Molecular Docking Studies and In Vitro Activity of Pancreatic Lipase Inhibitors from Yak Milk Cheese
Abstract
1. Introduction
2. Results and Discussion
2.1. Analysis of Physicochemical Properties of Peptides
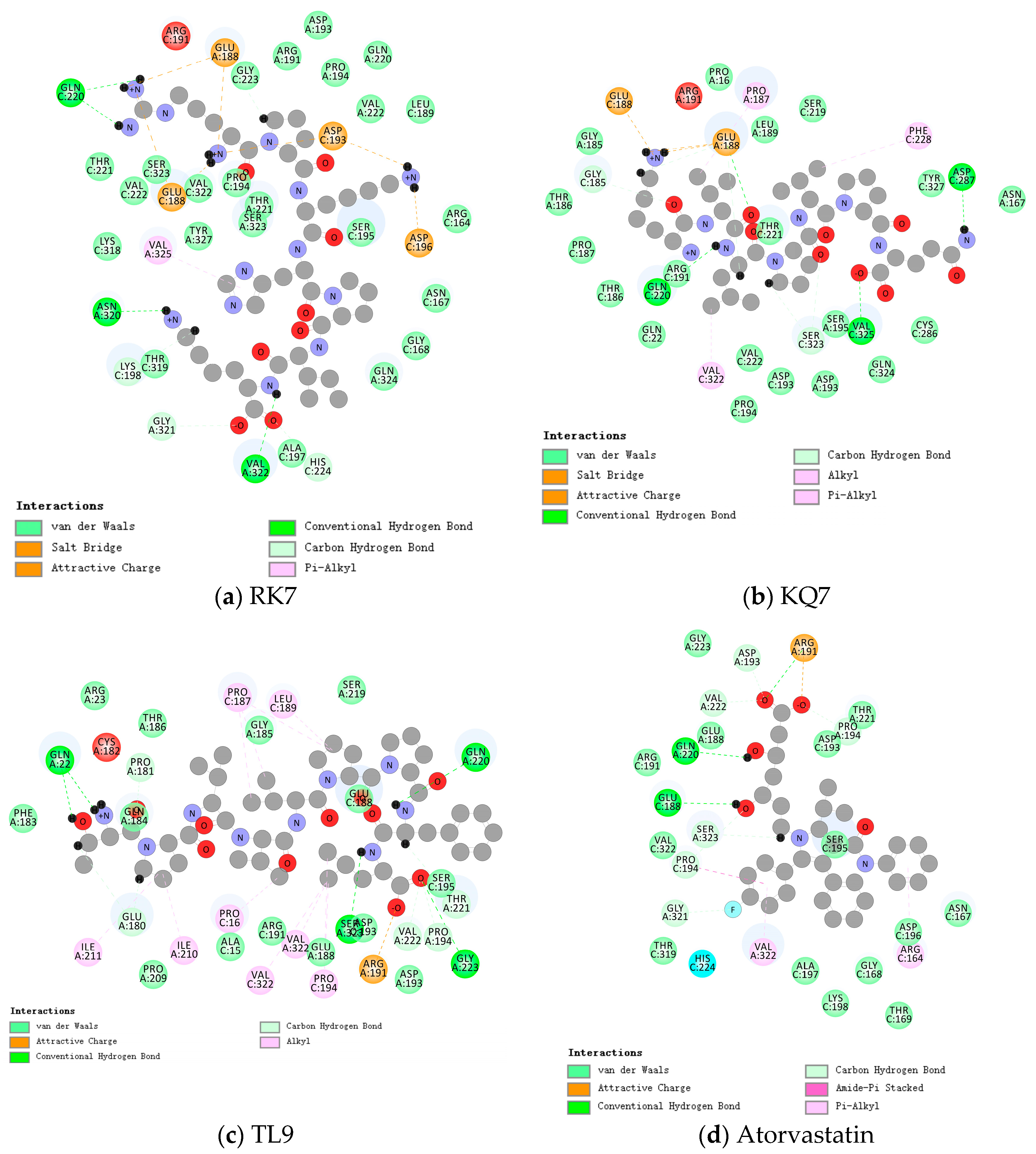
2.2. Molecular Docking Analysis
2.3. Molecular Dynamics Analysis
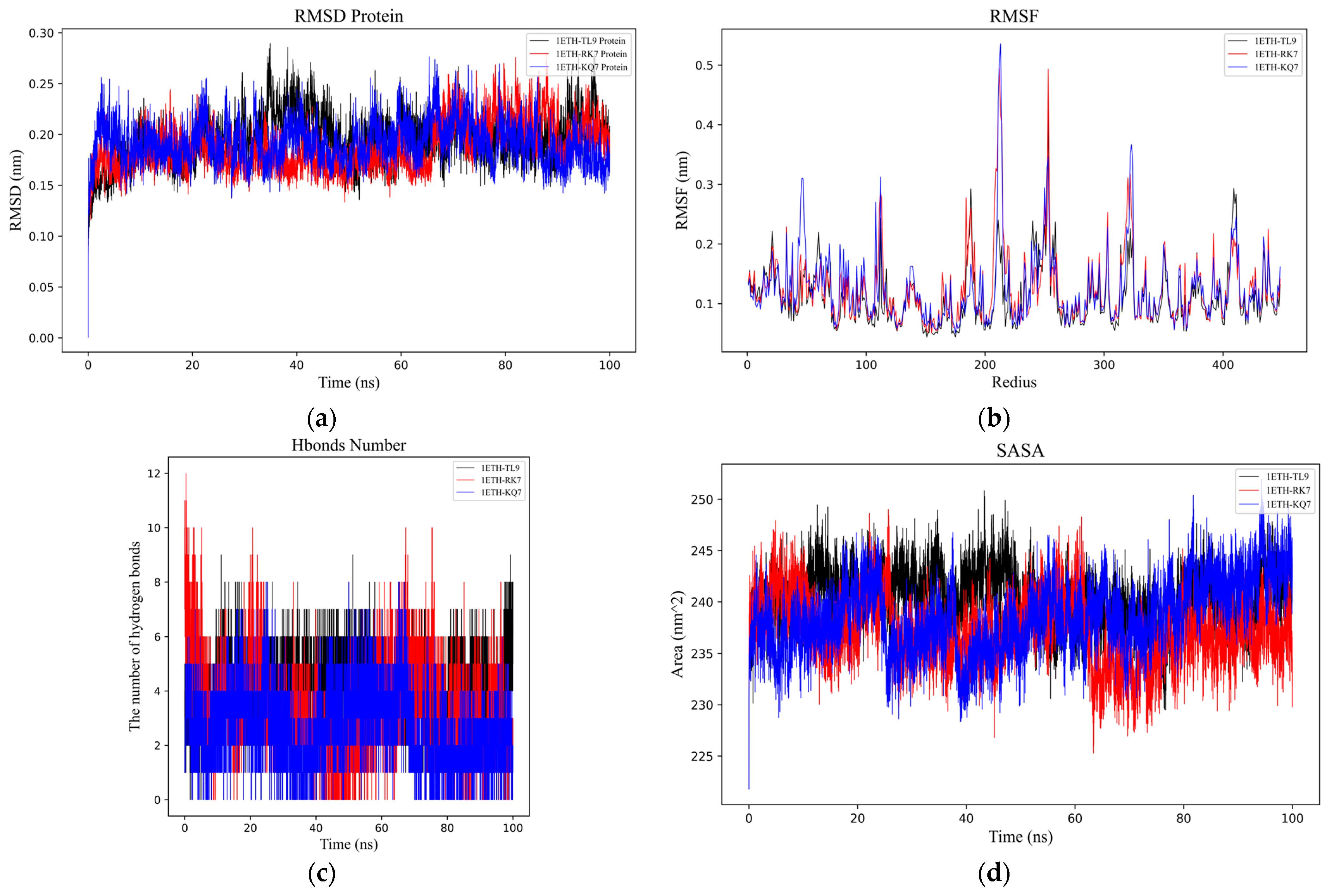
2.3.1. GROMACS Analysis of the RMSD, RMSF, Hydrogen Bonds, SASA, and Gibbs Free Energy Stability Analysis
2.3.2. MM/GBSA Calculations and Analyses
The MM/GBSA Approach for Free Energy Estimation in Protein-Ligand Complexes
2.3.3. Dynamic Interaction Analysis from 0 to 100 ns via Molecular Simulations
2.4. Synthesis and Validation of PL Inhibitory Activity of Peptides RK7 and KQ7
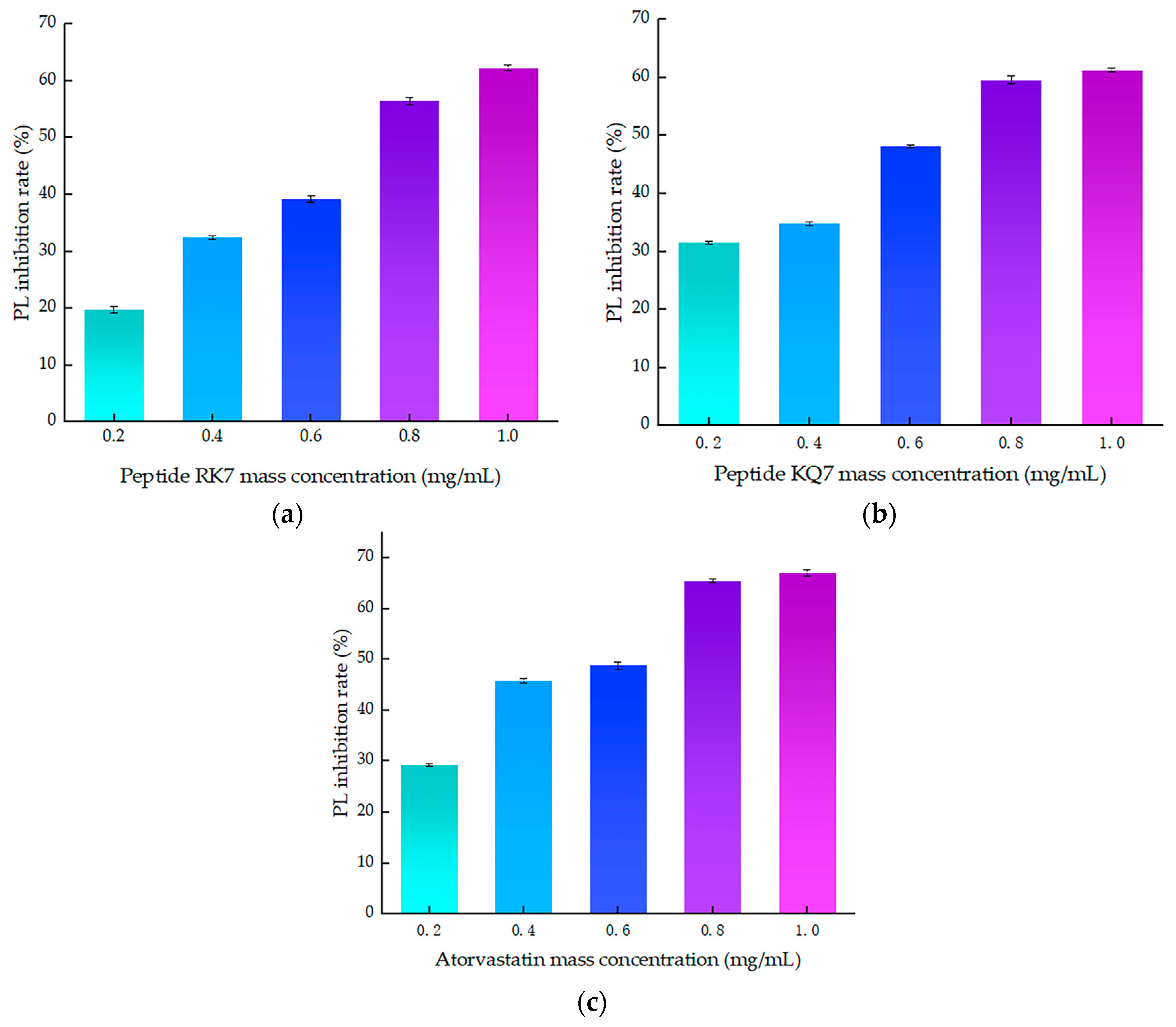
2.4.1. In Vitro Validation of RK7 and KQ7’s Inhibitory Efficacy
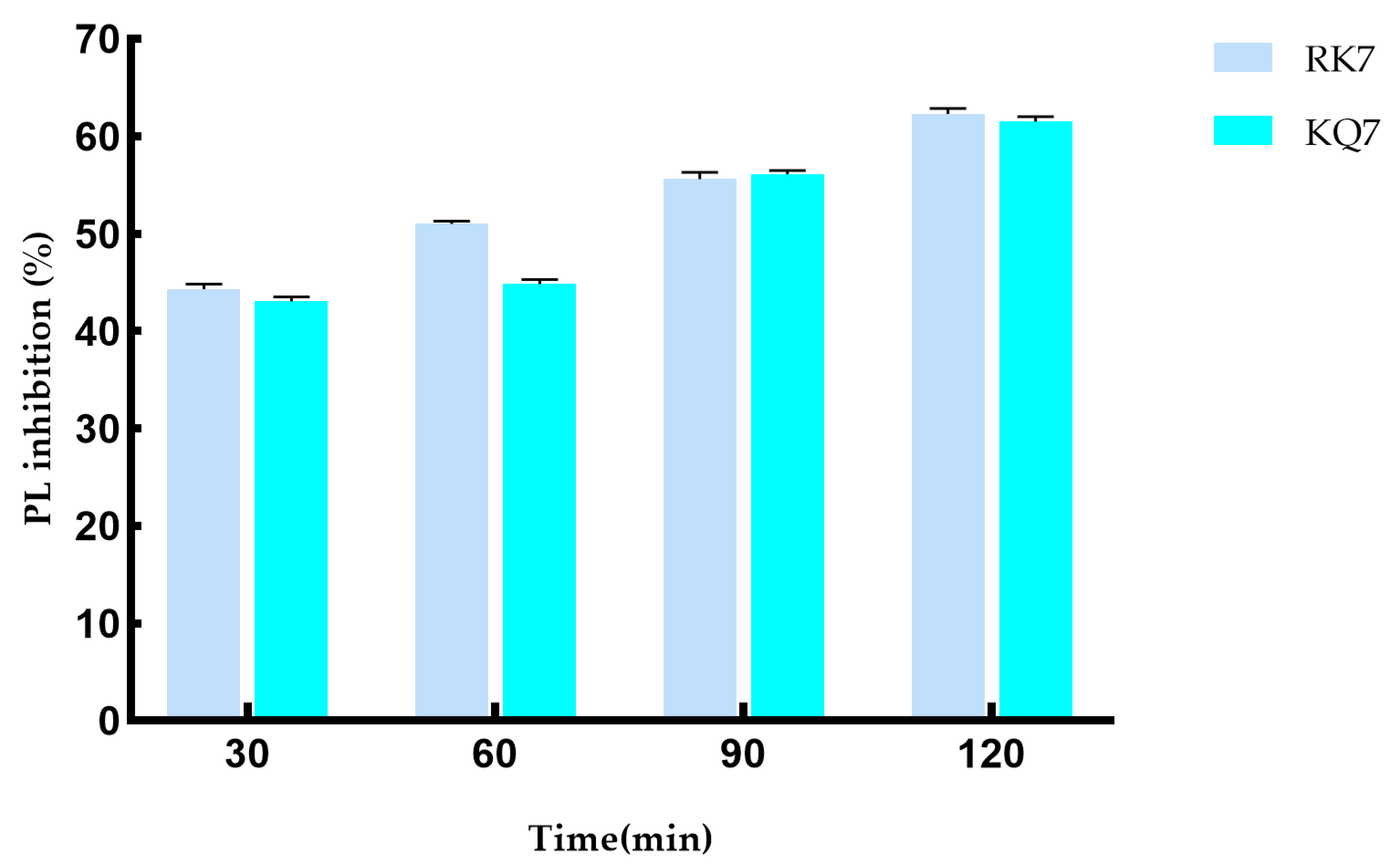
2.4.2. Simulating the Effect of Gastrointestinal Digestion on PL Inhibition Activity
3. Materials and Methods
3.1. Materials and Reagents
3.2. Instruments and Equipment
3.3. Experimental Methods
3.3.1. Forecasting the Physicochemical Traits of Peptides RK7, KQ7, and TL9
3.3.2. Structure Adjustment and Optimization of RK7, KQ7, TL9 and PL
3.3.3. Molecular Docking
3.3.4. GROMACS Molecular Dynamics Simulation
3.3.5. PL Inhibition Experiments with Peptides RK7 and KQ7
3.3.6. Simulating the Impact of Gastrointestinal Digestion on PL Inhibitory Activity
3.3.7. Data Processing
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mudgil, P.; Baby, B.; Ngoh, Y.Y.; Vijayan, R.; Gan, C.Y.; Maqsood, S. Identification and molecular docking study of novel cholesterol esterase inhibitory peptides from camel milk proteins. J. Dairy Sci. 2019, 102, 10748–10759. [Google Scholar] [CrossRef]
- Birari, R.B.; Bhutani, K.K. Pancreatic lipase inhibitors from natural sources: Unexplored potential. Drug Discov. Today 2007, 12, 879–889. [Google Scholar] [CrossRef]
- Sosnowska, D.; Podsędek, A.; Redzynia, M.; Kucharska, A.Z. Inhibitory effect of black chokeberry fruit polyphenols on pancreatic lipase—Searching for most active inhibitors. J. Funct. Foods 2018, 49, 196–204. [Google Scholar] [CrossRef]
- Sosnowska, D.; Podsędek, A.; Kucharska, A.Z. Proanthocyanidins as the main pancreatic lipase inhibitors in chokeberry fruits. Food Funct. 2022, 13, 5616–5625. [Google Scholar] [CrossRef]
- Huang, X.; Zhu, J.; Wang, L.; Jing, H.; Ma, C.; Kou, X.; Wang, H. Inhibitory mechanisms and interaction of tangeretin, 5-demethyltangeretin, nobiletin, and 5-demethylnobiletin from citrus peels on pancreatic lipase: Kinetics, spectroscopies, and molecular dynamics simulation. Int. J. Biol. Macromol. 2020, 164, 1927–1938. [Google Scholar] [CrossRef]
- Wu, M.; Zhao, H.; Tang, X.; Zhao, W.; Yi, X.; Li, Q.; Sun, X. Organization and Complexity of the Yak (Bos grunniens) Immunoglobulin Loci. Front. Immunol. 2022, 13, 876509. [Google Scholar] [CrossRef]
- Schormüller, J. The chemistry and biochemistry of cheese ripening. Adv. Food Res. 1968, 16, 231–334. [Google Scholar] [CrossRef]
- Song, X.; Zhang, Y.; Yang, M.; Liang, Q. Isolation and characterisation of peptides from yak milk cheese hard cheese. Food Sci. 2016, 37, 160–164. [Google Scholar] [CrossRef]
- Li, M.; Liang, Q.; Song, X. Investigating the α-Amylase Inhibitory Activity of Bitter Taste Peptides RK7 and KQ7 in Yak Milk Cheese through Molecular Docking Technology. Food Sci. 2023, 44, 132–138. [Google Scholar] [CrossRef]
- Yang, B.; Liang, Q.; Song, X. Research on the Antibacterial Activity Difference of Bitter Taste Peptides in Hard Yak Cheese Based on Computer Virtual Technology. J. Food Biotechnol. 2021, 40, 75–87. [Google Scholar] [CrossRef]
- Yang, B.; Liang, Q.; Song, X. Antioxidant Activity and Mechanism of Peptide RK7 from Yak Milk Cheese Hard Cheese. J. Chin. Inst. Food Sci. Technol. 2022, 22, 40–50. [Google Scholar] [CrossRef]
- Yang, B.; Liang, Q.; Song, X. Molecular Mechanisms Characterizing the ACE Inhibitory Activity of Yak Cheese Peptides. J. Chin. Food Sci. 2022, 22, 8–17. [Google Scholar] [CrossRef]
- Alnuaimi, A.; Ajayi, F.F.; Hamdi, M.; Mudgil, P.; Kamal, H.; Gan, C.Y.; Maqsood, S. A comparative analysis of anti-lipidemic potential of soybean (Glycine max) protein hydrolysates obtained from different ripening stages: Identification, and molecular interaction mechanisms of novel bioactive peptides. Food Chem. 2023, 402, 134192. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Liu, Z.; Wang, H.; Zhang, F.; Guo, S.; Shen, Q. Comparison of the generation of α-glucosidase inhibitory peptides derived from prolamins of raw and cooked foxtail millet: In vitro activity, de novo sequencing, and in silico docking. Food Chem. 2023, 411, 135378. [Google Scholar] [CrossRef] [PubMed]
- Urbizo-Reyes, U.; Liceaga, A.M.; Reddivari, L.; Kim, K.-H.; Anderson, J.M. Enzyme kinetics, molecular docking, and in silico characterization of canary seed (Phalaris canariensis L.) peptides with ACE and pancreatic lipase inhibitory activity. J. Funct. Foods 2022, 88, 104892. [Google Scholar] [CrossRef]
- Zhang, Z.; Ge, J.; Wei, L.; Shi, J.; Ji, Y.; Xing, X.; Shi, Y.; Dong, Y. Isolation and identification of novel hemp seed protein-derived pancreatic lipase inhibitory peptides. Food Biosci. 2025, 63, 105834. [Google Scholar] [CrossRef]
- Chen, H.; Dong, X.; Zhao, Y.; Zhou, X.; Zhou, M.; Xu, Z.; Xu, Z. Oyster-derived peptides as pancreatic lipase inhibitors: Kinetics, molecular interactions, and gastrointestinal stability. Food Biosci. 2025, 63, 105637. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. Software X 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Pagadala, N.S.; Syed, K.; Tuszynski, J. Software for molecular docking: A review. Biophys. Rev. 2017, 9, 91–102. [Google Scholar] [CrossRef]
- Esfandi, R.; Walters, M.E.; Tsopmo, A. Antioxidant properties and potential mechanisms of hydrolyzed proteins and peptides from cereals. Heliyon 2019, 5, e01538. [Google Scholar] [CrossRef]
- Ngoh, Y.Y.; Choi, S.; Gan, C.Y. The potential roles of Pinto bean (Phaseolus vulgaris cv. Pinto) bioactive peptides in regulating physiological functions: Protease activating, lipase inhibiting and bile acid binding activities. J. Funct. Foods 2017, 33, 67–75. [Google Scholar] [CrossRef]
- Martinez-Villaluenga, C.; Rupasinghe, S.G.; Schuler, M.A.; Gonzalez de Mejia, E. Peptides from purified soybean beta-conglycinin inhibit fatty acid synthase by interaction with the thioesterase catalytic domain. FEBS J. 2010, 277, 1481–1493. [Google Scholar] [CrossRef] [PubMed]
- Jafar, S.; Kamal, H.; Mudgil, P.; Hassan, H.M.; Maqsood, S. Camel whey protein hydrolysates displayed enhanced cholesteryl esterase and lipase inhibitory, anti-hypertensive and anti-haemolytic properties. LWT—Food Sci. Technol. 2018, 98, 212–218. [Google Scholar] [CrossRef]
- Khiari, Z.; Ndagijimana, M.; Betti, M. Low molecular weight bioactive peptides derived from the enzymatic hydrolysis of collagen after isoelectric solubilization/precipitation process of turkey by-products. Poult. Sci. 2014, 93, 2347–2362. [Google Scholar] [CrossRef]
- Luo, H.Q.; Shen, J.; Chen, C.P.; Ma, X.; Lin, C.; Ouyang, Q.; Xuan, C.X.; Liu, J.; Sun, H.B.; Liu, J. Lipid-lowering effects of oleanolic acid in hyperlipidemic patients. Chin. J. Nat. Med. 2018, 16, 339–346. [Google Scholar] [CrossRef]
- Xie, Z.; He, M.; Zhai, Y.; Xin, F.; Yu, S.; Yu, S.; Xiao, H.; Song, Y. Inhibitory kinetics and mechanism of oleanolic acid on α-glucosidase. Food Meas. Charact. 2021, 15, 3408–3418. [Google Scholar] [CrossRef]
- Shen, H.; Wang, J.; Ao, J.; Ye, L.; Shi, Y.; Liu, Y.; Li, M.; Luo, A. The inhibitory mechanism of pentacyclic triterpenoid acids on pancreatic lipase and cholesterol esterase. Food Biosci. 2023, 51, 102341. [Google Scholar] [CrossRef]
- Neveu, E.; Popov, P.; Hoffmann, A.; Migliosi, A.; Besseron, X.; Danoy, G.; Bouvry, P.; Grudinin, S. RapidRMSD: Rapid determination of RMSDs corresponding to motions of flexible molecules. Bioinformatics 2018, 34, 2757–2765. [Google Scholar] [CrossRef]
- Arshia, A.H.; Shadravan, S.; Solhjoo, A.; Sakhteman, A.; Sami, A. De novo design of novel protease inhibitor candidates in the treatment of SARS-CoV-2 using deep learning, docking, and molecular dynamic simulations. Comput. Biol. Med. 2021, 139, 104967. [Google Scholar] [CrossRef]
- Santos, L.H.S.; Ferreira, R.S.; Caffarena, E.R. Integrating Molecular Docking and Molecular Dynamics Simulations. Methods Mol. Biol. 2019, 2053, 13–34. [Google Scholar] [CrossRef]
- Mascoli, V.; Liguori, N.; Cupellini, L.; Elias, E.; Mennucci, B.; Croce, R. Uncovering the interactions driving carotenoid binding in light-harvesting complexes. Chem Sci. 2021, 12, 5113–5122. [Google Scholar] [CrossRef] [PubMed]
- Ragunathan, A.; Malathi, K.; Anbarasu, A. MurB as a target in an alternative approach to tackle the Vibrio cholerae resistance using molecular docking and simulation study. Cell Biochem. 2018, 119, 1726–1732. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.A.; Hassan, M.I.; Islam, A.; Ahmad, F.; Hassan, I. A review of methods available to estimate solvent-accessible surface areas of soluble proteinsin the folded and unfolded states. Curr. Protein Pept. Sci. 2014, 15, 456–476. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Zhong, J.; Wang, Y. A mutant T1 lipase homology modeling, and its molecular docking and molecular dynamics simulation with fatty acids. J. Biotechnol. 2021, 337, 24–34. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand–binding affinities. Expert. Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Chen, F.; Sun, H.; Wang, J.; Zhu, F.; Liu, H.; Wang, Z.; Lei, T.; Li, Y.; Hou, T. Assessing the performance of MM/PBSA and MM/GBSA methods. 8. Predicting binding free energies and poses of protein–RNA complexes. RNA 2018, 24, 1183–1194. [Google Scholar] [CrossRef]
- Gaillard, T.; Simonson, T. Full Protein Sequence Redesign with an MMGBSA Energy Function. Chem. Theory Comput. 2017, 13, 4932–4943. [Google Scholar] [CrossRef]
- Zhao, Q.; Fan, Y.; Zhao, L.; Zhu, Y.; Jiang, Y.; Gu, J.; Xue, Y.; Hao, Z.; Shen, Q. Identification and molecular binding mechanism of novel pancreatic lipase and cholesterol esterase inhibitory peptides from heat-treated adzuki bean protein hydrolysates. Food Chem. 2024, 439, 138129. [Google Scholar] [CrossRef]
- Siow, H.; Choi, S.; Gan, C. Structure–activity studies of protease activating, lipase inhibiting, bile acid binding and cholesterol-lowering effects of pre-screened cumin seed bioactive peptides. J. Funct. Foods 2016, 27, 600–611. [Google Scholar] [CrossRef]
- Lear, S.; Cobb, S.L. Pep-Calc.com: A set of web utilities for the calculation of peptide and peptoid properties and automatic mass spectral peak assignment. J. Comput. Aided. Mol. Des. 2016, 30, 271–277. [Google Scholar] [CrossRef]
- Soleymanzadeh, N.; Mirdamadi, S.; Mirzaei, M.; Kianirad, M. Novel β–casein derived antioxidant and ACE–inhibitory active peptide from camel milk fermented by Leuconostoc lactis PTCC1899: Identification and molecular docking. Int. Dairy J. 2019, 97, 201–208. [Google Scholar] [CrossRef]
- Gurung, D.; Danielson, J.A.; Tasnim, A.; Zhang, J.T.; Zou, Y.; Liu, J.Y. Proline Isomerization: From the Chemistry and Biology to Therapeutic Opportunities. Biology 2023, 12, 1008. [Google Scholar] [CrossRef] [PubMed]
- López-Martínez, C.; Flores-Morales, P.; Cruz, M.; González, T.; Feliz, M.; Diez, A.; Campanera, J.M. Proline cis-trans isomerization and its implications for the dimerization of analogues of cyclopeptide stylostatin 1: A combined computational and experimental study. Phys. Chem. Chem. Phys. 2016, 18, 12755–12767. [Google Scholar] [CrossRef] [PubMed]
- Glisan, S.L.; Grove, K.A.; Yennawar, N.H.; Lambert, J.D. Inhibition of pancreatic lipase by black tea theaflavins: Comparative enzymology and in silico modeling studies. Food Chem. 2017, 216, 296–300. [Google Scholar] [CrossRef]
- Jiang, F.; Liu, J.B.; Ma, S.T.; Yang, M.; Chen, Y.; Liu, B.; Zhang, T. Antioxidant Activity and Structural Characterization of Ovalbumin Digested in Vitro by Simulated Gastrointestinal Tract. J. Chin. Inst. Food Sci. Technol. 2021, 21, 10–18. [Google Scholar] [CrossRef]


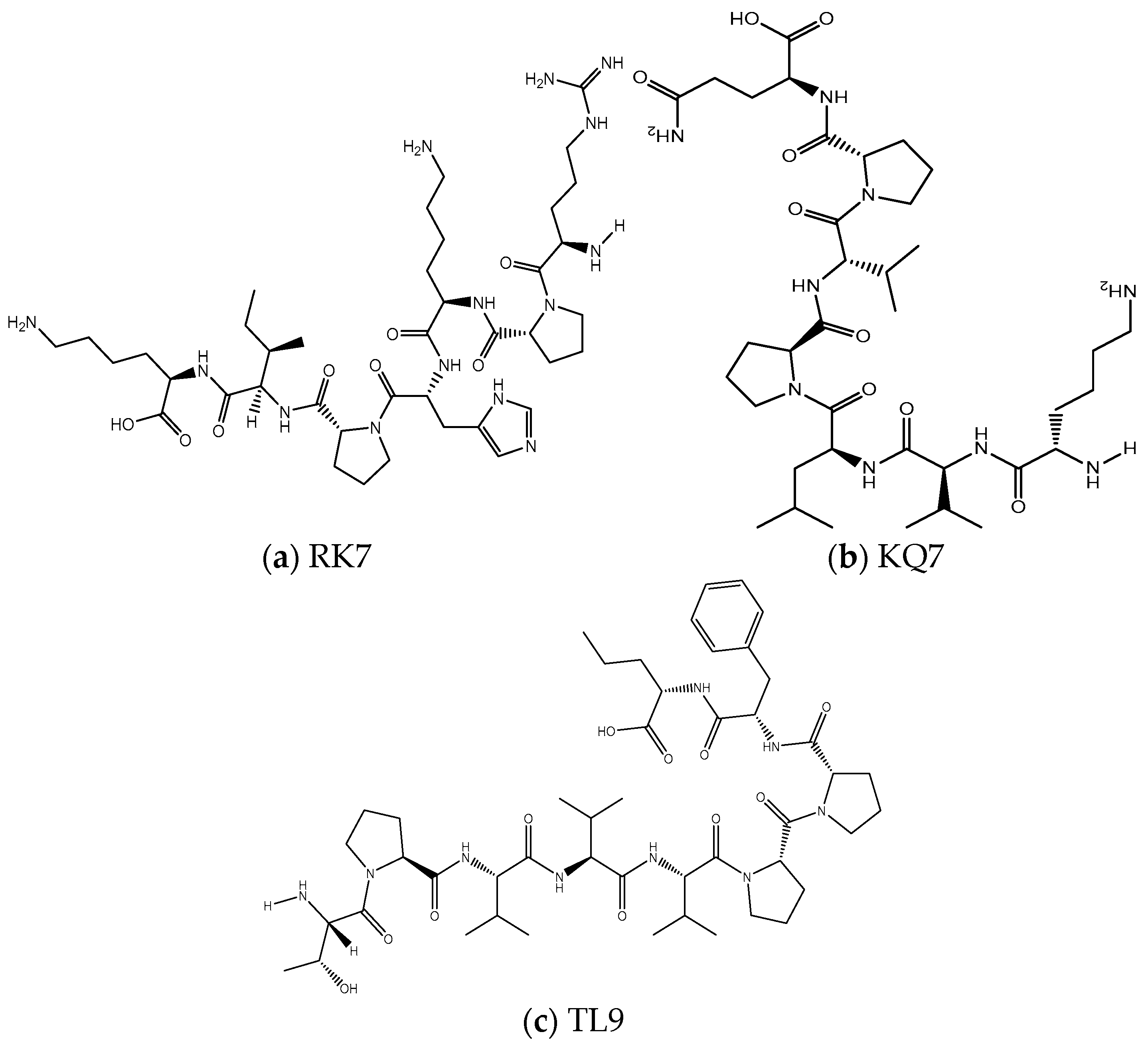
| Peptide Sequence | Molecular Weight/(Da) | Isoelectric Point | Net Charge | Hydrophobic Amino Acids | Proportion of Hydrophobic Amino Acids |
|---|---|---|---|---|---|
| RK7 | 874.90 | 11.57 | 3 | P, I | 42.86% |
| KQ7 | 779.50 | 9.84 | 1 | V, L, P | 71.42% |
| TL9 | 968.19 | 3.32 | 0 | P, V, F, L | 88.91% |
| RK7 | KQ7 | TL9 | Atorvastatin | |
|---|---|---|---|---|
| Hydrogen bonds | Val322, Asn320, Gln220, Glu188, Lys198, Gly321, His264 | Val325, Asp287, Glu188, Gln220, Ser323, Gly185 | Ser323, Gly223, Gln220, Gln22, Glu188, Val222, Pro194, Thr221 | Gln220, Glu188, Ser323, Pro194, Gly321, Pro194, Asp193, Val222, |
| Hydrophobic interactions | Val325 | Val322, Phe228, Pro187 | Ile211, Ile210, Pro16, Val322, Pro194, Pro187, Leu189 | Val322, Arg164, Pro194 |
| Van der Waals forces | Ala197, Gln324, Gly168, Asn167, Ser195, Arg164, Val222, Leu189, Gln220, Pro194, Gly223, Arg191, Asp193, Thr221, Ser323, Val322, Thr221, Tyr327, Lys318, Thr319 | Pro194, Asp193, Val222, Ser195, Gln324, Cys286, Tyr327, Asn167, Thr221, Ser219, Leu189, Pro16, Gly185, Thr186, Pro187, Thr186, Arg191, Gln22 | Pro209, Ala15, Arg191, Glu188, Asp193, Ser195, Ser219, Gly185, Thr186, Arg23, Gln184, Phe183, | Thr319, Ala197, Lys198, Gly168, Thr196, Asp196, Asn167, Ser195, Asp193, Thr221, Gly223, Glu188, A rg191, Val322, |
| Electrostatic interactions | Glu188 | Glu188 | Arg191 | Arg191 |
| (kcal/mol) | RK7 | KQ7 | TL9 |
|---|---|---|---|
| ΔVDWAALS | −71.11 ± 1.65 | −63.83 ± 2.46 | −49.54 ± 1.49 |
| ΔEEL | −95.60 ± 1.35 | −19.66 ± 2.15 | −43.28 ± 3.35 |
| ΔEGB | 65.11 ± 1.11 | 91.60 ± 3.06 | 70.12 ± 1.72 |
| ΔEsurf | −10.12 ± 0.06 | −8.39 ± 0.07 | −6.88 ± 0.04 |
| ΔGgas | −166.71 ± 2.13 | −83.49 ± 3.27 | −92.82 ± 3.67 |
| ΔGsolv | 111.05 ± 0.52 | 47.95 ± 0.09 | 63.23 ± 1.72 |
| ΔTotal | −55.66 ± 2.20 | −35.54 ± 3.27 | −29.59 ± 4.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Song, X.; Liang, Q. Molecular Docking Studies and In Vitro Activity of Pancreatic Lipase Inhibitors from Yak Milk Cheese. Int. J. Mol. Sci. 2025, 26, 756. https://doi.org/10.3390/ijms26020756
Wang P, Song X, Liang Q. Molecular Docking Studies and In Vitro Activity of Pancreatic Lipase Inhibitors from Yak Milk Cheese. International Journal of Molecular Sciences. 2025; 26(2):756. https://doi.org/10.3390/ijms26020756
Chicago/Turabian StyleWang, Peng, Xuemei Song, and Qi Liang. 2025. "Molecular Docking Studies and In Vitro Activity of Pancreatic Lipase Inhibitors from Yak Milk Cheese" International Journal of Molecular Sciences 26, no. 2: 756. https://doi.org/10.3390/ijms26020756
APA StyleWang, P., Song, X., & Liang, Q. (2025). Molecular Docking Studies and In Vitro Activity of Pancreatic Lipase Inhibitors from Yak Milk Cheese. International Journal of Molecular Sciences, 26(2), 756. https://doi.org/10.3390/ijms26020756





