Modulation of Nuclear Factor Kappa B Signaling and microRNA Profiles by Adalimumab in LPS-Stimulated Keratinocytes
Abstract
1. Introduction
2. Results
2.1. Cytotoxicity Results
2.2. Transcriptomic Profiling Reveals LPS-Induced NF-κB Activation and Its Attenuation by Adalimumab
2.3. Validation of NF-κB mRNA Expression Using RT-qPCR
2.4. Profiling of miRNA Expression Levels
2.5. Quantification of BCL2L1, CXCL2, MAP3K7, BIRC2, BIRC3, TNFAIP3, and XIAP Protein Levels
2.6. Protein–Protein Interaction (PPI) Network Analysis via STRING Database
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Keratinocyte Cell Culture
4.3. Total Ribonucleic Acid (RNA) Extraction
4.4. NF-κB Gene Expression Profiling via Oligonucleotide Microarrays
4.5. Identification of miRNAs Potentially Regulating NF-κB Gene Expression Using miRNA Microarrays
4.6. RT-qPCR
4.7. Protein Quantification by ELISA
4.8. Statistical Analysis
Protein–Protein Interaction (PPI) Networks and Gene Ontology (GO) Enrichment Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Abbreviation | Full Term |
| NF-κB | Nuclear factor kappa B |
| TNF-α | Tumor necrosis factor alpha |
| miRNA | MicroRNA |
| LPS | Lipopolysaccharide |
| DEG | Differentially expressed gene |
| RT-qPCR | Reverse transcription quantitative polymerase chain reaction |
| ELISA | Enzyme-linked immunosorbent assay |
| PPI | Protein–protein interaction |
| GO | Gene Ontology |
| HaCaT | Human immortalized keratinocyte |
| GEO | Gene Expression Omnibus |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| CXCL2 | C-X-C motif chemokine ligand 2 |
| IRAK1 | Interleukin-1 receptor–associated kinase 1 |
| MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 (TAK1) |
| BIRC2/3 | Baculoviral inhibitor of apoptosis repeat-containing protein 2/3 |
| XIAP | X-linked inhibitor of apoptosis protein |
| TNFAIP3 | Tumor necrosis factor alpha-induced protein 3 (A20) |
| IKBKB | Inhibitor of nuclear factor kappa-B kinase subunit beta (IKKβ) |
| SYK | Spleen tyrosine kinase |
| TAB1 | TGF-beta-activated kinase 1/MAP3K7 binding protein 1 |
| TAB2 | TGF-beta-activated kinase 1/MAP3K7 binding protein 2 |
| PASI | Psoriasis Area and Severity Index |
| JAK/STAT | Janus kinase/signal transducer and activator of transcription |
| MAPK | Mitogen-activated protein kinase |
| STAT3 | Signal transducer and activator of transcription 3 |
| AP-1 | Activator protein 1 |
| FDR | False discovery rate |
| DMEM | Dulbecco’s Modified Eagle Medium |
| FBS | Fetal bovine serum |
| PBS | Phosphate-buffered saline |
| RIPA | Radioimmunoprecipitation assay |
| STRING | Search Tool for the Retrieval of Interacting Genes/Proteins |
Appendix A
| ID | Term Description | Observed Gene Count | Background Gene Count | Strength | Signal | False Discovery Rate | Proteins in Network |
|---|---|---|---|---|---|---|---|
| GO:0038095 | Fc-epsilon receptor signaling pathway | 7 | 23 | 2.48 | 4.92 | 2.65 × 10−12 | PLCG1, MAP3K7, SYK, IKBKB, TRAF6, PLCG2, BTK |
| GO:0050851 | Antigen receptor-mediated signaling pathway | 8 | 141 | 1.75 | 3.08 | 6.22 × 10−10 | MAPK1, PLCG1, MAP3K7, SYK, IKBKB, TRAF6, PLCG2, BTK |
| GO:0038061 | NIK/NF-kappaB signaling | 6 | 36 | 2.22 | 3.67 | 1.16 × 10−9 | TRAF2, BIRC3, MAP3K7, IRAK1, TRAF6, BIRC2 |
| GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 7 | 68 | 2.01 | 3.55 | 5.05 × 10−10 | TRAF2, BIRC3, TAB2, MAP3K7, IRAK1,TNF, IKBKB, TRAF6, BIRC2, TNFAIP3, PLCG2 |
| GO:0002223 | Stimulatory C-type lectin receptor signaling pathway | 5 | 22 | 2.35 | 3.36 | 1.42 × 10−8 | MAP3K7, SYK, IKBKB, TRAF6, PLCG2 |
| GO:0043122 | Regulation of I-kappaB kinase/NF-kappaB signaling | 11 | 255 | 1.63 | 3.29 | 2.45 × 10−12 | TRAF2, BIRC3, TAB2, MAP3K7, IRAK1, TNF, IKBKB, TRAF6, BIRC2, TNFAIP3, PLCG2 |
| GO:0043123 | Positive regulation of I-kappaBkinase/NF-kappaB signaling | 9 | 191 | 1.67 | 3.05 | 1.51 × 10−10 | TRAF2, BIRC3, TAB2, MAP3K7, IRAK1, TNF, IKBKB, TRAF6, BIRC2 |
| GO:0033209 | Tumor necrosis factor-mediatedsignaling pathway | 6 | 56 | 2.02 | 3.16 | 9.38 × 10−9 | TRAF2, BIRC3, TNF, IKBKB, TRAF6, BIRC2 |
| GO:0002224 | Toll-like receptor signaling pathway | 6 | 61 | 1.99 | 3.06 | 1.38 × 10−8 | MAP3K7, IRAK1, TNF, TRAF6, PLCG2, BTK |
| GO:0002221 | Pattern recognition receptor signalingpathway | 7 | 101 | 1.83 | 3.04 | 3.66 × 10−9 | MAP3K7, IRAK1, TNF, TRAF6, TNFAIP3, PLCG2, BTK |
References
- Rendon, A.; Schäkel, K. Psoriasis Pathogenesis and Treatment. Int. J. Mol. Sci. 2019, 20, 1475. [Google Scholar] [CrossRef] [PubMed]
- Schön, M.P.; Wilsmann-Theis, D. Current Developments and Perspectives in Psoriasis. J. Dtsch. Dermatol. Ges. 2023, 21, 363–372. [Google Scholar] [CrossRef]
- Lee, H.-J.; Kim, M. Challenges and Future Trends in the Treatment of Psoriasis. Int. J. Mol. Sci. 2023, 24, 13313. [Google Scholar] [CrossRef]
- Prinz, J.C.; Choon, S.E.; Griffiths, C.E.M.; Merola, J.F.; Morita, A.; Ashcroft, D.M.; Viguier, M. Prevalence, Comorbidities and Mortality of Generalized Pustular Psoriasis: A Literature Review. J. Eur. Acad. Dermatol. Venereol. 2023, 37, 256–273. [Google Scholar] [CrossRef]
- Reich, A.; Adamski, Z.; Chodorowska, G.; Kaszuba, A.; Krasowska, D.; Lesiak, A.; Maj, J.; Narbutt, J.; Osmola-Mańkowska, A.J.; Owczarczyk-Saczonek, A. Łuszczyca. Rekomendacje Diagnostyczno-Terapeutyczne Polskiego Towarzystwa Dermatologicznego. Część 1. Dermatol. Rev./Prz. Dermatol. 2020, 107, 92. [Google Scholar] [CrossRef]
- Zhang, Q.; Xie, J.; Li, G.; Wang, F.; Lin, J.; Yang, M.; Du, A.; Zhang, D.; Han, L. Psoriasis Treatment Using Indigo Naturalis: Progress and Strategy. J. Ethnopharmacol. 2022, 297, 115522. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Chen, Y.; Yu, Q.; Shi, Y. Biologic and Small-Molecule Therapies for Moderate-to-Severe Psoriasis: Focus on Psoriasis Comorbidities. BioDrugs 2023, 37, 35–55. [Google Scholar] [CrossRef]
- Grabarek, B.O.; Wcisło-Dziadecka, D.L.; Mazurek, U. Signaling Pathways Associated with Interleukins 12 and 23 and Epigenetic Mechanism of Sequential-Specific Regulation of Expression of Related Genes by miRNAs as Molecular Markers for Assessing the Potential of Anti-Cytokine Therapies. Przegl. Dermatol. 2019, 106, 71–80. [Google Scholar] [CrossRef]
- Petit, R.G.; Cano, A.; Ortiz, A.; Espina, M.; Prat, J.; Muñoz, M.; Severino, P.; Souto, E.B.; García, M.L.; Pujol, M.; et al. Psoriasis: From Pathogenesis to Pharmacological and Nano-Technological-Based Therapeutics. Int. J. Mol. Sci. 2021, 22, 4983. [Google Scholar] [CrossRef]
- StrzaLka-Mrozik, B.; Krzaczyński, J. Farmakologiczne i Niefarmakologiczne Metody Terapii luszczycy Ze Szczególnym Uwzględnieniem Leków Biologicznych. Farm. Pol. 2020, 76, 333–343. [Google Scholar]
- Bellinato, F.; Gisondi, P.; Mason, E.; Ricci, P.; Maurelli, M.; Girolomoni, G. Real-Life Effectiveness of Adalimumab Biosimilars in Patients with Chronic Plaque Psoriasis. Dermatol. Ther. 2022, 12, 1303–1311. [Google Scholar] [CrossRef]
- Du, Y.; Yan, Q.; Chen, M.; Dong, Z.; Wang, F. Efficacy of Adalimumab in Pediatric Generalized Pustular Psoriasis: Case Series and Literature Review. J. Dermatol. Treat. 2022, 33, 2862–2868. [Google Scholar] [CrossRef]
- Wcisło-Dziadecka, D.; Grabarek, B.; Gola, J.; Plewka, A. The Comparison of Effectiveness of Therapy with Ustekinumab and Etanercept in Psoriatic Patients during 48 Weeks’ Observation. Postep. Dermatol. Alergol. 2021, 38, 173–175. [Google Scholar] [CrossRef]
- Ożóg, M.K.; Grabarek, B.O.; Wierzbik-Strońska, M.; Świder, M. Neurological Complications of Biological Treatment of Psoriasis. Life 2022, 12, 118. [Google Scholar] [CrossRef]
- Queiro, R.; Coto, P.; González-Lara, L.; Coto, E. Genetic Variants of the NF-κB Pathway: Unraveling the Genetic Architecture of Psoriatic Disease. Int. J. Mol. Sci. 2021, 22, 13004. [Google Scholar] [CrossRef] [PubMed]
- Goldminz, A.M.; Au, S.C.; Kim, N.; Gottlieb, A.B.; Lizzul, P.F. NF-κB: An Essential Transcription Factor in Psoriasis. J. Dermatol. Sci. 2013, 69, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Lin, L.; Zhang, Z.; Zhang, H.; Hu, H. Targeting NF-κB Pathway for the Therapy of Diseases: Mechanism and Clinical Study. Signal Transduct. Target. Ther. 2020, 5, 209. [Google Scholar] [CrossRef]
- Oeckinghaus, A.; Hayden, M.S.; Ghosh, S. Crosstalk in NF-κB Signaling Pathways. Nat. Immunol. 2011, 12, 695–708. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, Z.; Li, W.; Kang, Y.; Xu, Z.; Li, X.; Gao, Y.; Qi, Y. MAPKs/AP-1, Not NF-κB, Is Responsible for MCP-1 Production in TNF-α-Activated Adipocytes. Adipocyte 2022, 11, 477–486. [Google Scholar] [CrossRef]
- Ageeva, T.; Rizvanov, A.; Mukhamedshina, Y. NF-κB and JAK/STAT Signaling Pathways as Crucial Regulators of Neuroinflammation and Astrocyte Modulation in Spinal Cord Injury. Cells 2024, 13, 581. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, Y.; Cui, L.; Shi, Y.; Guo, C. Advances in the Pathogenesis of Psoriasis: From Keratinocyte Perspective. Cell Death Dis. 2022, 13, 81. [Google Scholar] [CrossRef]
- Ferrari, D.; Casciano, F.; Secchiero, P.; Reali, E. Purinergic Signaling and Inflammasome Activation in Psoriasis Pathogenesis. Int. J. Mol. Sci. 2021, 22, 9449. [Google Scholar] [CrossRef] [PubMed]
- Silvagni, E.; Missiroli, S.; Perrone, M.; Patergnani, S.; Boncompagni, C.; Bortoluzzi, A.; Govoni, M.; Giorgi, C.; Alivernini, S.; Pinton, P.; et al. From Bed to Bench and Back: TNF-α, IL-23/IL-17A, and JAK-Dependent Inflammation in the Pathogenesis of Psoriatic Synovitis. Front. Pharmacol. 2021, 12, 672515. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, J.; Lu, J.; Wu, J.; Zhang, X.; Ma, T.; Wu, X.; Zhu, Q.; Chen, Z.; Tai, Z. Interventions in Cytokine Signaling: Novel Horizons for Psoriasis Treatment. Front. Immunol. 2025, 16, 1573905. [Google Scholar] [CrossRef]
- García-Domínguez, M. The Role of IL-23 in the Development of Inflammatory Diseases. Biology 2025, 14, 347. [Google Scholar] [CrossRef]
- Liu, T.; Li, S.; Ying, S.; Tang, S.; Ding, Y.; Li, Y.; Qiao, J.; Fang, H. The IL-23/IL-17 Pathway in Inflammatory Skin Diseases: From Bench to Bedside. Front. Immunol. 2020, 11, 594735. [Google Scholar] [CrossRef]
- Ehst, B.; Wang, Z.; Leitenberger, J.; McClanahan, D.; De La Torre, R.; Sawka, E.; Ortega-Loayza, A.G.; Strunck, J.; Greiling, T.; Simpson, E.; et al. Synergistic Induction of IL-23 by TNFα, IL-17A, and EGF in Keratinocytes. Cytokine 2021, 138, 155357. [Google Scholar] [CrossRef] [PubMed]
- Krzysztofik, M.; Brzewski, P.; Kulbat, A.; Masajada, M.; Richter, K.; Wysocki, W.M. The IL-23/Th17 Pathway Inhibitors in the Treatment of Psoriasis and the Risk of Skin Malignancies: A Review. Postępy Dermatol. Alergol. 2024, 41, 552–559. [Google Scholar] [CrossRef]
- Nair, R.P.; Duffin, K.C.; Helms, C.; Ding, J.; Stuart, P.E.; Goldgar, D.; Gudjonsson, J.E.; Li, Y.; Tejasvi, T.; Feng, B.-J.; et al. Genome-Wide Scan Reveals Association of Psoriasis with IL-23 and NF-kappaB Pathways. Nat. Genet. 2009, 41, 199–204. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, S.; Zheng, G.; Huang, J.; Songyang, Z.; Zhao, X.; Lin, X. Gain-of-Function Mutation of Card14 Leads to Spontaneous Psoriasis-like Skin Inflammation through Enhanced Keratinocyte Response to IL-17A. Immunity 2018, 49, 66–79.e5. [Google Scholar] [CrossRef]
- Manils, J.; Webb, L.V.; Howes, A.; Janzen, J.; Boeing, S.; Bowcock, A.M.; Ley, S.C. CARD14E138A Signalling in Keratinocytes Induces TNF-Dependent Skin and Systemic Inflammation. eLife 2020, 9, e56720. [Google Scholar] [CrossRef]
- Xuan, T.-Q.; Gong, G.; Du, H.; Liu, C.; Wu, Y.; Bao, G.; Ma, Q.; Zhen, D. Protective Effect of Pteryxin on LPS-Induced Acute Lung Injury via Modulating MAPK/NF-κB Pathway and NLRP3 Inflammasome Activation. J. Ethnopharmacol. 2022, 286, 114924. [Google Scholar] [CrossRef]
- Romo-García, M.F.; Bastian, Y.; Zapata-Zuñiga, M.; Macías-Segura, N.; Castillo-Ortiz, J.D.; Lara-Ramírez, E.E.; Fernández-Ruiz, J.C.; Berlanga-Taylor, A.J.; González-Amaro, R.; Ramos-Remus, C. Identification of Putative miRNA Biomarkers in Early Rheumatoid Arthritis by Genome-Wide Microarray Profiling: A Pilot Study. Gene 2019, 720, 144081. [Google Scholar] [CrossRef]
- Sabina, S.; Panico, A.; Mincarone, P.; Leo, C.G.; Garbarino, S.; Grassi, T.; Bagordo, F.; De Donno, A.; Scoditti, E.; Tumolo, M.R. Expression and Biological Functions of miRNAs in Chronic Pain: A Review on Human Studies. Int. J. Mol. Sci. 2022, 23, 6016. [Google Scholar] [CrossRef] [PubMed]
- Mompeón, A.; Pérez-Cremades, D.; Paes, A.B.; Sanchis, J.; Ortega-Paz, L.; Andrea, R.; Brugaletta, S.; Sabate, M.; Novella, S.; Dantas, A.P. Circulating miRNA Fingerprint and Endothelial Function in Myocardial Infarction: Comparison at Acute Event and One-Year Follow-Up. Cells 2022, 11, 1823. [Google Scholar] [CrossRef] [PubMed]
- Alhamdan, F.; Greulich, T.; Daviaud, C.; Marsh, L.M.; Pedersen, F.; Thölken, C.; Pfefferle, P.I.; Bahmer, T.; Potaczek, D.P.; Tost, J.; et al. Identification of Extracellular Vesicle microRNA Signatures Specifically Linked to Inflammatory and Metabolic Mechanisms in Obesity-associated Low Type-2 Asthma. Allergy 2023, 78, 2944–2958. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Xu, W.; Doctor, A.; Driss, A.; Nezhat, C.; Sidell, N.; Taylor, R.N.; Thompson, W.E.; Chowdhury, I. TNFα-Induced Altered miRNA Expression Links to NF-κB Signaling Pathway in Endometriosis. Inflammation 2023, 46, 2055–2070. [Google Scholar] [CrossRef]
- Fei, M.; Li, Z.; Cao, Y.; Jiang, C.; Lin, H.; Chen, Z. MicroRNA-182 Improves Spinal Cord Injury in Mice by Modulating Apoptosis and the Inflammatory Response via IKKβ/NF-κB. Lab. Investig. 2021, 101, 1238–1253. [Google Scholar] [CrossRef]
- Han, P.; Sunada-Nara, K.; Kawashima, N.; Fujii, M.; Wang, S.; Kieu, T.Q.; Yu, Z.; Okiji, T. MicroRNA-146b-5p Suppresses Pro-Inflammatory Mediator Synthesis via Targeting TRAF6, IRAK1, and RELA in Lipopolysaccharide-Stimulated Human Dental Pulp Cells. Int. J. Mol. Sci. 2023, 24, 7433, Erratum in Int. J. Mol. Sci. 2024, 25, 2049. [Google Scholar] [CrossRef]
- Engel, A.; Ludwig, N.; Grandke, F.; Wagner, V.; Kern, F.; Fehlmann, T.; Schmartz, G.P.; Aparicio-Puerta, E.; Henn, D.; Walch-Rückheim, B.; et al. Skin Treatment with Non-Thermal Plasma Modulates the Immune System through miR-223-3p and Its Target Genes. RNA Biol. 2024, 21, 651–664. [Google Scholar] [CrossRef]
- Guelfi, G.; Capaccia, C.; Anipchenko, P.; Ciancabilla, F.; Oommen, O.P.; Bufalari, A.; Zerani, M.; Maranesi, M. Mimic miRNA and Anti-miRNA Activated Scaffolds as a Therapeutic Strategy to Promote Bone, Cartilage, and Skin Regeneration. Macromol 2024, 4, 165–189. [Google Scholar] [CrossRef]
- Trinidad-Barnech, J.M.; Fort, R.S.; Trinidad Barnech, G.; Garat, B.; Duhagon, M.A. Transcriptome-Wide Analysis of microRNA-mRNA Correlations in Tissue Identifies microRNA Targeting Determinants. Noncoding RNA 2023, 9, 15. [Google Scholar] [CrossRef]
- Jibing, C.; Weiping, L.; Yuwei, Y.; Bingzheng, F.; Zhiran, X. Exosomal microRNA-Based Therapies for Skin Diseases. Regen. Ther. 2024, 25, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Lopez, L.I.; Choudhary, V.; Bollag, W.B. Updated Perspectives on Keratinocytes and Psoriasis: Keratinocytes Are More Than Innocent Bystanders. PTT 2022, 12, 73–87. [Google Scholar] [CrossRef]
- Kamata, M.; Tada, Y. Crosstalk: Keratinocytes and Immune Cells in Psoriasis. Front. Immunol. 2023, 14, 1286344. [Google Scholar] [CrossRef]
- Wang, X.; Yao, Y.; Li, Y.; Guo, S.; Li, Y.; Zhang, G. Experimental Study on the Effect of Luteolin on the Proliferation, Apoptosis and Expression of Inflammation-Related Mediators in Lipopolysaccharide-Induced Keratinocytes. Int. J. Immunopathol. Pharmacol. 2023, 37, 3946320231169175. [Google Scholar] [CrossRef]
- Chen, L.; Liu, C.; Xiang, X.; Qiu, W.; Guo, K. miR-155 Promotes an Inflammatory Response in HaCaT Cells via the IRF2BP2/KLF2/NF-κB Pathway in Psoriasis. Int. J. Mol. Med. 2024, 54, 91. [Google Scholar] [CrossRef]
- Ciesielska, A.; Matyjek, M.; Kwiatkowska, K. TLR4 and CD14 Trafficking and Its Influence on LPS-Induced pro-Inflammatory Signaling. Cell. Mol. Life Sci. 2021, 78, 1233–1261. [Google Scholar] [CrossRef]
- Smith, R.L.; Hébert, H.L.; Massey, J.; Bowes, J.; Marzo-Ortega, H.; Ho, P.; McHugh, N.J.; Worthington, J.; Barton, A.; Griffiths, C.E.M.; et al. Association of Toll-like Receptor 4 (TLR4) with Chronic Plaque Type Psoriasis and Psoriatic Arthritis. Arch. Dermatol. Res. 2016, 308, 201–205. [Google Scholar] [CrossRef]
- Ye, Y.; Wang, P.; Zhou, F. miR-489-3p Inhibits TLR4/NF-κB Signaling to Prevent Inflammation in Psoriasis. Exp. Ther. Med. 2021, 22, 744. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Kim, B.-G.; Kim, H.-E.; Sun, Q.; Shi, S.; Ma, G.; Kim, Y.; Kim, O.-S.; Kim, O.-J. The Protective Role of Feruloylserotonin in LPS-Induced HaCaT Cells. Molecules 2019, 24, 3064. [Google Scholar] [CrossRef]
- Wójcik, M.; Plata-Babula, A.; Głowaczewska, A.; Sirek, T.; Orczyk, A.; Małecka, M.; Grabarek, B.O. Expression Profile of mRNAs and miRNAs Related to Mitogen-Activated Kinases in HaCaT Cell Culture Treated with Lipopolysaccharide a and Adalimumab. Cell Cycle 2024, 23, 385–404. [Google Scholar] [CrossRef]
- Kasela, T.; Dąbala, M.; Mistarz, M.; Wieczorek, W.; Wierzbik-Strońska, M.; Boroń, K.; Zawidlak-Węgrzyńska, B.; Oskar Grabarek, B. Effects of Cyclosporine A and Adalimumab on the Expression Profiles Histaminergic System-Associated Genes and microRNAs Regulating These Genes in HaCaT Cells. Cell Cycle 2022, 21, 2499–2516. [Google Scholar] [CrossRef]
- Grabarek, B.O.; Kasela, T.; Adwent, I.; Zawidlak-Węgrzyńska, B.; Brus, R. Evaluation of the Influence of Adalimumab on the Expression Profile of Leptin-Related Genes and Proteins in Keratinocytes Treated with Lipopolysaccharide A. Int. J. Mol. Sci. 2021, 22, 1595. [Google Scholar] [CrossRef] [PubMed]
- Wójcik, M.; Zmarzły, N.; Derkacz, A.; Kulpok-Bagiński, T.; Blek, N.; Grabarek, B.O. Gene Expression Profile of Mitogen-Activated Kinases and microRNAs Controlling Their Expression in HaCaT Cell Culture Treated with Lipopolysaccharide A and Cyclosporine A. Cell Cycle 2024, 23, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Adwent, I.; Grabarek, B.O.; Kojs-Mrożkiewicz, M.; Brus, R.; Staszkiewicz, R.; Plewka, A.; Stasiowski, M.; Lyssek-Boroń, A. The Influence of Adalimumab and Cyclosporine A on the Expression Profile of the Genes Related to TGFβ Signaling Pathways in Keratinocyte Cells Treated with Lipopolysaccharide A. Mediators Inflamm. 2020, 2020, 3821279. [Google Scholar] [CrossRef]
- Dopytalska, K.; Ciechanowicz, P.; Wiszniewski, K.; Szymańska, E.; Walecka, I. The Role of Epigenetic Factors in Psoriasis. Int. J. Mol. Sci. 2021, 22, 9294. [Google Scholar] [CrossRef]
- Chen, C.-H.; Wu, N.-L.; Tsai, T.-F. How Cells Die in Psoriasis? Int. J. Mol. Sci. 2025, 26, 3747. [Google Scholar] [CrossRef]
- Dai, F.-F.; Hu, M.; Zhang, Y.-W.; Zhu, R.-H.; Chen, L.-P.; Li, Z.-D.; Huang, Y.-J.; Hu, W.; Cheng, Y.-X. TNF-α/Anti-TNF-α Drugs and Its Effect on Pregnancy Outcomes. Expert Rev. Mol. Med. 2022, 24, e26. [Google Scholar] [CrossRef]
- Qian, S.; Wei, Z.; Yang, W.; Huang, J.; Yang, Y.; Wang, J. The Role of BCL-2 Family Proteins in Regulating Apoptosis and Cancer Therapy. Front. Oncol. 2022, 12, 985363. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, A.; Strzałka-Mrozik, B.; Wcisło-Dziadecka, D.; Grabarek, B.; Kimsa-Dudek, M.; Gola, J. Adalimumab Changes the Expression Profile of Selected BCL-2 Family Genes. Dermatol. Ther. 2020, 33, e13277. [Google Scholar] [CrossRef]
- Gogoleva, V.S.; Atretkhany, K.-S.N.; Dygay, A.P.; Yurakova, T.R.; Drutskaya, M.S.; Nedospasov, S.A. Current Perspectives on the Role of TNF in Hematopoiesis Using Mice With Humanization of TNF/LT System. Front. Immunol. 2021, 12, 661900. [Google Scholar] [CrossRef]
- Rivera-Yañez, C.R.; Ruiz-Hurtado, P.A.; Mendoza-Ramos, M.I.; Reyes-Reali, J.; García-Romo, G.S.; Pozo-Molina, G.; Reséndiz-Albor, A.A.; Nieto-Yañez, O.; Méndez-Cruz, A.R.; Méndez-Catalá, C.F.; et al. Flavonoids Present in Propolis in the Battle against Photoaging and Psoriasis. Antioxidants 2021, 10, 2014. [Google Scholar] [CrossRef]
- Agarwal, P.; Li, H.; Choi, K.; Hueneman, K.; He, J.; Welner, R.S.; Starczynowski, D.T.; Bhatia, R. TNF-α-Induced Alterations in Stromal Progenitors Enhance Leukemic Stem Cell Growth via CXCR2 Signaling. Cell Rep. 2021, 36, 109386. [Google Scholar] [CrossRef]
- Wang, H.; Chen, W.; Lie, C.; Zhang, Y.; Li, J.; Meng, J.; Zhang, N. ARG1 and CXCL2 Are Potential Biomarkers Target for Psoriasis Patients. Mol. Pain 2022, 18, 17448069221128423. [Google Scholar] [CrossRef]
- Lin, L.; Luo, Q.; Gao, X.; Li, Q.; Li, W.; Zhou, X.; Liu, W.; Zhong, X.; Yang, Y.; Zhang, X. Correlation and Clinical Significance of HBD-2 and CXCL-1/2 Levels at Skin Lesions with Psoriasis Vulgaris Severity. Immunol. Investig. 2024, 53, 1234–1249. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Chen, C.; Han, M.; Tian, C.; Song, F.; Feng, S.; Xu, M.; Zhao, Z.; Zhou, H.; Su, W.; et al. CXCL2: A Key Player in the Tumor Microenvironment and Inflammatory Diseases. Cancer Cell Int. 2025, 25, 133. [Google Scholar] [CrossRef]
- Fan, G.; Lu, J.; Zha, J.; Guo, W.; Zhang, Y.; Liu, Y.; Zhang, L. TAK1 in Vascular Signaling: “Friend or Foe”? J. Inflamm. Res. 2024, 17, 3031–3041. [Google Scholar] [CrossRef] [PubMed]
- Jang, D.-I.; Lee, A.-H.; Shin, H.-Y.; Song, H.-R.; Park, J.-H.; Kang, T.-B.; Lee, S.-R.; Yang, S.-H. The Role of Tumor Necrosis Factor Alpha (TNF-α) in Autoimmune Disease and Current TNF-α Inhibitors in Therapeutics. Int. J. Mol. Sci. 2021, 22, 2719. [Google Scholar] [CrossRef]
- Huang, K.; Zhang, Q.; Wan, H.; Ban, X.-X.; Chen, X.-Y.; Wan, X.-X.; Lu, R.; He, Y.; Xiong, K. TAK1 at the Crossroads of Multiple Regulated Cell Death Pathways: From Molecular Mechanisms to Human Diseases. FEBS J. 2025, 292, 3849–3877. [Google Scholar] [CrossRef] [PubMed]
- Cilek, M.Z.; de Vega, S.; Shiozawa, J.; Yoshinaga, C.; Miyamae, Y.; Chijiiwa, M.; Mochizuki, S.; Ito, M.; Kaneko, H.; Kaneko, K.; et al. Synergistic Upregulation of ADAMTS4 (Aggrecanase-1) by Cytokines and Its Suppression in Knee Osteoarthritic Synovial Fibroblasts. Lab. Investig. 2022, 102, 102–111. [Google Scholar] [CrossRef]
- Yang, X.; Yu, X.; Nie, H.; Jiang, W.; Zhou, J.; Ou, C.; He, X. Comprehensive Analysis of Prognostic Value and Immune Infiltration of IAPs Family in Hepatocellular Carcinoma. J. Cancer 2023, 14, 2848–2866. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Wei, K.; Shan, Y. E3 Ubiquitin Ligase Gene BIRC3 Modulates TNF-Induced Cell Death Pathways and Promotes Aberrant Proliferation in Rheumatoid Arthritis Fibroblast-like Synoviocytes. Front. Immunol. 2024, 15, 1433898. [Google Scholar] [CrossRef] [PubMed]
- Gosia, M.; Doshi, G.; Bagwe Parab, S.; Godad, A. Innovative Approaches to Psoriasis: Small Molecules Targeting Key Signaling Pathways. Immunol. Investig. 2025, 54, 457–493. [Google Scholar] [CrossRef]
- Pocino, K.; Carnazzo, V.; Stefanile, A.; Basile, V.; Guerriero, C.; Marino, M.; Rigante, D.; Basile, U. Tumor Necrosis Factor-Alpha: Ally and Enemy in Protean Cutaneous Sceneries. Int. J. Mol. Sci. 2024, 25, 7762. [Google Scholar] [CrossRef]
- Verstrepen, L.; Verhelst, K.; van Loo, G.; Carpentier, I.; Ley, S.C.; Beyaert, R. Expression, Biological Activities and Mechanisms of Action of A20 (TNFAIP3). Biochem. Pharmacol. 2010, 80, 2009–2020. [Google Scholar] [CrossRef] [PubMed]
- Pujari, R.; Hunte, R.; Khan, W.N.; Shembade, N. A20-Mediated Negative Regulation of Canonical NF-κB Signaling Pathway. Immunol. Res. 2013, 57, 166–171. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, W.; Zheng, X.; Jin, H. Immune Regulation of TNFAIP3 in Psoriasis through Its Association with Th1 and Th17 Cell Differentiation and P38 Activation. J. Immunol. Res. 2020, 2020, 5980190. [Google Scholar] [CrossRef]
- Urbano, P.C.M.; Aguirre-Gamboa, R.; Ashikov, A.; van Heeswijk, B.; Krippner-Heidenreich, A.; Tijssen, H.; Li, Y.; Azevedo, V.F.; Smits, L.J.T.; Hoentjen, F.; et al. TNF-α-Induced Protein 3 (TNFAIP3)/A20 Acts as a Master Switch in TNF-α Blockade-Driven IL-17A Expression. J. Allergy Clin. Immunol. 2018, 142, 517–529. [Google Scholar] [CrossRef]
- Wang, X.; Cheng, D.; Hu, G.; Liang, L.; Tan, F.; Xiao, T.; Xiao, S.; Xia, Y. Tumor Necrosis Factor (TNF) Receptor Expression Determines Keratinocyte Fate upon Stimulation with TNF-Like Weak Inducer of Apoptosis. Mediators Inflamm. 2019, 2019, 2945083. [Google Scholar] [CrossRef]
- Cao, T.; Yuan, X.; Fang, H.; Chen, J.; Xue, K.; Li, Z.; Dang, E.; Wang, G.; Shao, S. Neutrophil Extracellular Traps Promote Keratinocyte Inflammation via AIM2 Inflammasome and AIM2-XIAP in Psoriasis. Exp. Dermatol. 2023, 32, 368–378. [Google Scholar] [CrossRef]
- Hanifeh, M.; Ataei, F. XIAP as a Multifaceted Molecule in Cellular Signaling. Apoptosis 2022, 27, 441–453. [Google Scholar] [CrossRef]
- Vitale, I.; Pietrocola, F.; Guilbaud, E.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostini, M.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; et al. Apoptotic Cell Death in Disease-Current Understanding of the NCCD 2023. Cell Death Differ. 2023, 30, 1097–1154. [Google Scholar] [CrossRef]
- Schell, S.L.; Cong, Z.; Sennett, M.L.; Gettle, S.L.; Longenecker, A.L.; Goldberg, S.R.; Kirby, J.S.; Helm, M.F.; Nelson, A.M. Keratinocytes and Immune Cells in the Epidermis Are Key Drivers of Inflammation in Hidradenitis Suppurativa Providing a Rationale for Novel Topical Therapies. Br. J. Dermatol. 2023, 188, 407–419. [Google Scholar] [CrossRef]
- Wcisło-Dziadecka, D.; Grabarek, B.; Kaźmierczak, A.; Gola, J.; Kruszniewska-Rajs, C. The Influence of Adalimumab on the Expression Profile of mRNAs and miRNAs Related to the IL-12 and IL-23 Signal Paths. J. Eur. Acad. Dermatol. Venereol. 2019, 33, e198–e199. [Google Scholar] [CrossRef]
- Morizane, S.; Mukai, T.; Sunagawa, K.; Tachibana, K.; Kawakami, Y.; Ouchida, M. “Input/Output Cytokines” in Epidermal Keratinocytes and the Involvement in Inflammatory Skin Diseases. Front. Immunol. 2023, 14, 1239598. [Google Scholar] [CrossRef] [PubMed]
- Gunter, S.; Michel, F.S.; Fourie, S.S.; Singh, M.; le Roux, R.; Manilall, A.; Mokotedi, L.P.; Millen, A.M.E. The Effect of TNF-α Inhibitor Treatment on microRNAs and Endothelial Function in Collagen Induced Arthritis. PLoS ONE 2022, 17, e0264558. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Chen, Z.; Wang, C.; Huang, P.; Luo, M.; Zhou, R. STAT3/SH3PXD2A-AS1/miR-125b/STAT3 Positive Feedback Loop Affects Psoriasis Pathogenesis via Regulating Human Keratinocyte Proliferation. Cytokine 2021, 144, 155535. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, X.; Liu, C.; Chen, X.; Li, P.; Qiu, W.; Guo, K. Integrative Analysis of Gene and microRNA Expression Profiles Reveals Candidate Biomarkers and Regulatory Networks in Psoriasis. Medicine 2024, 103, e39002. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, H.; Huang, X.; Wu, C.; Zheng, K.; Deng, J.; Zheng, Y.; Wang, J.; Chi, X.; Ma, X.; et al. Innate Immune and Proinflammatory Signals Activate the Hippo Pathway via a Tak1-STRIPAK-Tao Axis. Nat. Commun. 2024, 15, 145. [Google Scholar] [CrossRef]
- Dżaman, K.; Czerwaty, K.; Reichert, T.E.; Szczepański, M.J.; Ludwig, N. Expression and Regulatory Mechanisms of MicroRNA in Cholesteatoma: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 12277. [Google Scholar] [CrossRef] [PubMed]
- Ciavarella, C.; Motta, I.; Vasuri, F.; Fittipaldi, S.; Valente, S.; Pollutri, D.; Ricci, F.; Gargiulo, M.; Pasquinelli, G. Involvement of miR-30a-5p and miR-30d in Endothelial to Mesenchymal Transition and Early Osteogenic Commitment under Inflammatory Stress in HUVEC. Biomolecules 2021, 11, 226. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zheng, J.; Han, X.; Zhao, J.; Cheng, S.; Li, A. Bovine Colostrum miR-30a-5p Targets the NF-κB Signaling Pathway to Alleviate Inflammation in Intestinal Epithelial Cells. J. Agric. Food Chem. 2024, 72, 9164–9178. [Google Scholar] [CrossRef]
- Egea Alonso, V. MicroRNA-Associated Regulatory Mechanisms in Human Mesenchymal Stem Cells and Endothelial Cells. Available online: https://edoc.ub.uni-muenchen.de/34285/ (accessed on 23 July 2025).
- Kim, S.-W.; Ramasamy, K.; Bouamar, H.; Lin, A.-P.; Jiang, D.; Aguiar, R.C.T. MicroRNAs miR-125a and miR-125b Constitutively Activate the NF-κB Pathway by Targeting the Tumor Necrosis Factor Alpha-Induced Protein 3 (TNFAIP3, A20). Proc. Natl. Acad. Sci. USA 2012, 109, 7865–7870. [Google Scholar] [CrossRef]
- Markopoulos, G.S.; Roupakia, E.; Tokamani, M.; Alabasi, G.; Sandaltzopoulos, R.; Marcu, K.B.; Kolettas, E. Roles of NF-κB Signaling in the Regulation of miRNAs Impacting on Inflammation in Cancer. Biomedicines 2018, 6, 40. [Google Scholar] [CrossRef]
- Dari, M.A.G.; Vahedian, V.; Kempisty, B.; Morenikeji, O.B.; Farzaneh, M.; Anbiyaiee, A. The Protective Role of miR-125b in Hepatocellular Carcinoma: Unraveling Tumor-Suppressive Mechanisms. Curr. Mol. Med. 2025, 25, 663–671. [Google Scholar] [CrossRef]
- Plawgo, K.; Raczynska, K.D. Context-Dependent Regulation of Gene Expression by Non-Canonical Small RNAs. Noncoding RNA 2022, 8, 29. [Google Scholar] [CrossRef] [PubMed]
- Hadi, N.; Seifati, S.M.; Nateghi, B.; Ravaghi, P.; Khosravian, F.; Namazi, F.; Fotouhi Firouzabad, M.; Shaygannejad, V.; Salehi, M. Study of The Correlation between miR-106a, miR-125b, and miR-330 on Multiple Sclerosis Patients by Targeting TNFSF4 and SP1 in NF-кb/TNF-α Pathway: A Case-Control Study. Cell J. 2022, 24, 403–409. [Google Scholar] [CrossRef]
- Raj, D.; Brash, D.E.; Grossman, D. Keratinocyte Apoptosis in Epidermal Development and Disease. J. Investig. Dermatol. 2006, 126, 243–257. [Google Scholar] [CrossRef]
- Jezernik, G.; Glavač, D.; Skok, P.; Krušič, M.; Potočnik, U.; Gorenjak, M. Discovery of Novel Biomarkers with Extended Non-Coding RNA Interactor Networks from Genetic and Protein Biomarkers. Int. J. Mol. Sci. 2024, 25, 10210. [Google Scholar] [CrossRef]
- Csók, Á.; Micsik, T.; Magyar, Z.; Tornóczky, T.; Kuthi, L.; Nishi, Y.; Szirák, K.; Csóka, M.; Ottóffy, G.; Soltész, B.; et al. Alterations of miRNA Expression in Diffuse Hyperplastic Perilobar Nephroblastomatosis: Mapping the Way to Understanding Wilms’ Tumor Development and Differential Diagnosis. Int. J. Mol. Sci. 2023, 24, 8793. [Google Scholar] [CrossRef]
- Iacobazzi, D.; Convertini, P.; Todisco, S.; Santarsiero, A.; Iacobazzi, V.; Infantino, V. New Insights into NF-κB Signaling in Innate Immunity: Focus on Immunometabolic Crosstalks. Biology 2023, 12, 776. [Google Scholar] [CrossRef]
- Garcia-Moreno, A.; López-Domínguez, R.; Villatoro-García, J.A.; Ramirez-Mena, A.; Aparicio-Puerta, E.; Hackenberg, M.; Pascual-Montano, A.; Carmona-Saez, P. Functional Enrichment Analysis of Regulatory Elements. Biomedicines 2022, 10, 590. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for Taxonomy-Based Analysis of Pathways and Genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Matsuura, Y.; Ishiguro-Watanabe, M. KEGG: Biological Systems Database as a Model of the Real World. Nucleic Acids Res. 2025, 53, D672–D677. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An Online Database for Prediction of Functional microRNA Targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, X. Prediction of Functional microRNA Targets by Integrative Modeling of microRNA Binding and Target Expression Data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING Database in 2023: Protein-Protein Association Networks and Functional Enrichment Analyses for Any Sequenced Genome of Interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]

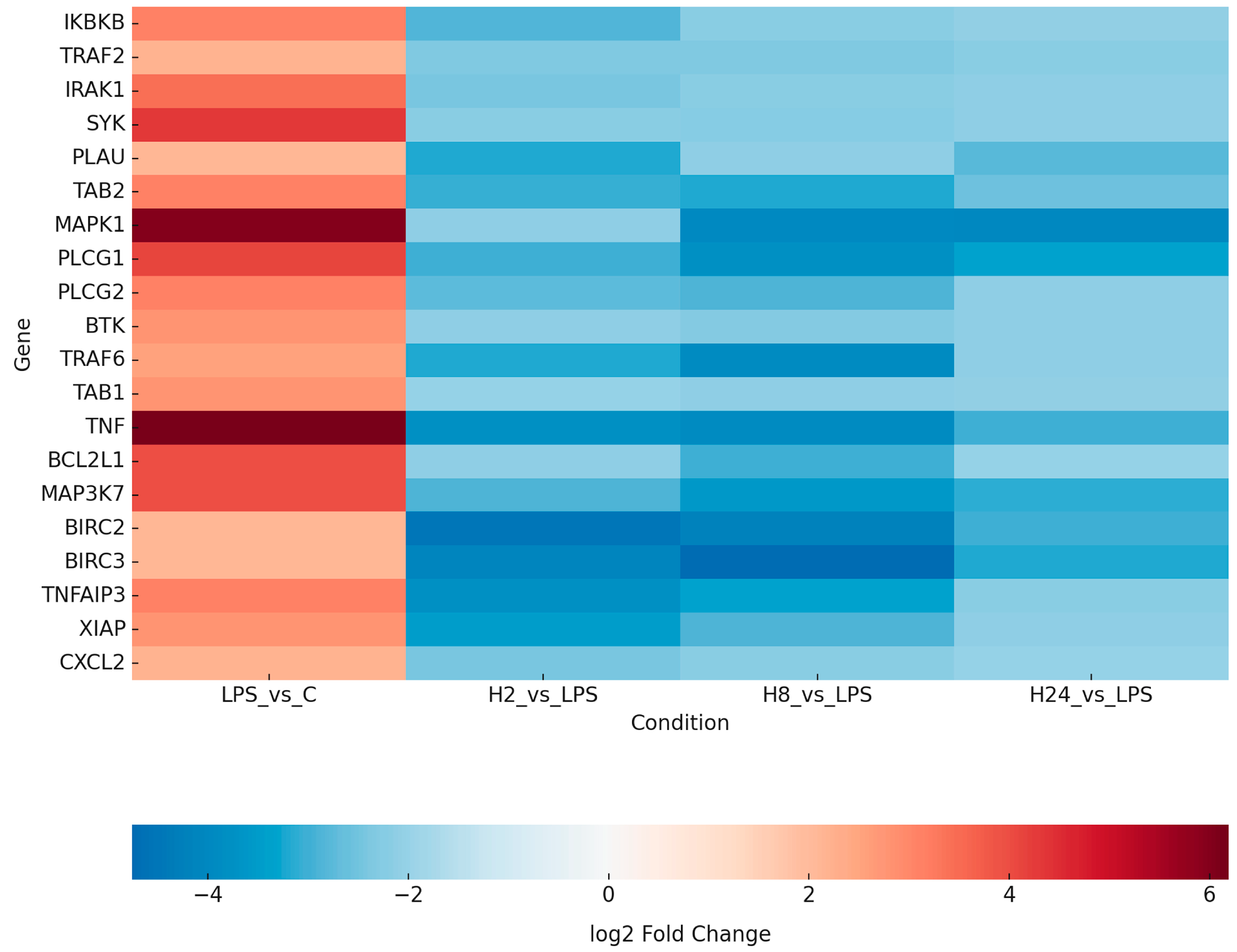
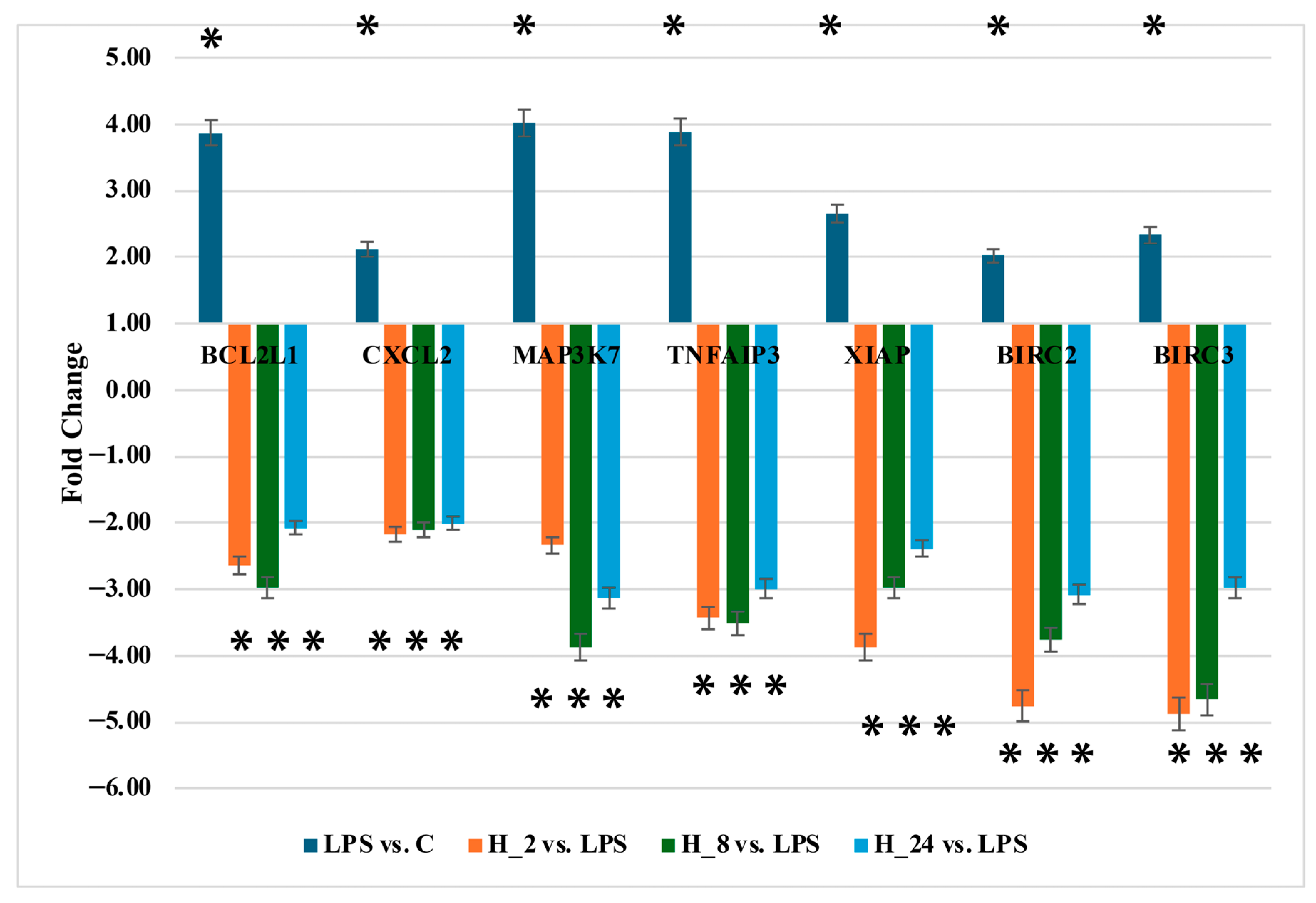
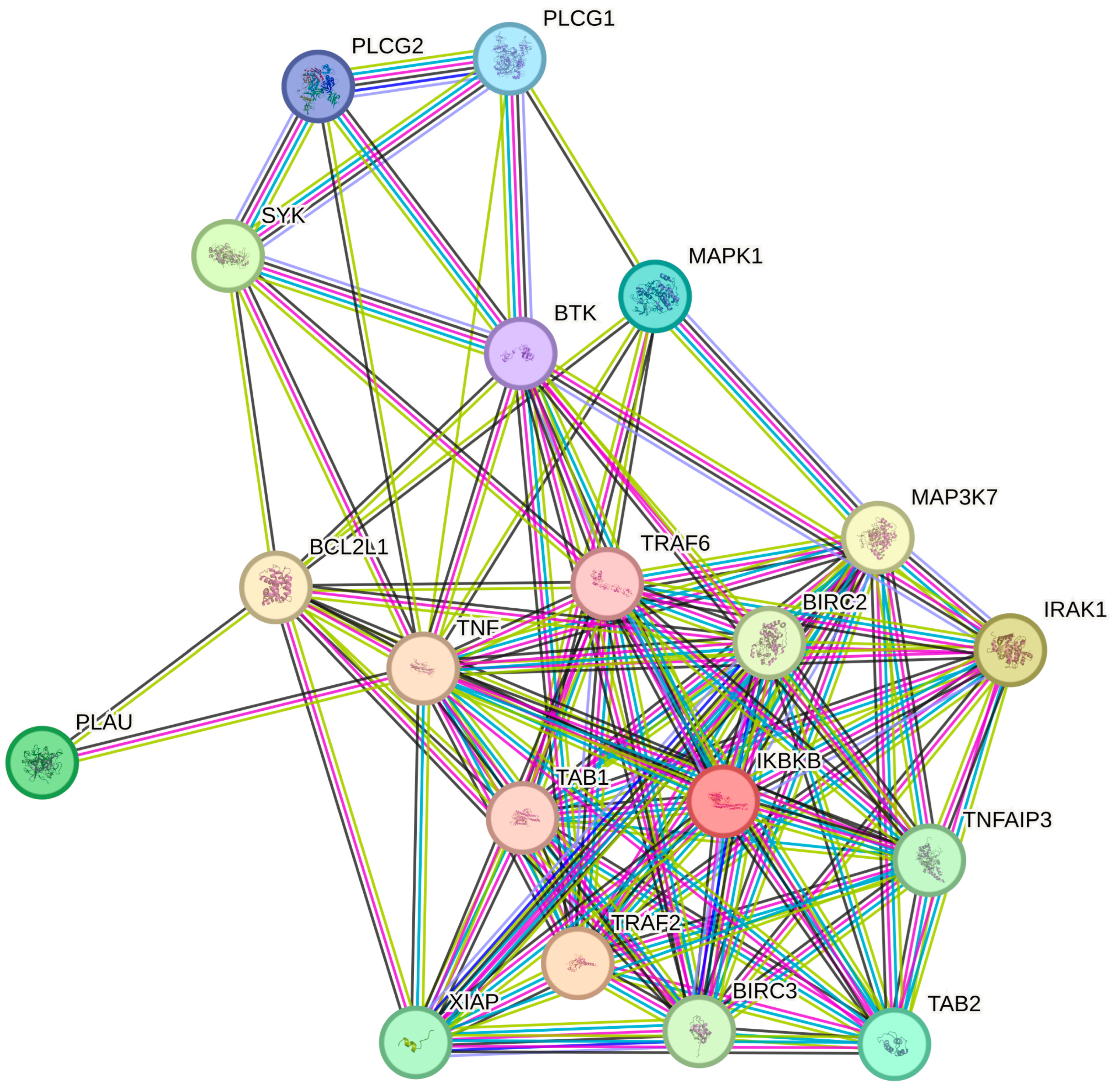

| ID | mRNA | LPS vs. C | H_2 vs. LPS | H_8 vs. LPS | H_24 vs. LPS |
|---|---|---|---|---|---|
| 1563357_at | (+)6.17 | (−)3.41 | (−)3.81 | (−)3.04 | |
| 206665_s_at | BCL2L1 | (+)3.98 | (−)2.11 | (−)2.98 | (−)2.01 |
| 212312_at | (+)3.28 | (−)2.12 | (−)2.71 | (−)2.09 | |
| 215037_s_at | (+)3.21 | (−)2.18 | (−)2.91 | (−)2.07 | |
| 231228_at | (+)3.18 | (−)2.13 | (−)3.01 | (−)2.11 | |
| 206853_s_at | MAP3K7 | (+)3.98 | (−)2.91 | (−)3.59 | (−)3.11 |
| 206854_s_at | (+)4.19 | (−)2.99 | (−)3.61 | (−)3.01 | |
| 211536_x_at | (+)3.99 | (−)2.91 | (−)3.44 | (−)3.17 | |
| 211537_x_at | (+)3.76 | (−)2.92 | (−)3.49 | (−)3.11 | |
| 202076_at | BIRC2 | (+)2.12 | (−)4.54 | (−)4.19 | (−)3.01 |
| 210538_s_at | BIRC3 | (+)2.12 | (−)4.14 | (−)4.76 | (−)3.22 |
| 230499_at | (+)2.11 | (−)3.17 | (−)4.91 | (−)3.19 | |
| 202643_s_at | TNFAIP3 | (+)3.09 | (−)3.81 | (−)3.31 | (−)2.19 |
| 202644_s_at | (+)3.01 | (−)3.89 | (−)3.48 | (−)2.18 | |
| 206536_s_at | XIAP | (+)2.81 | (−)3.44 | (−)2.91 | (−)2.11 |
| 206537_at | (+)2.76 | (−)3.43 | (−)2.99 | (−)2.18 | |
| 225858_s_at | (+)2.91 | (−)3.71 | (−)2.98 | (−)2.17 | |
| 225859_at | (+)2.81 | (−)3.32 | (−)2.78 | (−)2.16 | |
| 228363_at | (+)2.54 | (−)3.54 | (−)3.02 | (−)2.11 | |
| 235222_x_at | (+)2.79 | (−)3.45 | (−)2.89 | (−)2.24 | |
| 243026_x_at | (+)2.80 | (−)3.19 | (−)2.99 | (−)2.08 | |
| 209774_x_at | CXCL2 | (+)2.19 | (−)2.45 | (−)2.19 | (−)2.01 |
| 230101_at | (+)2.18 | (−)2.31 | (−)2.11 | (−)2.03 | |
| 1569203_at | (+)2.11 | (−)2.49 | (−)2.18 | (−)2.03 |
| mRNA | miRNA | Target Score | Fold Change | |||
|---|---|---|---|---|---|---|
| LPS vs. C | H_2 vs. LPS | H_8 vs. LPS | H_24 vs. LPS | |||
| MAP3K7 | miR-1297 | 90 | (−)2.13 ± 0.18 * | (+)2.18 ± 0.19 * | (+)2.11 ± 0.19 * | (+)2.87 ± 0.98 * |
| miR-30a | 83 | (−)2.17 ± 0.12 * | (+)2.17 ± 0.81 * | (+)2.45 ± 0.18 * | (+)2.54 ± 0.12 * | |
| CXCL2 | miR-95-5p | 96 | (−)3.12 ± 0.45 * | (+)2.87 ± 0.81 * | (+)3.12 ± 0.91 * | (+)2.10 ± 0.53 * |
| TNFAIP3 | miR-125b | 84 | (−)2.18 ± 0.13 * | (+)2.56 ± 0.43 * | (+)2.11 ± 0.71 * | (+)2.03 ± 0.71 * |
| XIAP | miR-4329 | 94 | (+)2.98 ± 0.76 * | (−)2.11 ± 0.71 * | (−)2.76 ± 0.76 * | (−)2.98 ± 0.13 * |
| BIRC3 | mir-20b-3p | 96 | (+)3.18 ± 0.55 * | (−)2.18 ± 0.12 * | (−)2.11 ± 0.43 * | (−)2.16 ± 0.41 * |
| Protein | C | LPS | H_2 | H_8 | H_24 |
|---|---|---|---|---|---|
| BCL2L1 [ng/mL] | 13.19 ± 0.23 | 87.12 ± 19 * | 18.13 ± 0.16 * | 17.87 ± 0.65 * | 17.19 ± 0.18 * |
| CXCL2 [ng/mL] | 1.56 ± 0.91 | 7.13 ± 1.09 * | 4.23 ± 0.65 * | 3.34 ± 0.51 * | 2.19 ± 0.81 * |
| MAP3K7 [ng/mL] | 2.91 ± 0.34 | 6.77 ± 1.12 * | 4.56 ± 0.91 * | 2.87 ± 0.51 * | 2.11 ± 0.13 * |
| BIRC2 [pg/mL] | 846.12 ± 5.71 | 2454.12 ± 105.91 * | 176.12 ± 3.67 * | 1056 ± 9.12 * | 1023 ± 9.81 * |
| BIRC3 [pg/mL] | 961.10 ± 32.98 | 3871.91 ± 176.65 * | 2098 ± 18.24 * | 2012 ± 14.81 * | 2019 ± 21.91 * |
| TNFAIP3 [ng/mL] | 6.18 ± 0.98 | 19.87 ± 1.76 * | 12.12 ± 2.17 * | 11.62 ± 0.98 * | 10.01 ± 0.12 * |
| XIAP [ng/mL] | 3.48 ± 0.91 | 7.65 ± 0.76 * | 5.57 ± 1.11 * | 6.12 ± 0.91 * | 6.80 ± 0.19 * |
| mRNA | RT-qPCR Primers (5′-3′) |
|---|---|
| BCL2L1 | Forward: GCCACTTACCTGAATGACCACC Reverse: AACCAGCGGTTGAAGCGTTCCT |
| CXCL2 | Forward: GGCAGAAAGCTTGTCTCAACCC Reverse: CTCCTTCAGGAACAGCCACCAA |
| MAP3K7 | Forward: CAGAGCAACTCTGCCACCAGTA Reverse: CATTTGTGGCAGGAACTTGCTCC |
| BIRC2 | Forward: CTGTGGTGGGAAGCTCAGTA Reverse: TCATTCGAGCTGCATGTGTC |
| BIRC3 | Forward: GGCTGTTACCGCTGAGAATG Reverse: C GGTGGCAGGAGAAACATCA |
| TNFAIP3 | Forward: CTCAACTGGTGTCGAGAAGTCC Reverse: TTCCTTGAGCGTGCTGAACAGC |
| XIAP | Forward: TGGCAGATTATGAAGCACGGATC Reverse: AGTTAGCCCTCCTCCACAGTGA |
| ACTB | Forward: TCACCCACACTGTGCCCATCTACGA Reverse: CAGCGGAACCGCTCATTGCCAATGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Plata-Babula, A.; Kulej, W.; Ordon, P.; Gajdeczka, J.; Stefaniak, M.; Chwalba, A.; Gościniewicz, P.; Kulpok, T.; Grabarek, B.O. Modulation of Nuclear Factor Kappa B Signaling and microRNA Profiles by Adalimumab in LPS-Stimulated Keratinocytes. Int. J. Mol. Sci. 2025, 26, 10035. https://doi.org/10.3390/ijms262010035
Plata-Babula A, Kulej W, Ordon P, Gajdeczka J, Stefaniak M, Chwalba A, Gościniewicz P, Kulpok T, Grabarek BO. Modulation of Nuclear Factor Kappa B Signaling and microRNA Profiles by Adalimumab in LPS-Stimulated Keratinocytes. International Journal of Molecular Sciences. 2025; 26(20):10035. https://doi.org/10.3390/ijms262010035
Chicago/Turabian StylePlata-Babula, Aleksandra, Wojciech Kulej, Paweł Ordon, Julia Gajdeczka, Martyna Stefaniak, Artur Chwalba, Piotr Gościniewicz, Tomasz Kulpok, and Beniamin Oskar Grabarek. 2025. "Modulation of Nuclear Factor Kappa B Signaling and microRNA Profiles by Adalimumab in LPS-Stimulated Keratinocytes" International Journal of Molecular Sciences 26, no. 20: 10035. https://doi.org/10.3390/ijms262010035
APA StylePlata-Babula, A., Kulej, W., Ordon, P., Gajdeczka, J., Stefaniak, M., Chwalba, A., Gościniewicz, P., Kulpok, T., & Grabarek, B. O. (2025). Modulation of Nuclear Factor Kappa B Signaling and microRNA Profiles by Adalimumab in LPS-Stimulated Keratinocytes. International Journal of Molecular Sciences, 26(20), 10035. https://doi.org/10.3390/ijms262010035





