Investigation of Exome-Wide Tumor Heterogeneity on Colorectal Tissue-Based Single Cells
Abstract
1. Introduction
2. Results
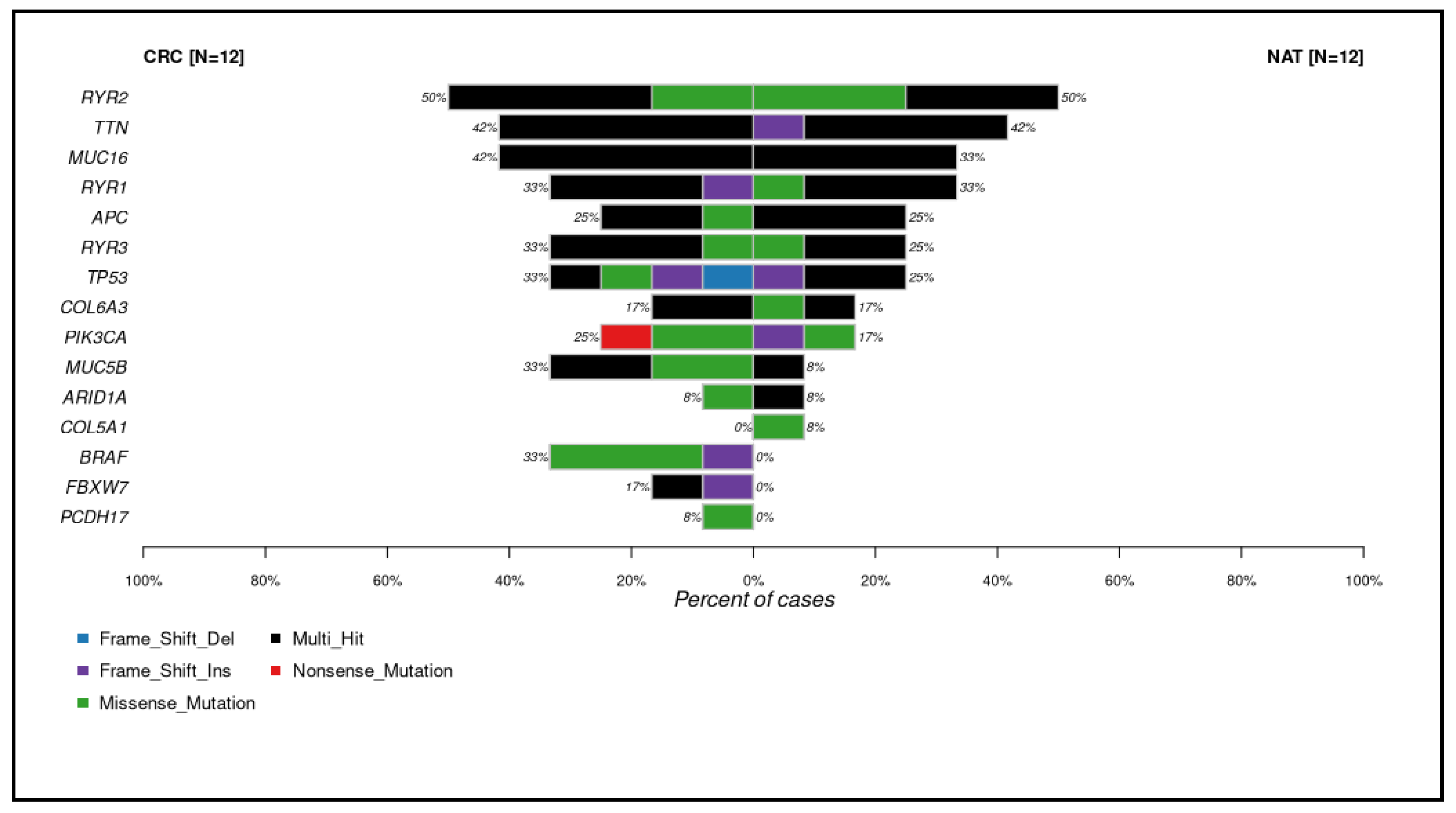
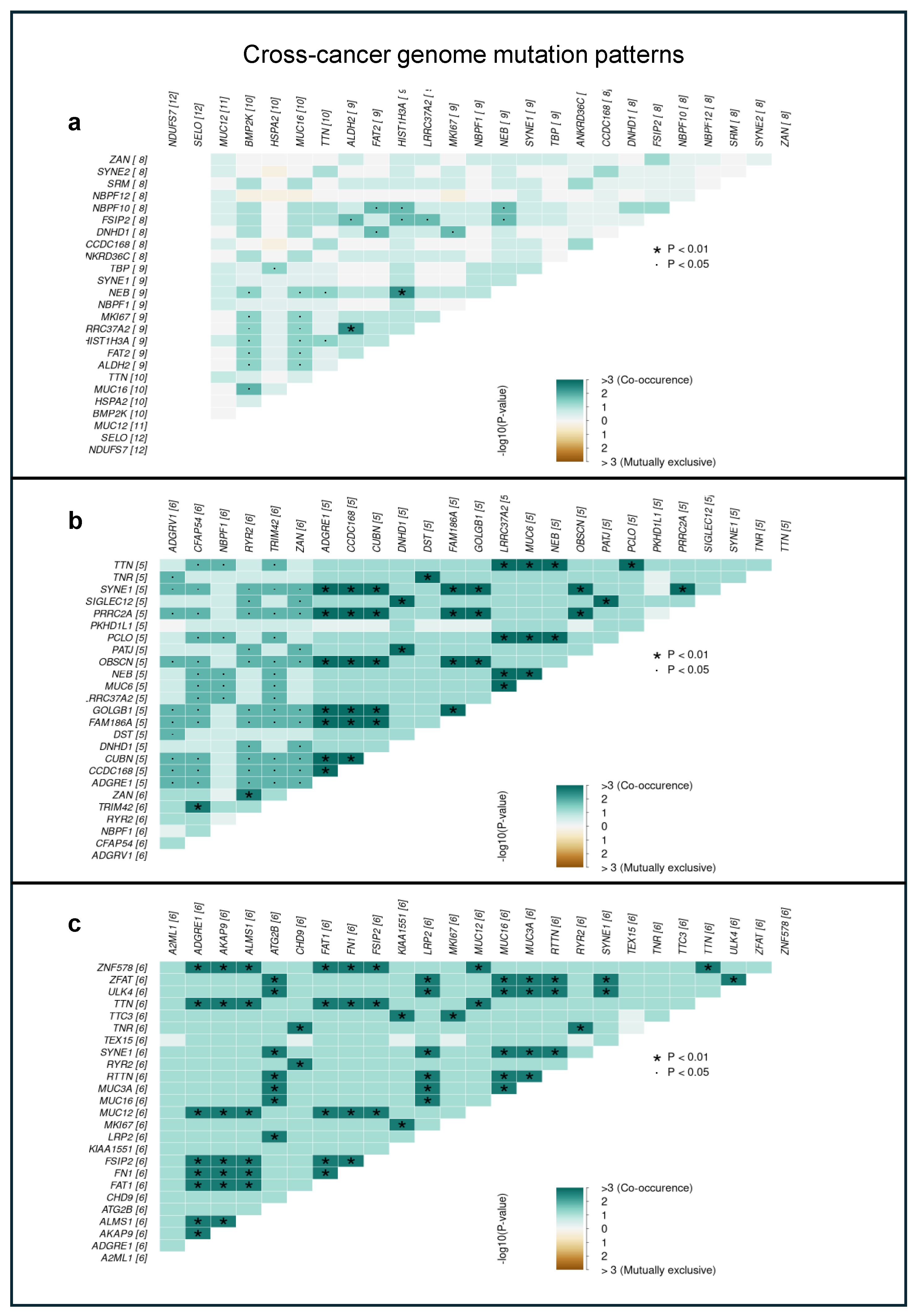
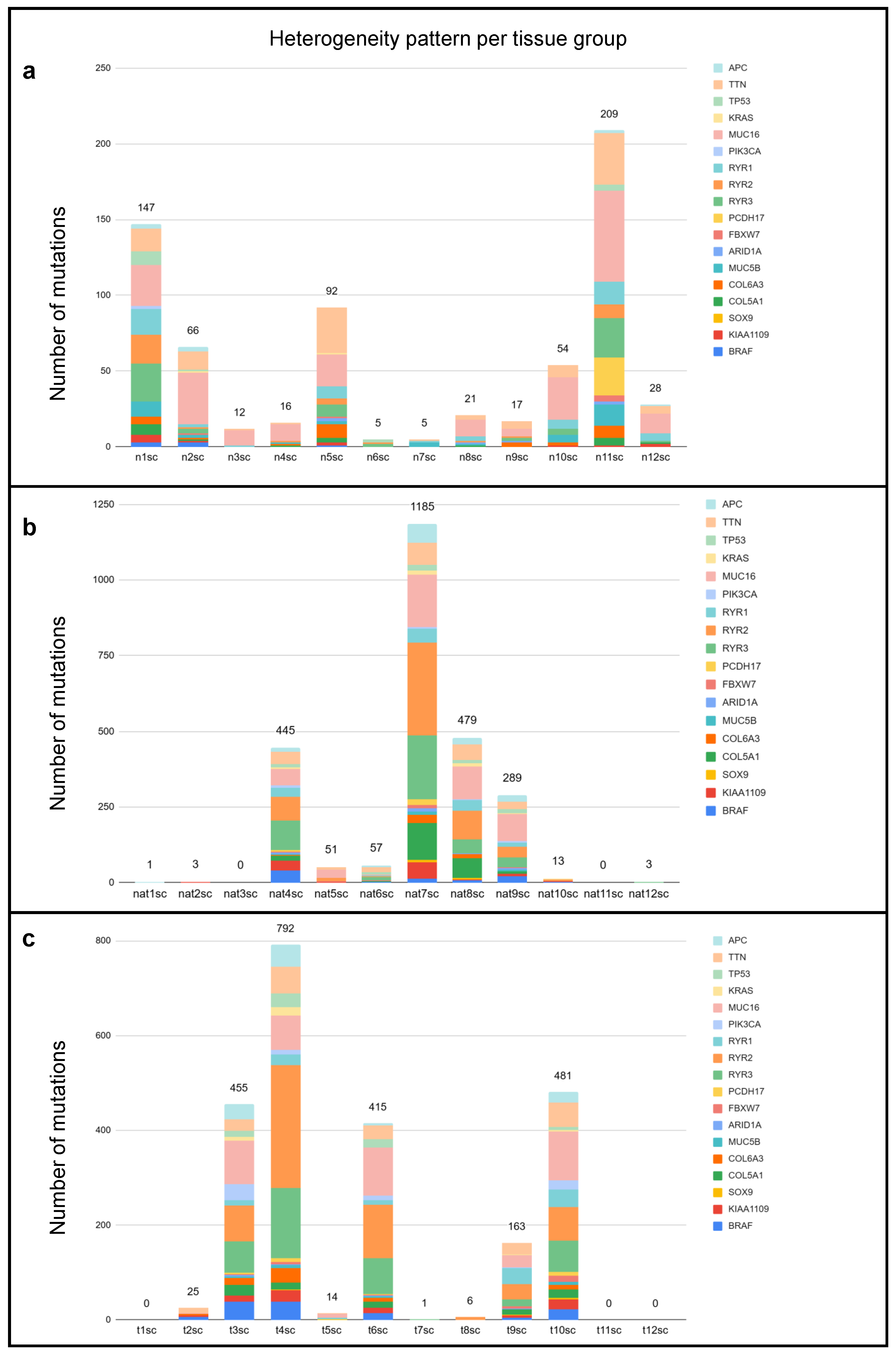
2.1. Investigation of Tumor Heterogeneity in Single-Cell Sequenced Samples
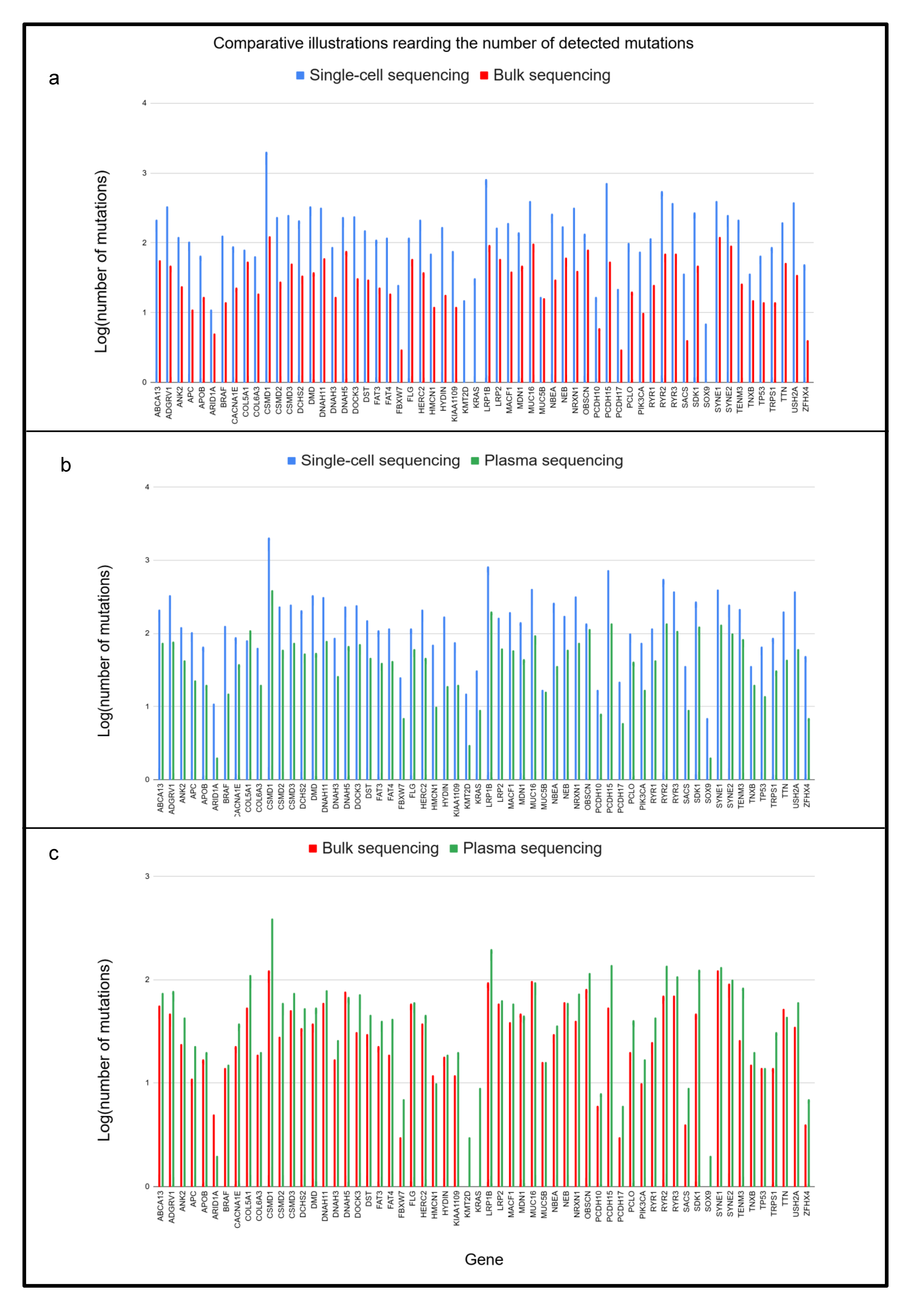
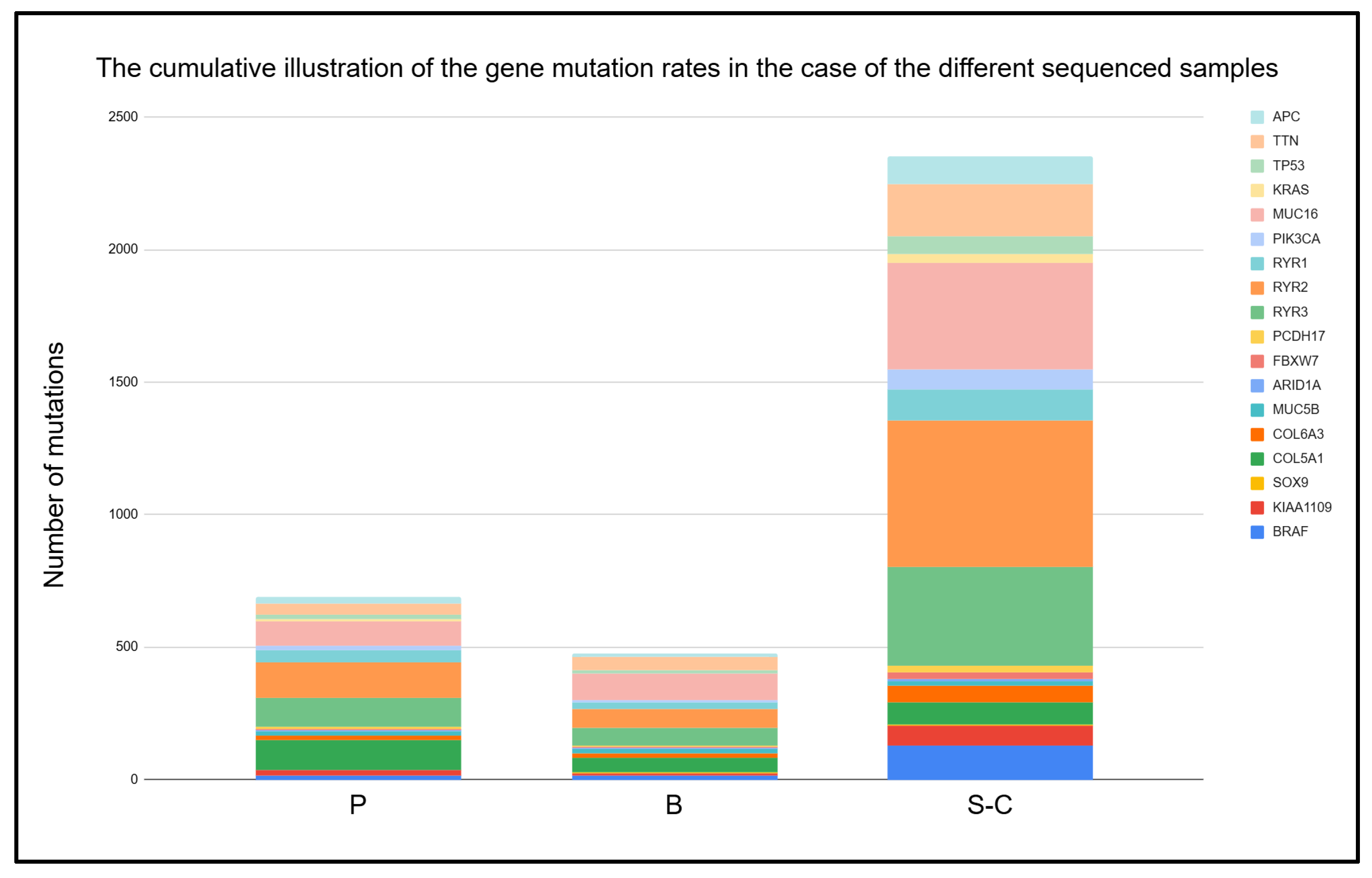
2.2. Comparison of Different Input Samples
3. Discussion
4. Materials and Methods
4.1. Clinical Samples
4.2. Single-Cell DNA Extraction, Library Preparation, and Next-Generation Sequencing
4.3. Bioinformatic Analyses
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Germline | Somatic | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| ID | Mean Region Coverage Depth | TMB | Median Fragment Length | SNP | Deletion | Insertion | SNP | Deletion | Insertion |
| NEG1 | 8.2 | 22.1 | 194 | 14,775 | 6 | 223 | |||
| NEG2 | 25.1 | 95.38 | 198 | 48,084 | 32 | 503 | |||
| NEG3 | 2.5 | 7.74 | 170 | 4280 | 16 | 422 | |||
| NEG4 | 4.2 | 6.04 | 179 | 5268 | 12 | 354 | |||
| NEG5 | 0.5 | 18.56 | 180 | 102 | 0 | 6 | |||
| NEG6 | 10.9 | 24.82 | 176 | 7490 | 188 | 340 | |||
| NEG7 | 76.6 | 39.8 | 191 | 60,524 | 1523 | 2158 | |||
| NEG8 | 126 | 178.72 | 191 | 183,465 | 137 | 3068 | |||
| NEG9 | 8.9 | 27.8 | 178 | 14,895 | 6 | 227 | |||
| NEG10 | 19.2 | 66.26 | 202 | 37,999 | 16 | 529 | |||
| NEG11 | 14.3 | 8.02 | 206 | 28,891 | 15 | 692 | |||
| NEG12 | 1.7 | 52.3 | 228 | 4359 | 0 | 62 | |||
| NAT1 | 37.2 | 73.94 | 264 | 92,880 | 101 | 1305 | 55,002 | 3501 | 5355 |
| NAT2 | 11.4 | 8.54 | 227 | 14,040 | 12 | 141 | 8906 | 636 | 1169 |
| NAT3 | 13.5 | 8.16 | 217 | 18,970 | 41 | 338 | 11,348 | 869 | 1178 |
| NAT4 | 125.5 | 226.52 | 198 | 261,094 | 54 | 110 | 137,773 | 7286 | 10,311 |
| NAT5 | 85.3 | 108.54 | 164 | 134,821 | 20 | 86 | 73,450 | 3753 | 5428 |
| NAT6 | 33.7 | 48.28 | 220 | 57,992 | 87 | 714 | 35,854 | 2470 | 3594 |
| NAT7 | 0.8 | 1.78 | 268 | 1892 | 12 | 87 | 1125 | 94 | 169 |
| NAT8 | 0.7 | 0.56 | 214 | 1518 | 2 | 19 | 979 | 75 | 126 |
| NAT9 | 5.6 | 1.88 | 205 | 3387 | 10 | 109 | 2196 | 164 | 266 |
| NAT10 | 0.9 | 0.18 | 224 | 970 | 3 | 25 | 717 | 47 | 62 |
| NAT11 | 1.6 | 0.54 | 239 | 950 | 2 | 24 | 630 | 47 | 80 |
| NAT12 | 0.2 | 0.08 | 257 | 230 | 0 | 5 | 159 | 8 | 22 |
| T1 | 141.2 | 294.7 | 241 | 232,133 | 148 | 1516 | 136,632 | 1355 | 9003 |
| T2 | 12.6 | 5.68 | 255 | 5510 | 16 | 168 | 3743 | 126 | 494 |
| T3 | 66 | 118.04 | 212 | 103,007 | 66 | 685 | 61,950 | 641 | 3810 |
| T4 | 0.3 | 0.64 | 266 | 270 | 3 | 7 | 220 | 4 | 42 |
| T5 | 1 | 0.86 | 255 | 665 | 1 | 23 | 505 | 15 | 53 |
| T6 | 14.1 | 51.66 | 259 | 24,802 | 48 | 444 | 15,029 | 349 | 1545 |
| T7 | 62 | 205.2 | 253 | 137,039 | 169 | 1724 | 82,054 | 1363 | 6703 |
| T8 | 0.8 | 1.16 | 259 | 1286 | 1 | 46 | 679 | 27 | 111 |
| T9 | 0.4 | 1.38 | 244 | 385 | 0 | 23 | 236 | 4 | 41 |
| T10 | 0.1 | 0.2 | 305 | 110 | 0 | 5 | 68 | 4 | 18 |
| T11 | 5 | 8.84 | 260 | 6608 | 14 | 126 | 3854 | 126 | 503 |
| T12 | 74.8 | 176.26 | 231 | 93,550 | 120 | 1121 | 57,145 | 1161 | 5684 |
| Gene | Sequence Variation | dbSNP ID | Mutation Classification |
|---|---|---|---|
| Mutations Detected by Single-Cell Sequencing Only | |||
| APC | n.112707592dupG | ||
| APC | c.135+5252G>C | ||
| APC | c.135+5253A>T | ||
| APC | c.135+5254G>C | ||
| APC | c.136-230C>A | rs2464805 | benign |
| APC | c.221-291C>A | ||
| APC | c.4853dupT | ||
| APC | c.6172G>T | ||
| APC | c.*433T>A | ||
| APC | c.*1958_*1959insTTAC | ||
| APC | c.*1965T>C | ||
| APC | c.*2220_*2221ins | ||
| APC | c.934-8_934-7ins | rs1561535860 | likely benign |
| APC | c.934-4dupA | ||
| APC | c.5613T>C | ||
| APC | c.*267_*273delCCATCCC | ||
| APC | c.*281_*283delTTT | rs42427 | benign |
| APC | c.*285A>G | rs866006 | benign |
| APC | c.*1098T>C | benign | |
| APC | c.*1556C>G | ||
| APC | c.560-12T>C | ||
| APC | c.1881-762G>A | rs41116 | benign |
| APC | c.*413_*414dupAA | rs2289484 | benign |
| APC | c.559+37C>A | rs1554084977 | uncertain significance |
| APC | c.559+223C>T | rs41115 | benign |
| APC | c.627_628insAGAAGATGAA | rs1580673845 | benign |
| APC | c.628_628+1ins | rs2229995 | benign |
| ARID1A | c.*421delA | ||
| ARID1A | c.*724C>A | ||
| ARID1A | c.2295-161delT | ||
| ARID1A | c.2496-130_2496-128delAAA | ||
| Mutations Detected by Single-Cell Sequencing Only | |||
| BRAF | c.1763T>C | rs1562954580 | uncertain significance |
| BRAF | c.1695-940C>A | ||
| BRAF | c.1695-1205G>A | ||
| BRAF | c.1695-5750C>G | ||
| BRAF | c.1694+8566G>A | ||
| BRAF | c.1694+3374T>G | ||
| BRAF | c.1694+3266A>G | ||
| BRAF | c.1694+2940C>T | ||
| BRAF | c.1140+3214G>A | ||
| BRAF | c.1140+2430C>T | ||
| BRAF | c.1140+2069A>G | ||
| BRAF | c.1140+1915G>T | ||
| BRAF | c.1140+1665dupG | ||
| BRAF | c.981-2296A>G | ||
| BRAF | c.980+2576T>A | ||
| BRAF | c.980+1801_980+1802delAA | ||
| BRAF | c.241-198G>C | ||
| BRAF | c.981-356dupA | ||
| BRAF | c.981-1080_981-1042del | ||
| BRAF | c.589G>A | ||
| BRAF | c.243C>A | ||
| BRAF | c.984-1276C>T | ||
| BRAF | c.1315-470C>T | ||
| BRAF | c.1315-479A>C | ||
| BRAF | c.1315-482A>T | ||
| BRAF | c.451-200C>T | ||
| BRAF | c.1251+48_1251+49delGA | ||
| BRAF | c.1178-1544T>C | ||
| BRAF | c.1178-1548A>G | ||
| BRAF | c.1178-1551C>T | ||
| BRAF | c.1178-1557C>T | ||
| BRAF | c.981-2446G>C | ||
| BRAF | c.160delC | ||
| BRAF | c.1394T>C | ||
| BRAF | c.328dupT | ||
| BRAF | c.1804-85C>A | ||
| BRAF | c.2138C>T | ||
| BRAF | c.814-174G>A | ||
| BRAF | c.814-77_814-76insAATA | ||
| BRAF | c.1059+170C>A | ||
| BRAF | c.1737A>G | ||
| BRAF | c.1141-1044C>T | ||
| BRAF | c.1141-1097A>G | ||
| BRAF | c.1141-1111G>A | rs373442098 | uncertain significance |
| BRAF | c.1141-1637_1141-1636delTT | ||
| BRAF | c.1140+633C>G | ||
| BRAF | c.1140+610_1140+615delAGCTAT | ||
| BRAF | c.861-75dupT | ||
| BRAF | c.860+457delC | ||
| BRAF | c.112-5717G>T | ||
| BRAF | c.112-5732A>G | ||
| BRAF | c.112-5778A>G | ||
| BRAF | c.112-6770_112-6769insA | ||
| BRAF | c.112-7499A>G | ||
| BRAF | c.112-7501A>T | ||
| BRAF | c.112-7503C>T | ||
| BRAF | c.*274+536A>G | ||
| BRAF | c.983+1398T>C | ||
| BRAF | c.983+1236_983+1237insCAAGAGGT | ||
| BRAF | c.983+1233_983+1234delTT | ||
| BRAF | c.983+1232T>C | ||
| BRAF | c.983+1186G>A | ||
| BRAF | c.876+630A>G | ||
| EGFR | c.88+48720_88+48721delTG | ||
| EGFR | c.322A>T | ||
| EGFR | c.2469+5015G>A | ||
| EGFR | c.2625+13C>G | ||
| EGFR | c.2947-203G>A | ||
| Mutations Detected by Single-Cell Sequencing Only | |||
| EGFR | c.3272-1104_3272-1092del | ||
| EGFR | c.3272-1071delG | ||
| EGFR | c.3272-1068delA | ||
| EGFR | c.3272-1064_3272-1063insAAAA | ||
| EGFR | c.3272-438T>C | ||
| EGFR | c.3272-408C>A | ||
| EGFR | c.*932dupA | ||
| EGFR | c.3271+191T>A | ||
| EGFR | c.3271+191T>G | ||
| EGFR | c.3271+1166C>A | ||
| EGFR | c.3272-1115_3272-1099del | ||
| EGFR | c.*781G>C | ||
| EGFR | c.*1151dupC | ||
| EGFR | c.*1382dupT | ||
| EGFR | c.*1957G>A | ||
| EGFR | c.1695-2105C>G | ||
| EGFR | c.1741+165A>G | rs10228436 | benign |
| EGFR | c.1695-1134A>G | rs2227984 | benign |
| EGFR | c.2469+108delG | ||
| EGFR | c.3271+188T>G | ||
| EGFR | c.3271+282T>A | rs2075110 | benign |
| EGFR | c.1721A>G | ||
| EGFR | c.1314+556_1314+557dupTT | rs10241451 | benign |
| EGFR | c.1314+394A>G | ||
| EGFR | c.1314+256G>A | ||
| EGFR | c.1178-443A>G | ||
| EGFR | c.1178-648G>A | ||
| EGFR | c.3271+809G>A | rs2072454 | benign |
| EGFR | c.3271+976G>C | ||
| EGFR | c.3272-611_3272-608delTACA | ||
| EGFR | n.140724136T>C | rs2075109 | benign |
| EGFR | n.140726106G>C | ||
| EGFR | c.2128-5dupT | ||
| EGFR | c.2128-27C>T | benign | |
| EGFR | c.1993-90G>T | rs2227983 | benign |
| EGFR | c.1993-93A>C | rs2227984 | benign |
| EGFR | c.1742-352C>G | rs2241055 | benign |
| EGFR | c.1742-353C>G | ||
| EGFR | c.1741+318A>G | ||
| EGFR | c.1695-53G>A | ||
| EGFR | c.1314+557dupT | ||
| FBXW7 | c.2001dupG | ||
| FBXW7 | c.*1125delT | ||
| FBXW7 | c.*1111A>C | ||
| FBXW7 | c.986-147A>C | ||
| FBXW7 | c.1728_1729insAAACAAC | ||
| FBXW7 | c.1727_1728ins | ||
| FBXW7 | c.1721_1722ins | ||
| FBXW7 | c.1716_1717ins | ||
| FBXW7 | c.933+18A>G | ||
| FBXW7 | c.934-191A>T | ||
| FBXW7 | c.934-15_934-14delTC | ||
| FBXW7 | c.934-12_934-11insAC | ||
| KRAS | c.876+408_876+409dupTT | ||
| KRAS | c.556-72_556-71delAA | ||
| KRAS | c.257C>T | ||
| KRAS | c.1308+459A>C | ||
| KRAS | c.1308+468C>T | ||
| KRAS | c.1308+472C>T | ||
| KRAS | c.1308+473A>G | ||
| NRAS | c.-2070dupT | ||
| NRAS | c.1251+52A>T | rs61758221 | benign |
| PIK3CA | c.*3631T>C | ||
| PIK3CA | c.*1606delA | ||
| PIK3CA | c.1251+54A>C | ||
| PIK3CA | c.1251+57G>C | ||
| PIK3CA | c.1251+58C>A | ||
| PIK3CA | c.*355G>T | ||
| PIK3CA | c.*480C>A | ||
| Mutations Detected by Single-Cell Sequencing Only | |||
| PIK3CA | c.*488_*490delTCC | ||
| PIK3CA | c.*494G>T | ||
| PIK3CA | c.1736_1737insAAAACAAA | ||
| PIK3CA | c.1735G>T | ||
| PIK3CA | c.1733C>A | ||
| PIK3CA | c.2496-124G>T | rs7623154 | benign |
| PIK3CA | c.2923A>T | rs17550640 | benign |
| PIK3CA | c.2936+21dupA | ||
| PIK3CA | c.*654T>G | rs3729676 | benign |
| PIK3CA | c.645+173A>G | ||
| PIK3CA | c.934-132delG | ||
| PIK3CA | c.3381G>C | ||
| PIK3CA | c.6200A>G | ||
| PIK3CA | c.6633C>T | ||
| RNF43 | c.*6514G>A | ||
| RNF43 | c.451-5T>C | ||
| RNF43 | c.376-90A>G | ||
| RNF43 | c.2936+19A>G | ||
| SMAD4 | c.692dupG | pathogenic | |
| SMAD4 | c.905-1G>C | ||
| SMAD4 | c.1139+385A>T | ||
| SMAD4 | c.*5005dupT | ||
| SMAD4 | c.*5116C>G | ||
| SMAD4 | c.*5131A>G | ||
| SMAD4 | c.*5191C>G | ||
| SMAD4 | c.*5535_*5536delAC | ||
| SMAD4 | c.*5541_*5552del | ||
| SMAD4 | c.*5863_*5867delGAAAA | benign | |
| SMAD4 | c.*5994A>C | benign | |
| SMAD4 | c.*6433G>A | ||
| SMAD4 | c.1060-42G>T | ||
| TP53 | c.984-1412delT | ||
| TP53 | c.983+1201A>G | ||
| TP53 | c.556-71delA | ||
| TP53 | c.556-149T>A | ||
| TP53 | c.424C>T | ||
| TP53 | c.259-161_259-158delAAAA | ||
| TP53 | c.259-162_259-158delAAAAA | ||
| TP53 | c.258+123dupT | ||
| TP53 | c.99dupC | ||
| TP53 | c.-22+41_-22+48delACCTGGAG | ||
| TP53 | c.-145-190C>A | ||
| TP53 | c.-145-1184T>C | ||
| TP53 | c.966dupG | ||
| TP53 | c.582+132T>C | ||
| TP53 | c.1060-69G>T | ||
| TP53 | c.1308+477T>C | ||
| TP53 | c.1308+478G>C | ||
| TP53 | c.1308+501C>T | ||
| TP53 | c.1308+521_1308+574del | ||
| TP53 | c.*3333C>T | ||
| Mutations Detected by Bulk Sequencing Only | |||
| APC | c.136-1428A>C | rs2464807 | other |
| APC | c.220+124C>G | rs76552546 | likely benign |
| APC | c.2413C>T | rs587779783 | pathogenic |
| APC | c.4666dupA | rs587783031 | pathogenic |
| ARID1A | c.1920+6177G>T | ||
| ARID1A | c.1921-1059A>T | ||
| ARID1A | c.2252-97A>T | rs113319329 | benign |
| ARID1A | c.2733-400A>G | ||
| ARID1A | c.3199-95A>G | rs76490152 | benign |
| BRAF | n.140726457T>C | ||
| BRAF | c.*1215A>T | ||
| BRAF | c.2128-16C>T | rs368721021 | benign/likely benign |
| BRAF | c.1177+146G>A | rs1267632 | benign |
| BRAF | c.1140+3180G>T | ||
| Mutations Detected by Bulk Sequencing Only | |||
| BRAF | c.505-6693T>C | ||
| BRAF | c.505-9562G>A | ||
| BRAF | c.504+3486T>C | ||
| BRAF | c.504+142G>A | benign | |
| BRAF | c.139-23483C>T | ||
| EGFR | c.88+37643T>C | ||
| EGFR | c.89-55393G>A | ||
| EGFR | c.89-29869C>A | ||
| EGFR | c.559+214G>T | rs2270427 | benign |
| EGFR | c.1498+22A>T | rs1558544 | benign |
| EGFR | c.1498+142C>T | rs759162 | benign |
| EGFR | c.1499-177A>G | rs11536635 | benign |
| EGFR | c.1880+733A>C | ||
| EGFR | c.2361G>A | benign | |
| EGFR | c.2469+4027T>C | ||
| EGFR | c.2508C>T | benign | |
| EGFR | c.2625+196A>G | rs6970262 | benign |
| EGFR | c.2709T>C | rs1140475 | benign |
| EGFR | c.2849-551T>G | ||
| EGFR | c.3162+200_3162+201insAG | rs34723095 | benign |
| EGFR | c.3272-123G>A | rs2692456 | benign |
| EGFR | c.3333_3334insTTTTTTTTTTTTT | ||
| EGFR | c.3337delC | ||
| EGFR | c.3339_3350delGCCTCTGAACCC | ||
| EGFR | c.3353C>G | ||
| EGFR | c.3355C>G | ||
| EGFR | c.3356C>A | ||
| EGFR | c.3368C>T | rs775317295 | uncertain significance |
| EGFR | c.*9367A>G | ||
| FBXW7 | c.*3466C>G | ||
| FBXW7 | c.-69-40817T>C | ||
| PIK3CA | c.1059+62C>A | rs2699895 | benign |
| PIK3CA | c.1060-17C>A | rs2699896 | benign |
| PIK3CA | c.1145+54A>G | rs3729679 | benign |
| PIK3CA | c.2016-27A>T | rs6443625 | benign |
| PIK3CA | c.*5631C>T | ||
| PIK3CA | c.*10339G>T | ||
| RNF43 | c.2057C>G | rs9652855 | benign |
| TP53 | c.*4020_*4049del | ||
| TP53 | c.*274+522T>G | ||
| TP53 | c.*274+31A>G | ||
| TP53 | c.877-1G>A | rs587782272 | pathogenic |
| TP53 | c.555+62A>G | benign | |
| TP53 | c.259-91G>A | benign | |
| TP53 | c.259-160_259-158delAAA | ||
| TP53 | c.98C>G | benign | |
| TP53 | c.-22+41_-21-54del | ||
| TP53 | c.-44+38C>G | benign | |
| TP53 | c.-14962_-14959dupGTTT | ||
| Mutations detected by both methods | |||
| APC | c.1458T>C | rs2229992 | benign |
| APC | c.4479G>A | rs41115 | benign |
| APC | c.5034G>A | rs42427 | benign |
| APC | c.5268T>G | rs866006 | benign |
| APC | c.5465T>A | rs459552 | benign |
| APC | c.5880G>A | rs465899 | benign |
| APC | c.7504G>A | rs2229995 | benign |
| BRAF | c.2128-54_2128-51dupCTTT | ||
| BRAF | c.1992+16G>C | rs3789806 | benign/likely benign |
| BRAF | c.1992+14A>G | ||
| BRAF | c.1929A>G | rs9648696 | benign |
| EGFR | c.474C>T | rs2072454 | benign |
| EGFR | c.560-84T>C | rs2075109 | benign |
| EGFR | c.628+104C>T | rs2075110 | benign |
| EGFR | c.629-62A>G | rs11506105 | benign |
| EGFR | c.1006+151T>C | rs3735059 | benign |
| EGFR | c.1562G>A | rs2227983 | benign |
| Mutations detected by both methods | |||
| EGFR | c.1881-600G>A | rs10228436 | benign |
| EGFR | c.1887T>A | rs2227984 | benign |
| EGFR | c.1920-215G>C | rs2241055 | benign |
| EGFR | c.2283+96A>G | rs2017000 | benign |
| EGFR | c.2284-60T>C | rs10241451 | benign |
| FBXW7 | c.1746G>A | ||
| NRAS | c.-3343C>T | ||
| PIK3CA | c.352+40A>G | rs3729674 | benign |
| PIK3CA | c.1173A>G | rs2230461 | benign |
| PIK3CA | c.2295-57C>G | rs2699889 | benign |
| PIK3CA | c.*10365T>C | ||
| RNF43 | c.350G>A | rs2257205 | benign |
| TP53 | c.665+92T>G | rs12951053 | benign |
| TP53 | c.665+72C>T | rs12947788 | benign |
References
- Galeano Nino, J.L.; Wu, H.; LaCourse, K.D.; Kempchinsky, A.G.; Baryiames, A.; Barber, B.; Futran, N.; Houlton, J.; Sather, C.; Sicinska, E.; et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature 2022, 611, 810–817. [Google Scholar] [CrossRef]
- Zhang, M.; Hu, S.; Min, M.; Ni, X.; Lu, Z.; Sun, X.; Wu, J.; Liu, B.; Ying, X.; Liu, Y. Dissecting transcriptional heterogeneity in primary gastric adenocarcinoma by single cell RNA sequencing. Gut 2021, 70, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Dang, N.; Tang, G.; Li, Z.; Li, X.; Shi, B.; Xu, Z.; Li, L.; Yang, X.; Xu, C.; et al. Integrating bulk and single-cell RNA sequencing reveals cellular heterogeneity and immune infiltration in hepatocellular carcinoma. Mol. Oncol. 2022, 16, 2195–2213. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Ramnarayanan, K.; Sundar, R.; Padmanabhan, N.; Srivastava, S.; Koiwa, M.; Yasuda, T.; Koh, V.; Huang, K.K.; Tay, S.T.; et al. Single-Cell Atlas of Lineage States, Tumor Microenvironment, and Subtype-Specific Expression Programs in Gastric Cancer. Cancer Discov. 2022, 12, 670–691. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wu, S.; Yang, Z.; Zhang, X.; Zheng, Q.; Lin, L.; Niu, Z.; Li, R.; Cai, Z.; Li, L. Single-cell exome sequencing identifies mutations in KCP, LOC440040, and LOC440563 as drivers in renal cell carcinoma stem cells. Cell Res. 2017, 27, 590–593. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Rowan, A.J.; Horsewell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 367, 976. [Google Scholar] [CrossRef] [PubMed]
- Ramón Y Cajal, S.; Sesé, M.; Capdevila, C.; Aasen, T.; De Mattos-Arruda, L.; Diaz-Cano, S.J.; Hernández-Losa, J.; Castellví, J. Clinical implications of intratumor heterogeneity: Challenges and opportunities. Mol. Med. 2020, 98, 161–177. [Google Scholar] [CrossRef]
- Zhu, L.; Jiang, M.; Wang, H.; Sun, H.; Zhu, J.; Zhao, W.; Fang, Q.; Yu, J.; Chen, P.; Wu, S.; et al. A narrative review of tumor heterogeneity and challenges to tumor drug therapy. Ann. Transl. Med. 2021, 9, 1351. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Guo, C.; Wang, C.; Xu, J.; Zheng, S.; Duan, J.; Li, Y.; Bai, H.; Xu, Q.; Ning, F.; et al. Single-cell RNA sequencing reveals tumor heterogeneity, microenvironment, and drug-resistance mechanisms of recurrent glioblastoma. Cancer Sci. 2023, 114, 2609–2621. [Google Scholar] [CrossRef]
- Walsh, E.M.; Halushka, M.K. A Comparison of Tissue Dissection Techniques for Diagnostic, Prognostic, and Theragnostic Analysis of Human Disease. Pathobiology 2023, 90, 199–208. [Google Scholar] [CrossRef]
- Peretz, C.A.C.; McGary, L.H.F.; Kumar, T.; Jackson, H.; Jacob, J.; Durruthy-Durruthy, R.; Levis, M.J.; Perl, A.; Huang, B.J.; Smith, C.C. Single-cell DNA sequencing reveals complex mechanisms of resistance to quizartinib. Blood Adv. 2021, 5, 1437–1441. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Tang, R.; Xu, J.; Wang, W.; Zhang, B.; Liu, J.; Yu, X.; Shi, S. Applications of single-cell sequencing in cancer research: Progress and perspectives. Hematol. Oncol. 2021, 14, 91. [Google Scholar] [CrossRef]
- Li, X.; Wang, C.Y. From bulk, single-cell to spatial RNA sequencing. Int. J. Oral. Sci. 2021, 13, 36. [Google Scholar] [CrossRef]
- Parikh, A.R.; Leshchiner, I.; Elagina, L.; Goyal, L.; Levovitz, C.; Siravegna, G.; Livitz, D.; Rhrissorrakrai, K.; Martin, E.E.; Van Seventer, E.E.; et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 2019, 25, 1415–1421. [Google Scholar] [CrossRef] [PubMed]
- Kalmár, A.; Galamb, O.; Szabó, G.; Pipek, O.; Medgyes-Horváth, A.; Barták, B.K.; Nagy, Z.B.; Szigeti, K.A.; Zsigrai, S.; Csabai, I.; et al. Patterns of Somatic Variants in Colorectal Adenoma and Carcinoma Tissue and Matched Plasma Samples from the Hungarian Oncogenome Program. Cancers 2023, 15, 907. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Mondal, S.; Martin, C.; DuPlissis, A.; Chizari, S.; Ma, K.Y.; Maiya, R.; Messing, R.O.; Jiang, N.; Ben-Yakar, A. Femtosecond laser microdissection for isolation of regenerating C. elegans neurons for single-cell RNA sequencing. Nat. Methods 2023, 20, 590–599. [Google Scholar] [CrossRef] [PubMed]
- Smajic, S.; Prada-Medina, C.A.; Landoulsi, Z.; Ghelfi, J.; Delcambre, S.; Dietrich, C.; Jarazo, J.; Henck, J.; Balachandran, S.; Pachchek, S.; et al. Single-cell sequencing of human midbrain reveals glial activation and a Parkinson-specific neuronal state. Brain 2022, 145, 964–978. [Google Scholar] [CrossRef]
- Turan, Z.G.; Richter, V.; Bochmann, J.; Parvizi, P.; Yapar, E.; Işıldak, U.; Waterholter, S.K.; Leclere-Turbant, S.; Son, C.D.; Duyckaerts, C.; et al. Somatic copy number variant load in neurons of healthy controls and Alzheimer’s disease patients. Acta Neuropath. Commun. 2022, 10, 175. [Google Scholar] [CrossRef] [PubMed]
- Massalha, H.; Bahar Halpern, K.; Abu-Gazala, S.; Jana, T.; Massasa, E.E.; Moor, A.E.; Buchauer, L.; Rozenberg, M.; Pikarsky, E.; Amit, I.; et al. A single cell atlas of the human liver tumor microenvironment. Mol. Syst. Biol. 2020, 16, e9682. [Google Scholar] [CrossRef] [PubMed]
- Paul, E.D.; Huraiová, B.; Valková, N.; Birknerova, N.; Gábrišová, D.; Gubova, S.; Ignačáková, H.; Ondris, T.; Bendíková, S.; Bíla, J.; et al. Multiplexed RNA-FISH-guided Laser Capture Microdissection RNA Sequencing Improves Breast Cancer Molecular Subtyping, Prognostic Classification, and Predicts Response to Antibody Drug Conjugates. medRxiv 2023. [Google Scholar] [CrossRef]
- Ikeda, H.; Miyao, S.; Nagaoka, S.; Takashima, T.; Law, S.M.; Yamamoto, T.; Kurimoto, K. High-quality single-cell transcriptomics from ovarian histological sections during folliculogenesis. Life Sci. Alliance 2023, 6, e202301929. [Google Scholar] [CrossRef] [PubMed]
- Pavlič, A.; Urh, K.; Boštjančič, E.; Zidar, N. Analyzing the invasive front of colorectal cancer—By punching tissue block or laser capture microdissection? Pathol. Res. Pract. 2023, 248, 154727. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.L.; Li, D.D.; Duan, J.Y.; Sheng, L.M.; Wang, X. Resistance to targeted therapy in metastatic colorectal cancer: Current status and new developments. World J. Gastroenterol. 2023, 29, 926–948. [Google Scholar] [CrossRef]
- Wang, Q.; Shen, X.; Chen, G.; Du, J. Drug Resistance in Colorectal Cancer: From Mechanism to Clinic. Cancers 2022, 14, 2928. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.M.; Qu, X.; Li, C.F. ARID1A mutations confer intrinsic and acquired resistance to cetuximab treatment in colorectal cancer. Nat. Commun. 2022, 13, 5478. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Fan, Y. Combined KRAS and TP53 mutation in patients with colorectal cancer enhance chemoresistance to promote postoperative recurrence and metastasis. BMC Cancer 2024, 24, 1155. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Wang, L.; Qiu, H.; Zhang, M.; Sun, L.; Peng, P.; Yu, Q.; Yuan, X. Mechanisms of resistance to anti-EGFR therapy in colorectal cancer. Oncotarget 2017, 8, 3980–4000. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.L.; Xin, H.Y.; Sun, R.Q.; Zhou, Z.J.; Hu, Z.Q.; Luo, C.B.; Wang, P.C.; Li, J.; Fan, J.; Zhou, J. Association of KRAS Variant Subtypes with Survival and Recurrence in Patients with Surgically Treated Intrahepatic Cholangiocarcinoma. JAMA Surg. 2022, 157, 59–65. [Google Scholar] [CrossRef]
- Sugihara, Y.; Taniguchi, H.; Kushima, R.; Tsuda, H.; Kubota, D.; Ichikawa, H.; Fujita, S.; Kondo, T. Laser microdissection and two-dimensional difference gel electrophoresis reveal proteomic intra-tumor heterogeneity in colorectal cancer. J. Proteom. 2013, 78, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ye, Y.; Shen, D.; Jiang, K.; Zhang, H.; Sun, W.; Zhang, J.; Xu, F.; Cui, Z.; Wang, S. Identification of transgelin-2 as a biomarker of colorectal cancer by laser capture microdissection and quantitative proteome analysis. Cancer Sci. 2010, 101, 523–529. [Google Scholar] [CrossRef]
- Lee-Six, H.; Olafsson, S.; Ellis, P.; Osborne, R.J.; Sanders, M.A.; Moore, L.; Georgakopoulos, N.; Torrente, F.; Noorani, A.; Goddard, M.; et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature 2019, 574, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.N.; Kopetz, E.S. BRAF mutant colorectal cancer as a distinct subset of colorectal cancer: Clinical characteristics, clinical behavior, and response to targeted therapies. J. Gastrointest. Oncol. 2015, 6, 660–667. [Google Scholar] [PubMed]
- Shang, W.; Yan, C.; Liu, R.; Chen, L.; Cheng, D.; Hao, L.; Yuan, W.; Chen, J.; Yang, H. Clinical significance of FBXW7 tumor suppressor gene mutations and expression in human colorectal cancer: A systemic review and meta-analysis. BMC Cancer. 2021, 21, 770. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Lin, H.; Wang, D.; Li, Q.; Luo, H.; Li, G.; Chen, X.; Li, Y.; Chen, P.; Zhai, B.; et al. PCDH17 increases the sensitivity of colorectal cancer to 5-fluorouracil treatment by inducing apoptosis and autophagic cell death. Signal Transduct. Target Ther. 2019, 4, 53. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Ma, H.M.; Huang, H.B.; Li, Y.W.; Zhang, P.; Huang, J.J.; Cheng, L.; Li, G.R. Overexpression of COL5A1 promotes tumor progression and metastasis and correlates with poor survival of patients with clear cell renal cell carcinoma. Cancer Manag. Res. 2019, 11, 1263–1274. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Han, J.; Liu, J.; Yang, R.; Wang, J.; Wang, X.; Chen, X. Genetic and Epigenetic Impact of Chronic Inflammation on Colon Mucosa Cells. Front. Genet. 2021, 12, 722835. [Google Scholar] [CrossRef]
- Valcz, G.; Buzás, E.; Gatenby, R.A.; Ujvari, B.; Molnár, B. Small extracellular vesicles from surviving cancer cells as multiparametric monitoring tools of measurable residual disease and therapeutic efficiency. Biochim. Biophys. Acta BBA Cancer 2024, 1879, 189088. [Google Scholar] [CrossRef] [PubMed]
- Dvir, A.; Camarda, R.; Odegaard, J.; Paik, H.; Oskotsky, B.; Krings, G.; Goga, A.; Sirota, M.; Butte, A. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat. Commun. 2017, 8, 1077. [Google Scholar]
- Kim, J.; Kim, H.; Lee, M.S.; Lee, H.; Kim, Y.J.; Lee, W.Y.; Yun, S.H.; Kim, H.C.; Hong, H.K.; Hannenhalli, S.; et al. Transcriptomes of the tumor-adjacent normal tissues are more informative than tumors in predicting recurrence in colorectal cancer patients. J. Transl. Med. 2023, 21, 304. [Google Scholar] [CrossRef] [PubMed]
- The Galaxy Community. The Galaxy platform for accessible, reproducible, and collaborative data analyses: 2024 update. Nucleic Acid Res. 2024, 52, W83–W94. [Google Scholar] [CrossRef] [PubMed]
- Kandoth, C.; Gao, J.; Mattioni, M.; Struck, A.; Boursin, Y.; Penson, A.; Chavan, S. mskcc/vcf2maf: vcf2maf v1.6.16. (v1.6.16), Zenodo: Genève, Switzerland, 2020. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Lee, M.J.; Riley, G.R.; Jang, W.; Rubinstein, W.S.; Church, D.M.; Maglott, D.R. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014, 42, D980-5. [Google Scholar] [CrossRef]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef] [PubMed]
- Hunter, J.D. Matplotlib is a 2D graphics package used for Python for application development, interactive scripting, and publication-quality image generation across user interfaces and operating systems. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]


| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | Mean | STD | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NEG | 8.2 | 25.1 | 2.5 | 4.2 | 0.5 | 10.9 | 76.6 | 126 | 8.9 | 19.2 | 14.3 | 1.7 | 24.8 | 37.9 |
| NAT | 37.2 | 11.4 | 13.5 | 125.5 | 85.3 | 33.7 | 0.8 | 0.7 | 5.6 | 0.9 | 1.6 | 0.2 | 26.4 | 39.9 |
| CRC | 141.2 | 12.6 | 66 | 0.3 | 1 | 14.1 | 62 | 0.8 | 0.4 | 0.1 | 5 | 74.8 | 31.5 | 44.9 |
| NAT | CRC | ||||||
|---|---|---|---|---|---|---|---|
| Gene | Number of Mutations | Samples Containing the Mutated Gene | Occurence Rate of the Mutated Genes | Gene | Number of Mutations | Samples Containing the Mutated Gene | Occurence Rate of the Mutated Genes |
| TTN | 174 | 6/6 | 100% | TTN | 197 | 7/7 | 100% |
| APC | 137 | 6/6 | 100% | APC | 197 | 5/7 | 71% |
| KRAS | 73 | 5/6 | 83% | KRAS | 45 | 5/7 | 71% |
| TP53 | 62 | 5/6 | 83% | TP53 | 66 | 4/7 | 57% |
| PIK3CA | 10 | 3/6 | 50% | PIK3CA | 38 | 5/7 | 71% |
| FBXW7 | 14 | 5/6 | 83% | FBXW7 | 25 | 5/7 | 71% |
| SOX9 | 7 | 2/6 | 33% | SOX9 | 7 | 4/7 | 57% |
| ∑ = 477 | = 477 | ∑ = 498 | = 427 | ||||
| Single-Cell Sequencing | |||
|---|---|---|---|
| Gene | Sequence Variation | dbSNP ID | Mutation Classification |
| BRAF | c.1763T>C | rs1562954580 | uncertain significance |
| BRAF | c.1141-1111G>A | rs373442098 | conflicting classifications of pathogenicity |
| TP53 | c.424C>T | rs1597371187 | uncertain significance |
| SMAD4 | c.692dupG | rs377767334 | pathogenic |
| APC | c.559+37C>A | rs1554084977 | uncertain significance |
| APC | c.560-84T>C | rs1561605775 | uncertain significance |
| Bulk Sequencing | |||
| Gene | Sequence Variation | dbSNP ID | Mutation Classification |
| APC | c.136-1428A>C | rs2464807 | other |
| APC | c.2413C>T | rs587779783 | pathogenic |
| APC | c.4666dupA | rs587783031 | pathogenic |
| EGFR | c.3368C>T | rs775317295 | uncertain significance |
| TP53 | c.877-1G>A | rs587782272 | pathogenic/likely pathogenic |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szakállas, N.; Kalmár, A.; Barták, B.K.; Nagy, Z.B.; Valcz, G.; Linkner, T.R.; Rada, K.R.; Takács, I.; Molnár, B. Investigation of Exome-Wide Tumor Heterogeneity on Colorectal Tissue-Based Single Cells. Int. J. Mol. Sci. 2025, 26, 737. https://doi.org/10.3390/ijms26020737
Szakállas N, Kalmár A, Barták BK, Nagy ZB, Valcz G, Linkner TR, Rada KR, Takács I, Molnár B. Investigation of Exome-Wide Tumor Heterogeneity on Colorectal Tissue-Based Single Cells. International Journal of Molecular Sciences. 2025; 26(2):737. https://doi.org/10.3390/ijms26020737
Chicago/Turabian StyleSzakállas, Nikolett, Alexandra Kalmár, Barbara Kinga Barták, Zsófia Brigitta Nagy, Gábor Valcz, Tamás Richárd Linkner, Kristóf Róbert Rada, István Takács, and Béla Molnár. 2025. "Investigation of Exome-Wide Tumor Heterogeneity on Colorectal Tissue-Based Single Cells" International Journal of Molecular Sciences 26, no. 2: 737. https://doi.org/10.3390/ijms26020737
APA StyleSzakállas, N., Kalmár, A., Barták, B. K., Nagy, Z. B., Valcz, G., Linkner, T. R., Rada, K. R., Takács, I., & Molnár, B. (2025). Investigation of Exome-Wide Tumor Heterogeneity on Colorectal Tissue-Based Single Cells. International Journal of Molecular Sciences, 26(2), 737. https://doi.org/10.3390/ijms26020737





