | R463H | 5 | 2.6 | MILD 39 (20.0%) |
| c.[155C>T] | p.Ser52Leu | S13L | 3 | 1.5 | |
| c.[160G>A] | p.Val54Met | V15M | 2 | 1.0 | |
| c.[455G>A] | p.Gly152Glu | G113E | 2 | 1.0 | |
| c.[485T>C] | p.Met162Thr | M123T | 2 | 1.0 | |
| c.[681T>A] | p.Asn227Lys | N188K | 2 | 1.0 | |
| c.[706C>T] | p.Leu236Phe | L197F | 2 | 1.0 | |
| c.[731A>G] | p.Tyr244Cys | Y205C | 2 | 1.0 | |
| c.[754T>A] | p.Phe252Ile | F213I | 2 | 1.0 | |
| c.[1193G>A] | p.Arg398Gln | R359Q | 2 | 1.0 | |
| c.[437C>T] | p.Ser146Leu | S107L | 1 | 0.5 | |
| c.[485T>A] | p.Met162Lys | M123K | 1 | 0.5 | |
| c.[496G>T] | p.Asp166Tyr | D127Y | 1 | 0.5 | |
| c.[508C>T] | p.Arg170Cys | R131C | 1 | 0.5 | |
| c.[625C>T] | p.Arg209Cys | R170C | 1 | 0.5 | |
| c.[695G>A] | p.Gly232Glu | G193E | 1 | 0.5 | |
| c.[700G>T] | p.Gly234Trp | G195T | 1 | 0.5 | |
| c.[709A>G] | p.Lys237Glu | K198E | 1 | 0.5 | |
| c.[746C>T] | p.Ala249Val | A210V | 1 | 0.5 | |
| c.[914C>T] | p.Pro305Leu | P266L | 1 | 0.5 | |
| c.[928A>C] | p.Ser310Arg | S271R | 1 | 0.5 | |
| c.[970C>T] | p.Arg324Cys | R285C | 1 | 0.5 | |
| c.[1207A>C] | p.Ser403Arg | S364R | 1 | 0.5 | |
| c.[1208G>A] | p.Ser403Asn | S364N | 1 | 0.5 | |
| c.[1348T>A] | p.Phe450Ile | F411I | 1 | 0.5 |
| Variable | Coef | Std_Err | z | p |
|---|---|---|---|---|
| Intercept | −0.634942 | 0.216538 | −2.932239 | 0.003365 |
| Age at Dx | 0.009794 | 0.006185 | 1.58356 | 0.113294 |
| S-MRI | 0.058678 | 0.022938 | 2.558053 | 0.010526 |
| Splenectomy | 0.678604 | 0.314432 | 2.158188 | 0.030913 |
| Family History of PD | 2.214037 | 0.388695 | 5.696077 | 1.23 × 10−8 |
| Gender | 0.636954 | 0.227262 | 2.802729 | 0.005067 |
| Genotype * | 0.455415 | 0.230146 | 1.97881 | 0.047837 |
| Variable | Coef | Std_Err | z | p |
|---|---|---|---|---|
| Intercept | −2.492958 | 0.328728 | −7.58364 | 3.35 × 10−14 |
| Age at Dx | 0.048291 | 0.008237 | 5.862482 | 4.56 × 10−9 |
| S-MRI | −0.046485 | 0.029223 | −1.590691 | 1.12 × 10−1 |
| Splenectomy | 1.293751 | 0.376888 | 3.432717 | 5.98 × 10−4 |
| Family History of PD | 2.214037 | 0.388695 | 5.696077 | 1.23 × 10−8 |
| Gender | −1.052176 | 0.29415 | −3.577003 | 3.48 × 10−4 |
| Genotype * | 2.146594 | 0.334419 | 6.418882 | 1.37 × 10−10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serrano-Gonzalo, I.; Bauza, F.; Lopez de Frutos, L.; Arevalo-Vargas, I.; Roca-Espiau, M.; Andrade-Campos, M.; Valero-Tena, E.; Roca-Esteve, S.; Iniguez, D.; Giraldo, P. Clinical Variability and Genotype–Phenotype Correlation in Spanish Patients with Type 1 Gaucher Disease: A Focus on Non-c.[1226A>G]; [1448T>C] Genotypes. Int. J. Mol. Sci. 2025, 26, 10088. https://doi.org/10.3390/ijms262010088
Serrano-Gonzalo I, Bauza F, Lopez de Frutos L, Arevalo-Vargas I, Roca-Espiau M, Andrade-Campos M, Valero-Tena E, Roca-Esteve S, Iniguez D, Giraldo P. Clinical Variability and Genotype–Phenotype Correlation in Spanish Patients with Type 1 Gaucher Disease: A Focus on Non-c.[1226A>G]; [1448T>C] Genotypes. International Journal of Molecular Sciences. 2025; 26(20):10088. https://doi.org/10.3390/ijms262010088
Chicago/Turabian StyleSerrano-Gonzalo, Irene, Francisco Bauza, Laura Lopez de Frutos, Isidro Arevalo-Vargas, Mercedes Roca-Espiau, Marcio Andrade-Campos, Esther Valero-Tena, Sonia Roca-Esteve, David Iniguez, and Pilar Giraldo. 2025. "Clinical Variability and Genotype–Phenotype Correlation in Spanish Patients with Type 1 Gaucher Disease: A Focus on Non-c.[1226A>G]; [1448T>C] Genotypes" International Journal of Molecular Sciences 26, no. 20: 10088. https://doi.org/10.3390/ijms262010088
APA StyleSerrano-Gonzalo, I., Bauza, F., Lopez de Frutos, L., Arevalo-Vargas, I., Roca-Espiau, M., Andrade-Campos, M., Valero-Tena, E., Roca-Esteve, S., Iniguez, D., & Giraldo, P. (2025). Clinical Variability and Genotype–Phenotype Correlation in Spanish Patients with Type 1 Gaucher Disease: A Focus on Non-c.[1226A>G]; [1448T>C] Genotypes. International Journal of Molecular Sciences, 26(20), 10088. https://doi.org/10.3390/ijms262010088






