Regulation of Flavonoid Biosynthesis by the MYB-bHLH-WDR (MBW) Complex in Plants and Its Specific Features in Cereals
Abstract
1. Introduction
2. Flavonoid Compounds and Flavonoid Pigments
3. Flavonoid Biosynthesis Pathway
4. General Mechanisms of Regulation of Flavonoid Biosynthesis by MBW Complexes
4.1. Features of MYB, bHLH, and WDR Transcription Factors Regulating Anthocyanin Biosynthesis
4.1.1. R2R3-MYB Transcription Factors
4.1.2. MYC-like bHLH Transcription Factors
4.1.3. WDR Proteins
4.2. Structure of MBW Complexes
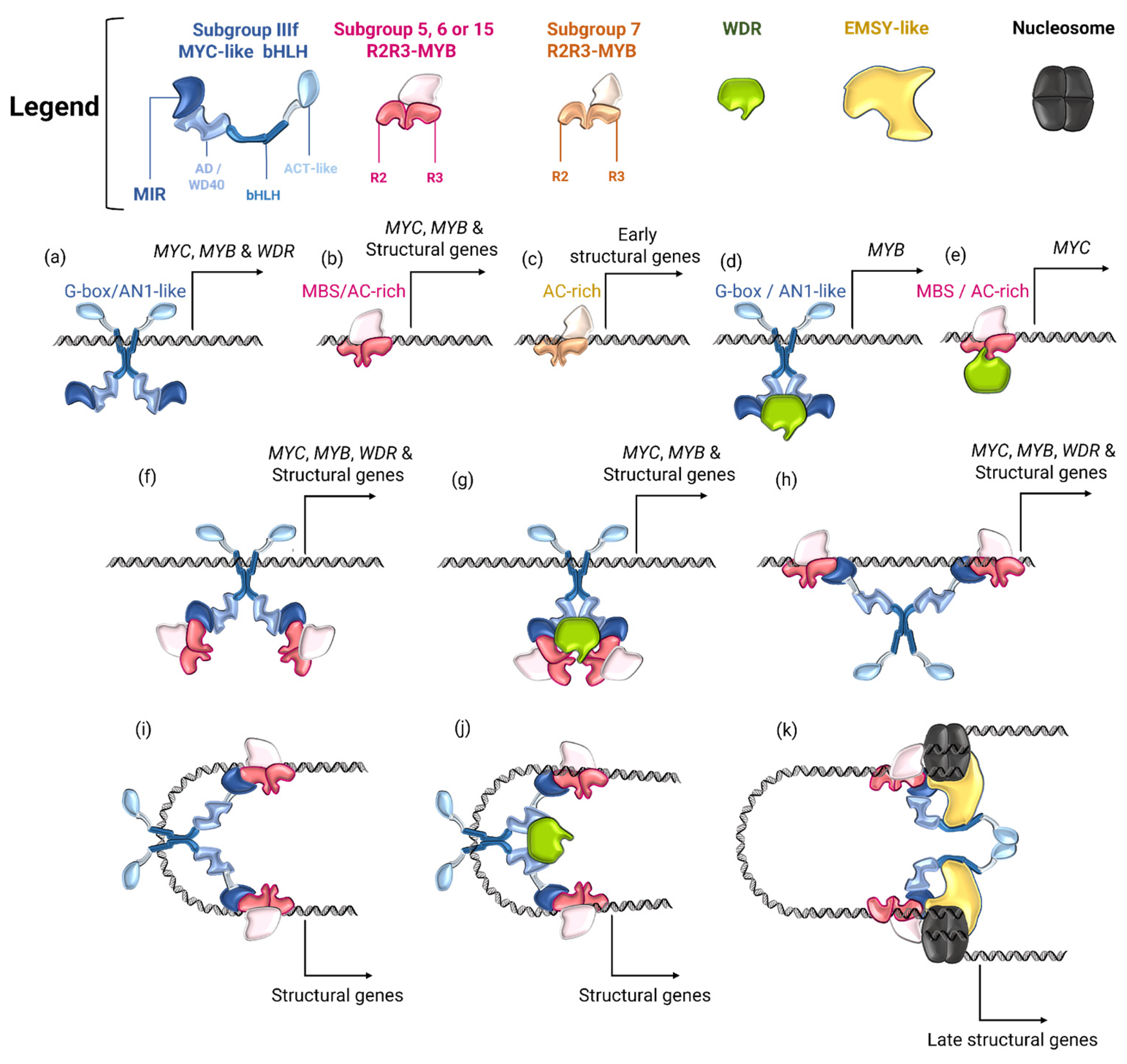
4.3. Regulatory Networks of MBW Complexes
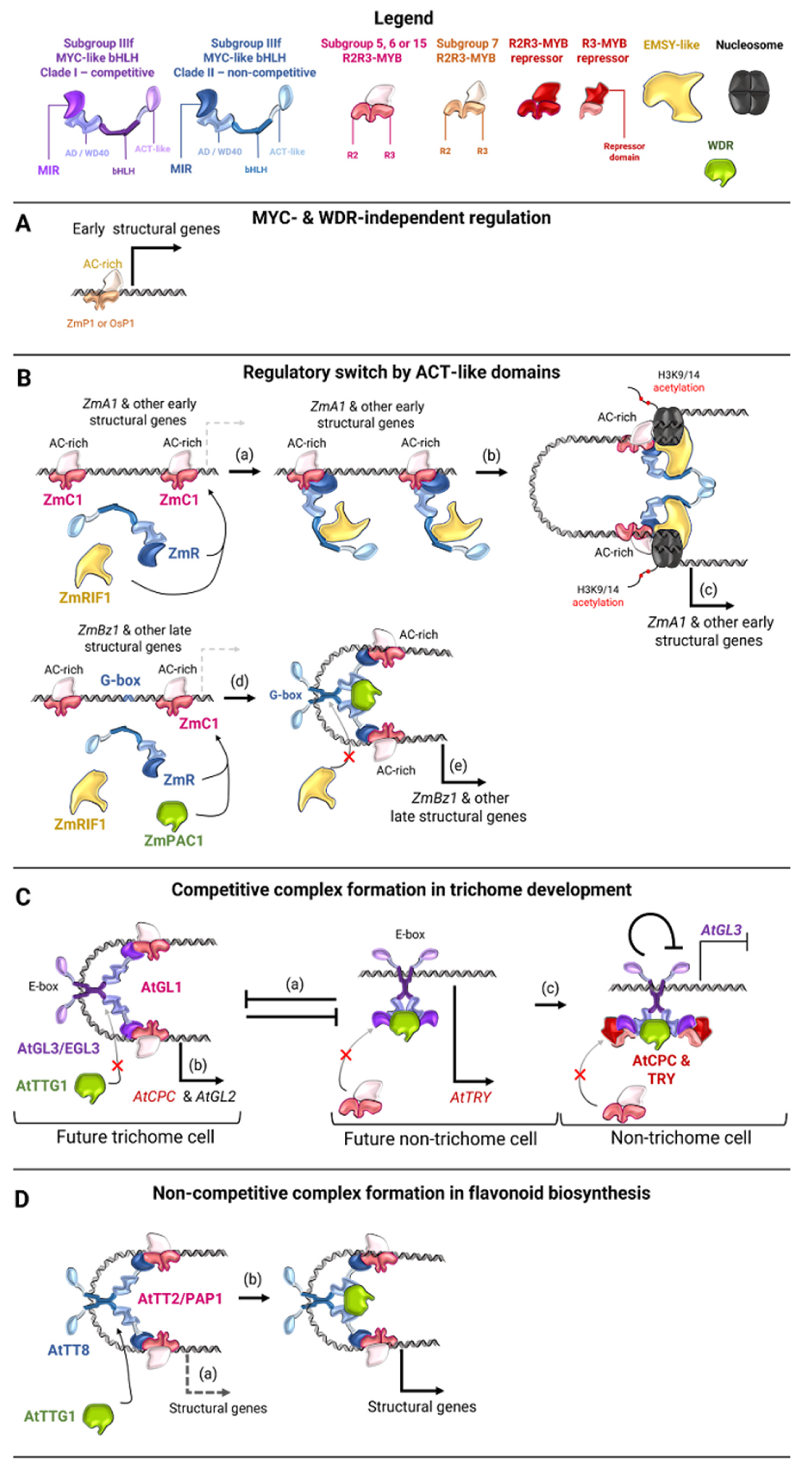
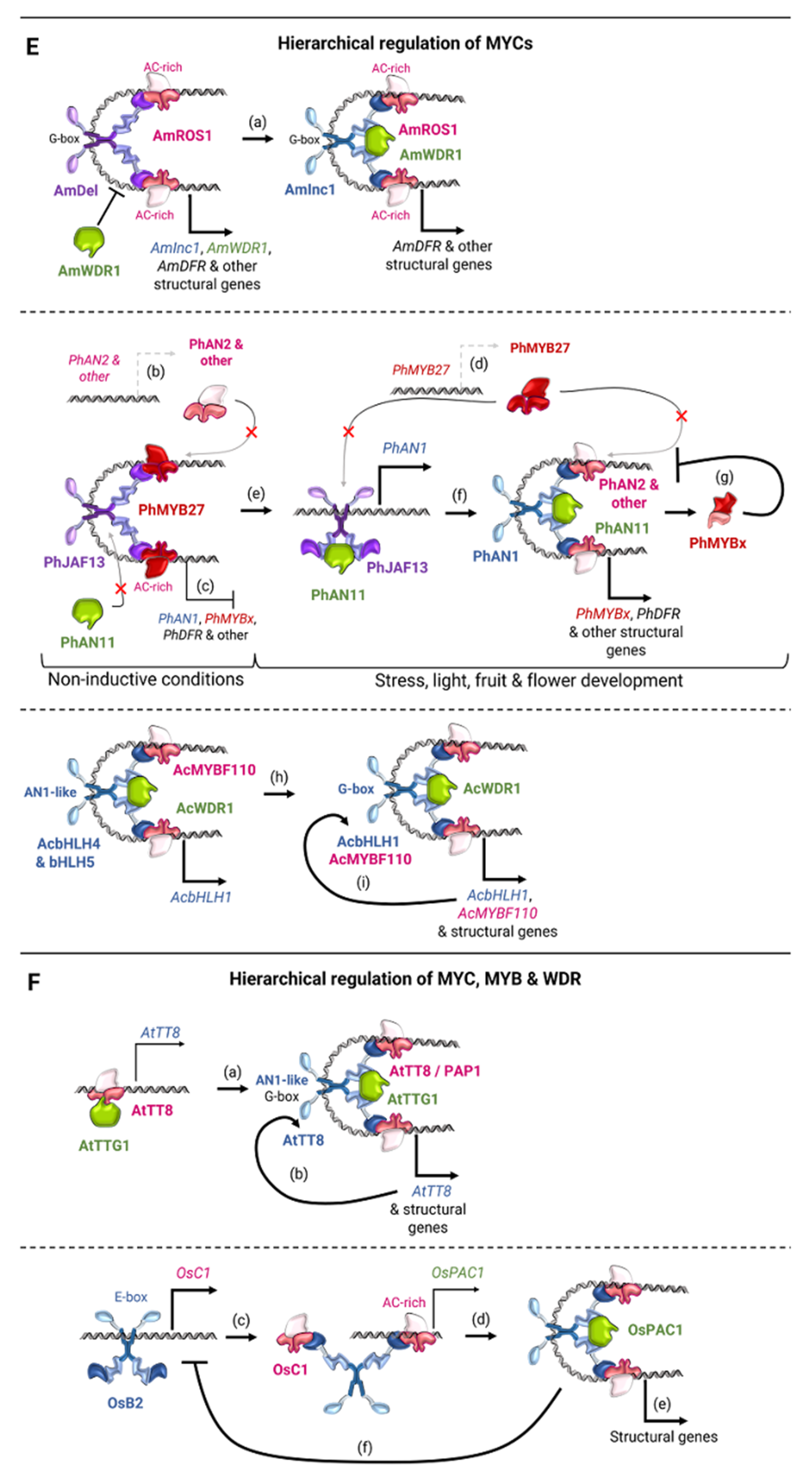
4.4. Genes of MBW Regulators of Flavonoid Biosynthesis in Cereal Plants
4.4.1. Diversity of MBW Genes in Maize (Zea mays L.)
| Family | Gene (Locus) | Chromosome | Function | MBW Complex Formation | Note | References |
|---|---|---|---|---|---|---|
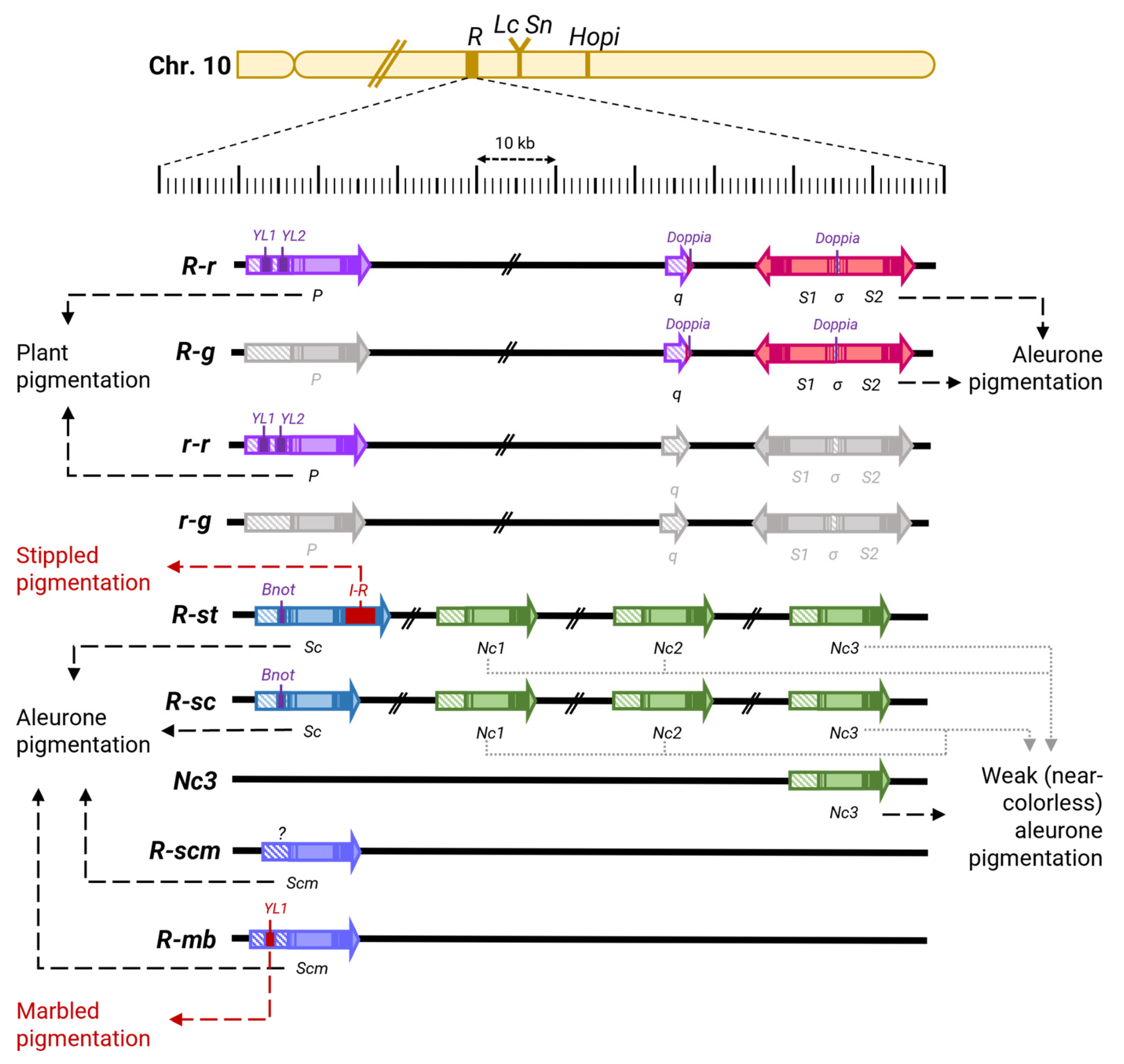
| MYC-like bHLH | ZmR (Red) | 10 | Positive regulator of anthocyanin pigmentation of almost all tissues and organs, depending on the allele (see Table 2) | Yes, with ZmC1/Pl and ZmPAC1 | A complex locus, different alleles contain varying numbers of copies of MYC genes and other sequences (see Table 2 and Figure 4) | [26,50,52,126,139,143,157,165,174,175,176,201,202,203,204,205,206,207,212,216] |
| ZmLc (Leaf color) | 10 | Positive regulator of anthocyanin pigmentation of leaf midrib, glumes, leaf auricles and pericarp | Yes, with ZmC1/Pl and, probably, ZmPAC1 | - | [15,207,208] | |
| ZmSn (Scuttelar node color) | 10 | Positive regulator of anthocyanin pigmentation of root, leaf, mesocotyl, scutellum and pericarp | Yes, with ZmC1/Pl and ZmPAC1 | Expression is induced by light in the mesocotyl and also in the early stages of embryo development in the scutellum and pericarp | [24,209,210] | |
| ZmHopi | 10 | Positive regulator of anthocyanin pigmentation of roots, leaves, mesocotyl, anthers, pericarp, scutellum and aleurone | Yes, with ZmC1/Pl and ZmPAC1 | Expression is induced by light in the late stages of embryo development in the scutellum and aleurone | [23] | |
| ZmB (Booster) | 2 | Positive regulator of anthocyanin pigmentation of vegetative tissues and pericarp, depending on the allele | Probably, with ZmC1/Pl and ZmPAC1 | Many different alleles with different promoter structures (see Table 3 and Figure 5) | [15,23,24,25,28,47,52,121,126,156,167,169,175,210,217,218] | |
| ZmPSH (Purple leaf sheath) | 10 | Positive regulator of anthocyanin pigmentation of leaf sheath, stem, leaves and root | Unknown | Expression in the root is activated by light | [211] | |
| ZmIN1 (Intensifier 1) | 7 | Negative regulator of anthocyanin pigmentation of aleurone | Probably, a competitor of C1/Pl–R/B–PAC1 complexes | IN1 likely competes with R family proteins for binding to MYB and WDR due to a defective structure | [26,219] | |
| R2R2-MYB | ZmC1 (Colorless 1) | 9 | Positive regulator of anthocyanin pigmentation of aleurone | Yes, with R/B family and ZmPAC1 | - | [15,16,22,23,26,28,47,126,156,157,164,169,220,221] |
| ZmPl (Purple leaf) | 6 | Positive regulator of anthocyanin pigmentation of vegetative tissues and pericarp | Yes, with R family and, probably, with ZmB and ZmPAC1 | - | [19,23,24,47,121,126,156,164,167,169,175,201,217,218] | |
| ZmP (Pericarp color) | 1 | Positive regulator of phlobaphene biosynthesis in pericarp, panicle, cob, silk and anthers | No | Contains two (or more) highly homologous P genes that activate the expression of early genes of flavonoid biosynthesis and activate the biosynthesis of phlobaphenes independently of MYC proteins | [46,64,128,129,130,131,222] | |
| WDR | ZmPAC1 (Pale aleurone color 1) | 5 | Positive regulator of anthocyanin pigmentation of roots and aleurone | Yes, with R/B family and ZmC1 | Expression is observed in many plant tissues, but mutations only result in the absence of root and aleurone pigmentation. | [19,26,164] |
| Allele | Structure of Allele * | Phenotype | References |
|---|---|---|---|
| R-r (Red-red) | Functional alleles of genes S1 and S2; promoter of R gene—q; functional allele of gene P | Uniform anthocyanin pigmentation of aleurone (R, Red); anthocyanin pigmentation of plant organs (-r, -red) | [50,52,165,201,202,203,206,207,212,216] |
| R-g/S (Red-green/Seed color) ** | Functional alleles of genes S1 and S2; promoter of R gene—q; non-functional allele of gene P, or absence of P | Uniform anthocyanin pigmentation of aleurone (R, Red); anthocyaninless plant organs (-g, -green) | |
| r-r/P (red-red/Plant color) ** | Non-functional alleles of genes S1 and S2; promoter of R gene—q, or their absence; functional allele of gene P | Uncolored aleurone (r, red); anthocyanin pigmentation of plant organs (-r, -red) | |
| r-g (red-green) ** | Non-functional alleles of genes S1 and S2; promoter of R gene—q; non-functional allele of P; or their absence | Uncolored aleurone (r, red); anthocyaninless vegetative organs (-g, -green) | |
| R-nj/Nj (Navajo aleurone color) | Functional allele of gene Nj | Anthocyanin pigmentation of aleurone in the grain apex; anthocyanin pigmentation of plant organs | [50,165,205,223] |
| R-st (Red-stippled) ** | Functional allele of Sc (Self-coloured) gene with insertion of mobile element I-R (Inhibitor of R) into exon 7; from 0 to 3 functional or non-functional alleles of Nc (Near-colorless) genes | Small spots of colored aleurone on a white background with variable anthocyanin pigmentation, increasing with a rise in the number of functional copies of the Nc genes; variable anthocyanin pigmentation of plant organs | [50,165,204,205,206,224,225] |
| R-sc (self-coloured aleurone) ** | Functional allele of Sc gene (Self-coloured); from 0 to 3 functional or non-functional alleles of Nc genes | Uniform anthocyanin pigmentation of aleurone; variable anthocyanin pigmentation of plant organs | |
| Nc3 (Near-colorless 3) | Functional allele of Nc3 gene | Uniform weak anthocyanin pigmentation of aleurone; lack of pigmentation of plant organs | |
| R-mb (marbled color) | Functional allele of Scm (Self-coloured marbled) gene with insertion of YL1 mobile element into promoter | “Marbled” anthocyanin pigmentation of aleurone with large spots of anthocyanin pigmentation on a white background of plant organs | [50,165,205] |
| R-scm (self-coloured marbled) | Functional allele of Scm (Self-coloured marbled) gene | Strong uniform anthocyanin pigmentation of aleurone, scutellum and plant organs |
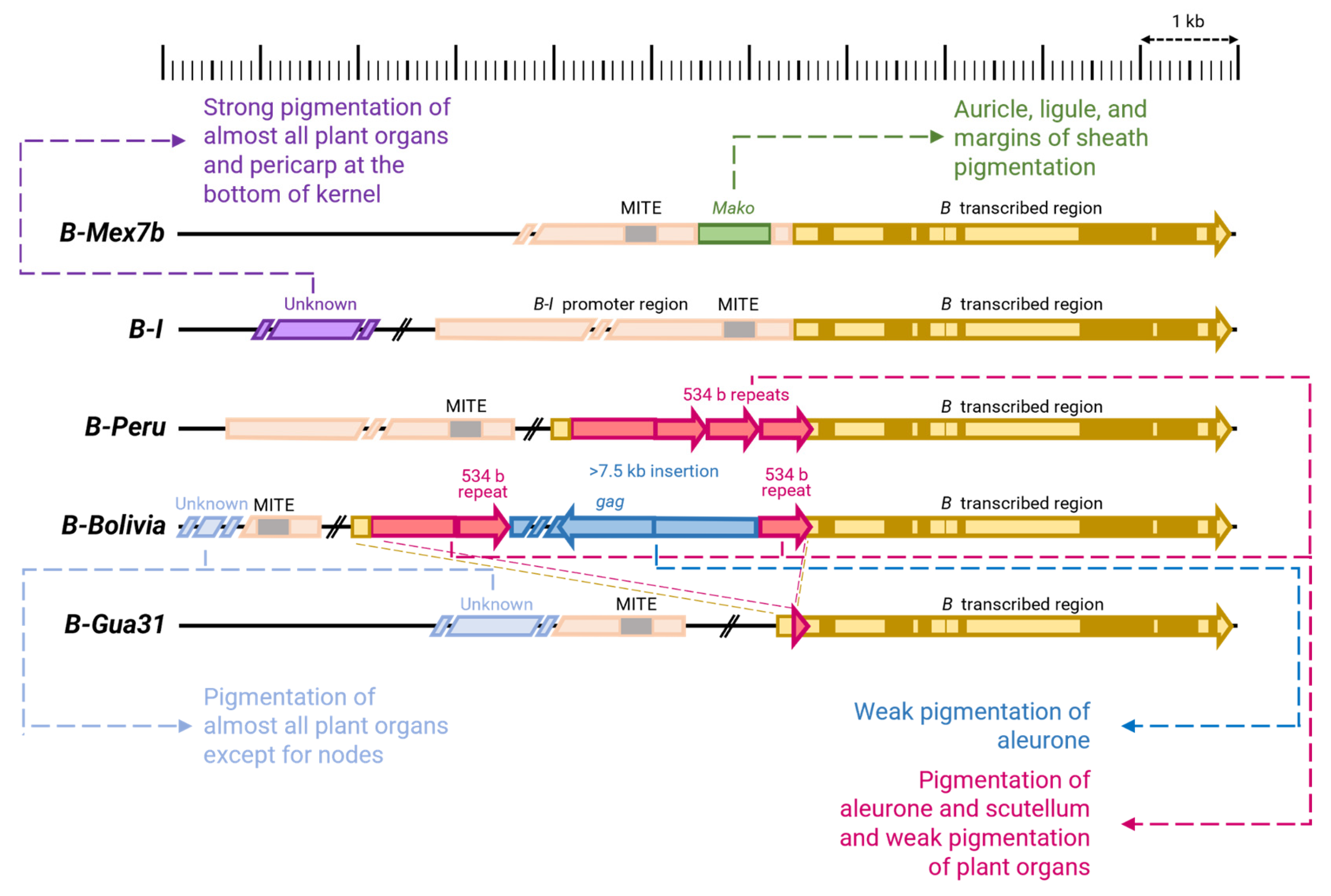
| Allele | Structure of Allele * | Phenotype | References |
|---|---|---|---|
| B-I (Intense color) | An unknown far upstream regulatory element is present, causing strong expression | Strong anthocyanin pigmentation of leaf sheaths, auricles, ligula, stem, panicle, cob husks and pericarp at the bottom of the kernels and weak pigmentation of leaves | [54,168,214,226] |
| B-Mex7b | The 5′UTR of the gene is similar in sequence to B-I, but an insertion of the Mako element, presumably a transposon, is present 220 bp upstream of the start codon, leading to weakening of gene expression | Anthocyanin pigmentation of auricles, ligula and margins of leaf sheaths | [54] |
| B-Peru | A 2.5 kb insertion in the 5 ′UTR with three tandem repeats of 534 bp results in the development of bright pigmentation in embryonic tissues | Weak anthocyanin pigmentation of nodes, stems, spotted pigmentation of leaf sheaths, panicles, ears, strong pigmentation of aleurone and scutellum | [25,54,156,167,168,214,226] |
| B-Bolivia | The same as in B-Peru, insertion with an additional insertion of a large (more than 7.5 kb) element with the retrotransposon sequence gag between two 534 bp repeats with the loss of the third one, which leads to weakening of the pigmentation of the embryonic tissues. An additional unknown regulatory element is present, promoting the pigmentation of plant organs. | Anthocyanin pigmentation of stems, leaf sheaths, panicles, cob sheaths, weak pigmentation of aleurone and scutellum | [25,54,214] |
| B-Gua31 | Presumably, the same as in B-bolivia, unknown regulatory element is present, promoting the pigmentation of plant organs | Anthocyanin pigmentation of stems, leaf sheaths, panicles, and cob husks | [25,54] |
4.4.2. Diversity of MBW Genes in Rice (Oryza sativa L.)
| Family | Gene (Locus) | Chromosome | Function | MBW Complex Formation | Note | References |
|---|---|---|---|---|---|---|
| MYC-like bHLH | OsPl (Purple leaf) | 4 | Positive regulator of anthocyanin pigmentation of leaf blades, sheaths and auricles, ligula, nodes, internodes, pericarp, stigma and glume tips, depending on the allele | Yes, with OsC1 and OsPAC1 | Contains two MYC genes with different expression patterns—OsB1 and OsB2 (Booster 1, 2). OsB2 activates the expression of its MYB partner gene OsC1, and in combination with it, activates the expression of the WDR partner OsPAC1 | [53,181,234,235,236,238,248] |
| OsPa (Purple apiculi) | 4 | Positive regulator of anthocyanin pigmentation of glume tips | Yes, with OsC1 and, probably, OsPAC1 | - | [238,239] | |
| OsPSH1 (Purple leaf sheath 1) | 1 | Positive regulator of anthocyanin pigmentation of leaf sheaths | Yes, with OsC1 and OsPAC1 | Contains two MYC genes, OsRb1 and OsRb2, that jointly regulate anthocyanin biosynthesis in leaf sheaths | [236,240] | |
| OsRc (Red c) | 7 | Positive regulator of proanthocyanidin biosynthesis in outer grain hulls | Possibly, with Kala3 | Ortholog of ZmIN1, however, performs the opposite function. | [235,241,252] | |
| R2R3-MYB | OsC1 (Colorless 1) | 6 | Positive regulator of anthocyanin pigmentation of almost all tissues and organs, except pericarp | Yes, with R/B family and OsPAC1 | Expression is activated by OsB2, which, in combination with it, activates the expression of the WDR partner OsPAC1 | [53,132,238,239,240,246,247,248] |
| OsKala3 (Key gene for black coloration by anthocyanin accumulation on chromosome 3) | 3 | Positive regulator of anthocyanin pigmentation of pericarp | Yes, with OsB2 and, probably, OsPAC1 | Combines the properties of R2R3-MYB transcription factors of both subgroups 5 and 6 | [127,181] | |
| OsPL (Purple Leaf) | 5 | Positive regulator of anthocyanin pigmentation of leaf blades, sheaths and stems | Unknown | - | [243] | |
| OsPL6 (Purple Leaf 6) | 6 | Positive regulator of anthocyanin pigmentation of leaf blades, including leaf midrib | Unknown | - | [244] | |
| OsP1 (Pericarp color 1) | 3 | Positive regulator of anthocyanin biosynthesis in leaves | No | Activates the expression of early genes of flavonoid biosynthesis independently of MYC proteins | [132] | |
| WDR | OsPAC1/TTG1/WA1 (Pale aleurone color 1/Transparent Testa Glabra 1/WD40 for Anthocyanin biosynthesis) | 2 | Positive regulator of anthocyanin pigmentation of almost all plant tissues | Yes, with R/B family and OsC1 | Expression is activated by the OsB2-OsC1 complex | [44,132,240,249] |
4.4.3. Diversity of MBW Genes in Barley (Hordeum vulgare L.)
| Family | Gene (Locus) | Chromosome | Function | MBW Complex Formation | Note | References |
|---|---|---|---|---|---|---|
| MYC-like bHLH | HvANT2/HvRc/HvMyc1 (Anthocyaninless 2/Red coleoptile) | 2H | Positive regulator of anthocyanin pigmentation of coleoptile, leaf sheaths and auricles, lemma, awns and pericarp | Yes, with HvANT1 and HvAnt13 | Mutually activates the expression of HvANT1 | [45,255,258,260,261,262] |
| HvMyc2 | 4H | Positive regulator of anthocyanin pigmentation of aleurone | Yes, with HvMpc1-2 and, probably, HvAnt13 | Closely colocalized with HvMpc2 and HvF3′5′H at the Blx1 locus and form a coordinately regulated unit with them | [75,255] | |
| R2R3-MYB | HvANT28/HvR-1/HvMyb10 (Anthocyaninless 28/Red 1) | 3H | Positive regulator of proanthocyanidin biosynthesis in outer grain coats | Possibly, no | - | [62,256] |
| HvANT1/HvMpc1-1 (Anthocyaninless 1/MYB protein c1 1) | 7H | Positive regulator of anthocyanin pigmentation of coleoptile, leaf sheaths and auricles, lemma, awns and pericarp | Yes, with HvANT2 and HvAnt13 | Mutually activates the expression of HvANT2 | [253,254,255,257,258] | |
| HvMpc1-2/HvMpc2/HvPL2 (MYB protein c1 2/Purple Leaf 2) | 4H | Positive regulator of anthocyanin pigmentation of lemma and aleurone | Yes, with HvMyc2 and, probably, HvAnt13 | Closely colocalizes with HvMyc2 and HvF3′5′H at the Blx1 locus and form a coordinately regulated unit with them. Likely represses HvMpc1-3 expression in the lemma but activates it in the aleurone | [57,75,255,264] | |
| HvMpc1-3 (MYB protein c1 3) | 4H | Positive regulator of anthocyanin pigmentation of aleurone | Unknown | Expression is repressed in the lemma but activated in the aleurone. Likely is a coregulator of HvMpc2 in the aleurone and regulated by it | [57] | |
| WDR | HvAnt13/HvWD40-140 (Anthocyaninless 13) | 6H | Positive regulator of flavonoid biosynthesis in aleurone, pericarp, lemma, stem and leaf sheaths | Yes, with HvANT1 and HvANT2, and, probably, with other Mpc1 and Myc proteins | Exhibits pleiotropic effects in plant growth | [245,255,263] |
4.4.4. Diversity of MBW Genes in Wheat (Triticum aestivum L.)
| Family | Gene (Locus) | Chromosome | Function | MBW Complex Formation | Note | References |
|---|---|---|---|---|---|---|
| MYC-like bHLH | TaPp-3/TaMyc-A1/TaMyc1.1/TaPpb1 (Purple pericarp 3/Purple pericarp bHLH 1) | 2A | Positive regulator of anthocyanin pigmentation in pericarp, coleoptile, leaf auricles and glumes | Yes, with TaPp-D1 and, probably, TaPp-A1 and TaPp-B1 | Expression is higher in the presence of the functional allele of TaPp-A1 than in the presence of both TaPp-A1 and TaPp-D1. In complex with TaPp-D1 activates expression of TaPp-D1. | [258,259,270,274,275] |
| TaMyc-A2/TaMyc1.2 | 2A | Probably a positive regulator of anthocyanin pigmentation in the coleoptile | Probably, with TaPp-1 and TaMpc1-D4 | Probably, is coregulator of TaPp-1-TaPp-3 complexes, potentially activating the biosynthesis of uncolored anthocyanin precursors in association with TaMpc1-D4 | [55,57] | |
| TaMyc-B1/TaMyc1.3 | 2B | Probably a positive regulator of anthocyanin pigmentation in the coleoptile | Probably, with TaPp-1 | Expression is increased in the presence of the functional allele of TaPp-A1. Potential coregulator of TaPp-3 | [55,57] | |
| TaMYC4D/TaMyc2.6 | 4D | Potential positive regulator of anthocyanin pigmentation of aleurone | Possibly, with TaMYB4D | Closely colocalizes with TaMYB4D and TaF3′5′H at the Ba1 (Blue aleurone 1) locus | [75,255] | |
| R2R3-MYB | TaR-1/TaMyb10 (Red 1/MYB protein c 10) | 3 | Positive regulator of proanthocyanidin biosynthesis in outer grain coats | Possibly, no | - | [62,256,276] |
| TaPp-A1/TaRc-A1/TaMpc1-A1/TaC1-A1 (A genome Red coleptile 1/Purple pericarp 1/MYB protein c1-1/Colorless 1) | 7A | Positive regulator of anthocyanin pigmentation in pericarp and coleoptile | Probably, with TaPp-3 and TaMyc-B1 | Increases TaMyc-B1 expression in coleoptile. Probably shares redundant functions with TaPp-D1. | [56,253,258,259,269,270,272,274,275] | |
| TaPp-B1/TaRc-B1/TaMpc1-B1/TaC-B1 (B genome Purple pericarp 1/Red coleptile 1/MYB protein c1-1/Colorless1-1) | 7B | Potential positive regulator of anthocyanin pigmentation in coleoptile and pericarp | Probably, with TaPp-3 | Expression significantly increases in the presence of the dominant TaPp-A1 allele. Potential coregulator of TaPp-A1 and TaPp-D1. | [56,253] | |
| TaPp-D1/TaRc-D1/TaMpc1-D1/TaC-D1/TaPpm1 (Purple pericarp MYB 1) | 7D | Positive regulator of anthocyanin pigmentation in coleoptile and pericarp | Yes, with TaPp-3 | Reduces TaPp-3 expression in pericarp. In complex with TaPp-3 activates its own expression. Probably shares redundant functions with TaPp-A1. | [56,253,258,259,269,275] | |
| TaMpc1-A2 (A genome MYB protein c1-1) | 5A | Probably, positive regulator of anthocyanin pigmentation in coleoptile and pericarp | Probably, with TaPp-3 | Probably, is coregulator of TaPp-1 genes, potentially activating the biosynthesis of uncolored anthocyanin precursors in association with TaPp-3 | [56] | |
| TaMpc1-D4/TaPL1-4D2 (D genome MYB protein c1-4/Purple Plant 1-4) | 4D | Probably, positive regulator of anthocyanin pigmentation in coleoptile and pericarp | Probably, with TaPp-3 and TaMyc-A2 | Probably, is coregulator of TaPp-1 genes, potentially activating the biosynthesis of uncolored anthocyanin precursors in association with TaPp-3 or TaMyc-A2 | [56,264] | |
| TaMYB4D/TaMpc1-D3/TaPL1–4D3 (D genome MYB protein c1-3/Purple Plant 1-3) | 4D | Potential positive regulator of anthocyanin pigmentation of aleurone | Possibly, with TaMY4D | Closely colocalizes with TaMYC4D and TaF3′5′H at the Ba1 (Blue aleurone 1) locus | [75,255] |
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ingham, J. A New Isoflavanone Phytoalexin from Medicago Rugosa. Planta Med. 1982, 45, 46–47. [Google Scholar] [CrossRef] [PubMed]
- Kodama, O.; Miyakawa, J.; Akatsuka, T.; Kiyosawa, S. Sakuranetin, a Flavanone Phytoalexin from Ultraviolet-Irradiated Rice Leaves. Phytochemistry 1992, 31, 3807–3809. [Google Scholar] [CrossRef]
- Weinstein, L.I.; Albersheim, P. Host-Pathogen Interactions: XXIII. The Mechanism of the Antibacterial Action of Glycinol, a Pterocarpan Phytoalexin Synthesized by Soybeans. Plant Physiol. 1983, 72, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Henry-Kirk, R.A.; Plunkett, B.; Hall, M.; McGhie, T.; Allan, A.C.; Wargent, J.J.; Espley, R.V. Solar UV Light Regulates Flavonoid Metabolism in Apple (Malus × Domestica). Plant Cell Environ. 2018, 41, 675–688. [Google Scholar] [CrossRef]
- Li, J.; Ou-Lee, T.-M.; Raba, R.; Amundson, R.G.; Last, R.L. Arabidopsis Flavonoid Mutants Are Hypersensitive to UV-B Irradiation. Plant Cell 1993, 5, 171. [Google Scholar] [CrossRef]
- Liu, Y.; Fang, S.; Yang, W.; Shang, X.; Fu, X. Light Quality Affects Flavonoid Production and Related Gene Expression in Cyclocarya Paliurus. J. Photochem. Photobiol. B 2018, 179, 66–73. [Google Scholar] [CrossRef]
- Merzlyak, M.N.; Chivkunova, O.B.; Solovchenko, A.E.; Naqvi, K.R. Light Absorption by Anthocyanins in Juvenile, Stressed, and Senescing Leaves. J. Exp. Bot. 2008, 59, 3903–3911. [Google Scholar] [CrossRef]
- Alappat, B.; Alappat, J. Anthocyanin Pigments: Beyond Aesthetics. Molecules 2020, 25, 5500. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of Plant Pigments: Anthocyanins, Betalains and Carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Flavonoid Biosynthesis. A Colorful Model for Genetics, Biochemistry, Cell Biology, and Biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef]
- Maj, D.; Wielbo, J.; Marek-Kozaczuk, M.; Skorupska, A. Response to Flavonoids as a Factor Influencing Competitiveness and Symbiotic Activity of Rhizobium leguminosarum. Microbiol. Res. 2010, 165, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Baudry, A.; Heim, M.A.; Dubreucq, B.; Caboche, M.; Weisshaar, B.; Lepiniec, L. TT2, TT8, and TTG1 Synergistically Specify the Expression of BANYULS and Proanthocyanidin Biosynthesis in Arabidopsis thaliana. Plant J. 2004, 39, 366–380. [Google Scholar] [CrossRef] [PubMed]
- Payne, C.T.; Zhang, F.; Lloyd, A.M. GL3 Encodes a bHLH Protein That Regulates Trichome Development in Arabidopsis through Interaction with GL1 and TTG1. Genetics 2000, 156, 1349–1362. [Google Scholar] [CrossRef]
- Zhang, F.; Gonzalez, A.; Zhao, M.; Payne, C.T.; Lloyd, A. A Network of Redundant bHLH Proteins Functions in All TTG1-Dependent Pathways of Arabidopsis. Development 2003, 130, 4859–4869. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, S.R.; Habera, L.F.; Dellaporta, S.L.; Wessler, S.R. Lc, a Member of the Maize R Gene Family Responsible for Tissue-Specific Anthocyanin Production, Encodes a Protein Similar to Transcriptional Activators and Contains the Myc-Homology Region. Proc. Natl. Acad. Sci. USA 1989, 86, 7092–7096. [Google Scholar] [CrossRef]
- Paz-Ares, J.; Wienand, U.; Peterson, P.A.; Saedler, H. Molecular Cloning of the c Locus of Zea mays: A Locus Regulating the Anthocyanin Pathway. Eur. Mol. Biol. Organ. J. 1986, 5, 829–833. [Google Scholar] [CrossRef]
- Pires, N.; Dolan, L. Origin and Diversification of Basic-Helix-Loop-Helix Proteins in Plants. Mol. Biol. Evol. 2010, 27, 862–874. [Google Scholar] [CrossRef]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB Gene Family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Zhang, B.; Hülskamp, M. Evolutionary Analysis of MBW Function by Phenotypic Rescue in Arabidopsis thaliana. Front. Plant Sci. 2019, 10, 375. [Google Scholar] [CrossRef]
- Zimmermann, I.M.; Heim, M.A.; Weisshaar, B.; Uhrig, J.F. Comprehensive Identification of Arabidopsis thaliana MYB Transcription Factors Interacting with R/B-like BHLH Proteins. Plant J. 2004, 40, 22–34. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the Anthocyanin Biosynthetic Pathway by the TTG1/bHLH/Myb Transcriptional Complex in Arabidopsis Seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Paz-Ares, J.; Ghosal, D.; Wienand, U.; Peterson, P.A.; Saedler, H. The Regulatory C1 Locus of Zea mays Encodes a Protein with Homology to Myb Proto-Oncogene Products and with Structural Similarities to Transcriptional Activators. Eur. Mol. Biol. Organ. J. 1987, 6, 3553–3558. [Google Scholar] [CrossRef] [PubMed]
- Petroni, K.; Cominelli, E.; Consonni, G.; Gusmaroli, G.; Gavazzi, G.; Tonelli, C. The Developmental Expression of the Maize Regulatory Gene Hopi Determines Germination-Dependent Anthocyanin Accumulation. Genetics 2000, 155, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Procissi, A.; Dolfini, S.; Ronchi, A.; Tonelli, C. Light-Dependent Spatial and Temporal Expression of Pigment Regulatory Genes in Developing Maize Seeds. Plant Cell 1997, 9, 1547–1557. [Google Scholar] [CrossRef]
- Selinger, D.A.; Chandler, V.L. B-Bolivia, an Allele of the Maize B1 Gene with Variable Expression, Contains a High Copy Retrotransposon-Related Sequence Immediately Upstream. Plant Physiol. 2001, 125, 1363–1379. [Google Scholar] [CrossRef]
- Carey, C.C.; Strahle, J.T.; Selinger, D.A.; Chandler, V.L. Mutations in the Pale Aleurone Color1 Regulatory Gene of the Zea mays Anthocyanin Pathway Have Distinct Phenotypes Relative to the Functionally Similar TRANSPARENT TESTA GLABRA1 Gene in Arabidopsis thaliana. Plant Cell 2004, 16, 450–464. [Google Scholar] [CrossRef]
- Pesch, M.; Schultheiß, I.; Klopffleisch, K.; Uhrig, J.F.; Koegl, M.; Clemen, C.S.; Simon, R.; Weidtkamp-Peters, S.; Hülskamp, M. TRANSPARENT TESTA GLABRA1 and GLABRA1 Compete for Binding to GLABRA3 in Arabidopsis. Plant Physiol. 2015, 168, 584–597. [Google Scholar] [CrossRef]
- Zhang, B.; Chopra, D.; Schrader, A.; Hülskamp, M. Evolutionary Comparison of Competitive Protein-Complex Formation of MYB, bHLH, and WDR Proteins in Plants. J. Exp. Bot. 2019, 70, 3197–3209. [Google Scholar] [CrossRef]
- Muhlemann, J.K.; Younts, T.L.B.; Muday, G.K. Flavonols Control Pollen Tube Growth and Integrity by Regulating ROS Homeostasis during High-Temperature Stress. Proc. Natl. Acad. Sci. USA 2018, 115, E11188–E11197. [Google Scholar] [CrossRef]
- Wang, L.; Ying Lam, L.P.; Lui, A.C.W.; Zhu, F.-Y.; Chen, M.-X.; Liu, H.; Zhang, J.; Lo, C. Flavonoids Are Indispensable for Complete Male Fertility in Rice. J. Exp. Bot. 2020, 71, 4715–4728. [Google Scholar] [CrossRef]
- Agarwal, P.; Holland, T.M.; Wang, Y.; Bennett, D.A.; Morris, M.C. Association of Strawberries and Anthocyanidin Intake with Alzheimer’s Dementia Risk. Nutrients 2019, 11, 3060. [Google Scholar] [CrossRef] [PubMed]
- Arora, A.; Byrem, T.M.; Nair, M.G.; Strasburg, G.M. Modulation of Liposomal Membrane Fluidity by Flavonoids and Isoflavonoids. Arch. Biochem. Biophys. 2000, 373, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Cotelle, N. Antioxidant Properties of Hydroxy-Flavones. Free Radic. Biol. Med. 1996, 20, 35–43. [Google Scholar] [CrossRef]
- Jovanovic, S.V.; Steenken, S.; Tosic, M.; Marjanovic, B.; Simic, M.G. Flavonoids as Antioxidants. J. Am. Chem. Soc. 1994, 116, 4846–4851. [Google Scholar] [CrossRef]
- Markovics, A.; Biró, A.; Kun-Nemes, A.; Fazekas, M.É.; Rácz, A.A.; Paholcsek, M.; Lukács, J.; Stündl, L.; Remenyik, J. Effect of Anthocyanin-Rich Extract of Sour Cherry for Hyperglycemia-Induced Inflammatory Response and Impaired Endothelium-Dependent Vasodilation. Nutrients 2020, 12, 3373. [Google Scholar] [CrossRef]
- Mattioli, R.; Francioso, A.; Mosca, L.; Silva, P. Anthocyanins: A Comprehensive Review of Their Chemical Properties and Health Effects on Cardiovascular and Neurodegenerative Diseases. Molecules 2020, 25, 3809. [Google Scholar] [CrossRef]
- Rice-Evans, C.A.; Miller, N.J.; Paganga, G. Structure-Antioxidant Activity Relationships of Flavonoids and Phenolic Acids. Free Radic. Biol. Med. 1996, 20, 933–956. [Google Scholar] [CrossRef]
- Abdel-Aal, E.-S.M.; Young, J.C.; Rabalski, I. Anthocyanin Composition in Black, Blue, Pink, Purple, and Red Cereal Grains. J. Agric. Food Chem. 2006, 54, 4696–4704. [Google Scholar] [CrossRef]
- Birla, D.S.; Malik, K.; Sainger, M.; Chaudhary, D.; Jaiwal, R.; Jaiwal, P.K. Progress and Challenges in Improving the Nutritional Quality of Rice (Oryza sativa L.). Crit. Rev. Food Sci. Nutr. 2017, 57, 2455–2481. [Google Scholar] [CrossRef]
- Sang, T.; Li, J. Molecular Genetic Basis of the Domestication Syndrome in Cereals. In Cereal Genomics II; Springer: Dordrecht, The Netherlands, 2013; pp. 319–340. ISBN 9789400764002. [Google Scholar]
- Shewry, P.R.; Hawkesford, M.J.; Piironen, V.; Lampi, A.-M.; Gebruers, K.; Boros, D.; Andersson, A.A.M.; Åman, P.; Rakszegi, M.; Bedo, Z.; et al. Natural Variation in Grain Composition of Wheat and Related Cereals. J. Agric. Food Chem. 2013, 61, 8295–8303. [Google Scholar] [CrossRef]
- Zykin, P.A.; Andreeva, E.A.; Lykholay, A.N.; Tsvetkova, N.V.; Voylokov, A.V. Anthocyanin Composition and Content in Rye Plants with Different Grain Color. Molecules 2018, 23, 948. [Google Scholar] [CrossRef] [PubMed]
- Casas, M.I.; Duarte, S.; Doseff, A.I.; Grotewold, E. Flavone-Rich Maize: An Opportunity to Improve the Nutritional Value of an Important Commodity Crop. Front. Plant Sci. 2014, 5, 440. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhang, Z.; Li, J.; Zhang, H.; Peng, Y.; Li, Z. Uncovering Hierarchical Regulation among MYB-bHLH-WD40 Proteins and Manipulating Anthocyanin Pigmentation in Rice. Int. J. Mol. Sci. 2022, 23, 8203. [Google Scholar] [CrossRef]
- Zhou, C.; Zeng, Z.; Suo, J.; Li, X.; Bian, H.; Wang, J.; Zhu, M.; Han, N. Manipulating a Single Transcription Factor, Ant1, Promotes Anthocyanin Accumulation in Barley Grains. J. Agric. Food Chem. 2021, 69, 5306–5317. [Google Scholar] [CrossRef]
- Casas, M.I.; Falcone-Ferreyra, M.L.; Jiang, N.; Mejía-Guerra, M.K.; Rodríguez, E.; Wilson, T.; Engelmeier, J.; Casati, P.; Grotewold, E. Identification and Characterization of Maize Salmon Silks Genes Involved in Insecticidal Maysin Biosynthesis. Plant Cell 2016, 28, 1297–1309. [Google Scholar] [CrossRef]
- Cone, K.C.; Cocciolone, S.M.; Burr, F.A.; Burr, B. Maize Anthocyanin Regulatory Gene Pl Is a Duplicate of C1 That Functions in the Plant. Plant Cell 1993, 5, 1795–1805. [Google Scholar] [CrossRef]
- Harris, L.J.; Currie, K.; Chandler, V.L. Large Tandem Duplication Associated with a Mu2 Insertion in Zea mays B-Peru Gene. Plant Mol. Biol. 1994, 25, 817–828. [Google Scholar] [CrossRef]
- Hu, J.; Reddy, V.S.; Wessler, S.R. The rice R gene family: Two distinct subfamilies containing several miniature inverted-repeat transposable elements. Plant Mol. Biol. 2000, 42, 667–678. [Google Scholar] [CrossRef]
- Li, Y.; Bernot, J.P.; Illingworth, C.; Lison, W.; Bernot, K.M.; Eggleston, W.B.; Fogle, K.J.; DiPaola, J.E.; Kermicle, J.; Alleman, M. Gene Conversion within Regulatory Sequences Generates Maize r Alleles with Altered Gene Expression. Genetics 2001, 159, 1727–1740. [Google Scholar] [CrossRef]
- Ludwig, S.R.; Wessler, S.R. Maize R Gene Family: Tissue-Specific Helix-Loop-Helix Proteins. Cell 1990, 62, 849–851. [Google Scholar] [CrossRef]
- Robbins, T.P.; Walker, E.L.; Kermicle, J.L.; Alleman, M.; Dellaporta, S.L. Meiotic Instability of the R-r Complex Arising from Displaced Intragenic Exchange and Intrachromosomal Rearrangement. Genetics 1991, 129, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, W.; Ohmori, T.; Kageyama, K.; Miyazaki, C.; Saito, A.; Murata, M.; Noda, K.; Maekawa, M. The Purple Leaf (Pl) Locus of Rice: The Plw Allele Has a Complex Organization and Includes Two Genes Encoding Basic Helix-Loop-Helix Proteins Involved in Anthocyanin Biosynthesis. Plant Cell Physiol. 2001, 42, 982–991. [Google Scholar] [CrossRef] [PubMed]
- Selinger, D.A.; Chandler, V.L. Major Recent and Independent Changes in Levels and Patterns of Expression Have Occurred at the b Gene, a Regulatory Locus in Maize. Proc. Natl. Acad. Sci. USA 1999, 96, 15007–15012. [Google Scholar] [CrossRef] [PubMed]
- Strygina, K.V.; Khlestkina, E.K. MYC Gene Family in Cereals: Transformations during Evolution of Hexaploid Bread Wheat and Its Relatives. Mol. Biol. 2017, 51, 674–680. [Google Scholar] [CrossRef]
- Strygina, K.V.; Khlestkina, E.K. Structural and Functional Divergence of the Mpc1 Genes in Wheat and Barley. BMC Evol. Biol. 2019, 19, 45. [Google Scholar] [CrossRef]
- Strygina, K.V.; Khlestkina, E.K. Myc-like Transcriptional Factors in Wheat: Structural and Functional Organization of the Subfamily I Members. BMC Plant Biol. 2019, 19, 50. [Google Scholar] [CrossRef]
- Williams, C.A.; Grayer, R.J. Anthocyanins and Other Flavonoids. Nat. Prod. Rep. 2004, 21, 539. [Google Scholar] [CrossRef]
- Pourcel, L.; Routaboul, J.; Cheynier, V.; Lepiniec, L.; Debeaujon, I. Flavonoid Oxidation in Plants: From Biochemical Properties to Physiological Functions. Trends Plant Sci. 2007, 12, 29–36. [Google Scholar] [CrossRef]
- Dixon, R.A.; Xie, D.-Y.; Sharma, S.B. Proanthocyanidins—A Final Frontier in Flavonoid Research? New Phytol. 2005, 165, 9–28. [Google Scholar] [CrossRef]
- Vaughan, S.P.; Baker, J.M.; Primavesi, L.F.; Patil, A.; King, R.; Hassani-Pak, K.; Kulasekaran, S.; Coghill, J.; Ward, J.L.; Huttly, A.K.; et al. Proanthocyanidin Biosynthesis in the Developing Wheat Seed Coat Investigated by Chemical and RNA-Seq Analysis. Plant Direct 2022, 6, e453. [Google Scholar] [CrossRef]
- Himi, E.; Maekawa, M.; Miura, H.; Noda, K. Development of PCR Markers for Tamyb10 Related to R-1, Red Grain Color Gene in Wheat. Züchter Genet. Breed. Res. 2011, 122, 1561–1576. [Google Scholar] [CrossRef] [PubMed]
- Francavilla, A.; Joye, I.J. Anthocyanins in Whole Grain Cereals and Their Potential Effect on Health. Nutrients 2020, 12, 2922. [Google Scholar] [CrossRef] [PubMed]
- Grotewold, E.; Athma, P.; Peterson, T. Alternatively Spliced Products of the Maize P Gene Encode Proteins with Homology to the DNA-Binding Domain of Myb-like Transcription Factors. Proc. Natl. Acad. Sci. USA 1991, 88, 4587–4591. [Google Scholar] [CrossRef] [PubMed]
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and Anthocyanins: Colored Pigments as Food, Pharmaceutical Ingredients, and the Potential Health Benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef] [PubMed]
- Saigo, T.; Wang, T.; Watanabe, M.; Tohge, T. Diversity of Anthocyanin and Proanthocyanin Biosynthesis in Land Plants. Curr. Opin. Plant Biol. 2020, 55, 93–99. [Google Scholar] [CrossRef]
- Alfenito, M.R.; Souer, E.; Goodman, C.D.; Buell, R.; Mol, J.; Koes, R.; Walbot, V. Functional Complementation of Anthocyanin Sequestration in the Vacuole by Widely Divergent Glutathione S-Transferases. Plant Cell 1998, 10, 1135–1149. [Google Scholar] [CrossRef]
- Jiang, S.; Chen, M.; He, N.; Chen, X.; Wang, N.; Sun, Q.; Zhang, T.; Xu, H.; Fang, H.; Wang, Y.; et al. MdGSTF6, Activated by MdMYB1, Plays an Essential Role in Anthocyanin Accumulation in Apple. Hortic. Res. 2019, 6, 40. [Google Scholar] [CrossRef]
- Kou, M.; Liu, Y.-J.; Li, Z.-Y.; Zhang, Y.-G.; Tang, W.; Yan, H.; Wang, X.; Chen, X.-G.; Su, Z.-X.; Arisha, M.H.; et al. A Novel Glutathione S-Transferase Gene from Sweetpotato, IbGSTF4, Is Involved in Anthocyanin Sequestration. Plant Physiol. Biochem. 2019, 135, 395–403. [Google Scholar] [CrossRef]
- Sasaki, N.; Nakayama, T. Achievements and Perspectives in Biochemistry Concerning Anthocyanin Modification for Blue Flower Coloration. Plant Cell Physiol. 2015, 56, 28–40. [Google Scholar] [CrossRef]
- Petroni, K.; Pilu, R.; Tonelli, C. Anthocyanins in Corn: A Wealth of Genes for Human Health. Planta 2014, 240, 901–911. [Google Scholar] [CrossRef]
- Dedio, W.; Hill, R.D.; Evans, L.E. Anthocyanins in the Pericarp and Coleoptiles of Purple-Seeded Rye. Can. J. Plant Sci. 1972, 52, 981–983. [Google Scholar] [CrossRef][Green Version]
- Glagoleva, A.; Kukoeva, T.; Mursalimov, S.; Khlestkina, E.; Shoeva, O. Effects of Combining the Genes Controlling Anthocyanin and Melanin Synthesis in the Barley Grain on Pigment Accumulation and Plant Development. Agronomy 2022, 12, 112. [Google Scholar] [CrossRef]
- Gordeeva, E.; Shoeva, O.; Mursalimov, S.; Adonina, I.; Khlestkina, E. Fine Points of Marker-Assisted Pyramiding of Anthocyanin Biosynthesis Regulatory Genes for the Creation of Black-Grained Bread Wheat (Triticum aestivum L.) Lines. Agronomy 2022, 12, 2934. [Google Scholar] [CrossRef]
- Jia, Y.; Selva, C.; Zhang, Y.; Li, B.; McFawn, L.A.; Broughton, S.; Zhang, X.; Westcott, S.; Wang, P.; Tan, C.; et al. Uncovering the Evolutionary Origin of Blue Anthocyanins in Cereal Grains. Plant J. 2020, 101, 1057–1074. [Google Scholar] [CrossRef]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive Oxygen Species, Oxidative Damage, and Antioxidative Defense Mechanism in Plants under Stressful Conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef]
- Ben Abdallah, S.; Aung, B.; Amyot, L.; Lalin, I.; Lachâal, M.; Karray-Bouraoui, N.; Hannoufa, A. Salt Stress (NaCl) Affects Plant Growth and Branch Pathways of Carotenoid and Flavonoid Biosyntheses in Solanum nigrum. Acta Physiol. Plant 2016, 38, 72. [Google Scholar] [CrossRef]
- Borghesi, E.; González-Miret, M.L.; Escudero-Gilete, M.L.; Malorgio, F.; Heredia, F.J.; Meléndez-Martínez, A.J. Effects of Salinity Stress on Carotenoids, Anthocyanins, and Color of Diverse Tomato Genotypes. J. Agric. Food Chem. 2011, 59, 11676–11682. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of Oxidative and Drought Tolerance in Arabidopsis by Overaccumulation of Antioxidant Flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef]
- Ouhibi, C.; Attia, H.; Rebah, F.; Msilini, N.; Chebbi, M.; Aarrouf, J.; Urban, L.; Lachaal, M. Salt Stress Mitigation by Seed Priming with UV-C in Lettuce Plants: Growth, Antioxidant Activity and Phenolic Compounds. Plant Physiol. Biochem. 2014, 83, 126–133. [Google Scholar] [CrossRef]
- Petrussa, E.; Braidot, E.; Zancani, M.; Peresson, C.; Bertolini, A.; Patui, S.; Vianello, A. Plant Flavonoids—Biosynthesis, Transport and Involvement in Stress Responses. Int. J. Mol. Sci. 2013, 14, 14950–14973. [Google Scholar] [CrossRef]
- Schenke, D.; Utami, H.P.; Zhou, Z.; Gallegos, M.-T.; Cai, D. Suppression of UV-B Stress Induced Flavonoids by Biotic Stress: Is There Reciprocal Crosstalk? Plant Physiol. Biochem. 2019, 134, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, K.; Matsuoka, H.; Hayashi, H. Isolation and Identification of a Phosphate Deficiency-Induced C-Glycosylflavonoid That Stimulates Arbuscular Mycorrhiza Formation in Melon Roots. Mol. Plant. Microbe. Interact. 2002, 15, 334–340. [Google Scholar] [CrossRef]
- Lagrange, H.; Jay-Allgmand, C.; Lapeyrie, F. Rutin, the Phenolglycoside from Eucalyptus Root Exudates, Stimulates Pisolithus Hyphal Growth at Picomolar Concentrations. New Phytol. 2001, 149, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Nair, M.G.; Safir, G.R.; Siqueira, J.O. Isolation and Identification of Vesicular-Arbuscular Mycorrhiza-Stimulatory Compounds from Clover (Trifolium Repens) Roots. Appl. Environ. Microbiol. 1991, 57, 434–439. [Google Scholar] [CrossRef]
- Ponce, M.A.; Scervino, J.M.; Erra-Balsells, R.; Ocampo, J.A.; Godeas, A.M. Flavonoids from Shoots and Roots of Trifolium Repens (White Clover) Grown in Presence or Absence of the Arbuscular Mycorrhizal Fungus Glomus Intraradices. Phytochemistry 2004, 65, 1925–1930. [Google Scholar] [CrossRef]
- Schliemann, W.; Ammer, C.; Strack, D. Metabolite Profiling of Mycorrhizal Roots of Medicago Truncatula. Phytochemistry 2008, 69, 112–146. [Google Scholar] [CrossRef]
- Tsai, S.M.; Phillips, D.A. Flavonoids Released Naturally from Alfalfa Promote Development of Symbiotic Glomus Spores in Vitro. Appl. Environ. Microbiol. 1991, 57, 1485–1488. [Google Scholar] [CrossRef]
- Hartwig, U.A.; Maxwell, C.A.; Joseph, C.M.; Phillips, D.A. Chrysoeriol and Luteolin Released from Alfalfa Seeds Induce Nod Genes in Rhizobium Meliloti. Plant Physiol. 1990, 92, 116–122. [Google Scholar] [CrossRef]
- Webster, G.; Jain, V.; Davey, M.R.; Gough, C.; Vasse, J.; Dénarié, J.; Cocking, E.C. The Flavonoid Naringenin Stimulates the Intercellular Colonization of Wheat Roots by Azorhizobium caulinodans. Plant Cell Environ. 1998, 21, 373–383. [Google Scholar] [CrossRef]
- de Castro, C.C.B.; Costa, P.S.; Laktin, G.T.; de Carvalho, P.H.D.; Geraldo, R.B.; de Moraes, J.; Pinto, P.L.S.; Couri, M.R.C.; Pinto, P.d.F.; Da Silva Filho, A.A. Cardamonin, a Schistosomicidal Chalcone from Piper aduncum L. (Piperaceae) That Inhibits Schistosoma Mansoni ATP Diphosphohydrolase. Phytomedicine 2015, 22, 921–928. [Google Scholar] [CrossRef]
- Shaik, A.; Bhandare, R.R.; Palleapati, K.; Nissankararao, S.; Kancharlapalli, V.; Shaik, S. Antimicrobial, Antioxidant, and Anticancer Activities of Some Novel Isoxazole Ring Containing Chalcone and Dihydropyrazole Derivatives. Molecules 2020, 25, 1047. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.-X.; Wang, S.; Wei, L.; Cui, Y.-Y.; Chen, Y.-H. Proanthocyanidins: Components, Pharmacokinetics and Biomedical Properties. Am. J. Chin. Med. 2020, 48, 813–869. [Google Scholar] [CrossRef] [PubMed]
- Mallikarjuna, N.; Kranthi, K.R.; Jadhav, D.R.; Kranthi, S.; Chandra, S. Influence of Foliar Chemical Compounds on the Development of Spodoptera litura (Fab.) in Interspecific Derivatives of Groundnut. J. Appl. Entomol. 2004, 128, 321–328. [Google Scholar] [CrossRef]
- Sosa, T.; Chaves, N.; Alias, J.C.; Escudero, J.C.; Henao, F.; Gutiérrez-Merino, C. Inhibition of Mouth Skeletal Muscle Relaxation by Flavonoids of Cistus ladanifer L.: A Plant Defense Mechanism against Herbivores. J. Chem. Ecol. 2004, 30, 1087–1101. [Google Scholar] [CrossRef]
- Star, A.E. Frond Exudate Flavonoids as Allelopathic Agents in Pityrogramma. Bull. Torrey Bot. Club 1980, 107, 146. [Google Scholar] [CrossRef]
- Zhang, Y.-C.; He, R.-R.; Lian, J.-P.; Zhou, Y.-F.; Zhang, F.; Li, Q.-F.; Yu, Y.; Feng, Y.-Z.; Yang, Y.-W.; Lei, M.-Q.; et al. OsmiR528 Regulates Rice-Pollen Intine Formation by Targeting an Uclacyanin to Influence Flavonoid Metabolism. Proc. Natl. Acad. Sci. USA 2020, 117, 727–732. [Google Scholar] [CrossRef]
- Bowtell, J.L.; Aboo-Bakkar, Z.; Conway, M.E.; Adlam, A.-L.R.; Fulford, J. Enhanced Task-Related Brain Activation and Resting Perfusion in Healthy Older Adults after Chronic Blueberry Supplementation. Appl. Physiol. Nutr. Metab. 2017, 42, 773–779. [Google Scholar] [CrossRef]
- Nomi, Y.; Iwasaki-Kurashige, K.; Matsumoto, H. Therapeutic Effects of Anthocyanins for Vision and Eye Health. Molecules 2019, 24, 3311. [Google Scholar] [CrossRef]
- Huang, W.-Y.; Wu, H.; Li, D.-J.; Song, J.-F.; Xiao, Y.-D.; Liu, C.-Q.; Zhou, J.-Z.; Sui, Z.-Q. Protective Effects of Blueberry Anthocyanins against H2O2-Induced Oxidative Injuries in Human Retinal Pigment Epithelial Cells. J. Agric. Food Chem. 2018, 66, 1638–1648. [Google Scholar] [CrossRef]
- Lee, S.; Keirsey, K.I.; Kirkland, R.; Grunewald, Z.I.; Fischer, J.G.; de La Serre, C.B. Blueberry Supplementation Influences the Gut Microbiota, Inflammation, and Insulin Resistance in High-Fat-Diet–Fed Rats. J. Nutr. 2018, 148, 209–219. [Google Scholar] [CrossRef]
- Muscarà, C.; Molonia, M.S.; Speciale, A.; Bashllari, R.; Cimino, F.; Occhiuto, C.; Saija, A.; Cristani, M. Anthocyanins Ameliorate Palmitate-induced Inflammation and Insulin Resistance in 3T3-L1 Adipocytes. Phytother. Res. 2019, 33, 1888–1897. [Google Scholar] [CrossRef] [PubMed]
- Le Phuong Nguyen, T.; Fenyvesi, F.; Remenyik, J.; Homoki, J.R.; Gogolák, P.; Bácskay, I.; Fehér, P.; Ujhelyi, Z.; Vasvári, G.; Vecsernyés, M.; et al. Protective Effect of Pure Sour Cherry Anthocyanin Extract on Cytokine-Induced Inflammatory Caco-2 Monolayers. Nutrients 2018, 10, 861. [Google Scholar] [CrossRef] [PubMed]
- Paramanantham, A.; Kim, M.J.; Jung, E.J.; Nagappan, A.; Yun, J.W.; Kim, H.J.; Shin, S.C.; Kim, G.S.; Lee, W.S. Pretreatment of Anthocyanin from the Fruit of Vitis Coignetiae Pulliat Acts as a Potent Inhibitor of TNF-α Effect by Inhibiting NF-κB-Regulated Genes in Human Breast Cancer Cells. Molecules 2020, 25, 2396. [Google Scholar] [CrossRef]
- Wang, X.; Yang, D.-Y.; Yang, L.-Q.; Zhao, W.-Z.; Cai, L.-Y.; Shi, H.-P. Anthocyanin Consumption and Risk of Colorectal Cancer: A Meta-Analysis of Observational Studies. J. Am. Coll. Nutr. 2019, 38, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Aschenbach, J.R.; Borau, T.; Gäbel, G. Glucose Uptake via SGLT-1 Is Stimulated by Β2-Adrenoceptors in the Ruminal Epithelium of Sheep. J. Nutr. 2002, 132, 1254–1257. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Dadmohammadi, Y.; Lee, M.C.; Abbaspourrad, A. Combination of Copigmentation and Encapsulation Strategies for the Synergistic Stabilization of Anthocyanins. Compr. Rev. Food Sci. Food Saf. 2021, 20, 3164–3191. [Google Scholar] [CrossRef]
- Rosler, J.; Krekel, F.; Amrhein, N.; Schmid, J. Maize Phenylalanine Ammonia-Lyase Has Tyrosine Ammonia-Lyase Activity. Plant Physiol. 1997, 113, 175–179. [Google Scholar] [CrossRef]
- Holton, T.A.; Cornish, E.C. Genetics and Biochemistry of Anthocyanin Biosynthesis. Plant Cell 1995, 7, 1071–1083. [Google Scholar] [CrossRef]
- Kreuzaler, F.; Ragg, H.; Fautz, E.; Kuhn, D.N.; Hahlbrock, K. UV-Induction of Chalcone Synthase mRNA in Cell Suspension Cultures of Petroselinum hortense. Proc. Natl. Acad. Sci. USA 1983, 80, 2591–2593. [Google Scholar] [CrossRef]
- Koes, R.E.; Spelt, C.E.; Mol, J.N.M. The Chalcone Synthase Multigene Family of Petunia hybrida (V30): Differential, Light-Regulated Expression during Flower Development and UV Light Induction. Plant Mol. Biol. 1989, 12, 213–225. [Google Scholar] [CrossRef]
- van Tunen, A.J.; Hartman, S.A.; Mur, L.A.; Mol, J.N.M. Regulation of Chalcone Flavanone Isomerase (CHI) Gene Expression in Petunia hybrida: The Use of Alternative Promoters in Corolla, Anthers and Pollen. Plant Mol. Biol. 1989, 12, 539–551. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.-H.; Shih, M.-C.; Li, W.-H. Transcription Factor Families Have Much Higher Expansion Rates in Plants than in Animals. Plant Physiol. 2005, 139, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Klempnauer, K.-H.; Gonda, T.J.; Michael Bishop, J. Nucleotide Sequence of the Retroviral Leukemia Gene V-Myb and Its Cellular Progenitor c-Myb: The Architecture of a Transduced Oncogene. Cell 1982, 31, 453–463. [Google Scholar] [CrossRef]
- Kranz, H.; Scholz, K.; Weisshaar, B. c-MYB Oncogene-like Genes Encoding Three MYB Repeats Occur in All Major Plant Lineages. Plant J. 2000, 21, 231–235. [Google Scholar] [CrossRef]
- Kanei-Ishii, C.; Sarai, A.; Sawazaki, T.; Nakagoshi, H.; He, D.N.; Ogata, K.; Nishimura, Y.; Ishii, S. The Tryptophan Cluster: A Hypothetical Structure of the DNA-Binding Domain of the Myb Protooncogene Product. J. Biol. Chem. 1990, 265, 19990–19995. [Google Scholar] [CrossRef]
- Ogata, K.; Kanei-Ishii, C.; Sasaki, M.; Hatanaka, H.; Nagadoi, A.; Enari, M.; Nakamura, H.; Nishimura, Y.; Ishii, S.; Sarai, A. The Cavity in the Hydrophobic Core of Myb DNA-Binding Domain Is Reserved for DNA Recognition and Trans-Activation. Nat. Struct. Biol. 1996, 3, 178–187. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.-Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis Transcription Factors: Genome-Wide Comparative Analysis among Eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, K.; Qi, Y.; Lv, G.; Ren, X.; Liu, Z.; Ma, F. Transcriptional Regulation of Anthocyanin Synthesis by MYB-bHLH-WDR Complexes in Kiwifruit (Actinidia chinensis). J. Agric. Food Chem. 2021, 69, 3677–3691. [Google Scholar] [CrossRef]
- Patzlaff, A.; Newman, L.J.; Dubos, C.; Whetten, R.W.; Smith, C.; McInnis, S.; Bevan, M.W.; Sederoff, R.R.; Campbell, M.M. Characterisation of PtMYB1, an R2R3-MYB from Pine Xylem. Plant Mol. Biol. 2003, 53, 597–608. [Google Scholar] [CrossRef]
- Sainz, M.B.; Grotewold, E.; Chandler, V.L. Evidence for Direct Activation of an Anthocyanin Promoter by the Maize C1 Protein and Comparison of DNA Binding by Related Myb Domain Proteins. Plant Cell 1997, 9, 611–625. [Google Scholar] [CrossRef]
- Xu, W.; Grain, D.; Bobet, S.; Le Gourrierec, J.; Thévenin, J.; Kelemen, Z.; Lepiniec, L.; Dubos, C. Complexity and Robustness of the Flavonoid Transcriptional Regulatory Network Revealed by Comprehensive Analyses of MYB–bHLH–WDR Complexes and Their Targets in Arabidopsis Seed. New Phytol. 2014, 202, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Grain, D.; Le Gourrierec, J.; Harscoët, E.; Berger, A.; Jauvion, V.; Scagnelli, A.; Berger, N.; Bidzinski, P.; Kelemen, Z.; et al. Regulation of Flavonoid Biosynthesis Involves an Unexpected Complex Transcriptional Regulation of TT8 Expression, in Arabidopsis. New Phytol. 2013, 198, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Kranz, H.D.; Denekamp, M.; Greco, R.; Jin, H.; Leyva, A.; Meissner, R.C.; Petroni, K.; Urzainqui, A.; Bevan, M.; Martin, C.; et al. Towards Functional Characterisation of the Members of the R2R3-MYB Gene Family from Arabidopsis thaliana. Plant J. 1998, 16, 263–276. [Google Scholar] [CrossRef]
- Rabinowicz, P.D.; Braun, E.L.; Wolfe, A.D.; Bowen, B.; Grotewold, E. Maize R2R3 Myb Genes: Sequence Analysis Reveals Amplification in the Higher Plants. Genetics 1999, 153, 427–444. [Google Scholar] [CrossRef]
- Grotewold, E.; Sainz, M.B.; Tagliani, L.; Hernandez, J.M.; Bowen, B.; Chandler, V.L. Identification of the Residues in the Myb Domain of Maize C1 That Specify the Interaction with the bHLH Cofactor R. Proc. Natl. Acad. Sci. USA 2000, 97, 13579–13584. [Google Scholar] [CrossRef]
- Kim, D.-H.; Yang, J.; Ha, S.-H.; Kim, J.K.; Lee, J.-Y.; Lim, S.-H. An OsKala3, R2R3 MYB TF, Is a Common Key Player for Black Rice Pericarp as Main Partner of an OsKala4, bHLH TF. Front. Plant Sci. 2021, 12, 765049. [Google Scholar] [CrossRef]
- Byrne, P.F.; McMullen, M.D.; Snook, M.E.; Musket, T.A.; Theuri, J.M.; Widstrom, N.W.; Wiseman, B.R.; Coe, E.H. Quantitative Trait Loci and Metabolic Pathways: Genetic Control of the Concentration of Maysin, a Corn Earworm Resistance Factor, in Maize Silks. Proc. Natl. Acad. Sci. USA 1996, 93, 8820–8825. [Google Scholar] [CrossRef]
- Grotewold, E.; Drummond, B.J.; Bowen, B.; Peterson, T. The Myb-Homologous P Gene Controls Phlobaphene Pigmentation in Maize Floral Organs by Directly Activating a Flavonoid Biosynthetic Gene Subset. Cell 1994, 76, 543–553. [Google Scholar] [CrossRef]
- Sharma, M.; Chai, C.; Morohashi, K.; Grotewold, E.; Snook, M.E.; Chopra, S. Expression of Flavonoid 3′-Hydroxylase Is Controlled by P1, the Regulator of 3-Deoxyflavonoid Biosynthesis in Maize. BMC Plant Biol. 2012, 12, 196. [Google Scholar] [CrossRef]
- Zhang, P.; Chopra, S.; Peterson, T. A Segmental Gene Duplication Generated Differentially Expressed Myb-Homologous Genes in Maize. Plant Cell 2000, 12, 2311. [Google Scholar] [CrossRef]
- Zheng, J.; Wu, H.; Zhu, H.; Huang, C.; Liu, C.; Chang, Y.; Kong, Z.; Zhou, Z.; Wang, G.; Lin, Y.; et al. Determining Factors, Regulation System, and Domestication of Anthocyanin Biosynthesis in Rice Leaves. New Phytol. 2019, 223, 705–721. [Google Scholar] [CrossRef] [PubMed]
- Sheiness, D.; Bishop, J.M. DNA and RNA from Uninfected Vertebrate Cells Contain Nucleotide Sequences Related to the Putative Transforming Gene of Avian Myelocytomatosis Virus. J. Virol. 1979, 31, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Martínez-García, J.F.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-Wide Classification and Evolutionary Analysis of the bHLH Family of Transcription Factors in Arabidopsis, Poplar, Rice, Moss, and Algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef] [PubMed]
- Pires, N.; Dolan, L. Early Evolution of bHLH Proteins in Plants. Plant Signal. Behav. 2010, 5, 911–912. [Google Scholar] [CrossRef]
- Murre, C.; McCaw, P.S.; Baltimore, D. A New DNA Binding and Dimerization Motif in Immunoglobulin Enhancer Binding, Daughterless, MyoD, and Myc Proteins. Cell 1989, 56, 777–783. [Google Scholar] [CrossRef]
- Atchley, W.R.; Terhalle, W.; Dress, A. Positional Dependence, Cliques, and Predictive Motifs in the bHLH Protein Domain. J. Mol. Evol. 1999, 48, 501–516. [Google Scholar] [CrossRef]
- Ferré-D’Amaré, A.R.; Prendergast, G.C.; Ziff, E.B.; Burley, S.K. Recognition by Max of Its Cognate DNA through a Dimeric b/HLH/Z Domain. Nature 1993, 363, 38–45. [Google Scholar] [CrossRef]
- Kong, Q.; Pattanaik, S.; Feller, A.; Werkman, J.R.; Chai, C.; Wang, Y.; Grotewold, E.; Yuan, L. Regulatory Switch Enforced by Basic Helix-Loop-Helix and ACT-Domain Mediated Dimerizations of the Maize Transcription Factor R. Proc. Natl. Acad. Sci. USA 2012, 109, E2091–E2097. [Google Scholar] [CrossRef]
- Montefiori, M.; Brendolise, C.; Dare, A.P.; Lin-Wang, K.; Davies, K.M.; Hellens, R.P.; Allan, A.C. In the Solanaceae, a Hierarchy of bHLHs Confer Distinct Target Specificity to the Anthocyanin Regulatory Complex. J. Exp. Bot. 2015, 66, 1427–1436. [Google Scholar] [CrossRef]
- Heim, M.A. The Basic Helix-Loop-Helix Transcription Factor Family in Plants: A Genome-Wide Study of Protein Structure and Functional Diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef]
- Li, Y.; Shan, X.; Gao, R.; Yang, S.; Wang, S.; Gao, X.; Wang, L. Two IIIf Clade-bHLHs from Freesia hybrida Play Divergent Roles in Flavonoid Biosynthesis and Trichome Formation When Ectopically Expressed in Arabidopsis. Sci. Rep. 2016, 6, 30514. [Google Scholar] [CrossRef] [PubMed]
- Feller, A.; Hernandez, J.M.; Grotewold, E. An ACT-like Domain Participates in the Dimerization of Several Plant Basic-Helix-Loop-Helix Transcription Factors. J. Biol. Chem. 2006, 281, 28964–28974. [Google Scholar] [CrossRef] [PubMed]
- Morohashi, K.; Zhao, M.; Yang, M.; Read, B.; Lloyd, A.; Lamb, R.; Grotewold, E. Participation of the Arabidopsis bHLH Factor GL3 in Trichome Initiation Regulatory Events. Plant Physiol. 2007, 145, 736–746. [Google Scholar] [CrossRef] [PubMed]
- Tao, R.; Yu, W.; Gao, Y.; Ni, J.; Yin, L.; Zhang, X.; Li, H.; Wang, D.; Bai, S.; Teng, Y. Light-Induced Basic/Helix-Loop-Helix64 Enhances Anthocyanin Biosynthesis and Undergoes CONSTITUTIVELY PHOTOMORPHOGENIC1-Mediated Degradation in Pear. Plant Physiol. 2020, 184, 1684–1701. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, H.; Luo, T.; Liu, Y.; Nie, X.; Li, H. Characteristics and Expression Pattern of MYC Genes in Triticum aestivum, Oryza sativa, and Brachypodium distachyon. Plants 2019, 8, 274. [Google Scholar] [CrossRef]
- Fernández-Calvo, P.; Chini, A.; Fernández-Barbero, G.; Chico, J.-M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M.; et al. The Arabidopsis bHLH Transcription Factors MYC3 and MYC4 Are Targets of JAZ Repressors and Act Additively with MYC2 in the Activation of Jasmonate Responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef]
- Gong, Z.-Z.; Yamagishi, E.; Yamazaki, M.; Saito, K. A constitutively expressed Myc-like gene involved in anthocyanin biosynthesis from Perilla frutescens: Molecular characterization, heterologous expression in transgenic plants and transactivation in yeast cells. Plant Mol. Biol. 1999, 41, 33–44. [Google Scholar] [CrossRef]
- Ma, H.; Pooler, M.; Griesbach, R. Ratio of Myc and Myb Transcription Factors Regulates Anthocyanin Production in Orchid Flowers. J. Am. Soc. Hortic. Sci. 2008, 133, 133–138. [Google Scholar] [CrossRef]
- Pattanaik, S.; Xie, C.H.; Yuan, L. The Interaction Domains of the Plant Myc-like bHLH Transcription Factors Can Regulate the Transactivation Strength. Planta 2008, 227, 707–715. [Google Scholar] [CrossRef]
- Swinnen, G.; De Meyer, M.; Pollier, J.; Molina-Hidalgo, F.J.; Ceulemans, E.; Venegas-Molina, J.; De Milde, L.; Fernández-Calvo, P.; Ron, M.; Pauwels, L.; et al. The Basic Helix–Loop–Helix Transcription Factors MYC1 and MYC2 Have a Dual Role in the Regulation of Constitutive and Stress-inducible Specialized Metabolism in Tomato. New Phytol. 2022, 236, 911–928. [Google Scholar] [CrossRef]
- Lu, X.; Yang, L.; Yu, M.; Lai, J.; Wang, C.; McNeil, D.; Zhou, M.; Yang, C. A Novel Zea Mays Ssp. Mexicana L. MYC-Type ICE-like Transcription Factor Gene ZmmICE1, Enhances Freezing Tolerance in Transgenic Arabidopsis Thaliana. Plant Physiol. Biochem. 2017, 113, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.-H.; Hong, X.; Agarwal, M.; Zhu, J.-K. ICE1: A Regulator of Cold-Induced Transcriptome and Freezing Tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) Function as Transcriptional Activators in Abscisic Acid Signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Purugganan, M.D.; Wessler, S.R. Molecular Evolution of the Plant R Regulatory Gene Family. Genetics 1994, 138, 849–854. [Google Scholar] [CrossRef]
- Goff, S.A.; Cone, K.C.; Chandler, V.L. Functional Analysis of the Transcriptional Activator Encoded by the Maize B Gene: Evidence for a Direct Functional Interaction between Two Classes of Regulatory Proteins. Genes Dev. 1992, 6, 864–875. [Google Scholar] [CrossRef]
- Hernandez, J.M.; Heine, G.F.; Irani, N.G.; Feller, A.; Kim, M.-G.; Matulnik, T.; Chandler, V.L.; Grotewold, E. Different Mechanisms Participate in the R-Dependent Activity of the R2R3 MYB Transcription Factor C1. J. Biol. Chem. 2004, 279, 48205–48213. [Google Scholar] [CrossRef]
- Nesi, N.; Debeaujon, I.; Jond, C.; Pelletier, G.; Caboche, M.; Lepiniec, L. The TT8 Gene Encodes a Basic Helix-Loop-Helix Domain Protein Required for Expression of DFR and BAN Genes in Arabidopsis Siliques. Plant Cell 2000, 12, 1863–1878. [Google Scholar] [CrossRef]
- Nesi, N.; Jond, C.; Debeaujon, I.; Caboche, M.; Lepiniec, L. The Arabidopsis TT2 Gene Encodes and R2R3 MYB Domain Protein That Acts as a Key Determinant for Proanthocyanidin Accumulation in Developing Seed. Plant Cell 2001, 13, 2099. [Google Scholar] [CrossRef]
- Walker, A.R.; Davison, P.A.; Bolognesi-Winfield, A.C.; James, C.M.; Srinivasan, N.; Blundell, T.L.; Esch, J.J.; Marks, M.D.; Gray, J.C. The TRANSPARENT TESTA GLABRA1 Locus, Which Regulates Trichome Differentiation and Anthocyanin Biosynthesis in Arabidopsis, Encodes a WD40 Repeat Protein. Plant Cell 1999, 11, 1337–1349. [Google Scholar] [CrossRef]
- Neer, E.J.; Schmidt, C.J.; Nambudripad, R.; Smith, T.F. The Ancient Regulatory-Protein Family of WD-Repeat Proteins. Nature 1994, 371, 297–300. [Google Scholar] [CrossRef]
- Fong, H.K.; Hurley, J.B.; Hopkins, R.S.; Miake-Lye, R.; Johnson, M.S.; Doolittle, R.F.; Simon, M.I. Repetitive Segmental Structure of the Transducin Beta Subunit: Homology with the CDC4 Gene and Identification of Related mRNAs. Proc. Natl. Acad. Sci. USA 1986, 83, 2162–2166. [Google Scholar] [CrossRef] [PubMed]
- de Vetten, N.; Quattrocchio, F.; Mol, J.; Koes, R. The An11 Locus Controlling Flower Pigmentation in Petunia Encodes a Novel WD-Repeat Protein Conserved in Yeast, Plants, and Animals. Genes Dev. 1997, 11, 1422–1434. [Google Scholar] [CrossRef] [PubMed]
- Selinger, D.A.; Chandler, V.L. A Mutation in the Pale Aleurone Color1 Gene Identifies a Novel Regulator of the Maize Anthocyanin Pathway. Plant Cell 1999, 11, 5–14. [Google Scholar] [CrossRef]
- Coe, E.H., Jr.; Neuffer, M.G.; Hoisington, D.A. The Genetics of Corn. In Agronomy Monographs; American Society of Agronomy, Crop Science Society of America; Soil Science Society of America: Madison, WI, USA, 2015; pp. 81–258. ISBN 9780891182122. [Google Scholar]
- Perrot, G.H.; Cone, K.C. Nucleotide Sequence of the Maize R-S Gene. Nucleic Acids Res. 1989, 17, 8003. [Google Scholar] [CrossRef][Green Version]
- Radicella, J.P.; Turks, D.; Chandler, V.L. Cloning and Nucleotide Sequence of a cDNA encoding B-Peru, a Regulatory Protein of the Anthocyanin Pathway in Maize. Plant Mol. Biol. 1991, 17, 127–130. [Google Scholar] [CrossRef]
- Goff, S.A.; Klein, T.M.; Roth, B.A.; Fromm, M.E.; Cone, K.C.; Radicella, J.P.; Chandler, V.L. Transactivation of Anthocyanin Biosynthetic Genes Following Transfer of B Regulatory Genes into Maize Tissues. Eur. Mol. Biol. Organ. J. 1990, 9, 2517–2522. [Google Scholar] [CrossRef]
- Roth, B.A.; Goff, S.A.; Klein, T.M.; Fromm, M.E. C1- and R-Dependent Expression of the Maize Bz1 Gene Requires Sequences with Homology to Mammalian Myb and Myc Binding Sites. Plant Cell 1991, 3, 317–325. [Google Scholar] [CrossRef][Green Version]
- Oppenheimer, D.G.; Herman, P.L.; Sivakumaran, S.; Esch, J.; Marks, M.D. A Myb Gene Required for Leaf Trichome Differentiation in Arabidopsis Is Expressed in Stipules. Cell 1991, 67, 483–493. [Google Scholar] [CrossRef]
- Borevitz, J.O.; Xia, Y.; Blount, J.; Dixon, R.A.; Lamb, C. Activation Tagging Identifies a Conserved MYB Regulator of Phenylpropanoid Biosynthesis. Plant Cell 2000, 12, 2383–2393. [Google Scholar] [CrossRef]
- Kaspar, P.; Pajer, P.; Sedlak, D.; Tamaoki, T.; Dvorak, M. C-Myb Inhibits Myogenic Differentiation through Repression of MyoD. Exp. Cell Res. 2005, 309, 419–428. [Google Scholar] [CrossRef]
- Albert, N.W.; Butelli, E.; Moss, S.M.A.; Piazza, P.; Waite, C.N.; Schwinn, K.E.; Davies, K.M.; Martin, C. Discrete bHLH Transcription Factors Play Functionally Overlapping Roles in Pigmentation Patterning in Flowers of Antirrhinum majus. New Phytol. 2021, 231, 849–863. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Herrera-Tequia, A.; Silwal, J.; Geiger, J.H.; Grotewold, E. A Hydrophobic Residue Stabilizes Dimers of Regulatory ACT-like Domains in Plant Basic Helix–Loop–Helix Transcription Factors. J. Biol. Chem. 2021, 296, 100708. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, J.M.; Feller, A.; Morohashi, K.; Frame, K.; Grotewold, E. The Basic Helix–Loop–Helix Domain of Maize R Links Transcriptional Regulation and Histone Modifications by Recruitment of an EMSY-Related Factor. Proc. Natl. Acad. Sci. USA 2007, 104, 17222–17227. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Li, W.; Jiang, N.; Yu, H.; Morohashi, K.; Ouma, W.Z.; Morales-Mantilla, D.E.; Gomez-Cano, F.A.; Mukundi, E.; Prada-Salcedo, L.D.; et al. A Maize Gene Regulatory Network for Phenolic Metabolism. Mol. Plant 2017, 10, 498–515. [Google Scholar] [CrossRef]
- Tuerck, J.A.; Fromm, M.E. Elements of the Maize A1 Promoter Required for Transactivation by the Anthocyanin B/C1 or Phlobaphene P Regulatory Genes. Plant Cell 1994, 6, 1655–1663. [Google Scholar] [CrossRef]
- Albert, N.W.; Davies, K.M.; Lewis, D.H.; Zhang, H.; Montefiori, M.; Brendolise, C.; Boase, M.R.; Ngo, H.; Jameson, P.E.; Schwinn, K.E. A Conserved Network of Transcriptional Activators and Repressors Regulates Anthocyanin Pigmentation in Eudicots. Plant Cell 2014, 26, 962–980. [Google Scholar] [CrossRef]
- Baudry, A.; Caboche, M.; Lepiniec, L. TT8 Controls Its Own Expression in a Feedback Regulation Involving TTG1 and Homologous MYB and bHLH Factors, Allowing a Strong and Cell-specific Accumulation of Flavonoids in Arabidopsis thaliana. Plant J. 2006, 46, 768–779. [Google Scholar] [CrossRef]
- Morohashi, K.; Grotewold, E. A Systems Approach Reveals Regulatory Circuitry for Arabidopsis Trichome Initiation by the GL3 and GL1 Selectors. PLoS Genet. 2009, 5, e1000396. [Google Scholar] [CrossRef]
- Oikawa, T.; Maeda, H.; Oguchi, T.; Yamaguchi, T.; Tanabe, N.; Ebana, K.; Yano, M.; Ebitani, T.; Izawa, T. The Birth of a Black Rice Gene and Its Local Spread by Introgression. Plant Cell 2015, 27, 2401–2414. [Google Scholar] [CrossRef]
- An, J.-P.; Wang, X.-F.; Zhang, X.-W.; Xu, H.-F.; Bi, S.-Q.; You, C.-X.; Hao, Y.-J. An Apple MYB Transcription Factor Regulates Cold Tolerance and Anthocyanin Accumulation and Undergoes MIEL1-mediated Degradation. Plant Biotechnol. J. 2020, 18, 337–353. [Google Scholar] [CrossRef]
- An, J.-P.; Zhang, X.-W.; Liu, Y.-J.; Wang, X.-F.; You, C.-X.; Hao, Y.-J. ABI5 Regulates ABA-Induced Anthocyanin Biosynthesis by Modulating the MYB1-bHLH3 Complex in Apple. J. Exp. Bot. 2021, 72, 1460–1472. [Google Scholar] [CrossRef] [PubMed]
- An, J.-P.; Xu, R.-R.; Liu, X.; Zhang, J.-C.; Wang, X.-F.; You, C.-X.; Hao, Y.-J. Jasmonate Induces Biosynthesis of Anthocyanin and Proanthocyanidin in Apple by Mediating the JAZ1–TRB1–MYB9 Complex. Plant J. 2021, 106, 1414–1430. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Liu, L.; Liu, Q.; Shen, Q.; Wang, C.; Yang, P.; Zhu, C.; Wang, Q. ZmMYC2 Exhibits Diverse Functions and Enhances JA Signaling in Transgenic Arabidopsis. Plant Cell Rep. 2020, 39, 273–288. [Google Scholar] [CrossRef]
- Gupta, O.P.; Karkute, S.G.; Banerjee, S.; Meena, N.L.; Dahuja, A. Contemporary Understanding of miRNA-Based Regulation of Secondary Metabolites Biosynthesis in Plants. Front. Plant Sci. 2017, 8, 374. [Google Scholar] [CrossRef]
- Hou, Q.; Zhao, W.; Lu, L.; Wang, L.; Zhang, T.; Hu, B.; Yan, T.; Qi, Y.; Zhang, F.; Chao, N.; et al. Overexpression of HLH4 Inhibits Cell Elongation and Anthocyanin Biosynthesis in Arabidopsis thaliana. Cells 2022, 11, 1087. [Google Scholar] [CrossRef]
- Kim, J.; Kim, D.-H.; Lee, J.-Y.; Lim, S.-H. The R3-Type MYB Transcription Factor BrMYBL2.1 Negatively Regulates Anthocyanin Biosynthesis in Chinese Cabbage (Brassica rapa L.) by Repressing MYB–bHLH–WD40 Complex Activity. Int. J. Mol. Sci. 2022, 23, 3382. [Google Scholar] [CrossRef]
- LaFountain, A.M.; Yuan, Y.-W. Repressors of Anthocyanin Biosynthesis. New Phytol. 2021, 231, 933–949. [Google Scholar] [CrossRef]
- Li, C.; Yu, W.; Xu, J.; Lu, X.; Liu, Y. Anthocyanin Biosynthesis Induced by MYB Transcription Factors in Plants. Int. J. Mol. Sci. 2022, 23, 11701. [Google Scholar] [CrossRef]
- Ma, Y.; Ma, X.; Gao, X.; Wu, W.; Zhou, B. Light Induced Regulation Pathway of Anthocyanin Biosynthesis in Plants. Int. J. Mol. Sci. 2021, 22, 11116. [Google Scholar] [CrossRef]
- Mackon, E.; Jeazet Dongho Epse Mackon, G.C.; Ma, Y.; Haneef Kashif, M.; Ali, N.; Usman, B.; Liu, P. Recent Insights into Anthocyanin Pigmentation, Synthesis, Trafficking, and Regulatory Mechanisms in Rice (Oryza sativa L.) Caryopsis. Biomolecules 2021, 11, 394. [Google Scholar] [CrossRef]
- Mao, W.; Han, Y.; Chen, Y.; Sun, M.; Feng, Q.; Li, L.; Liu, L.; Zhang, K.; Wei, L.; Han, Z.; et al. Low Temperature Inhibits Anthocyanin Accumulation in Strawberry Fruit by Activating FvMAPK3-Induced Phosphorylation of FvMYB10 and Degradation of Chalcone Synthase 1. Plant Cell 2022, 34, 1226–1249. [Google Scholar] [CrossRef] [PubMed]
- Peniche-Pavía, H.A.; Guzmán, T.J.; Magaña-Cerino, J.M.; Gurrola-Díaz, C.M.; Tiessen, A. Maize Flavonoid Biosynthesis, Regulation, and Human Health Relevance: A Review. Molecules 2022, 27, 5166. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Sun, L.; Wei, L.; Yuan, J.; Kong, F.; Zhang, Y.; Miao, X.; Xia, G.; Liu, S. Maize SRO1e Represses Anthocyanin Synthesis through Regulating the MBW Complex in Response to Abiotic Stress. Plant J. 2021, 105, 1010–1025. [Google Scholar] [CrossRef]
- Singh, P.K.; Rawal, H.C.; Panda, A.K.; Roy, J.; Mondal, T.K.; Sharma, T.R. Pan-Genomic, Transcriptomic, and miRNA Analyses to Decipher Genetic Diversity and Anthocyanin Pathway Genes among the Traditional Rice Landraces. Genomics 2022, 114, 110436. [Google Scholar] [CrossRef]
- Xie, T.; Zan, X.; Chen, X.; Zhu, H.; Rong, H.; Wang, Y.; Jiang, J. An R3-MYB Repressor, BnCPC Forms a Feedback Regulation with MBW Complex to Modulate Anthocyanin Biosynthesis in Brassica napus. Biotechnol. Biofuels Bioprod. 2022, 15, 133. [Google Scholar] [CrossRef]
- Yang, J.; Chen, Y.; Xiao, Z.; Shen, H.; Li, Y.; Wang, Y. Multilevel Regulation of Anthocyanin-Promoting R2R3-MYB Transcription Factors in Plants. Front. Plant Sci. 2022, 13, 1008829. [Google Scholar] [CrossRef]
- Zheng, T.; Tan, W.; Yang, H.; Zhang, L.; Li, T.; Liu, B.; Zhang, D.; Lin, H. Regulation of Anthocyanin Accumulation via MYB75/HAT1/TPL-Mediated Transcriptional Repression. PLoS Genet. 2019, 15, e1007993. [Google Scholar] [CrossRef]
- Zheng, T.; Li, Y.; Lei, W.; Qiao, K.; Liu, B.; Zhang, D.; Lin, H. SUMO E3 Ligase SIZ1 Stabilizes MYB75 to Regulate Anthocyanin Accumulation under High Light Conditions in Arabidopsis. Plant Sci. 2020, 292, 110355. [Google Scholar] [CrossRef]
- Dooner, H.K.; Kermicle, J.L. Reconstruction of the Rr compound allele in maize. Genetics 1974, 78, 691–701. [Google Scholar] [CrossRef]
- Dooner, H.K.; Kermicle, J.L. Structure of the Rr tandem duplication in maize. Genetics 1971, 67, 427–436. [Google Scholar] [CrossRef]
- Walker, E.L.; Robbins, T.P.; Bureau, T.E.; Kermicle, J.; Dellaporta, S.L. Transposon-Mediated Chromosomal Rearrangements and Gene Duplications in the Formation of the Maize R-r Complex. Eur. Mol. Biol. Organ. J. 1995, 14, 2350–2363. [Google Scholar] [CrossRef] [PubMed]
- Eggleston, W.B.; Alleman, M.; Kermicle, J.L. Molecular Organization and Germinal Instability of R-Stippled Maize. Genetics 1995, 141, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Kermicle, J.L. Somatic and Meiotic Instability of R-Stippled, an Aleurone Spotting Factor in Maize. Genetics 1970, 64, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Kermicle, J.L. Recombination between Components of a Mutable Gene System in Maize. Genetics 1984, 107, 489–500. [Google Scholar] [CrossRef] [PubMed]
- Dooner, H.K.; Kermicle, J.L. Displaced and Tandem Duplications in the Long Arm of Chromosome 10 in Maize. Genetics 1976, 82, 309–322. [Google Scholar] [CrossRef]
- Li, T.; Wang, Y.; Dong, Q.; Wang, F.; Kong, F.; Liu, G.; Lei, Y.; Yang, H.; Zhou, Y.; Li, C. Weighted Gene Co-Expression Network Analysis Reveals Key Module and Hub Genes Associated with the Anthocyanin Biosynthesis in Maize Pericarp. Front. Plant Sci. 2022, 13, 1013412. [Google Scholar] [CrossRef]
- Consonni, G.; Viotti, A.; Dellaporta, S.L.; Tonelli, C. cDNA Nucleotide Sequence of Sn, a Regulatory Gene in Maize. Nucleic Acids Res. 1992, 20, 373. [Google Scholar] [CrossRef]
- Tonelli, C.; Consonni, G.; Dolfini, S.F.; Dellaporta, S.L.; Viotti, A.; Gavazzi, G. Genetic and Molecular Analysis of Sn, a Light-Inducible, Tissue Specific Regulatory Gene in Maize. Mol. Gen. Genet. 1991, 225, 401–410. [Google Scholar] [CrossRef]
- Li, P.; Du, C.; Zhang, Y.; Yin, S.; Zhang, E.; Fang, H.; Lin, D.; Xu, C.; Yang, Z. Combined Bulked Segregant Sequencing and Traditional Linkage Analysis for Identification of Candidate Gene for Purple Leaf Sheath in Maize. PLoS ONE 2018, 13, e0190670. [Google Scholar] [CrossRef]
- Consonni, G.; Geuna, F.; Gavazzi, G.; Tonelli, C. Molecular Homology among Members of the R Gene Family in Maize. Plant J. 1993, 3, 335–346. [Google Scholar] [CrossRef]
- Chandler, V.L.; Radicella, J.P.; Robbins, T.P.; Chen, J.; Turks, D. Two Regulatory Genes of the Maize Anthocyanin Pathway Are Homologous: Isolation of B Utilizing R Genomic Sequences. Plant Cell 1989, 1, 1175–1183. [Google Scholar] [CrossRef] [PubMed]
- Radicella, J.P.; Brown, D.; Tolar, L.A.; Chandler, V.L. Allelic Diversity of the Maize B Regulatory Gene: Different Leader and Promoter Sequences of Two B Alleles Determine Distinct Tissue Specificities of Anthocyanin Production. Genes Dev. 1992, 6, 2152–2164. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, L.; Wessler, S.R. Role of mRNA Secondary Structure in Translational Repression of the Maize Transcriptional ActivatorLC. Plant Physiol. 2001, 125, 1380–1387. [Google Scholar] [CrossRef] [PubMed]
- Taylor, L.P.; Briggs, W.R. Genetic Regulation and Photocontrol of Anthocyanin Accumulation in Maize Seedlings. Plant Cell 1990, 2, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Racchi, M.L. Effect of the Genes B and Pl on Anthocyanin Synthesis in Maize Endosperm Culture. Plant Cell Rep. 1985, 4, 184–187. [Google Scholar] [CrossRef]
- Yonemaru, J.-I.; Miki, K.; Choi, S.; Kiyosawa, A.; Goto, K. A Genomic Region Harboring the Pl1 Allele from the Peruvian Cultivar JC072A Confers Purple Cob on Japanese Flint Corn (Zea mays L.). Breed. Sci. 2018, 68, 582–586. [Google Scholar] [CrossRef]
- Burr, F.A.; Burr, B.; Scheffler, B.E.; Blewitt, M.; Wienand, U.; Matz, E.C. The Maize Repressor-like Gene Intensifier1 Shares Homology with the R1/B1 Multigene Family of Transcription Factors and Exhibits Missplicing. Plant Cell 1996, 8, 1249–1259. [Google Scholar] [CrossRef]
- Cone, K.C.; Burr, F.A.; Burr, B. Molecular Analysis of the Maize Anthocyanin Regulatory Locus C1. Proc. Natl. Acad. Sci. USA 1986, 83, 9631–9635. [Google Scholar] [CrossRef]
- Goff, S.A.; Cone, K.C.; Fromm, M.E. Identification of Functional Domains in the Maize Transcriptional Activator C1: Comparison of Wild-Type and Dominant Inhibitor Proteins. Genes Dev. 1991, 5, 298–309. [Google Scholar] [CrossRef]
- Morohashi, K.; Casas, M.I.; Falcone Ferreyra, M.L.; Mejía-Guerra, M.K.; Pourcel, L.; Yilmaz, A.; Feller, A.; Carvalho, B.; Emiliani, J.; Rodriguez, E.; et al. A Genome-Wide Regulatory Framework Identifies Maize Pericarp Color1 Controlled Genes. Plant Cell 2012, 24, 2745–2764. [Google Scholar] [CrossRef]
- Dellaporta, S.L.; Greenblatt, I.; Kermicle, J.L.; Hicks, J.B.; Wessler, S.R. Molecular Cloning of the Maize R-Nj Allele by Transposon Tagging with Ac; Stadler Genetics Symposia Series; Springer: Boston, MA, USA, 1988; pp. 263–282. ISBN 9781461283041. [Google Scholar]
- Kermicle, J.L.; Eggleston, W.B.; Alleman, M. Organization of Paramutagenicity in R-Stippled Maize. Genetics 1995, 141, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Alleman, M.; Wessler, S.R. A Ds Insertion Alters the Nuclear Localization of the Maize Transcriptional Activator R. Proc. Natl. Acad. Sci. USA 1996, 93, 7816–7820. [Google Scholar] [CrossRef] [PubMed]
- Patterson, G.I.; Kubo, K.M.; Shroyer, T.; Chandler, V.L. Sequences Required for Paramutation of the Maize b Gene Map to a Region Containing the Promoter and Upstream Sequences. Genetics 1995, 140, 1389–1406. [Google Scholar] [CrossRef] [PubMed]
- Bureau, T.E.; Wessler, S.R. Mobile Inverted-Repeat Elements of the Tourist Familyare Associated with the Genes of Many Cereal Grasses. Proc. Natl. Acad. Sci. USA 1994, 91, 1411–1415. [Google Scholar] [CrossRef]
- Li, Y.; Fang, X.; Lin, Z. Convergent Loss of Anthocyanin Pigments Is Controlled by the Same MYB Gene in Cereals. J. Exp. Bot. 2022, 73, 6089–6102. [Google Scholar] [CrossRef]
- Chen, C.; Liu, X.; Li, S.; Liu, C.; Zhang, Y.; Luo, L.; Miao, L.; Yang, W.; Xiao, Z.; Zhong, Y.; et al. Co-expression of Transcription Factors ZmC1 and ZmR2 Establishes an Efficient and Accurate Haploid Embryo Identification System in Maize. Plant J. 2022, 111, 1296–1307. [Google Scholar] [CrossRef]
- Paz-Ares, J.; Ghosal, D.; Saedler, H. Molecular Analysis of the C1-I Allele from Zea mays: A Dominant Mutant of the Regulatory C1 Locus. Eur. Mol. Biol. Organ. J. 1990, 9, 315–321. [Google Scholar] [CrossRef]
- Zou, T.; Wang, X.; Sun, T.; Rong, H.; Wu, L.; Deng, J.; Guo, T.; Wang, H.; Wang, J.; Huang, M. MYB Transcription Factor OsC1PLSr Involves the Regulation of Purple Leaf Sheath in Rice. Int. J. Mol. Sci. 2023, 24, 6655. [Google Scholar] [CrossRef]
- Pilu, R.; Piazza, P.; Petroni, K.; Ronchi, A.; Martin, C.; Tonelli, C. Pl-bol3, a Complex Allele of the Anthocyanin Regulatory Pl1 Locus That Arose in a Naturally Occurring Maize Population. Plant J. 2003, 36, 510–521. [Google Scholar] [CrossRef]
- Chopra, S.; Athma, P.; Li, X.; Peterson, T. A Maize Myb Homolog Is Encoded by a Multicopy Gene Complex. Mol. Gen. Genet. 1998, 260, 372–380. [Google Scholar] [CrossRef]
- Kinoshita, T.; Maekawa, M. Inheritance of purple leaf color found in Indica Rice. J. Fac. Agric. 1986, 62, 453–466. [Google Scholar]
- Lachagari, V.B.R.; Gupta, R.; Lekkala, S.P.; Mahadevan, L.; Kuriakose, B.; Chakravartty, N.; Mohan Katta, A.V.S.K.; Santhosh, S.; Reddy, A.R.; Thomas, G. Whole Genome Sequencing and Comparative Genomic Analysis Reveal Allelic Variations Unique to a Purple Colored Rice Landrace (Oryza sativa ssp. Indica cv. Purpleputtu). Front. Plant Sci. 2019, 10, 513. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Anderson, B.; Wessler, S.R. Isolation and Characterization of Rice R Genes: Evidence for Distinct Evolutionary Paths in Rice and Maize. Genetics 1996, 142, 1021–1031. [Google Scholar] [CrossRef]
- Wang, C.; Shu, Q. Fine Mapping and Candidate Gene Analysis of Purple Pericarp Gene Pb in Rice (Oryza sativa L.). Chin. Sci. Bull. 2007, 52, 3097–3104. [Google Scholar] [CrossRef]
- Meng, L.; Qi, C.; Wang, C.; Wang, S.; Zhou, C.; Ren, Y.; Cheng, Z.; Zhang, X.; Guo, X.; Zhao, Z.; et al. Determinant Factors and Regulatory Systems for Anthocyanin Biosynthesis in Rice Apiculi and Stigmas. Rice 2021, 14, 37. [Google Scholar] [CrossRef]
- Qiao, W.; Wang, Y.; Xu, R.; Yang, Z.; Sun, Y.; Su, L.; Zhang, L.; Wang, J.; Huang, J.; Zheng, X.; et al. A Functional Chromogen Gene C from Wild Rice Is Involved in a Different Anthocyanin Biosynthesis Pathway in Indica and Japonica. Züchter Genet. Breed. Res. 2021, 134, 1531–1543. [Google Scholar] [CrossRef]
- Hu, W.; Zhou, T.; Han, Z.; Tan, C.; Xing, Y. Dominant Complementary Interaction between OsC1 and Two Tightly Linked Genes, Rb1 and Rb2, Controls the Purple Leaf Sheath in Rice. Züchter Genet. Breed. Res. 2020, 133, 2555–2566. [Google Scholar] [CrossRef]
- Sweeney, M.T.; Thomson, M.J.; Pfeil, B.E.; McCouch, S. Caught Red-Handed: Rc Encodes a Basic Helix-Loop-Helix Protein Conditioning Red Pericarp in Rice. Plant Cell 2006, 18, 283–294. [Google Scholar] [CrossRef]
- Maeda, H.; Yamaguchi, T.; Omoteno, M.; Takarada, T.; Fujita, K.; Murata, K.; Iyama, Y.; Kojima, Y.; Morikawa, M.; Ozaki, H.; et al. Genetic Dissection of Black Grain Rice by the Development of a near Isogenic Line. Breed. Sci. 2014, 64, 134–141. [Google Scholar] [CrossRef]
- Akhter, D.; Qin, R.; Nath, U.K.; Eshag, J.; Jin, X.; Shi, C. A Rice Gene, OsPL, Encoding a MYB Family Transcription Factor Confers Anthocyanin Synthesis, Heat Stress Response and Hormonal Signaling. Gene 2019, 699, 62–72. [Google Scholar] [CrossRef]
- Khan, A.; Jalil, S.; Cao, H.; Tsago, Y.; Sunusi, M.; Chen, Z.; Shi, C.; Jin, X. The Purple Leaf (Pl6) Mutation Regulates Leaf Color by Altering the Anthocyanin and Chlorophyll Contents in Rice. Plants 2020, 9, 1477. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Cui, Y.; Yao, Y.; An, L.; Bai, Y.; Li, X.; Yao, X.; Wu, K. Genome-Wide Identification of WD40 Transcription Factors and Their Regulation of the MYB-bHLH-WD40 (MBW) Complex Related to Anthocyanin Synthesis in Qingke (Hordeum vulgare L. var. nudum Hook. f.). BMC Genom. 2023, 24, 166. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.S.; Scheffler, B.E.; Wienand, U.; Wessler, S.R.; Reddy, A.R. Cloning and characterization of the rice homologue of the maize C1 anthocyanin regulatory gene. Plant Mol. Biol. 1998, 36, 497–498. [Google Scholar] [CrossRef]
- Shih, C.H.; Chu, H.; Tang, L.K.; Sakamoto, W.; Maekawa, M.; Chu, I.K.; Wang, M.; Lo, C. Functional Characterization of Key Structural Genes in Rice Flavonoid Biosynthesis. Planta 2008, 228, 1043–1054. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, Z.; Chen, C.; Wu, W.; Ren, N.; Jiang, C.; Yu, J.; Zhao, Y.; Zheng, X.; Yang, Q.; et al. The C–S–A Gene System Regulates Hull Pigmentation and Reveals Evolution of Anthocyanin Biosynthesis Pathway in Rice. J. Exp. Bot. 2018, 69, 1485–1498. [Google Scholar] [CrossRef]
- Yang, X.; Wang, J.; Xia, X.; Zhang, Z.; He, J.; Nong, B.; Luo, T.; Feng, R.; Wu, Y.; Pan, Y.; et al. OsTTG1, a WD40 Repeat Gene, Regulates Anthocyanin Biosynthesis in Rice. Plant J. 2021, 107, 198–214. [Google Scholar] [CrossRef]
- Lee, C.; Lee, Y.-S.; Hong, H.-C.; Hong, W.-J.; Koh, H.-J.; Jung, K.-H. Reinterpretation of Anthocyanins Biosynthesis in Developing Black Rice Seeds through Gene Expression Analysis. PLoS ONE 2023, 18, e0286539. [Google Scholar] [CrossRef]
- Ke, S.; Jiang, Y.; Zhou, M.; Li, Y. Genome-Wide Identification, Evolution, and Expression Analysis of the WD40 Subfamily in Oryza Genus. Int. J. Mol. Sci. 2023, 24, 15776. [Google Scholar] [CrossRef]
- Chen, M.-H.; Pinson, S.R.M.; Jackson, A.K.; Edwards, J.D. Genetic Loci Regulating the Concentrations of Anthocyanins and Proanthocyanidins in the Pericarps of Purple and Red Rice. Plant Genome 2023, 16, e20338. [Google Scholar] [CrossRef]
- Himi, E.; Taketa, S. Isolation of Candidate Genes for the Barley Ant1 and Wheat Rc Genes Controlling Anthocyanin Pigmentation in Different Vegetative Tissues. Mol. Genet. Genom. 2015, 290, 1287–1298. [Google Scholar] [CrossRef]
- Shoeva, O.Y.; Kukoeva, T.V.; Börner, A.; Khlestkina, E.K. Barley Ant1 Is a Homolog of Maize C1 and Its Product Is Part of the Regulatory Machinery Governing Anthocyanin Synthesis in the Leaf Sheath. Plant Breed. 2015, 134, 400–405. [Google Scholar] [CrossRef]
- Strygina, K.V.; Börner, A.; Khlestkina, E.K. Identification and Characterization of Regulatory Network Components for Anthocyanin Synthesis in Barley Aleurone. BMC Plant Biol. 2017, 17, 184. [Google Scholar] [CrossRef] [PubMed]
- Himi, E.; Yamashita, Y.; Haruyama, N.; Yanagisawa, T.; Maekawa, M.; Taketa, S. Ant28 Gene for Proanthocyanidin Synthesis Encoding the R2R3 MYB Domain Protein (Hvmyb10) Highly Affects Grain Dormancy in Barley. Euphytica 2012, 188, 141–151. [Google Scholar] [CrossRef]
- Jende-Strid, B.; Lundqvist, U. Diallelic Tests of Anthocyanin-deficient Mutants. Barley Genet. Newsl. 1978, 8, 57–59. [Google Scholar]
- Gordeeva, E.I.; Glagoleva, A.Y.; Kukoeva, T.V.; Khlestkina, E.K.; Shoeva, O.Y. Purple-Grained Barley (Hordeum vulgare L.): Marker-Assisted Development of NILs for Investigating Peculiarities of the Anthocyanin Biosynthesis Regulatory Network. BMC Plant Biol. 2019, 19, 49–57. [Google Scholar] [CrossRef]
- Jiang, W.; Liu, T.; Nan, W.; Jeewani, D.C.; Niu, Y.; Li, C.; Wang, Y.; Shi, X.; Wang, C.; Wang, J.; et al. Two Transcription Factors TaPpm1 and TaPpb1 Co-Regulate Anthocyanin Biosynthesis in Purple Pericarps of Wheat. J. Exp. Bot. 2018, 69, 2555–2567. [Google Scholar] [CrossRef]
- Cockram, J.; White, J.; Zuluaga, D.L.; Smith, D.; Comadran, J.; Macaulay, M.; Luo, Z.; Kearsey, M.J.; Werner, P.; Harrap, D.; et al. Genome-Wide Association Mapping to Candidate Polymorphism Resolution in the Unsequenced Barley Genome. Proc. Natl. Acad. Sci. USA 2010, 107, 21611–21616. [Google Scholar] [CrossRef]
- Shoeva, O.Y.; Mock, H.-P.; Kukoeva, T.V.; Börner, A.; Khlestkina, E.K. Regulation of the Flavonoid Biosynthesis Pathway Genes in Purple and Black Grains of Hordeum vulgare. PLoS ONE 2016, 11, e0163782. [Google Scholar] [CrossRef]
- Zhang, X.-W.; Jiang, Q.-T.; Wei, Y.-M.; Liu, C. Inheritance Analysis and Mapping of Quantitative Trait Loci (QTL) Controlling Individual Anthocyanin Compounds in Purple Barley (Hordeum vulgare L.) Grains. PLoS ONE 2017, 12, e0183704. [Google Scholar] [CrossRef]
- Shoeva, O.Y.; Mukhanova, M.A.; Zakhrabekova, S.; Hansson, M. Ant13 Encodes Regulatory Factor WD40 Controlling Anthocyanin and Proanthocyanidin Synthesis in Barley (Hordeum vulgare L.). J. Agric. Food Chem. 2023, 71, 6967–6977. [Google Scholar] [CrossRef]
- Shin, D.H.; Choi, M.-G.; Kang, C.-S.; Park, C.-S.; Choi, S.-B.; Park, Y.-I. A Wheat R2R3-MYB Protein PURPLE PLANT1 (TaPL1) Functions as a Positive Regulator of Anthocyanin Biosynthesis. Biochem. Biophys. Res. Commun. 2016, 469, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Li, W.L.; Faris, J.D.; Chittoor, J.M.; Leach, J.E.; Hulbert, S.H.; Liu, D.J.; Chen, P.D.; Gill, B.S. Genomic Mapping of Defense Response Genes in Wheat. Züchter Genet. Breed. Res. 1999, 98, 226–233. [Google Scholar] [CrossRef]
- Khlestkina, E.K.; Röder, M.S.; Salina, E.A. Relationship between Homoeologous Regulatory and Structural Genes in Allopolyploid Genome—A Case Study in Bread Wheat. BMC Plant Biol. 2008, 8, 88. [Google Scholar] [CrossRef] [PubMed]
- Khlestkina, E.; Pshenichnikova, T.; Röder, M.; Börner, A. Clustering Anthocyanin Pigmentation Genes in Wheat Group 7 Chromosomes. Cereal Res. Commun. 2009, 37, 391–398. [Google Scholar] [CrossRef]
- Khlestkina, E.K.; Gordeeva, E.I.; Arbuzova, V.S. Molecular and Functional Characterization of Wheat Near-isogenic Line ‘i:S29Ra’ Having Intensive Anthocyanin Pigmentation of the Coleoptile, Culm, Leaves and Auricles. Plant Breed. 2014, 133, 454–458. [Google Scholar] [CrossRef]
- Tereshchenko, O.; Gordeeva, E.; Arbuzova, V.; Börner, A.; Khlestkina, E. The D Genome Carries a Gene Determining Purple Grain Colour in Wheat. Cereal Res. Commun. 2012, 40, 334–341. [Google Scholar] [CrossRef]
- Tereshchenko, O.Y.; Arbuzova, V.S.; Khlestkina, E.K. Allelic State of the Genes Conferring Purple Pigmentation in Different Wheat Organs Predetermines Transcriptional Activity of the Anthocyanin Biosynthesis Structural Genes. J. Cereal Sci. 2013, 57, 10–13. [Google Scholar] [CrossRef]
- Khlestkina, E.K.; Röder, M.S.; Pshenichnikova, T.A.; Börner, A. Functional Diversity at the Rc (Red coleoptile) Gene in Bread Wheat. Mol. Breed. 2010, 25, 125–132. [Google Scholar] [CrossRef]
- Khlestkina, E.K.; Tereshchenko, O.Y.; Salina, E.A. Anthocyanin Biosynthesis Genes Location and Expression in Wheat–Rye Hybrids. Mol. Genet. Genom. 2009, 282, 475–485. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Hou, X.J.; Zhang, B.; Chen, W.J.; Liu, D.C.; Liu, B.L.; Zhang, H.G. Identification of a Candidate Gene for Rc-D1, a Locus Controlling Red Coleoptile Colour in Wheat. Cereal Res. Commun. 2016, 44, 35–46. [Google Scholar] [CrossRef]
- Shoeva, O.Y. Complex Regulation of the TaMyc1 Gene Expression in Wheat Grain Synthesizing Anthocyanin Pigments. Mol. Biol. Rep. 2018, 45, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Shoeva, O.; Gordeeva, E.; Khlestkina, E. The Regulation of Anthocyanin Synthesis in the Wheat Pericarp. Molecules 2014, 19, 20266–20279. [Google Scholar] [CrossRef] [PubMed]
- Himi, E.; Nisar, A.; Noda, K. Colour Genes (R and Rc) for Grain and Coleoptile Upregulate Flavonoid Biosynthesis Genes in Wheat. Genome 2005, 48, 747–754. [Google Scholar] [CrossRef] [PubMed]
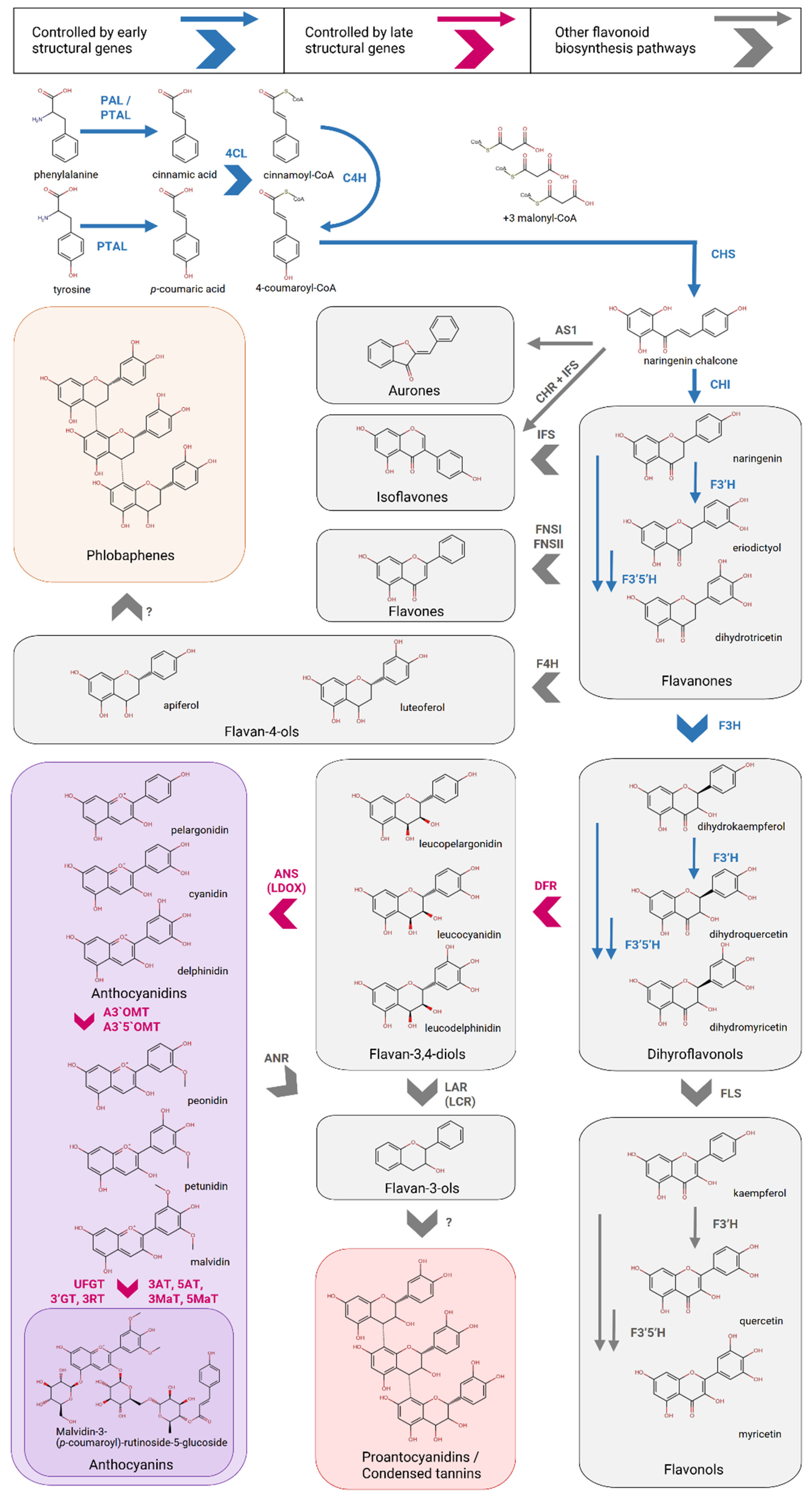
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bulanov, A.N.; Andreeva, E.A.; Tsvetkova, N.V.; Zykin, P.A. Regulation of Flavonoid Biosynthesis by the MYB-bHLH-WDR (MBW) Complex in Plants and Its Specific Features in Cereals. Int. J. Mol. Sci. 2025, 26, 734. https://doi.org/10.3390/ijms26020734
Bulanov AN, Andreeva EA, Tsvetkova NV, Zykin PA. Regulation of Flavonoid Biosynthesis by the MYB-bHLH-WDR (MBW) Complex in Plants and Its Specific Features in Cereals. International Journal of Molecular Sciences. 2025; 26(2):734. https://doi.org/10.3390/ijms26020734
Chicago/Turabian StyleBulanov, Andrey N., Elena A. Andreeva, Natalia V. Tsvetkova, and Pavel A. Zykin. 2025. "Regulation of Flavonoid Biosynthesis by the MYB-bHLH-WDR (MBW) Complex in Plants and Its Specific Features in Cereals" International Journal of Molecular Sciences 26, no. 2: 734. https://doi.org/10.3390/ijms26020734
APA StyleBulanov, A. N., Andreeva, E. A., Tsvetkova, N. V., & Zykin, P. A. (2025). Regulation of Flavonoid Biosynthesis by the MYB-bHLH-WDR (MBW) Complex in Plants and Its Specific Features in Cereals. International Journal of Molecular Sciences, 26(2), 734. https://doi.org/10.3390/ijms26020734









