Genetic and Pathogenic Overlaps Between Autism Spectrum Disorder and Alzheimer’s Disease: Evolutionary Features and Opportunities for Drug Repurposing
Abstract
1. Introduction
2. Results
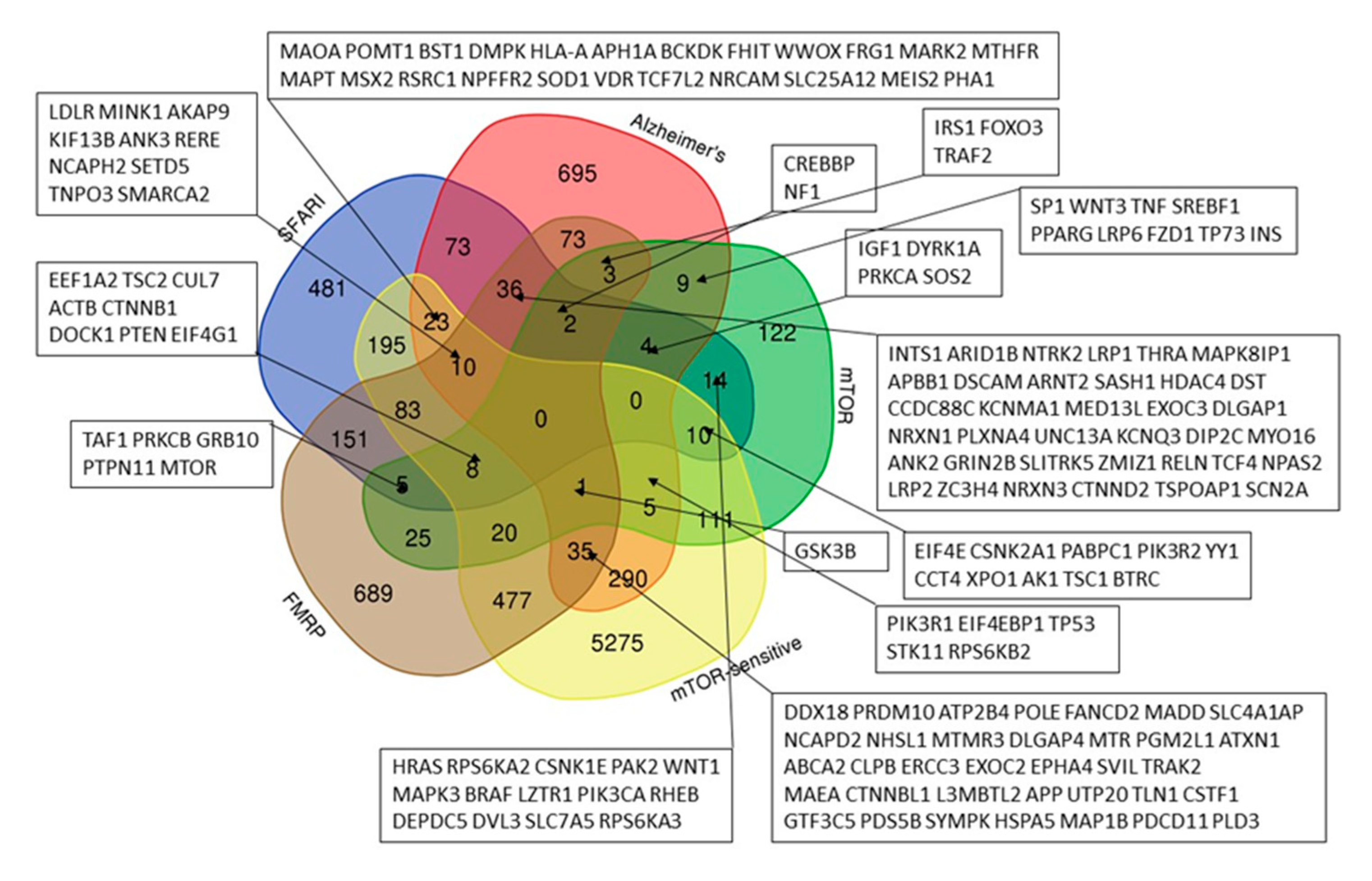
2.1. SFARI Gene Database and AD Genes Comparative Gene-Set and Pathway Analysis
2.2. Analysis of Evolutionary Characteristics of Gene Sets
2.2.1. Phylostratigraphic Age of Genes (PAI-Based Analysis)
2.2.2. Evolutionary Variability of Genes (DI-Based Analysis)
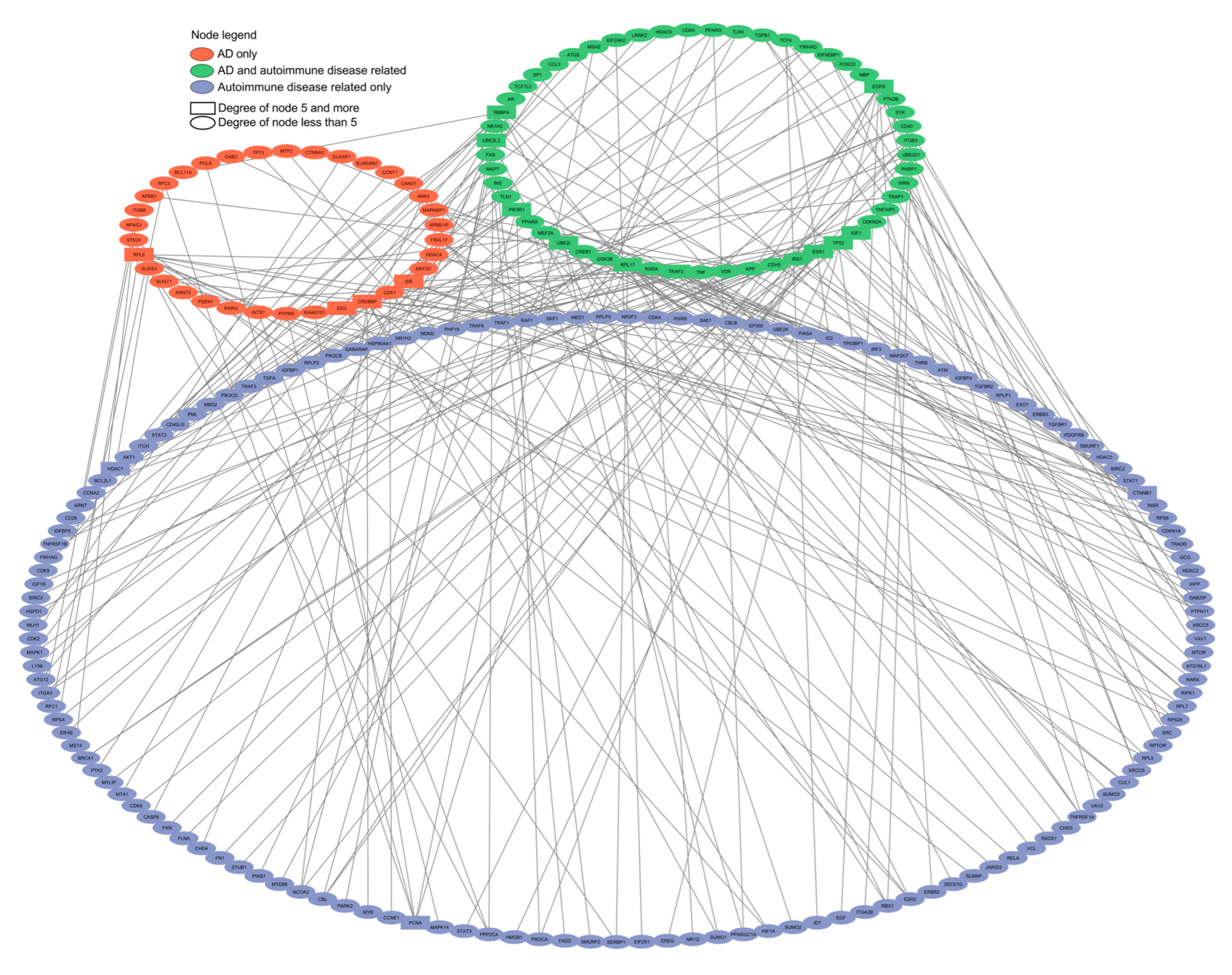
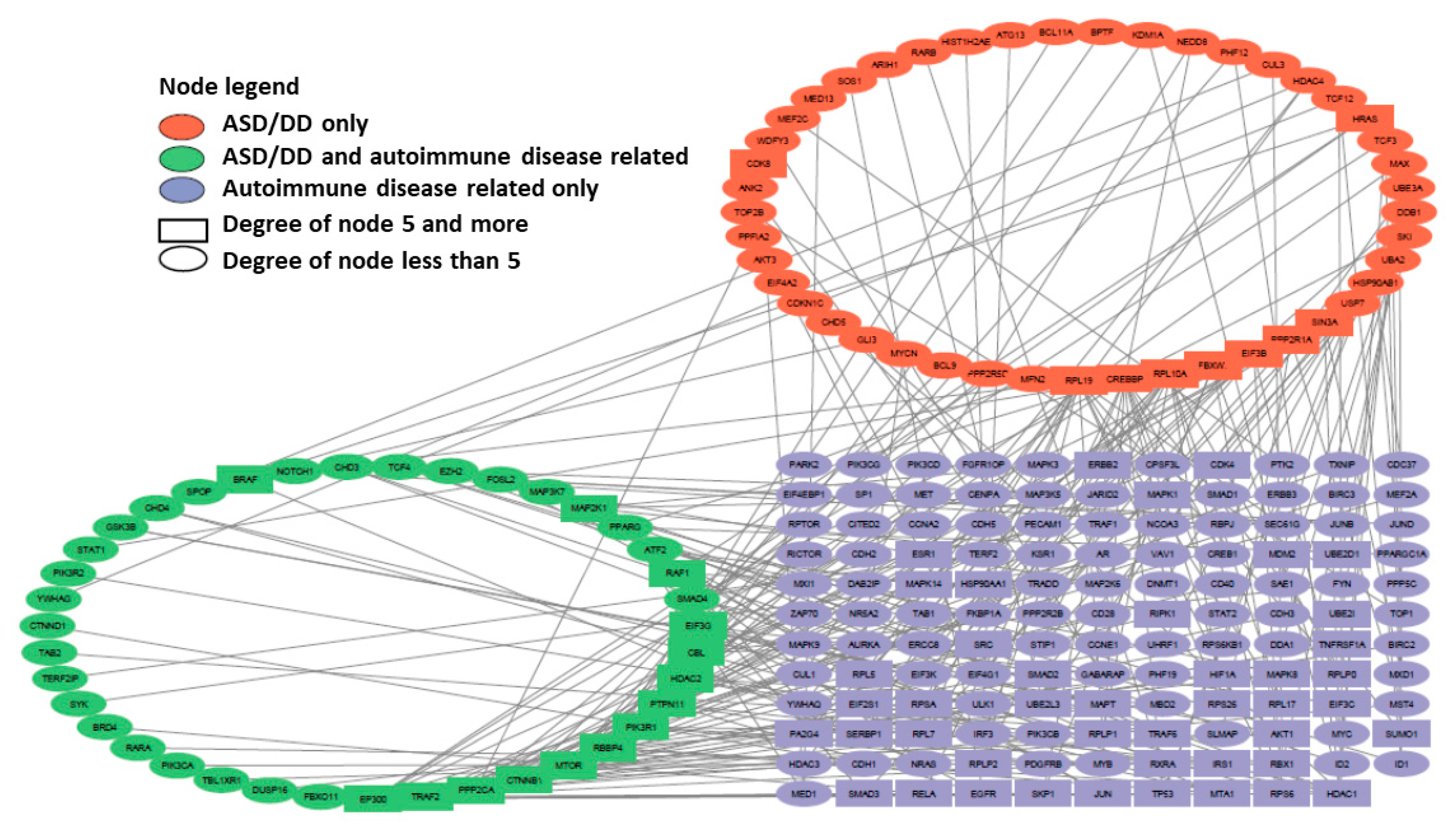
2.3. Comparative Network Analysis of Genes Predisposing to Autism and Alzheimer’s Disease with Genes of Autoimmune Diseases
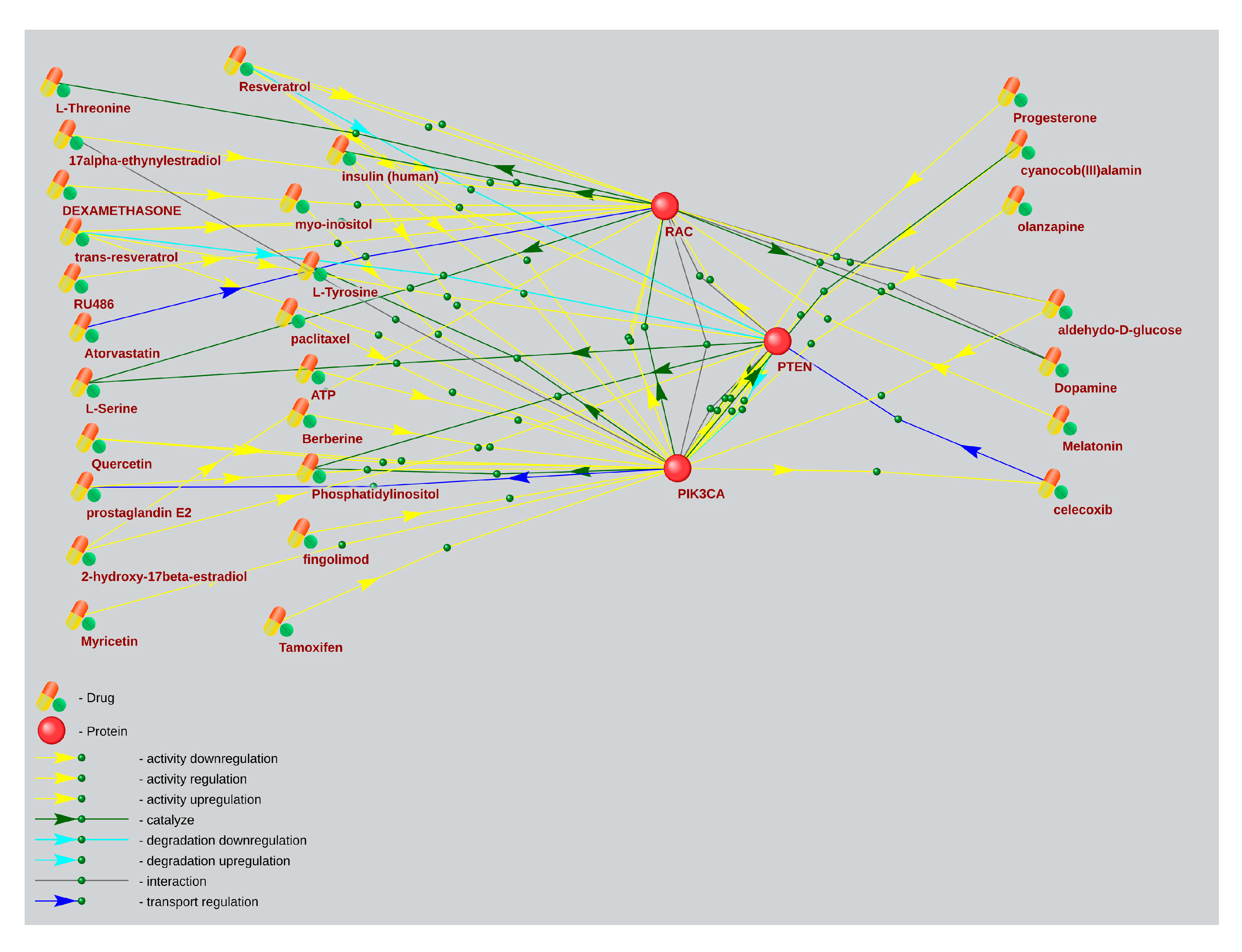
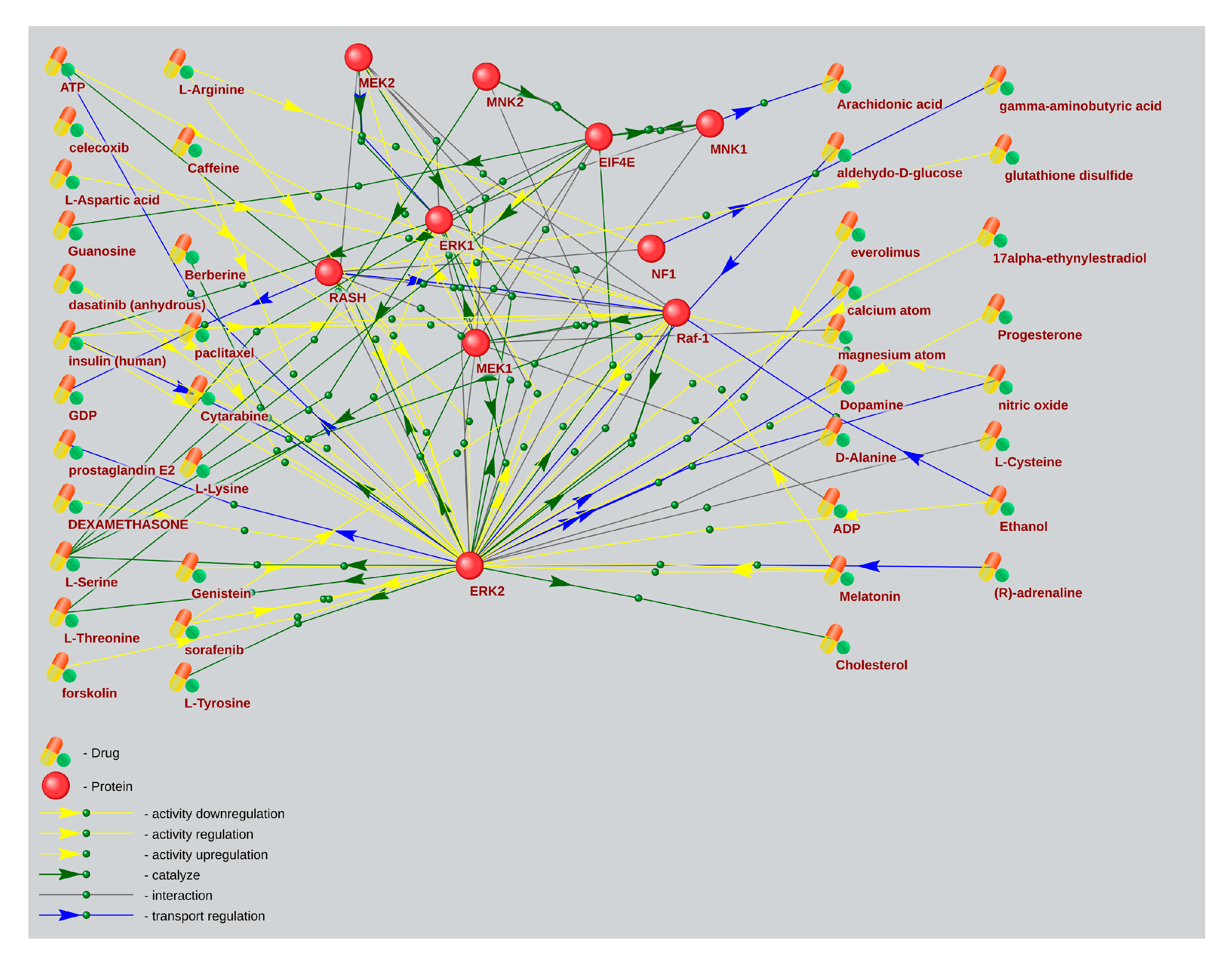
2.4. The Associative Network Analysis of the Main Elements of the mTOR Pathway and Substances Regulating Their Activity Using for ASD and AD Treatment
3. Discussion
4. Materials and Methods
4.1. Extracting Genes from Diverse Data Sources and Gene-Set Analysis
- Genes implicated in autism susceptibility (from SFARI Gene database released 20 October 2022 [22])—1095 genes;
- Autism predisposition genes (41588_2022_1104_MOESM3_ESM.xlsx [20])—185 genes;
- Autism and developmental delay predisposition genes (ASD/DD) (41588_2022_1104_MOESM3_ESM.xlsx [20])—664 genes;
- Genes predisposing to Alzheimer’s disease (13195_2017_252_MOESM2_ESM.doc [55])—430 genes;
- Alzheimer’s disease predisposition genes (from the ADVP database [21])—956 genes;
- Genes included in the mTOR signaling network (Table S1.xlsx [23]—248 genes and KEGG database [58]—153 genes)—341 genes;
- mTOR-sensitive genes (mTOR-sensitive 5UTR.xlsx [59])—6543 genes;
- Genes associated with autoimmune diseases (from the GAAD [60])—4186 genes.
- All protein-coding genes of the human genome for which PAI and DI values were calculated [24]—19478 genes.
4.2. Phylostratigraphic Analysis and Divergence Analysis
4.3. Network Construction
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ASD | Autism Spectrum Disorder |
| AD | Alzheimer’s disease |
| SFARI | Simon’s Foundation Autism Research Initiative |
| mTOR | mechanistic target of rapamycin |
| FMRP | fragile X mental retardation protein |
| PAI | Phylostratigraphic Age Index |
| DI | Divergence Index |
| DD | developmental delay |
References
- Center for Disease Control and Prevention. Available online: https://www.cdc.gov/autism/data-research/?CDC_AAref_Val=https://www.cdc.gov/ncbddd/autism/data.html (accessed on 27 May 2025).
- Soysal, P.; Tan, S.G. The prevalence and co-incidence of geriatric syndromes in older patients with early-stage Alzheimer’s disease and dementia with Lewy bodies. Aging Clin. Exp. Res. 2021, 33, 2599–2603. [Google Scholar] [CrossRef] [PubMed]
- Rajan, K.B.; Weuve, J.; Barnes, L.L.; McAninch, E.A.; Wilson, R.S.; Evans, D.A. Population estimate of people with clinical Alzheimer’s disease and mild cognitive impairment in the United States (2020–2060). Alzheimers. Dement. 2021, 17, 1966–1975. [Google Scholar] [CrossRef]
- Mencer, S.; Kartawy, M.; Lendenfeld, F.; Soluh, H.; Tripathi, M.K.; Khaliulin, I.; Amal, H. Proteomics of autism and Alzheimer’s mouse models reveal common alterations in mTOR signaling pathway. Transl. Psychiatry 2021, 11, 480. [Google Scholar] [CrossRef]
- Hughes, H.K.; Mills Ko, E.; Rose, D.; Ashwood, P. Immune Dysfunction and Autoimmunity as Pathological Mechanisms in Autism Spectrum Disorders. Front. Cell. Neurosci. 2018, 12, 405. [Google Scholar] [CrossRef]
- Ashwood, P.; van deWater, J. Is autism an autoimmune disease? Autoimmun. Rev. 2004, 3, 557–562. [Google Scholar] [CrossRef]
- Edmiston, E.; Ashwood, P.; van de Water, J. Autoimmunity, Autoantibodies, and Autism Spectrum Disorder. Biol. Psychiatry 2017, 81, 383–390. [Google Scholar] [CrossRef]
- DiStasio, M.M.; Nagakura, I.; Nadler, M.J.; Anderson, M.P. T lymphocytes and cytotoxic astrocyte blebs correlate across autism brains. Ann. Neurol. 2019, 86, 885–898. [Google Scholar] [CrossRef]
- Chen, X.; Holtzman, D.M. Emerging roles of innate and adaptive immunity in Alzheimer’s disease. Immunity 2022, 55, 2236–2254. [Google Scholar] [CrossRef] [PubMed]
- Meier-Stephenson, F.S.; Meier-Stephenson, V.C.; Carter, M.D.; Meek, A.R.; Wang, Y.; Pan, L.; Chen, Q.; Jacobo, S.; Wu, F.; Lu, E.; et al. Alzheimer’s disease as an autoimmune disorder of innate immunity endogenously modulated by tryptophan metabolites. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2022, 8, e12283. [Google Scholar] [CrossRef] [PubMed]
- Shuid, A.N.; Jayusman, P.A.; Shuid, N.; Ismail, J.; Kamal, N.N.; Naina Mohamed, I. Update on Atypicalities of Central Nervous System in Autism Spectrum Disorder. Brain Sci. 2020, 10, 309. [Google Scholar] [CrossRef]
- Delgoffe, G.M.; Pollizzi, K.N.; Waickman, A.T.; Heikamp, E.; Meyers, D.J.; Horton, M.R.; Xiao, B.; Worley, P.F.; Powell, J.D. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat. Immunol. 2011, 12, 295–303. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, D.T.; Liu, X.G. mTOR signaling in T cell immunity and autoimmunity. Int. Rev. Immunol. 2015, 34, 50–66. [Google Scholar] [CrossRef]
- Ortiz-González, X.R. Mitochondrial Dysfunction: A Common Denominator in Neurodevelopmental Disorders? Dev. Neurosci. 2021, 43, 222–229. [Google Scholar] [CrossRef]
- Lenzi, P.; Ferese, R.; Biagioni, F.; Fulceri, F.; Busceti, C.L.; Falleni, A.; Gambardella, S.; Frati, A.; Fornai, F. Rapamycin Ameliorates Defects in Mitochondrial Fission and Mitophagy in Glioblastoma Cells. Int. J. Mol. Sci. 2021, 22, 5379. [Google Scholar] [CrossRef]
- Thellung, S.; Corsaro, A.; Nizzari, M.; Barbieri, F.; Florio, T. Autophagy Activator Drugs: A New Opportunity in Neuroprotection from Misfolded Protein Toxicity. Int. J. Mol. Sci. 2019, 20, 901. [Google Scholar] [CrossRef]
- Richardson, A.; Galvan, V.; Lin, A.L.; Oddo, S. How longevity research can lead to therapies for Alzheimer’s disease: The rapamycin story. Exp. Gerontol. 2015, 68, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Caccamo, A.; Majumder, S.; Richardson, A.; Strong, R.; Oddo, S. Molecular interplay between mammalian target of rapamycin (mTOR), amyloid-beta, and Tau: Effects on cognitive impairments. J. Biol. Chem. 2010, 285, 13107–13120. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, U.; Nalavadi, V.; Nakamoto, M.; Thomas, G.; Ceman, S.; Bassell, G.J.; Warren, S.T. S6K1 phosphorylates and regulates fragile X mental retardation protein (FMRP) with the neuronal protein synthesis-dependent mammalian target of rapamycin (mTOR) signaling cascade. J. Biol. Chem. 2008, 283, 18478–18482. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.M.; Satterstrom, F.K.; Peng, M.; Brand, H.; Collins, R.L.; Dong, S.; Wamsley, B.; Klei, L.; Wang, L.; Hao, S.P.; et al. Rare coding variation provides insight into the genetic architecture and phenotypic context of autism. Nat. Genet. 2022, 54, 1320–1331. [Google Scholar] [CrossRef]
- Kuksa, P.P.; Liu, C.L.; Fu, W.; Qu, L.; Zhao, Y.; Katanic, Z.; Clark, K.; Kuzma, A.B.; Ho, P.C.; Tzeng, K.T.; et al. Alzheimer’s Disease Variant Portal: A Catalog of Genetic Findings for Alzheimer’s Disease. J. Alzheimers. Dis. 2022, 86, 461–477. [Google Scholar] [CrossRef]
- Abrahams, B.S.; Arking, D.E.; Campbell, D.B.; Meord, H.C.; Morrow, E.M.; Weiss, L.A.; Menashe, I.; Wadkins, T.; Banerjee-Basu, S.; Packer, A. SFARI Gene 2.0: A community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism 2013, 4, 36. [Google Scholar] [CrossRef] [PubMed]
- Caron, E.; Ghosh, S.; Matsuoka, Y.; Ashton-Beaucage, D.; Therrien, M.; Lemieux, S.; Perreault, C.; Roux, P.P.; Kitano, H. A comprehensive map of the mTOR signaling network. Mol. Syst. Biol. 2010, 6, 453. [Google Scholar] [CrossRef]
- Piovesan, A.; Antonaros, F.; Vitale, L.; Strippoli, P.; Pelleri, M.C.; Caracausi, M. Human protein-coding genes and gene feature statistics in 2019. BMC Res. Notes 2019, 12, 315. [Google Scholar] [CrossRef]
- Feng, L.; Wang, G.; Song, Q.; Feng, X.; Su, J.; Ji, G.; Li, M. Proteomics revealed an association between ribosome-associated proteins and amyloid beta deposition in Alzheimer’s disease. Metab. Brain Dis. 2024, 39, 263–282. [Google Scholar] [CrossRef]
- Liu, P.P.; Han, X.; Li, X.; Dai, S.K.; Xu, Y.J.; Jiao, L.F.; Du, H.Z.; Zhao, L.H.; Li, R.F.; Teng, Z.Q.; et al. An EED/PRC2-H19 Loop Regulates Cerebellar Development. Adv. Sci. 2025, 12, e2403591. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yuan, S.; Niu, X.; Kelleher, R.; Sheridan, H. ESR1 dysfunction triggers neuroinflammation as a critical upstream causative factor of the Alzheimer’s disease process. Aging 2022, 14, 8595–8614. [Google Scholar] [CrossRef] [PubMed]
- Barral, S.; Reitz, C.; Small, S.A.; Mayeux, R. Genetic variants in a ‘cAMP element binding protein’ (CREB)-dependent histone acetylation pathway influence memory performance in cognitively healthy elderly individuals. Neurobiol. Aging 2014, 35, 2881.e7–2881.e10. [Google Scholar] [CrossRef]
- Zhu, Y.; Huang, Y.; Tang, T.; Xie, Y. HDAC1 and HDAC2 orchestrate Wnt signaling to regulate neural progenitor transition during brain development. iScience 2024, 27, 110600. [Google Scholar] [CrossRef]
- Lewerissa, E.I.; Nadif Kasri, N.; Linda, K. Epigenetic regulation of autophagy-related genes: Implications for neurodevelopmental disorders. Autophagy 2024, 20, 15–28. [Google Scholar] [CrossRef]
- Xie, P.-L.; Zheng, M.-Y.; Han, R.; Chen, W.-X.; Mao, J.-H. Pharmacological mTOR inhibitors in ameliorating Alzheimer’s disease: Current review and perspectives. Front. Pharmacol. 2024, 15, 1366061. [Google Scholar] [CrossRef]
- Ehninger, D.; Han, S.; Shilyansky, C.; Zhou, Y.; Li, W.; Kwiatkowski, D.J.; Ramesh, V.; Silva, A.J. Reversalof learning deficits in a Tsc2+/− mouse model of tuberoussclerosis. Nat. Med. 2008, 14, 843–848. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cano, L.; Azidane Chenlo, S.; Sabido-Vera, R.; Sirci, F.; Durham, L.; Guney, E. Translating precision medicine for autism spectrum disorder: A pressing need. Drug Discov. Today 2023, 28, 103486. [Google Scholar] [CrossRef]
- Mustafin, Z.S.; Lashin, S.A.; Matushkin, Y.G. Phylostratigraphic analysis of gene networks of human diseases. Vavilovskii Zhurnal Genet. Selektsii 2021, 25, 46–56. [Google Scholar] [CrossRef]
- Nagy, P.L.; Griesenbeck, J.; Kornberg, R.D.; Cleary, M.L. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc. Natl. Acad. Sci. USA 2002, 99, 90–94. [Google Scholar] [CrossRef]
- Li, S.; Qiao, Y.; Di, Q.; Le, X.; Zhang, L.; Zhang, X.; Zhang, C.; Cheng, J.; Zong, S.; Koide, S.S.; et al. Interaction of SH3P13 and DYDC1 protein: A germ cell component that regulates acrosome biogenesis during spermiogenesis. Eur. J. Cell Biol. 2009, 88, 509–520, Erratum in Eur. J. Cell Biol. 2009, 98, 51–52. [Google Scholar] [CrossRef]
- Demenkov, P.S.; Ivanisenko, T.V.; Kolchanov, N.A.; Ivanisenko, V.A. ANDVisio: A new tool for graphic visualization and analysis of literature mined associative gene networks in the ANDSystem. Silico Biol. 2011, 11, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Ivanisenko, V.A.; Saik, O.V.; Ivanisenko, N.V.; Tiys, E.S.; Ivanisenko, T.V.; Demenkov, P.S.; Kolchanov, N.A. ANDSystem: An Associative Network Discovery System for automated literature mining in the field of biology. BMC Syst. Biol. 2015, 9 (Suppl. S2), S2. [Google Scholar] [CrossRef]
- Wang, Z.; Cao, B.; Ji, P.; Yao, F. Propofol inhibits tumor angiogenesis through targeting VEGF/VEGFR and mTOR/eIF4E signaling. Biochem. Biophys. Res. Commun. 2021, 555, 13–18. [Google Scholar] [CrossRef]
- Bossmann, M.; Ackermann, B.W.; Thome, U.H.; Laube, M. Signaling Cascade Involved in Rapid Stimulation of Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) by Dexamethasone. Int. J. Mol. Sci. 2017, 18, 1807. [Google Scholar] [CrossRef]
- Morita, M.; Suyama, H.; Igishi, T.; Shigeoka, Y.; Kodani, M.; Hashimoto, K.; Takeda, K.; Sumikawa, T.; Shimizu, E. Dexamethasone inhibits paclitaxel-induced cytotoxic activity through retinoblastoma protein dephosphorylation in non-small cell lung cancer cells. Int. J. Oncol. 2007, 30, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; He, D.; Song, E.; Jiang, M.; Song, Y. Celecoxib enhances the sensitivity of non-small-cell lung cancer cells to radiation-induced apoptosis through downregulation of the Akt/mTOR signaling pathway and COX-2 expression. PLoS ONE 2019, 14, e0223760, Erratum in PLoS ONE 2019, 14, e0224843. [Google Scholar] [CrossRef]
- Asadabadi, M.; Mohammadi, M.R.; Ghanizadeh, A.; Modabbernia, A.; Ashrafi, M.; Hassanzadeh, E.; Forghani, S.; Akhondzadeh, S. Celecoxib as adjunctive treatment to risperidone in children with autistic disorder: A randomized, double-blind, placebo-controlled trial. Psychopharmacology 2013, 225, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Lashgari, N.A.; Roudsari, N.M.; Zadeh, S.S.T.; Momtaz, S.; Abbasifard, M.; Reiner, Ž.; Abdolghaffari, A.H.; Sahebkar, A. Statins block mammalian target of rapamycin pathway: A possible novel therapeutic strategy for inflammatory, malignant and neurodegenerative diseases. Inflammopharmacology 2023, 31, 57–75. [Google Scholar] [CrossRef]
- Okubo, S.; Uto, T.; Goto, A.; Tanaka, H.; Nishioku, T.; Yamada, K.; Shoyama, Y. Berberine Induces Apoptotic Cell Death via Activation of Caspase-3 and -8 in HL-60 Human Leukemia Cells: Nuclear Localization and Structure-Activity Relationships. Am. J. Chin. Med. 2017, 45, 1497–1511. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhang, Y.; Wang, J.; Wei, L.; Li, H.; Hanley, G.; Zhao, M.; Li, Y.; Yin, D. Beta-arrestin 2 modulates resveratrol-induced apoptosis and regulation of Akt/GSK3ß pathways. Biochim. Biophys. Acta 2010, 1800, 912–918. [Google Scholar] [CrossRef]
- Russo, M.; Milito, A.; Spagnuolo, C.; Carbone, V.; Rosén, A.; Minasi, P.; Lauria, F.; Russo, G.L. CK2 and PI3K are direct molecular targets of quercetin in chronic lymphocytic leukaemia. Oncotarget 2017, 8, 42571–42587. [Google Scholar] [CrossRef]
- Lim, J.Y.; Lee, J.Y.; Byun, B.J.; Kim, S.H. Fisetin targets phosphatidylinositol-3-kinase and induces apoptosis of human B lymphoma Raji cells. Toxicol. Rep. 2015, 2, 984–989. [Google Scholar] [CrossRef]
- Marchezan, J.; Deckmann, I.; da Fonseca, G.C.; Margis, R.; Riesgo, R.; Gottfried, C. Resveratrol Treatment of Autism Spectrum Disorder-A Pilot Study. Clin. Neuropharmacol. 2022, 45, 122–127. [Google Scholar] [CrossRef]
- Alvarez-Arellano, L.; Salazar-García, M.; Corona, J.C. Neuroprotective Effects of Quercetin in Pediatric Neurological Diseases. Molecules 2020, 25, 5597. [Google Scholar] [CrossRef]
- Song, X.; Tan, L.; Wang, M.; Ren, C.; Guo, C.; Yang, B.; Ren, Y.; Cao, Z.; Li, Y.; Pei, J. Myricetin: A review of the most recent research. Biomed. Pharmacother. 2021, 134, 111017. [Google Scholar] [CrossRef] [PubMed]
- Xiong, D.; Pan, J.; Zhang, Q.; Szabo, E.; Miller, M.S.; Lubet, R.A.; You, M.; Wang, Y. Bronchial airway gene expression signatures in mouse lung squamous cell carcinoma and their modulation by cancer chemopreventive agents. Oncotarget 2017, 8, 18885–18900. [Google Scholar] [CrossRef]
- Rameh, L.E.; York, J.D.; Blind, R.D. Inositol phosphates dynamically enhance stability, solubility, and catalytic activity of mTOR. J. Biol. Chem. 2025, 301, 108095. [Google Scholar] [CrossRef]
- van Sadelhoff, J.H.J.; Perez Pardo, P.; Wu, J.; Garssen, J.; van Bergenhenegouwen, J.; Hogenkamp, A.; Hartog, A.; Kraneveld, A.D. The Gut-Immune-Brain Axis in Autism Spectrum Disorders; A Focus on Amino Acids. Front. Endocrinol. 2019, 10, 247. [Google Scholar] [CrossRef]
- Hu, Y.S.; Xin, J.; Hu, Y.; Zhang, L.; Wang, J. Analyzing the genes related to Alzheimer’s disease via a network and pathway-based approach. Alzheimers Res. Ther. 2017, 9, 29. [Google Scholar] [CrossRef]
- Darnell, J.C.; Van Driesche, S.J.; Zhang, C.; Hung, K.Y.; Mele, A.; Fraser, C.E.; Stone, E.F.; Chen, C.; Fak, J.J.; Chi, S.W.; et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 2011, 146, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Jansen, A.; Dieleman, G.C.; Smit, A.B.; Verhage, M.; Verhulst, F.C.; Polderman, T.J.C.; Posthuma, D. Gene-set analysis shows association between FMRP targets and autism spectrum disorder. Eur. J. Hum. Genet. 2017, 25, 863–868. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Gandin, V.; Masvidal, L.; Hulea, L.; Gravel, S.P.; Cargnello, M.; McLaughlan, S.; Cai, Y.; Balanathan, P.; Morita, M.; Rajakumar, A.; et al. nanoCAGE reveals 5′ UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res. 2016, 26, 636–648. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Hao, X.; Chen, W.H.; Mu, S. GAAD: A Gene and Autoimmiune Disease Association Database. Genom. Proteom. Bioinform. 2018, 16, 252–261. [Google Scholar] [CrossRef]
- Trifonova, E.A.; Klimenko, A.I.; Mustafin, Z.S.; Lashin, S.A.; Kochetov, A.V. The mTOR Signaling Pathway Activity and Vitamin D Availability Control the Expression of Most Autism Predisposition Genes. Int. J. Mol. Sci. 2019, 20, 6332. [Google Scholar] [CrossRef]
- Domazet-Loso, T.; Brajković, J.; Tautz, D. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 2007, 23, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Domazet-Lošo, M.; Široki, T.; Šimičević, K.; Domazet-Lošo, T. Macroevolutionary dynamics of gene family gain and loss along multicellular eukaryotic lineages. Nat. Commun. 2024, 15, 2663. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Ivanov, R.A.; Mukhin, A.M.; Kazantsev, F.V.; Mustafin, Z.S.; Afonnikov, D.A.; Matushkin, Y.G.; Lashin, S.A. Orthoweb: A Software Package for Evolutionary Analysis of Gene Networks. Vavilovskii Zhurnal Genet. Selektsii 2025, 28, 874–881. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
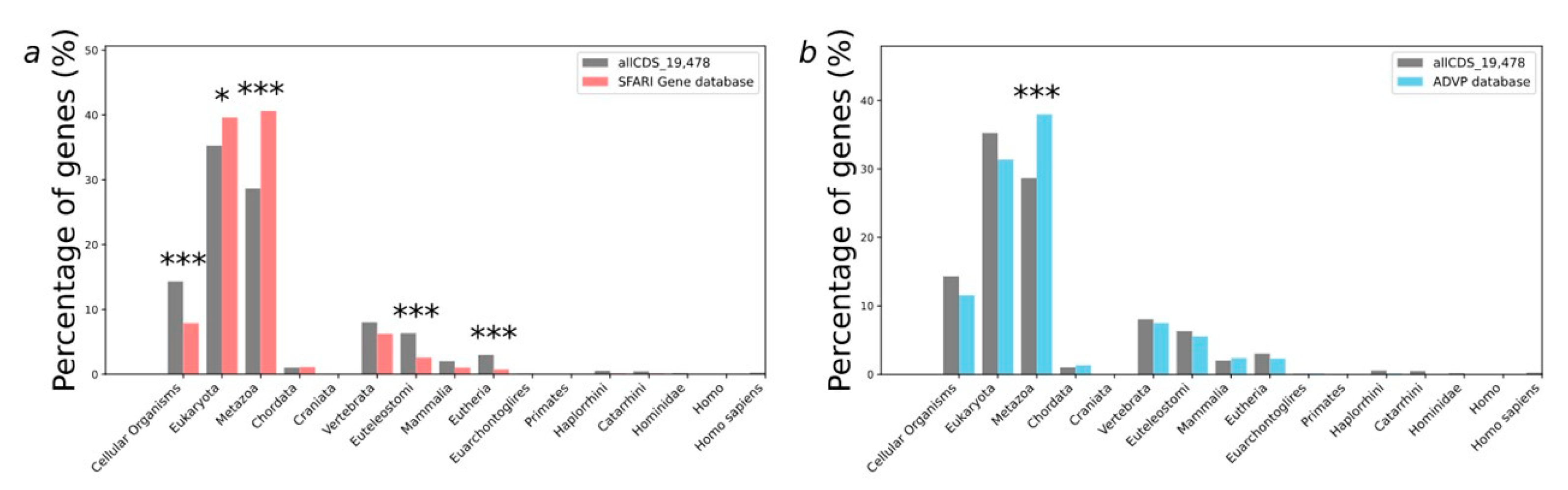



| Gene Set Number | Gene Sets | PAI | DI |
|---|---|---|---|
| 1 | Genes implicated in autism susceptibility (from SFARI Gene database) [22] | 2.86 | 0.24 |
| 2 | Autism predisposition genes [20] | 2.80 | 0.16 |
| 3 | Autism and developmental delay predisposition genes (ASD/DD) [20] | 2.59 | 0.15 |
| 5 | Alzheimer’s disease predisposition genes (from the ADVP database) [21] | 3.18 | 0.33 |
| 7 | Genes included in the mTOR signaling network [23] | 2.29 | 0.18 |
| 10 | All protein-coding human genes [24] | 3.29 | 0.38 |
| PAI | Phylostratum |
|---|---|
| 1 | Cellular Organisms |
| 2 | Eukaryota |
| 3 | Metazoa |
| 4 | Chordata |
| 5 | Craniata |
| 6 | Vertebrata |
| 7 | Euteleostomi |
| 8 | Mammalia |
| 9 | Eutheria |
| 10 | Euarchontoglires |
| 11 | Primates |
| 12 | Haplorrhini |
| 13 | Catarrhini |
| 14 | Hominidae |
| 15 | Homo |
| 16 | Homo sapiens |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trifonova, E.A.; Pashchenko, A.A.; Ivanov, R.A.; Kochetov, A.V.; Lashin, S.A. Genetic and Pathogenic Overlaps Between Autism Spectrum Disorder and Alzheimer’s Disease: Evolutionary Features and Opportunities for Drug Repurposing. Int. J. Mol. Sci. 2025, 26, 10066. https://doi.org/10.3390/ijms262010066
Trifonova EA, Pashchenko AA, Ivanov RA, Kochetov AV, Lashin SA. Genetic and Pathogenic Overlaps Between Autism Spectrum Disorder and Alzheimer’s Disease: Evolutionary Features and Opportunities for Drug Repurposing. International Journal of Molecular Sciences. 2025; 26(20):10066. https://doi.org/10.3390/ijms262010066
Chicago/Turabian StyleTrifonova, Ekaterina A., Anna A. Pashchenko, Roman A. Ivanov, Alex V. Kochetov, and Sergey A. Lashin. 2025. "Genetic and Pathogenic Overlaps Between Autism Spectrum Disorder and Alzheimer’s Disease: Evolutionary Features and Opportunities for Drug Repurposing" International Journal of Molecular Sciences 26, no. 20: 10066. https://doi.org/10.3390/ijms262010066
APA StyleTrifonova, E. A., Pashchenko, A. A., Ivanov, R. A., Kochetov, A. V., & Lashin, S. A. (2025). Genetic and Pathogenic Overlaps Between Autism Spectrum Disorder and Alzheimer’s Disease: Evolutionary Features and Opportunities for Drug Repurposing. International Journal of Molecular Sciences, 26(20), 10066. https://doi.org/10.3390/ijms262010066







