Cellular Signaling of Amino Acid Metabolism in Prostate Cancer
Abstract
1. Introduction to Prostate Cancer (PCa)
2. Amino Acid Metabolism in PCa
2.1. Glutamine
2.2. Tryptophan
2.3. Leucine
2.4. Arginine
2.5. Methionine
2.6. Histidine
2.7. Serine
2.8. Glycine
| Amino Acid | Gene Symbol | Full Name | Biological Function | Alteration in PCa | Effects on PCa | Reference |
|---|---|---|---|---|---|---|
| Arginine | CKM | Creatine kinase, M-type | Catalyses the reversible transfer of a phosphate group between ATP and various phosphogens | ↓ a | Decrease in both trancription and protein levels; high expression was correlated with prolonged relapse-free survival | [106] |
| Aspartate | FOLH1 | Folate hydrolase 1 | Acts as a glutamate carboxypeptidase and plays a key role in folate metabolism by breaking down folate and folate derivatives | ↑ b | Known as the prostate-specific membrane antigen and highly expressed; a diagnostic biomarker | [107,108] |
| Leucine | MCCC2 | Methylcrotonyl-CoA carboxylase subunit 2 | Engaged in leucine and isovaleric acid catabolism | ↑ | As an androgen-regulated gene; involved in carcinogenesis by upregulating GLUD1 in the GLUD1-p38 MAPK signalling axis | [109,110] |
| Lysine | EZH2 | Enhancer of zeste 2 polycomb repressive complex 2 subunit | Responsible for trimethylating histone H3 on lysine 27, i.e., yielding H3K27me3, a histone modification that represses gene transcription | ↑ | Overexpression promoted tumour proliferation, metastasis, drug resistance and poor prognosis | [111,112,113,114] |
| MECOM | MDS1 and EVI1 complex locus | Encodes an oncoprotein (transcription factor) known as Evi-1, regulating the gene expression during hematopoiesis and development | ↓ | Overexpression contributed to docetaxel resistance in PC3 cells | [115] | |
| Proline | OAT | Ornithine aminotransferase | Catalyses the reversible interconversion of L-ornithine and 2-oxoglutarate into L-glutamate semialdehyde and L-glutamate | ↓ | Repressed by AR knockdown in C4-2B cells | [116] |
| PRODH | Proline dehydrogenase 1 | A mitochondrial protein that catalyses the oxidation of proline and its conversion into P5C in a FAD-dependent manner | ↓ | Upregulation in both mRNA and protein levels; overexpression promoted PCa tumour growth and reduced T-cell infiltration in mice | [117] | |
| PYCR1 | Pyrroline-5-carboxylate reductase 1 | One of the isoforms of the pyrroline-5-carboxylate reductase, which is involved in proline biosynthesis | ↑ | PYCR1 knockdown suppressed the growth of DU145 and PC-3 cells | [118,119] | |
| Tryptophan | INMT | Indolethylamine N-methyltransferase | Involved in the methylation of indolethylamines, such as tryptamine and serotonin, converting them into N-methylated derivatives | ↓ | Low expression in primary PCa but elevated in CRPC.; INMT knockdown in DU145 cells was inhibitory to cell proliferation | [120,121,122] |
| Multiple | AOX1 | Aldehyde oxidase 1 | Exhibits a broad substrate specificity and involved in cell redox homeostasis by regulating the production of reactive oxygen species | ↓ | Hypermethylated and downregulated; overexpression inhibited the proliferation and invasion of PC-3 cells | [123,124] |
| ASPA | Aspartoacylase | Crucial to the breakdown of N-acetylaspartate into aspartate and acetate | ↓ | Inhibited the phosphorylation of LYN and suppressed the proliferation, migration and invasion of PC-3 cells | [125] | |
| CBS | Cystathionine beta-synthase | The rate-limiting enzyme in the first step of the transsulfuration pathway, catalysing the conversion of homocysteine into cystathionine; SAM is required for the allosteric activation of its homotetramer | ↑ | Elevated in the early stages but drops during tumour metastasis | [126] | |
| GNMT | Glycine N-methyltransferase | Catalyses the transfer of a methyl group from SAM to glycine, producing S-adenosylhomocysteine and sarcosine | ↑ | Regulated in an AR-dependent manner and contributed to the survival of PCa cells | [127,128,129] | |
| MAOB | Monoamine oxidase B | Located at the mitochondrial outer membrane and involved in the breakdown of monoamines, including neurotransmitters like dopamine and serotonin | ↓ | Decreased expression could enhance the proliferation of PC-3 cells and was associated with a poor prognosis in PCa patients | [130,131] | |
| SMS | Spermine synthase | Catalyses the production of spermine from spermidine and decarboxylated SAM | ↑ | Sensitive to androgen exposure | [132,133,134] |
3. Signalling Pathways Affected by an Altered AAM in PCa
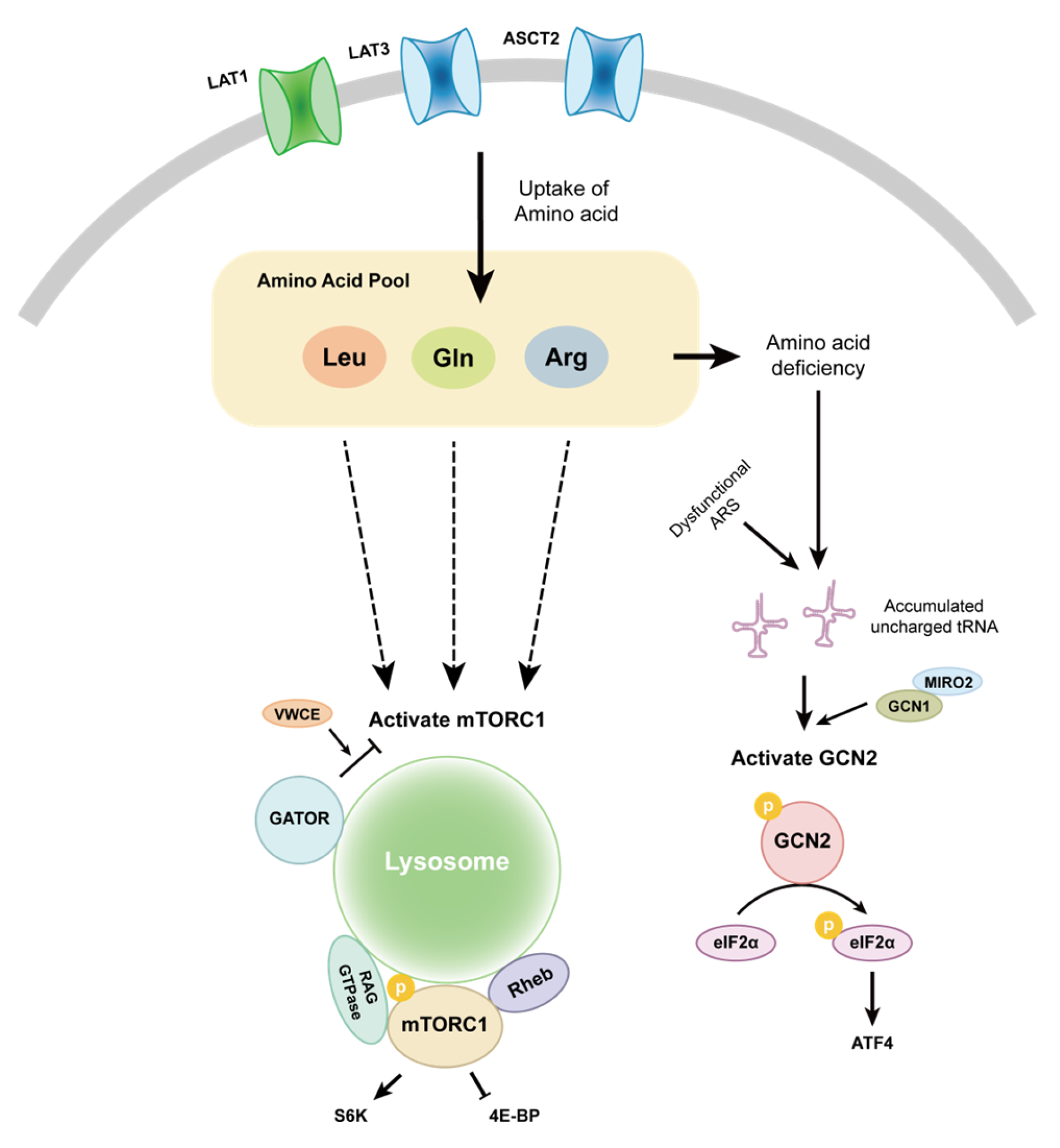
3.1. mTOR
3.2. GCN2
4. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Rebello, R.J.; Oing, C.; Knudsen, K.E.; Loeb, S.; Johnson, D.C.; Reiter, R.E.; Gillessen, S.; Van Der Kwast, T.; Bristow, R.G. Prostate Cancer. Nat. Rev. Dis. Primer 2021, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Desai, K.; McManus, J.M.; Sharifi, N. Hormonal Therapy for Prostate Cancer. Endocr. Rev. 2021, 42, 354–373. [Google Scholar] [CrossRef]
- Wasim, S.; Lee, S.-Y.; Kim, J. Complexities of Prostate Cancer. Int. J. Mol. Sci. 2022, 23, 14257. [Google Scholar] [CrossRef]
- Butler, M.; Van Der Meer, L.T.; Van Leeuwen, F.N. Amino Acid Depletion Therapies: Starving Cancer Cells to Death. Trends Endocrinol. Metab. 2021, 32, 367–381. [Google Scholar] [CrossRef]
- Bhowmick, N.; Posadas, E.; Ellis, L.; Freedland, S.J.; Vizio, D.D.; Freeman, M.R.; Theodorescu, D.; Figlin, R.; Gong, J. Targeting Glutamine Metabolism in Prostate Cancer. Front. Biosci.-Elite 2023, 15, 2. [Google Scholar] [CrossRef]
- Moon, D.; Hauck, J.S.; Jiang, X.; Quang, H.; Xu, L.; Zhang, F.; Gao, X.; Wild, R.; Everitt, J.I.; Macias, E.; et al. Targeting Glutamine Dependence with DRP-104 Inhibits Proliferation and Tumor Growth of Castration-Resistant Prostate Cancer. Prostate 2024, 84, 349–357. [Google Scholar] [CrossRef]
- Fu, Y.-M.; Yu, Z.-X.; Li, Y.-Q.; Ge, X.; Sanchez, P.J.; Fu, X.; Meadows, G.G. Specific Amino Acid Dependency Regulates Invasiveness and Viability of Androgen-Independent Prostate Cancer Cells. Nutr. Cancer 2003, 45, 60–73. [Google Scholar] [CrossRef]
- Poirson-Bichat, F.; Gonfalone, G.; Bras-Gonçalves, R.A.; Dutrillaux, B.; Poupon, M.F. Growth of Methionine-Dependent Human Prostate Cancer (PC-3) Is Inhibited by Ethionine Combined with Methionine Starvation. Br. J. Cancer 1997, 75, 1605–1612. [Google Scholar] [CrossRef]
- Delage, B.; Fennell, D.A.; Nicholson, L.; McNeish, I.; Lemoine, N.R.; Crook, T.; Szlosarek, P.W. Arginine Deprivation and Argininosuccinate Synthetase Expression in the Treatment of Cancer. Int. J. Cancer 2010, 126, 2762–2772. [Google Scholar] [CrossRef]
- Zheng, H.; Dong, B.; Ning, J.; Shao, X.; Zhao, L.; Jiang, Q.; Ji, H.; Cai, A.; Xue, W.; Gao, H. NMR-Based Metabolomics Analysis Identifies Discriminatory Metabolic Disturbances in Tissue and Biofluid Samples for Progressive Prostate Cancer. Clin. Chim. Acta 2020, 501, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Wang, J.; Chen, L.; Wang, H.; Liang, C.-Z.; Huang, J.; Xu, L.-F. The Role of Glutamine Metabolism in Castration-Resistant Prostate Cancer. Asian J. Androl. 2023, 25, 192–197. [Google Scholar] [CrossRef]
- Thiruvalluvan, M.; Billet, S.; Bhowmick, N.A. Antagonizing Glutamine Bioavailability Promotes Radiation Sensitivity in Prostate Cancer. Cancers 2022, 14, 2491. [Google Scholar] [CrossRef] [PubMed]
- Dereziński, P.; Klupczynska, A.; Sawicki, W.; Pałka, J.A.; Kokot, Z.J. Amino Acid Profiles of Serum and Urine in Search for Prostate Cancer Biomarkers: A Pilot Study. Int. J. Med. Sci. 2017, 14, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Mahmud, I.; Marchica, J.; Dereziński, P.; Qi, F.; Wang, F.; Joshi, P.; Valerio, F.; Rivera, I.; Patel, V.; et al. Integrated RNA and Metabolite Profiling of Urine Liquid Biopsies for Prostate Cancer Biomarker Discovery. Sci. Rep. 2020, 10, 3716. [Google Scholar] [CrossRef]
- Costello, L.C.; Franklin, R.B. The Intermediary Metabolism of the Prostate: A Key to Understanding the Pathogenesis and Progression of Prostate Malignancy. Oncology 2000, 59, 269–282. [Google Scholar] [CrossRef]
- Ahmad, F.; Cherukuri, M.K.; Choyke, P.L. Metabolic Reprogramming in Prostate Cancer. Br. J. Cancer 2021, 125, 1185–1196. [Google Scholar] [CrossRef]
- Costello, L.C.; Franklin, R.B. A Comprehensive Review of the Role of Zinc in Normal Prostate Function and Metabolism and Its Implications in Prostate Cancer. Arch. Biochem. Biophys. 2016, 611, 100–112. [Google Scholar] [CrossRef]
- Singh, K.K.; Desouki, M.M.; Franklin, R.B.; Costello, L.C. Mitochondrial Aconitase and Citrate Metabolism in Malignant and Nonmalignant Human Prostate Tissues. Mol. Cancer 2006, 5, 14. [Google Scholar] [CrossRef]
- Pereira-Nunes, A.; Simões-Sousa, S.; Pinheiro, C.; Miranda-Gonçalves, V.; Granja, S.; Baltazar, F. Targeting Lactate Production and Efflux in Prostate Cancer. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165894. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, H.J.; Carvalho, T.M.A.; Fonseca, L.R.S.; Figueira, M.I.; Vaz, C.V.; Socorro, S. Revisiting Prostate Cancer Metabolism: From Metabolites to Disease and Therapy. Med. Res. Rev. 2021, 41, 1499–1538. [Google Scholar] [CrossRef]
- Chetta, P.; Sriram, R.; Zadra, G. Lactate as Key Metabolite in Prostate Cancer Progression: What Are the Clinical Implications? Cancers 2023, 15, 3473. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Yin, Y.; Li, Y.; Chen, X.; Chang, Y.; Zhang, H.; Liu, J.; Beasley, J.; McCaw, P.; Zhang, H.; et al. A Glutaminase Isoform Switch Drives Therapeutic Resistance and Disease Progression of Prostate Cancer. Proc. Natl. Acad. Sci. USA 2021, 118, e2012748118. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Le, A. Glutamine Metabolism in Cancer. In The Heterogeneity of Cancer Metabolism; Le, A., Ed.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2018; Volume 1063, pp. 13–32. ISBN 978-3-319-77735-1. [Google Scholar]
- Shinka, T.; Onodera, D.; Tanaka, T.; Shoji, N.; Miyazaki, T.; Moriuchi, T.; Fukumoto, T. Serotonin Synthesis and Metabolism-Related Molecules in a Human Prostate Cancer Cell Line. Oncol. Lett. 2011, 2, 211–215. [Google Scholar] [CrossRef]
- Zang, X.; Jones, C.M.; Long, T.Q.; Monge, M.E.; Zhou, M.; Walker, L.D.; Mezencev, R.; Gray, A.; McDonald, J.F.; Fernández, F.M. Feasibility of Detecting Prostate Cancer by Ultraperformance Liquid Chromatography-Mass Spectrometry Serum Metabolomics. J. Proteome Res. 2014, 13, 3444–3454. [Google Scholar] [CrossRef]
- Mandarano, M.; Orecchini, E.; Bellezza, G.; Vannucci, J.; Ludovini, V.; Baglivo, S.; Tofanetti, F.R.; Chiari, R.; Loreti, E.; Puma, F.; et al. Kynurenine/Tryptophan Ratio as a Potential Blood-Based Biomarker in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2021, 22, 4403. [Google Scholar] [CrossRef]
- Tappenden, D.M.; Hwang, H.J.; Yang, L.; Thomas, R.S.; Lapres, J.J. The Aryl-Hydrocarbon Receptor Protein Interaction Network (AHR-PIN) as Identified by Tandem Affinity Purification (TAP) and Mass Spectrometry. J. Toxicol. 2013, 2013, 279829. [Google Scholar] [CrossRef]
- Perez-Castro, L.; Garcia, R.; Venkateswaran, N.; Barnes, S. Tryptophan and Its Metabolites in Normal Physiology and Cancer Etiology. FEBS J. 2023, 290, 7–27. [Google Scholar] [CrossRef]
- Li, Z.; Ding, B.; Ali, M.R.K.; Zhao, L.; Zang, X.; Lv, Z. Dual Effect of Tryptamine on Prostate Cancer Cell Growth Regulation: A Pilot Study. Int. J. Mol. Sci. 2022, 23, 11087. [Google Scholar] [CrossRef]
- Sivanand, S.; Vander Heiden, M.G. Emerging Roles for Branched-Chain Amino Acid Metabolism in Cancer. Cancer Cell 2020, 37, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Ananieva, E.A.; Wilkinson, A.C. Branched-Chain Amino Acid Metabolism in Cancer. Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, E.; Marin De Mas, I.; Zodda, E.; Marin, S.; Morrish, F.; Selivanov, V.; Meca-Cortés, Ó.; Delowar, H.; Pons, M.; Izquierdo, I.; et al. Metabolic Reprogramming and Dependencies Associated with Epithelial Cancer Stem Cells Independent of the Epithelial-Mesenchymal Transition Program. Stem Cells 2016, 34, 1163–1176. [Google Scholar] [CrossRef] [PubMed]
- Schcolnik-Cabrera, A.; Juárez-López, D. Dual Contribution of the mTOR Pathway and of the Metabolism of Amino Acids in Prostate Cancer. Cell. Oncol. 2022, 45, 831–859. [Google Scholar] [CrossRef]
- Morris, S.M. Regulation of Enzymes of the Urea Cycle and Arginine Metabolism. Annu. Rev. Nutr. 2002, 22, 87–105. [Google Scholar] [CrossRef]
- Matos, A.; Carvalho, M.; Bicho, M.; Ribeiro, R. Arginine and Arginases Modulate Metabolism, Tumor Microenvironment and Prostate Cancer Progression. Nutrients 2021, 13, 4503. [Google Scholar] [CrossRef]
- Gannon, P.O.; Godin-Ethier, J.; Hassler, M.; Delvoye, N.; Aversa, M.; Poisson, A.O.; Péant, B.; Alam Fahmy, M.; Saad, F.; Lapointe, R.; et al. Androgen-Regulated Expression of Arginase 1, Arginase 2 and Interleukin-8 in Human Prostate Cancer. PLoS ONE 2010, 5, e12107. [Google Scholar] [CrossRef]
- Wang, J.; Torbenson, M.; Wang, Q.; Ro, J.Y.; Becich, M. Expression of Inducible Nitric Oxide Synthase in Paired Neoplastic and Non-Neoplastic Primary Prostate Cell Cultures and Prostatectomy Specimen. Urol. Oncol. Semin. Orig. Investig. 2003, 21, 117–122. [Google Scholar] [CrossRef]
- Bronte, V.; Kasic, T.; Gri, G.; Gallana, K.; Borsellino, G.; Marigo, I.; Battistini, L.; Iafrate, M.; Prayer-Galetti, T.; Pagano, F.; et al. Boosting Antitumor Responses of T Lymphocytes Infiltrating Human Prostate Cancers. J. Exp. Med. 2005, 201, 1257–1268. [Google Scholar] [CrossRef]
- Changou, C.A.; Chen, Y.-R.; Xing, L.; Yen, Y.; Chuang, F.Y.S.; Cheng, R.H.; Bold, R.J.; Ann, D.K.; Kung, H.-J. Arginine Starvation-Associated Atypical Cellular Death Involves Mitochondrial Dysfunction, Nuclear DNA Leakage, and Chromatin Autophagy. Proc. Natl. Acad. Sci. USA 2014, 111, 14147–14152. [Google Scholar] [CrossRef]
- Hsu, S.-C.; Chen, C.-L.; Cheng, M.-L.; Chu, C.-Y.; Changou, C.A.; Yu, Y.-L.; Yeh, S.-D.; Kuo, T.-C.; Kuo, C.-C.; Chuu, C.-P.; et al. Arginine Starvation Elicits Chromatin Leakage and cGAS-STING Activation via Epigenetic Silencing of Metabolic and DNA-Repair Genes. Theranostics 2021, 11, 7527–7545. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-L.; Hsu, S.-C.; Chung, T.-Y.; Chu, C.-Y.; Wang, H.-J.; Hsiao, P.-W.; Yeh, S.-D.; Ann, D.K.; Yen, Y.; Kung, H.-J. Arginine Is an Epigenetic Regulator Targeting TEAD4 to Modulate OXPHOS in Prostate Cancer Cells. Nat. Commun. 2021, 12, 2398. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.-T.; Qi, Y.; Wang, Y.-C.; Chi, K.K.; Chung, Y.; Ouyang, C.; Chen, Y.-R.; Oh, M.E.; Sheng, X.; Tang, Y.; et al. Arginine Starvation Kills Tumor Cells through Aspartate Exhaustion and Mitochondrial Dysfunction. Commun. Biol. 2018, 1, 178. [Google Scholar] [CrossRef] [PubMed]
- Mecham, J.O.; Rowitch, D.; Wallace, C.D.; Stern, P.H.; Hoffman, R.M. The Metabolic Defect of Methionine Dependence Occurs Frequently in Human Tumor Cell Lines. Biochem. Biophys. Res. Commun. 1983, 117, 429–434. [Google Scholar] [CrossRef]
- Chaturvedi, S.; Hoffman, R.M.; Bertino, J.R. Exploiting Methionine Restriction for Cancer Treatment. Biochem. Pharmacol. 2018, 154, 170–173. [Google Scholar] [CrossRef]
- Lu, S.; Epner, D.E. Molecular Mechanisms of Cell Cycle Block by Methionine Restriction in Human Prostate Cancer Cells. Nutr. Cancer 2000, 38, 123–130. [Google Scholar] [CrossRef]
- Liu, X.; Fu, Y.-M.; Meadows, G.G. Differential Effects of Specific Amino Acid Restriction on Glucose Metabolism, Reduction/Oxidation Status and Mitochondrial Damage in DU145 and PC3 Prostate Cancer Cells. Oncol. Lett. 2011, 2, 349–355. [Google Scholar] [CrossRef]
- Fu, Y.; Zhang, H.; Ding, M.; Li, Y.; Fu, X.; Yu, Z.; Meadows, G.G. Selective Amino Acid Restriction Targets Mitochondria to Induce Apoptosis of Androgen-independent Prostate Cancer Cells. J. Cell. Physiol. 2006, 209, 522–534. [Google Scholar] [CrossRef]
- Fu, Y.-M.; Yu, Z.-X.; Lin, H.; Fu, X.; Meadows, G.G. Selective Amino Acid Restriction Differentially Affects the Motility and Directionality of DU145 and PC3 Prostate Cancer Cells. J. Cell. Physiol. 2008, 217, 184–193. [Google Scholar] [CrossRef]
- Han, Q.; Hoffman, R.M. Lowering and Stabilizing PSA Levels in Advanced-Prostate Cancer Patients With Oral Methioninase. Anticancer Res. 2021, 41, 1921. [Google Scholar] [CrossRef]
- Han, Q.; Tan, Y.; Hoffman, R.M. Oral Dosing of Recombinant Methioninase Is Associated With a 70% Drop in PSA in a Patient With Bone-Metastatic Prostate Cancer and 50% Reduction in Circulating Methionine in a High-Stage Ovarian Cancer Patient. Anticancer Res. 2020, 40, 2813. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Shao, Y.; Zhao, X.; Hong, C.S.; Wang, F.; Lu, X.; Li, J.; Ye, G.; Yan, M.; Zhuang, Z.; et al. Integration of Metabolomics and Transcriptomics Reveals Major Metabolic Pathways and Potential Biomarker Involved in Prostate Cancer. Mol. Cell. Proteom. 2016, 15, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Tomasi, M.L.; Cossu, C.; Spissu, Y.; Floris, A.; Ryoo, M.; Iglesias-Ara, A.; Wang, Q.; Pandol, S.J.; Bhowmick, N.A.; Seki, E.; et al. S-Adenosylmethionine and Methylthioadenosine Inhibit Cancer Metastasis by Targeting microRNA 34a/b-Methionine Adenosyltransferase 2A/2B Axis. Oncotarget 2017, 8, 78851–78869. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, L.Y.; Arsene, D.; Mato, J.M.; Lu, S.C. Methionine Adenosyltransferases in Cancers: Mechanisms of Dysregulation and Implications for Therapy. Exp. Biol. Med. 2018, 243, 107–117. [Google Scholar] [CrossRef]
- Cacciatore, A.; Shinde, D.; Musumeci, C.; Sandrini, G.; Guarrera, L.; Albino, D.; Civenni, G.; Storelli, E.; Mosole, S.; Federici, E.; et al. Epigenome-Wide Impact of MAT2A Sustains the Androgen-Indifferent State and Confers Synthetic Vulnerability in ERG Fusion-Positive Prostate Cancer. Nat. Commun. 2024, 15, 6672. [Google Scholar] [CrossRef]
- Schmidt, T.; Leha, A.; Salinas-Riester, G. Treatment of Prostate Cancer Cells with S-Adenosylmethionine Leads to Genome-Wide Alterations in Transcription Profiles. Gene 2016, 595, 161–167. [Google Scholar] [CrossRef]
- Schmidt, T. S-Adenosylmethionine Affects ERK1/2 and STAT3 Pathway in Androgen-Independent Prostate Cancer Cells. Mol. Biol. Rep. 2022, 49, 4805–4817. [Google Scholar] [CrossRef]
- Shukeir, N.; Pakneshan, P.; Chen, G.; Szyf, M.; Rabbani, S.A. Alteration of the Methylation Status of Tumor-Promoting Genes Decreases Prostate Cancer Cell Invasiveness and Tumorigenesis In Vitro and In Vivo. Cancer Res. 2006, 66, 9202–9210. [Google Scholar] [CrossRef]
- Mathes, A.; Duman, M.B.; Neumann, A.; Dobreva, G.; Schmidt, T. S-Adenosylmethionine Treatment Affects Histone Methylation in Prostate Cancer Cells. Gene 2024, 893, 147915. [Google Scholar] [CrossRef]
- Branco, A.C.C.C.; Yoshikawa, F.S.Y.; Pietrobon, A.J.; Sato, M.N. Role of Histamine in Modulating the Immune Response and Inflammation. Mediat. Inflamm. 2018, 2018, 9524075. [Google Scholar] [CrossRef]
- Matsushita, M.; Fujita, K.; Hatano, K.; Hayashi, T.; Kayama, H.; Motooka, D.; Hase, H.; Yamamoto, A.; Uemura, T.; Yamamichi, G.; et al. High-Fat Diet Promotes Prostate Cancer Growth through Histamine Signaling. Int. J. Cancer 2022, 151, 623–636. [Google Scholar] [CrossRef] [PubMed]
- Labuschagne, C.F.; van den Broek, N.J.F.; Mackay, G.M.; Vousden, K.H.; Maddocks, O.D.K. Serine, but Not Glycine, Supports One-Carbon Metabolism and Proliferation of Cancer Cells. Cell Rep. 2014, 7, 1248–1258. [Google Scholar] [CrossRef]
- Holeček, M. Serine Metabolism in Health and Disease and as a Conditionally Essential Amino Acid. Nutrients 2022, 14, 1987. [Google Scholar] [CrossRef]
- Wang, W.; He, Z.; Kong, Y.; Liu, Z.; Gong, L. GC-MS-Based Metabolomics Reveals New Biomarkers to Assist the Differentiation of Prostate Cancer and Benign Prostatic Hyperplasia. Clin. Chim. Acta 2021, 519, 10–17. [Google Scholar] [CrossRef]
- Falegan, O.S.; Jarvi, K.; Vogel, H.J.; Hyndman, M.E. Seminal Plasma Metabolomics Reveals Lysine and Serine Dysregulation as Unique Features Distinguishing between Prostate Cancer Tumors of Gleason Grades 6 and 7. Prostate 2021, 81, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Saruta, M.; Takahara, K.; Yoshizawa, A.; Niimi, A.; Takeuchi, T.; Nukaya, T.; Takenaka, M.; Zennami, K.; Ichino, M.; Sasaki, H.; et al. Alanine-Serine-Cysteine Transporter 2 Inhibition Suppresses Prostate Cancer Cell Growth In Vitro. J. Clin. Med. 2022, 11, 5466. [Google Scholar] [CrossRef]
- Buqué, A.; Galluzzi, L.; Montrose, D.C. Targeting Serine in Cancer: Is Two Better Than One? Trends Cancer 2021, 7, 668–670. [Google Scholar] [CrossRef] [PubMed]
- Reina-Campos, M.; Linares, J.F.; Duran, A.; Cordes, T.; L’Hermitte, A.; Badur, M.G.; Bhangoo, M.S.; Thorson, P.K.; Richards, A.; Rooslid, T.; et al. Increased Serine and One-Carbon Pathway Metabolism by PKCλ/ι Deficiency Promotes Neuroendocrine Prostate Cancer. Cancer Cell 2019, 35, 385–400.e9. [Google Scholar] [CrossRef]
- Sun, W.; Zhao, E.; Cui, H. Target Enzymes in Serine-glycine-one-carbon Metabolic Pathway for Cancer Therapy. Int. J. Cancer 2023, 152, 2446–2463. [Google Scholar] [CrossRef]
- Li, A.M.; Ye, J. Reprogramming of Serine, Glycine and One-Carbon Metabolism in Cancer. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165841. [Google Scholar] [CrossRef]
- Chandrika, M.; Chua, P.J.; Muniasamy, U.; Huang, R.Y.J.; Thike, A.A.; Ng, C.T.; Tan, P.H.; Yip, G.W.; Bay, B.H. Prognostic Significance of Phosphoglycerate Dehydrogenase in Breast Cancer. Breast Cancer Res. Treat. 2021, 186, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Zhang, J.; Zheng, Z.; Yang, F.; Liu, S.; Wu, Y.; Chen, Y.; Xu, T.; Mao, S.; Yan, Y.; et al. PHGDH Inhibits Ferroptosis and Promotes Malignant Progression by Upregulating SLC7A11 in Bladder Cancer. Int. J. Biol. Sci. 2022, 18, 5459–5474. [Google Scholar] [CrossRef] [PubMed]
- Rathore, R.; Schutt, C.R.; Van Tine, B.A. PHGDH as a Mechanism for Resistance in Metabolically-Driven Cancers. Cancer Drug Resist. 2020, 3, 762–774. [Google Scholar] [CrossRef]
- Wang, J.; Zeng, L.; Wu, N.; Liang, Y.; Jin, J.; Fan, M.; Lai, X.; Chen, Z.-S.; Pan, Y.; Zeng, F.; et al. Inhibition of Phosphoglycerate Dehydrogenase Induces Ferroptosis and Overcomes Enzalutamide Resistance in Castration-Resistant Prostate Cancer Cells. Drug Resist. Updat. 2023, 70, 100985. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.Y.; Haverty, P.M.; Li, L.; Kljavin, N.M.; Bourgon, R.; Lee, J.; Stern, H.; Modrusan, Z.; Seshagiri, S.; Zhang, Z.; et al. Comparative Oncogenomics Identifies PSMB4 and SHMT2 as Potential Cancer Driver Genes. Cancer Res. 2014, 74, 3114–3126. [Google Scholar] [CrossRef]
- Zeng, Y.; Zhang, J.; Xu, M.; Chen, F.; Zi, R.; Yue, J.; Zhang, Y.; Chen, N.; Chin, Y.E. Roles of Mitochondrial Serine Hydroxymethyltransferase 2 (SHMT2) in Human Carcinogenesis. J. Cancer 2021, 12, 5888–5894. [Google Scholar] [CrossRef]
- Chen, L.; Liu, H.; Ji, Y.; Ma, Z.; Shen, K.; Shangguan, X.; Qian, H.; Zhao, Y.; Pan, C.-W.; Xue, W. Downregulation of SHMT2 Promotes the Prostate Cancer Proliferation and Metastasis by Inducing Epithelial-Mesenchymal Transition. Exp. Cell Res. 2022, 415, 113138. [Google Scholar] [CrossRef]
- Marrocco, I.; Altieri, F.; Rubini, E.; Paglia, G.; Chichiarelli, S.; Giamogante, F.; Macone, A.; Perugia, G.; Magliocca, F.M.; Gurtner, A.; et al. Shmt2: A Stat3 Signaling New Player in Prostate Cancer Energy Metabolism. Cells 2019, 8, 1048. [Google Scholar] [CrossRef]
- Jain, M.; Nilsson, R.; Sharma, S.; Madhusudhan, N.; Kitami, T.; Souza, A.L.; Kafri, R.; Kirschner, M.W.; Clish, C.B.; Mootha, V.K. Metabolite Profiling Identifies a Key Role for Glycine in Rapid Cancer Cell Proliferation. Science 2012, 336, 1040–1044. [Google Scholar] [CrossRef]
- Kumar, D.; Gupta, A.; Mandhani, A.; Sankhwar, S.N. NMR Spectroscopy of Filtered Serum of Prostate Cancer: A New Frontier in Metabolomics: Metabolomics of Prostate Cancer. Prostate 2016, 76, 1106–1119. [Google Scholar] [CrossRef]
- Kumar, D.; Gupta, A.; Mandhani, A.; Sankhwar, S.N. Metabolomics-Derived Prostate Cancer Biomarkers: Fact or Fiction? J. Proteome Res. 2015, 14, 1455–1464. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Rambla, C.; Puchades-Carrasco, L.; García-Flores, M.; Rubio-Briones, J.; López-Guerrero, J.A.; Pineda-Lucena, A. Non-Invasive Urinary Metabolomic Profiling Discriminates Prostate Cancer from Benign Prostatic Hyperplasia. Metabolomics 2017, 13, 52. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Zhang, C.; Cheng, S.; Li, G.; Griebel, J.; Neuhaus, J. Novel Metabolic Signatures of Prostate Cancer Revealed by 1H-NMR Metabolomics of Urine. Diagnostics 2021, 11, 149. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Cebrián, N.; García-Flores, M.; Rubio-Briones, J.; López-Guerrero, J.A.; Pineda-Lucena, A.; Puchades-Carrasco, L. Targeted Metabolomics Analyses Reveal Specific Metabolic Alterations in High-Grade Prostate Cancer Patients. J. Proteome Res. 2020, 19, 4082–4092. [Google Scholar] [CrossRef] [PubMed]
- Amelio, I.; Cutruzzolá, F.; Antonov, A.; Agostini, M.; Melino, G. Serine and Glycine Metabolism in Cancer. Trends Biochem. Sci. 2014, 39, 191–198. [Google Scholar] [CrossRef]
- Liu, R.; Zeng, L.-W.; Gong, R.; Yuan, F.; Shu, H.-B.; Li, S. mTORC1 Activity Regulates Post-Translational Modifications of Glycine Decarboxylase to Modulate Glycine Metabolism and Tumorigenesis. Nat. Commun. 2021, 12, 4227. [Google Scholar] [CrossRef]
- Yu, H.; Hu, X.; Zhang, Y.; Wang, J.; Ni, Z.; Wang, Y.; Zhu, H. GLDC Promotes Colorectal Cancer Metastasis through Epithelial–Mesenchymal Transition Mediated by Hippo Signaling Pathway. Med. Oncol. 2023, 40, 293. [Google Scholar] [CrossRef]
- Meiser, J.; Vazquez, A. Give It or Take It: The Flux of One-carbon in Cancer Cells. FEBS J. 2016, 283, 3695–3704. [Google Scholar] [CrossRef]
- Mukha, D.; Fokra, M.; Feldman, A.; Sarvin, B.; Sarvin, N.; Nevo-Dinur, K.; Besser, E.; Hallo, E.; Aizenshtein, E.; Schug, Z.T.; et al. Glycine Decarboxylase Maintains Mitochondrial Protein Lipoylation to Support Tumor Growth. Cell Metab. 2022, 34, 775–782.e9. [Google Scholar] [CrossRef]
- Kikuchi, G.; Motokawa, Y.; Yoshida, T.; Hiraga, K. Glycine Cleavage System: Reaction Mechanism, Physiological Significance, and Hyperglycinemia. Proc. Jpn. Acad. Ser. B 2008, 84, 246–263. [Google Scholar] [CrossRef]
- Woo, C.C.; Kaur, K.; Chan, W.X.; Teo, X.Q.; Lee, T.H.P. Inhibiting Glycine Decarboxylase Suppresses Pyruvate-to-Lactate Metabolism in Lung Cancer Cells. Front. Oncol. 2018, 8, 196. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.C.; Shyh-Chang, N.; Yang, H.; Rai, A.; Umashankar, S.; Ma, S.; Soh, B.S.; Sun, L.L.; Tai, B.C.; Nga, M.E.; et al. Glycine Decarboxylase Activity Drives Non-Small Cell Lung Cancer Tumor-Initiating Cells and Tumorigenesis. Cell 2012, 148, 259–272. [Google Scholar] [CrossRef]
- Xie, H.; Yan, T.; Lu, X.; Du, Y.; Xu, S.; Kong, Y.; Yu, L.; Sun, J.; Zhou, L.; Ma, J. GLDC Mitigated by miR-30e Regulates Cell Proliferation and Tumor Immune Infiltration in TNBC. Front. Immunol. 2022, 13, 1033367. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Xiao, Z.-Y.; Huang, Z.-P.; Xue, K.-Y.; Xia, H.; Zhou, J.-W.; Liao, D.-Y.; Liang, Z.-J.; Xie, X.; Wei, Q.-Z.; et al. Glycine Decarboxylase (GLDC) Plays a Crucial Role in Regulating Energy Metabolism, Invasion, Metastasis and Immune Escape for Prostate Cancer. Int. J. Biol. Sci. 2023, 19, 4726–4743. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.C.; Han, J.M.; Kim, S. Aminoacyl-tRNA Synthetases and Amino Acid Signaling. Biochim. Biophys. Acta BBA Mol. Cell Res. 2021, 1868, 118889. [Google Scholar] [CrossRef]
- Zhou, Z.; Sun, B.; Nie, A.; Yu, D.; Bian, M. Roles of Aminoacyl-tRNA Synthetases in Cancer. Front. Cell Dev. Biol. 2020, 8, 599765. [Google Scholar] [CrossRef]
- Kwon, N.H.; Fox, P.L.; Kim, S. Aminoacyl-tRNA Synthetases as Therapeutic Targets. Nat. Rev. Drug Discov. 2019, 18, 629–650. [Google Scholar] [CrossRef]
- Vellaichamy, A.; Sreekumar, A.; Strahler, J.R.; Rajendiran, T.; Yu, J.; Varambally, S.; Li, Y.; Omenn, G.S.; Chinnaiyan, A.M.; Nesvizhskii, A.I. Proteomic Interrogation of Androgen Action in Prostate Cancer Cells Reveals Roles of Aminoacyl tRNA Synthetases. PLoS ONE 2009, 4, e7075. [Google Scholar] [CrossRef]
- Sangha, A.K.; Kantidakis, T. The Aminoacyl-tRNA Synthetase and tRNA Expression Levels Are Deregulated in Cancer and Correlate Independently with Patient Survival. Curr. Issues Mol. Biol. 2022, 44, 3001–3019. [Google Scholar] [CrossRef]
- Khosh Kish, E.; Gamallat, Y.; Choudhry, M.; Ghosh, S.; Seyedi, S.; Bismar, T.A. Glycyl-tRNA Synthetase (GARS) Expression Is Associated with Prostate Cancer Progression and Its Inhibition Decreases Migration, and Invasion In Vitro. Int. J. Mol. Sci. 2023, 24, 4260. [Google Scholar] [CrossRef]
- Nie, J.; Liu, T.; Mao, T.; Yang, H.; Deng, W.; Liu, X.; Fu, B. Transcriptome Sequencing and Single-Cell Sequencing Analysis Identify GARS1 as a Potential Prognostic and Immunotherapeutic Biomarker for Multiple Cancers, Including Bladder Cancer. Front. Immunol. 2023, 14, 1169588. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xu, C.; Lee, H.J.; Ren, S.; Zi, X.; Zhang, Z.; Wang, H.; Yu, Y.; Yang, C.; Gao, X.; et al. A Genomic and Epigenomic Atlas of Prostate Cancer in Asian Populations. Nature 2020, 580, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015, 163, 1011–1025. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, G.; Yao, X.; Fang, Y.; Lin, Q.; Zhou, M.; Yang, Y.; Meng, Q.; Zhang, Q.; Wang, S. Proteomic Profiling of Prostate Cancer Reveals Molecular Signatures under Antiandrogen Treatment. Clin. Proteom. 2024, 21, 44. [Google Scholar] [CrossRef]
- Jeong, S.-H.; Kwak, C. Prostate-Specific Membrane Antigen-Mediated Theragnostics in Prostate Cancer. Investig. Clin. Urol. 2021, 62, 497–499. [Google Scholar] [CrossRef]
- Stamatakos, P.V.; Fragkoulis, C.; Leventi, A.; Gklinos, K.; Kontolatis, N.; Papatsoris, A.; Dellis, A. PSMA-Based Therapeutics for Prostate Cancer. Expert. Opin. Pharmacother. 2024, 25, 1405–1419. [Google Scholar] [CrossRef]
- Marques, R.B.; Dits, N.F.; Erkens-Schulze, S.; van Ijcken, W.F.J.; van Weerden, W.M.; Jenster, G. Modulation of Androgen Receptor Signaling in Hormonal Therapy-Resistant Prostate Cancer Cell Lines. PLoS ONE 2011, 6, e23144. [Google Scholar] [CrossRef]
- He, J.; Mao, Y.; Huang, W.; Li, M.; Zhang, H.; Qing, Y.; Lu, S.; Xiao, H.; Li, K. Methylcrotonoyl-CoA Carboxylase 2 Promotes Proliferation, Migration and Invasion and Inhibits Apoptosis of Prostate Cancer Cells Through Regulating GLUD1-P38 MAPK Signaling Pathway. OncoTargets Ther. 2020, 13, 7317–7327. [Google Scholar] [CrossRef]
- Park, S.H.; Fong, K.-W.; Mong, E.; Martin, M.C.; Schiltz, G.E.; Yu, J. Going beyond Polycomb: EZH2 Functions in Prostate Cancer. Oncogene 2021, 40, 5788–5798. [Google Scholar] [CrossRef]
- Kaur, P.; Shankar, E.; Gupta, S. EZH2-Mediated Development of Therapeutic Resistance in Cancer. Cancer Lett. 2024, 586, 216706. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Wu, Z.J.; Groner, A.C.; He, H.H.; Cai, C.; Lis, R.T.; Wu, X.; Stack, E.C.; Loda, M.; Liu, T.; et al. EZH2 Oncogenic Activity in Castration-Resistant Prostate Cancer Cells Is Polycomb-Independent. Science 2012, 338, 1465–1469. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, X.; Kim, M.; He, D.; Wang, C.; Fong, K.W.; Liu, X. Downregulation of EZH2 Inhibits Epithelial-Mesenchymal Transition in Enzalutamide-Resistant Prostate Cancer. Prostate 2023, 83, 1458–1469. [Google Scholar] [CrossRef] [PubMed]
- Queisser, A.; Hagedorn, S.; Wang, H.; Schaefer, T.; Konantz, M.; Alavi, S.; Deng, M.; Vogel, W.; von Mässenhausen, A.; Kristiansen, G.; et al. Ecotropic Viral Integration Site 1, a Novel Oncogene in Prostate Cancer. Oncogene 2017, 36, 1573–1584. [Google Scholar] [CrossRef]
- Jariwala, U.; Prescott, J.; Jia, L.; Barski, A.; Pregizer, S.; Cogan, J.P.; Arasheben, A.; Tilley, W.D.; Scher, H.I.; Gerald, W.L.; et al. Identification of Novel Androgen Receptor Target Genes in Prostate Cancer. Mol. Cancer 2007, 6, 39. [Google Scholar] [CrossRef]
- Yan, Y.; Chang, L.; Tian, H.; Wang, L.; Zhang, Y.; Yang, T.; Li, G.; Hu, W.; Shah, K.; Chen, G.; et al. 1-Pyrroline-5-Carboxylate Released by Prostate Cancer Cell Inhibit T Cell Proliferation and Function by Targeting SHP1/Cytochrome c Oxidoreductase/ROS Axis. J. Immunother. Cancer 2018, 6, 148. [Google Scholar] [CrossRef]
- Xu, H.; He, Y.; Lin, L.; Li, M.; Zhou, Z.; Yang, Y. MiR-1207-5p Targets PYCR1 to Inhibit the Progression of Prostate Cancer. Biochem. Biophys. Res. Commun. 2021, 575, 56–64. [Google Scholar] [CrossRef]
- Zeng, T.; Zhu, L.; Liao, M.; Zhuo, W.; Yang, S.; Wu, W.; Wang, D. Knockdown of PYCR1 Inhibits Cell Proliferation and Colony Formation via Cell Cycle Arrest and Apoptosis in Prostate Cancer. Med. Oncol. 2017, 34, 27. [Google Scholar] [CrossRef]
- Jianfeng, W.; Yutao, W.; Jianbin, B. Indolethylamine-N-Methyltransferase Inhibits Proliferation and Promotes Apoptosis of Human Prostate Cancer Cells: A Mechanistic Exploration. Front. Cell Dev. Biol. 2022, 10, 805402. [Google Scholar] [CrossRef]
- Zhong, S.; Jeong, J.-H.; Huang, C.; Chen, X.; Dickinson, S.I.; Dhillon, J.; Yang, L.; Luo, J.-L. Targeting INMT and Interrupting Its Methylation Pathway for the Treatment of Castration Resistant Prostate Cancer. J. Exp. Clin. Cancer Res. 2021, 40, 307. [Google Scholar] [CrossRef]
- Larkin, S.E.T.; Holmes, S.; Cree, I.A.; Walker, T.; Basketter, V.; Bickers, B.; Harris, S.; Garbis, S.D.; Townsend, P.A.; Aukim-Hastie, C. Identification of Markers of Prostate Cancer Progression Using Candidate Gene Expression. Br. J. Cancer 2012, 106, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Middha, M.; Bicak, M.; Sjoberg, D.D.; Vertosick, E.; Dahlin, A.; Häggström, C.; Hallmans, G.; Rönn, A.-C.; Stattin, P.; et al. Genome-Wide Scan Identifies Role for AOX1 in Prostate Cancer Survival. Eur. Urol. 2018, 74, 710–719. [Google Scholar] [CrossRef]
- Wu, J.; Wei, Y.; Li, T.; Lin, L.; Yang, Z.; Ye, L. DNA Methylation-Mediated Lowly Expressed AOX1 Promotes Cell Migration and Invasion of Prostate Cancer. Urol. Int. 2023, 107, 517–525. [Google Scholar] [CrossRef]
- Weng, H.; Xiong, K.-P.; Wang, W.; Qian, K.-Y.; Yuan, S.; Wang, G.; Yu, F.; Luo, J.; Lu, M.-X.; Yang, Z.-H.; et al. Aspartoacylase Suppresses Prostate Cancer Progression by Blocking LYN Activation. Mil. Med. Res. 2023, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Ascenção, K.; Szabo, C. Emerging Roles of Cystathionine β-Synthase in Various Forms of Cancer. Redox Biol. 2022, 53, 102331. [Google Scholar] [CrossRef] [PubMed]
- Zabala-Letona, A.; Arruabarrena-Aristorena, A.; Fernandez-Ruiz, S.; Viera, C.; Carlevaris, O.; Ercilla, A.; Mendizabal, I.; Martin, T.; Macchia, A.; Camacho, L.; et al. PI3K-Regulated Glycine N-Methyltransferase Is Required for the Development of Prostate Cancer. Oncogenesis 2022, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.H.; Shiota, M.; Kuroiwa, K.; Naito, S.; Oda, Y. The Important Role of Glycine N-Methyltransferase in the Carcinogenesis and Progression of Prostate Cancer. Mod. Pathol. 2011, 24, 1272–1280. [Google Scholar] [CrossRef]
- Ottaviani, S.; Brooke, G.N.; O’Hanlon-Brown, C.; Waxman, J.; Ali, S.; Buluwela, L. Characterisation of the Androgen Regulation of Glycine N-Methyltransferase in Prostate Cancer Cells. J. Mol. Endocrinol. 2013, 51, 301–312. [Google Scholar] [CrossRef]
- Pu, T.; Wang, J.; Wei, J.; Zeng, A.; Zhang, J.; Chen, J.; Yin, L.; Li, J.; Lin, T.-P.; Melamed, J.; et al. Stromal-Derived MAOB Promotes Prostate Cancer Growth and Progression. Sci. Adv. 2024, 10, eadi4935. [Google Scholar] [CrossRef]
- Huang, H.-C.; Hsieh, Y.-H.; Hsiao, C.-H.; Lin, C.-Y.; Wang, S.-S.; Ho, K.-H.; Chang, L.-C.; Huang, H.-M.; Yang, S.-F.; Chien, M.-H. MAOB Expression Correlates with a Favourable Prognosis in Prostate Cancer, and Its Genetic Variants Are Associated with the Metastasis of the Disease. J. Cell. Mol. Med. 2024, 28, e18229. [Google Scholar] [CrossRef]
- Shukla-Dave, A.; Castillo-Martin, M.; Chen, M.; Lobo, J.; Gladoun, N.; Collazo-Lorduy, A.; Khan, F.M.; Ponomarev, V.; Yi, Z.; Zhang, W.; et al. Ornithine Decarboxylase Is Sufficient for Prostate Tumorigenesis via Androgen Receptor Signaling. Am. J. Pathol. 2016, 186, 3131–3145. [Google Scholar] [CrossRef] [PubMed]
- DePrimo, S.E.; Diehn, M.; Nelson, J.B.; Reiter, R.E.; Matese, J.; Fero, M.; Tibshirani, R.; Brown, P.O.; Brooks, J.D. Transcriptional Programs Activated by Exposure of Human Prostate Cancer Cells to Androgen. Genome Biol. 2002, 3, research0032.1. [Google Scholar] [CrossRef]
- Giskeødegård, G.F.; Bertilsson, H.; Selnæs, K.M.; Wright, A.J.; Bathen, T.F.; Viset, T.; Halgunset, J.; Angelsen, A.; Gribbestad, I.S.; Tessem, M.-B. Spermine and Citrate as Metabolic Biomarkers for Assessing Prostate Cancer Aggressiveness. PLoS ONE 2013, 8, e62375. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [PubMed]
- Sancak, Y.; Peterson, T.R.; Shaul, Y.D.; Lindquist, R.A.; Thoreen, C.C.; Bar-Peled, L.; Sabatini, D.M. The Rag GTPases Bind Raptor and Mediate Amino Acid Signaling to mTORC1. Science 2008, 320, 1496–1501. [Google Scholar] [CrossRef]
- Chantranupong, L.; Scaria, S.M.; Saxton, R.A.; Gygi, M.P.; Shen, K.; Wyant, G.A.; Wang, T.; Harper, J.W.; Gygi, S.P.; Sabatini, D.M. The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell 2016, 165, 153–164. [Google Scholar] [CrossRef]
- Jewell, J.L.; Kim, Y.C.; Russell, R.C.; Yu, F.-X.; Park, H.W.; Plouffe, S.W.; Tagliabracci, V.S.; Guan, K.-L. Differential Regulation of mTORC1 by Leucine and Glutamine. Science 2015, 347, 194–198. [Google Scholar] [CrossRef]
- Wolfson, R.L.; Chantranupong, L.; Saxton, R.A.; Shen, K.; Scaria, S.M.; Cantor, J.R.; Sabatini, D.M. Sestrin2 Is a Leucine Sensor for the mTORC1 Pathway. Science 2016, 351, 43–48. [Google Scholar] [CrossRef]
- Chen, J.; Ou, Y.; Luo, R.; Wang, J.; Wang, D.; Guan, J.; Li, Y.; Xia, P.; Chen, P.R.; Liu, Y. SAR1B Senses Leucine Levels to Regulate mTORC1 Signalling. Nature 2021, 596, 281–284. [Google Scholar] [CrossRef]
- Han, J.M.; Jeong, S.J.; Park, M.C.; Kim, G.; Kwon, N.H.; Kim, H.K.; Ha, S.H.; Ryu, S.H.; Kim, S. Leucyl-tRNA Synthetase Is an Intracellular Leucine Sensor for the mTORC1-Signaling Pathway. Cell 2012, 149, 410–424. [Google Scholar] [CrossRef]
- Rebsamen, M.; Pochini, L.; Stasyk, T.; De Araújo, M.E.G.; Galluccio, M.; Kandasamy, R.K.; Snijder, B.; Fauster, A.; Rudashevskaya, E.L.; Bruckner, M.; et al. SLC38A9 Is a Component of the Lysosomal Amino Acid Sensing Machinery That Controls mTORC1. Nature 2015, 519, 477–481. [Google Scholar] [CrossRef]
- Wang, Q.; Bailey, C.G.; Ng, C.; Tiffen, J.; Thoeng, A.; Minhas, V.; Lehman, M.L.; Hendy, S.C.; Buchanan, G.; Nelson, C.C.; et al. Androgen Receptor and Nutrient Signaling Pathways Coordinate the Demand for Increased Amino Acid Transport during Prostate Cancer Progression. Cancer Res. 2011, 71, 7525–7536. [Google Scholar] [CrossRef] [PubMed]
- Otsuki, H.; Kimura, T.; Yamaga, T.; Kosaka, T.; Suehiro, J.; Sakurai, H. Prostate Cancer Cells in Different Androgen Receptor Status Employ Different Leucine Transporters. Prostate 2017, 77, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Tiffen, J.; Bailey, C.G.; Lehman, M.L.; Ritchie, W.; Fazli, L.; Metierre, C.; Feng, Y.; Li, E.; Gleave, M.; et al. Targeting Amino Acid Transport in Metastatic Castration-Resistant Prostate Cancer: Effects on Cell Cycle, Cell Growth, and Tumor Development. JNCI J. Natl. Cancer Inst. 2013, 105, 1463–1473. [Google Scholar] [CrossRef] [PubMed]
- Rii, J.; Sakamoto, S.; Mizokami, A.; Xu, M.; Fujimoto, A.; Saito, S.; Koike, H.; Tamura, T.; Arai, T.; Yamada, Y.; et al. L-type Amino Acid Transporter 1 Inhibitor JPH203 Prevents the Growth of Cabazitaxel-resistant Prostate Cancer by Inhibiting Cyclin-dependent Kinase Activity. Cancer Sci. 2024, 115, 937–953. [Google Scholar] [CrossRef]
- Saito, S.; Ando, K.; Sakamoto, S.; Xu, M.; Yamada, Y.; Rii, J.; Kanaoka, S.; Wei, J.; Zhao, X.; Pae, S.; et al. The LAT1 Inhibitor JPH203 Suppresses the Growth of Castration-resistant Prostate Cancer through a CD24 -mediated Mechanism. Cancer Sci. 2024, 115, 2461–2472. [Google Scholar] [CrossRef]
- Dasgupta, S.; Putluri, N.; Long, W.; Zhang, B.; Wang, J.; Kaushik, A.K.; Arnold, J.M.; Bhowmik, S.K.; Stashi, E.; Brennan, C.A.; et al. Coactivator SRC-2-Dependent Metabolic Reprogramming Mediates Prostate Cancer Survival and Metastasis. J. Clin. Investig. 2015, 125, 1174–1188. [Google Scholar] [CrossRef]
- Wang, Q.; Hardie, R.-A.; Hoy, A.J.; van Geldermalsen, M.; Gao, D.; Fazli, L.; Sadowski, M.C.; Balaban, S.; Schreuder, M.; Nagarajah, R.; et al. Targeting ASCT2-mediated glutamine uptake blocks prostate cancer growth and tumour development. J. Pathol. 2015, 236, 278–289. [Google Scholar] [CrossRef]
- Dennis, P.B.; Jaeschke, A.; Saitoh, M.; Fowler, B.; Kozma, S.C.; Thomas, G. Mammalian TOR: A Homeostatic ATP Sensor. Science 2001, 294, 1102–1105. [Google Scholar] [CrossRef]
- Xu, J.; Ji, J.; Yan, X.-H. Cross-Talk between AMPK and mTOR in Regulating Energy Balance. Crit. Rev. Food Sci. Nutr. 2012, 52, 373–381. [Google Scholar] [CrossRef]
- Lodi, A.; Saha, A.; Lu, X.; Wang, B.; Sentandreu, E.; Collins, M.; Kolonin, M.G.; DiGiovanni, J.; Tiziani, S. Combinatorial Treatment with Natural Compounds in Prostate Cancer Inhibits Prostate Tumor Growth and Leads to Key Modulations of Cancer Cell Metabolism. NPJ Precis. Oncol. 2017, 1, 18. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Kusnadi, E.P.; Furic, L.; Selth, L.A. Regulation of mRNA Translation by Hormone Receptors in Breast and Prostate Cancer. Cancers 2021, 13, 3254. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, E.C.; Knebel, S.M.; Lo, W.-H.; Leung, Y.-C.; Cheng, P.N.-M.; Hsueh, C.-T. Deprivation of Arginine by Recombinant Human Arginase in Prostate Cancer Cells. J. Hematol. Oncol. 2012, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Guan, Y.; Xu, C.; Wang, D.; Guan, J.; Liu, Y. VWCE Modulates Amino Acid-Dependent mTOR Signaling and Coordinates with KICSTOR to Recruit GATOR1 to the Lysosomes. Nat. Commun. 2023, 14, 8464. [Google Scholar] [CrossRef]
- Gallinetti, J.; Harputlugil, E.; Mitchell, J.R. Amino Acid Sensing in Dietary-Restriction-Mediated Longevity: Roles of Signal-Transducing Kinases GCN2 and TOR. Biochem. J. 2013, 449, 1–10. [Google Scholar] [CrossRef]
- Masson, G.R. Towards a Model of GCN2 Activation. Biochem. Soc. Trans. 2019, 47, 1481–1488. [Google Scholar] [CrossRef]
- Ye, J.; Kumanova, M.; Hart, L.S.; Sloane, K.; Zhang, H.; De Panis, D.N.; Bobrovnikova-Marjon, E.; Diehl, J.A.; Ron, D.; Koumenis, C. The GCN2-ATF4 Pathway Is Critical for Tumour Cell Survival and Proliferation in Response to Nutrient Deprivation. EMBO J. 2010, 29, 2082–2096. [Google Scholar] [CrossRef]
- Furnish, M.; Boulton, D.P.; Genther, V.; Grofova, D.; Ellinwood, M.L.; Romero, L.; Lucia, M.S.; Cramer, S.D.; Caino, M.C. MIRO2 Regulates Prostate Cancer Cell Growth via GCN1-Dependent Stress Signaling. Mol. Cancer Res. 2022, 20, 607–621. [Google Scholar] [CrossRef]
- Gold, L.T.; Masson, G.R. GCN2: Roles in Tumour Development and Progression. Biochem. Soc. Trans. 2022, 50, 737–745. [Google Scholar] [CrossRef]
- Cordova, R.A.; Misra, J.; Amin, P.H.; Klunk, A.J.; Damayanti, N.P.; Carlson, K.R.; Elmendorf, A.J.; Kim, H.-G.; Mirek, E.T.; Elzey, B.D.; et al. GCN2 eIF2 Kinase Promotes Prostate Cancer by Maintaining Amino Acid Homeostasis. eLife 2022, 11, e81083. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, P.; Cao, S.; Zhu, Z.; Wen, Y.; Guo, Y.; Liang, W.; Xie, J. Cellular Signaling of Amino Acid Metabolism in Prostate Cancer. Int. J. Mol. Sci. 2025, 26, 776. https://doi.org/10.3390/ijms26020776
Yao P, Cao S, Zhu Z, Wen Y, Guo Y, Liang W, Xie J. Cellular Signaling of Amino Acid Metabolism in Prostate Cancer. International Journal of Molecular Sciences. 2025; 26(2):776. https://doi.org/10.3390/ijms26020776
Chicago/Turabian StyleYao, Ping, Shiqi Cao, Ziang Zhu, Yunru Wen, Yawen Guo, Wenken Liang, and Jianling Xie. 2025. "Cellular Signaling of Amino Acid Metabolism in Prostate Cancer" International Journal of Molecular Sciences 26, no. 2: 776. https://doi.org/10.3390/ijms26020776
APA StyleYao, P., Cao, S., Zhu, Z., Wen, Y., Guo, Y., Liang, W., & Xie, J. (2025). Cellular Signaling of Amino Acid Metabolism in Prostate Cancer. International Journal of Molecular Sciences, 26(2), 776. https://doi.org/10.3390/ijms26020776






