Genome-Wide Identification of CONSTANS-like (COL) Gene Family and the Potential Function of ApCOL08 Under Salt Stress in Andrographis paniculata
Abstract
1. Introduction
2. Results
2.1. Distribution, Characterization, and Expansion of the Nine Identified ApCOL Genes
2.2. Phylogenetic Analysis and Well-Defined Classification of Andrographis paniculata COL Genes
2.3. Gene Structure and Conserved Motifs Analysis of ApCOLs
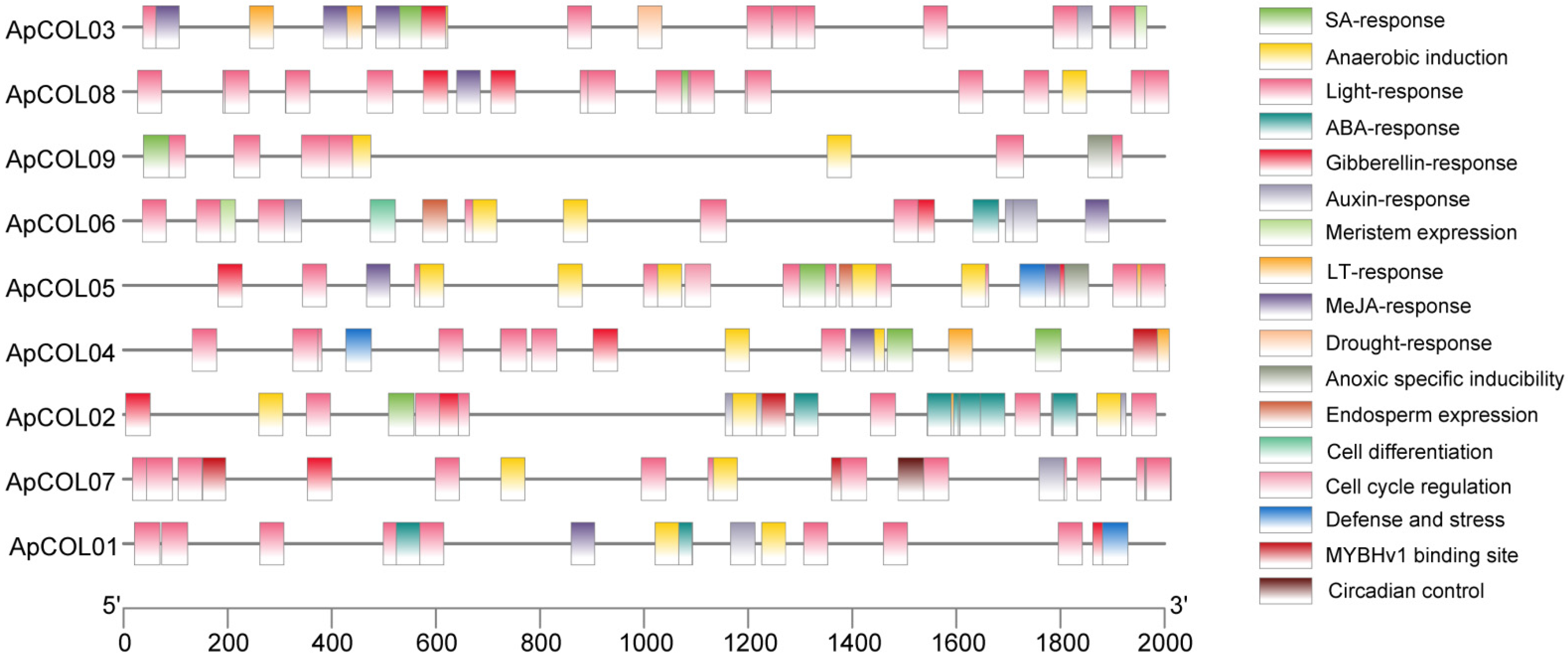
2.4. Cis-Regulatory Elements and Functional Analysis of the ApCOL Promoter Regions
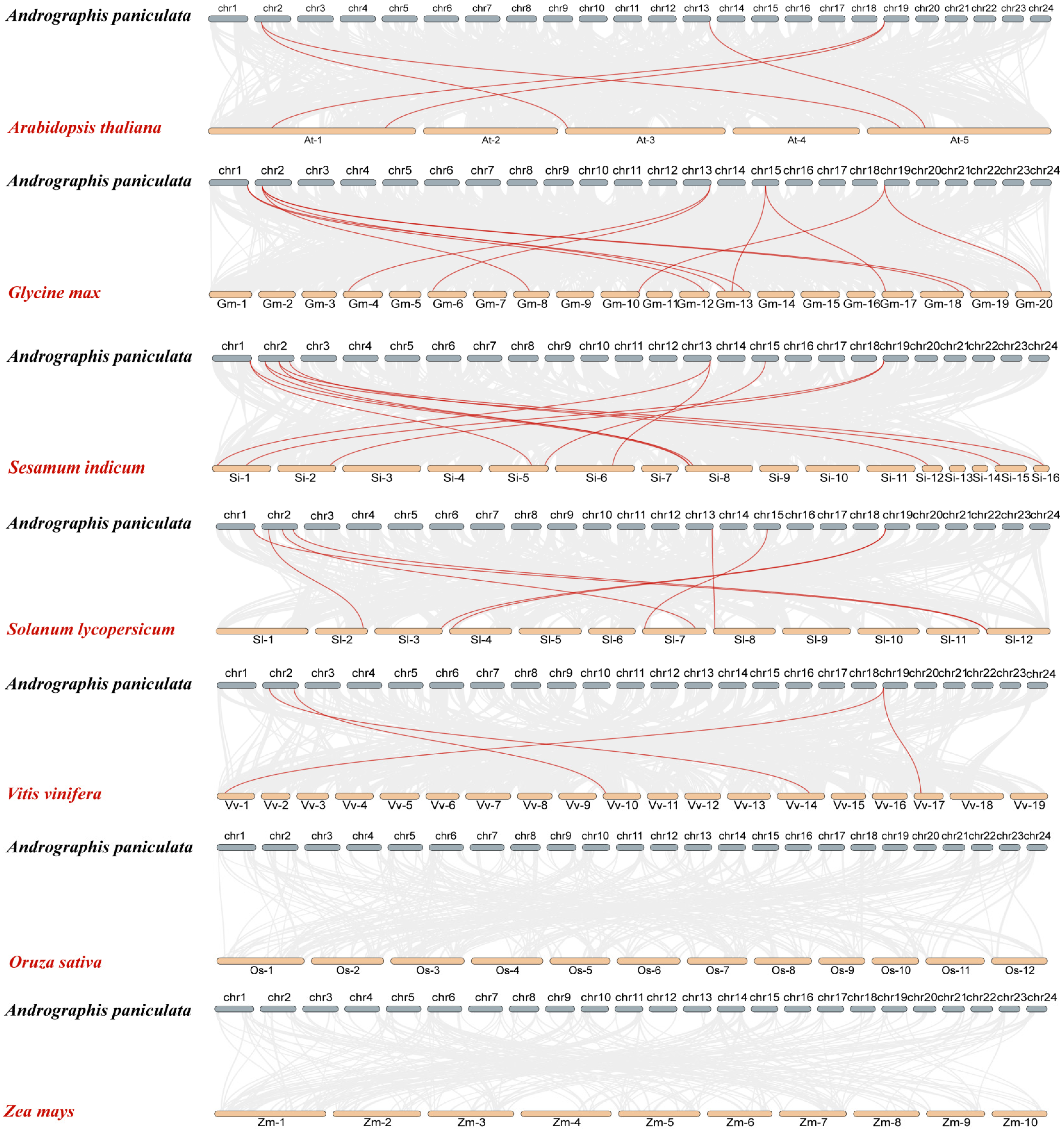
2.5. Interspecies Syntenic Analysis
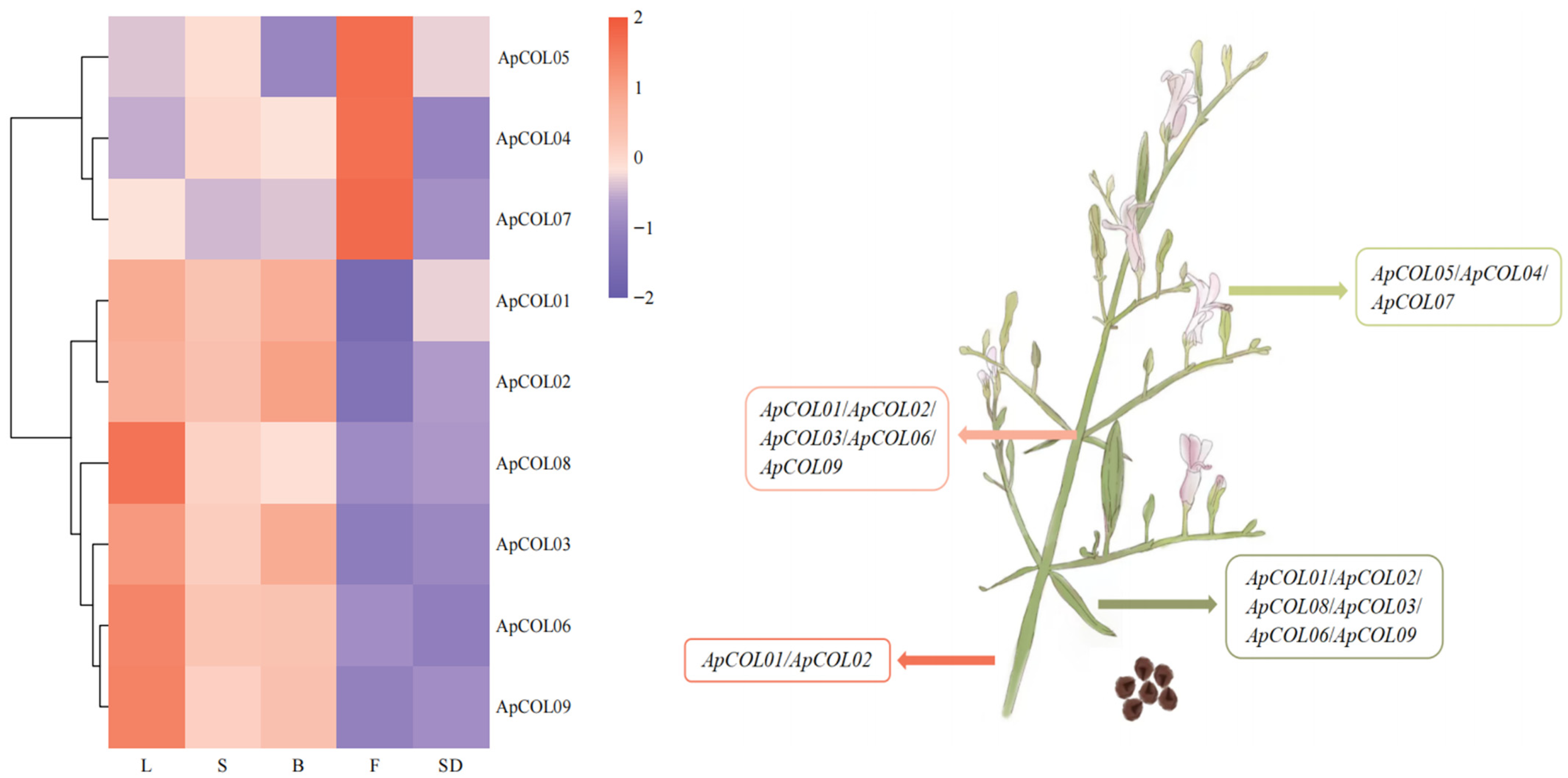
2.6. Expression Profiles of ApCOL Genes in Different Tissues
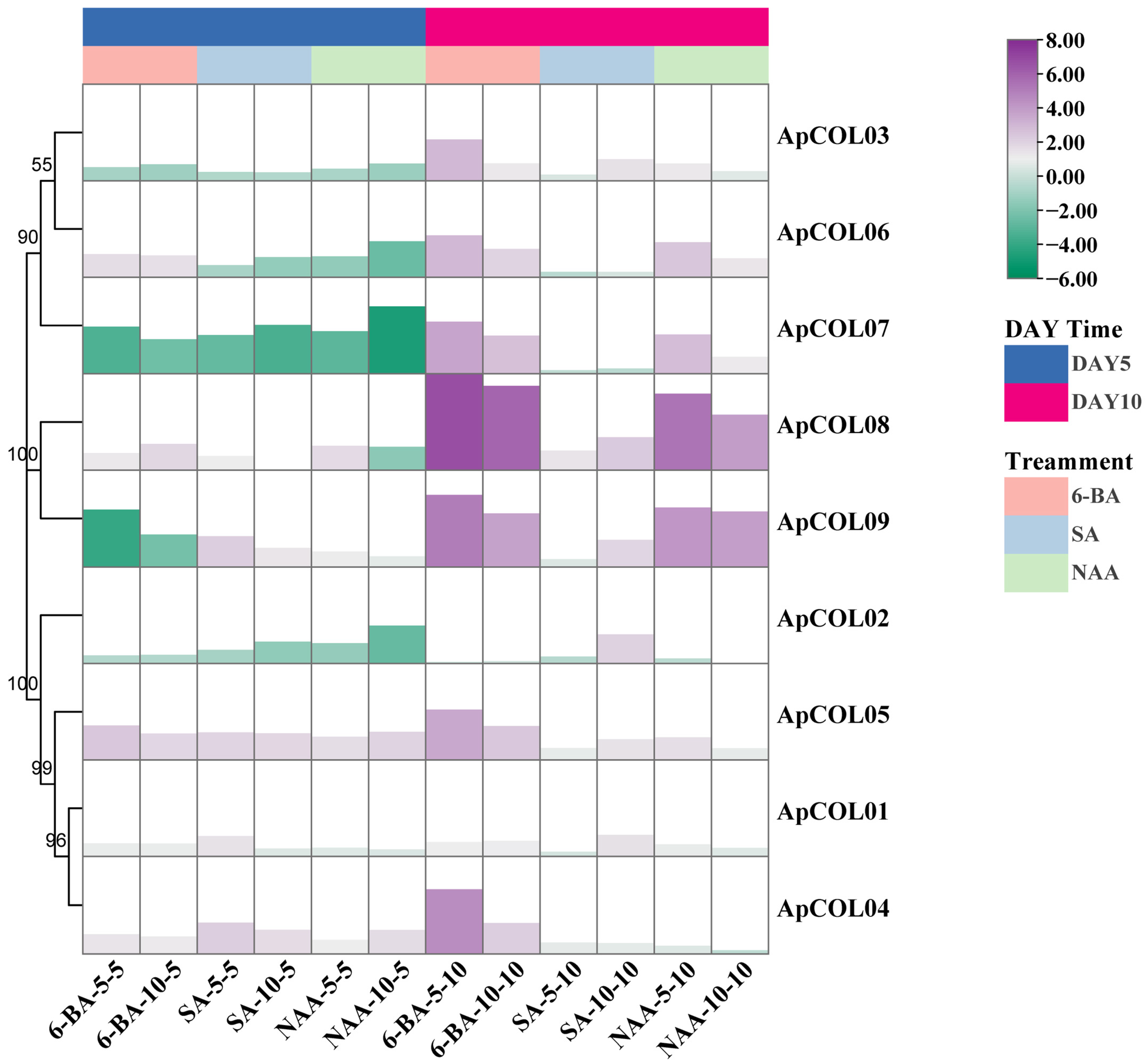
2.7. Expression Profiles of ApCOL Genes Under Hormone Treatment
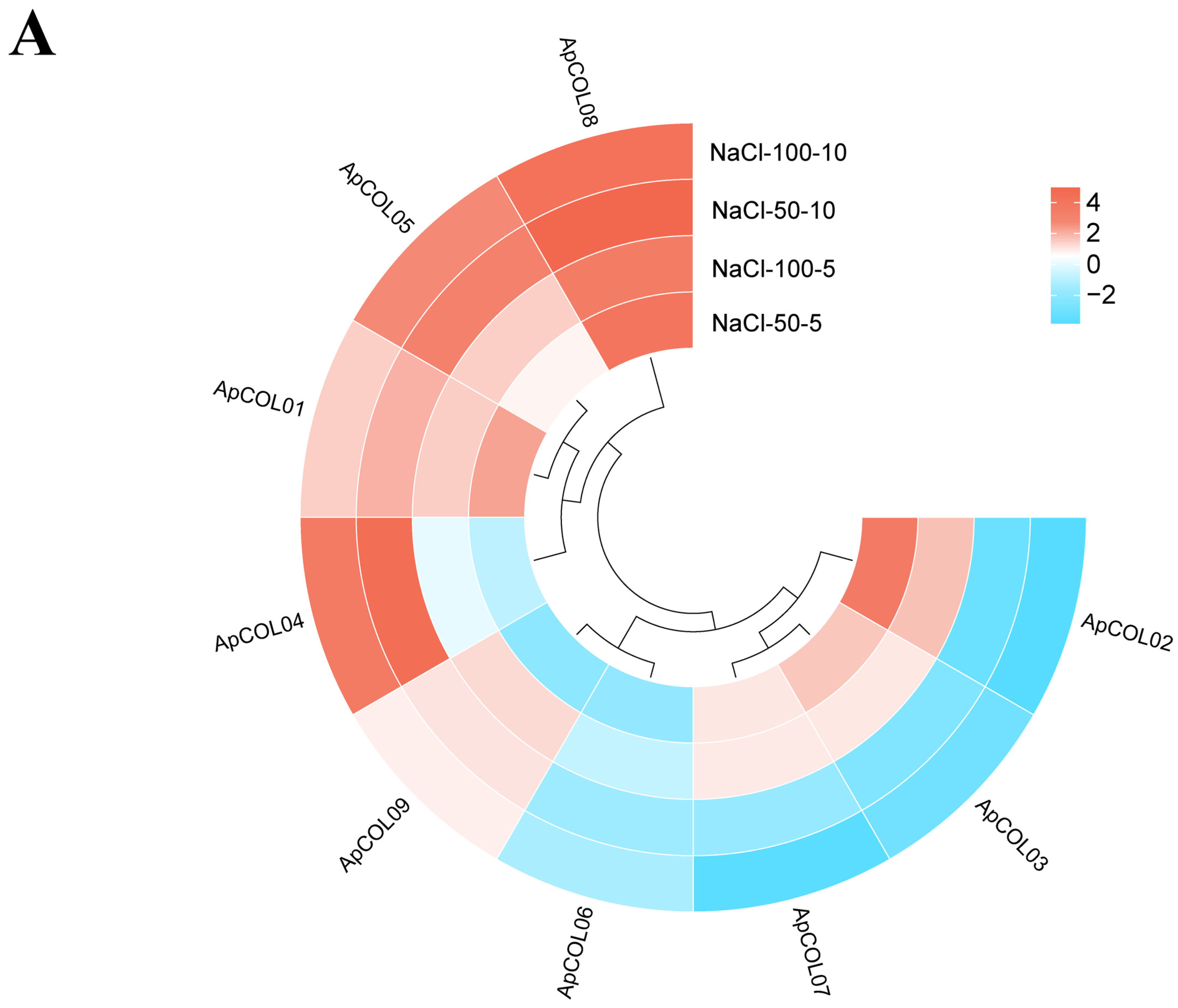
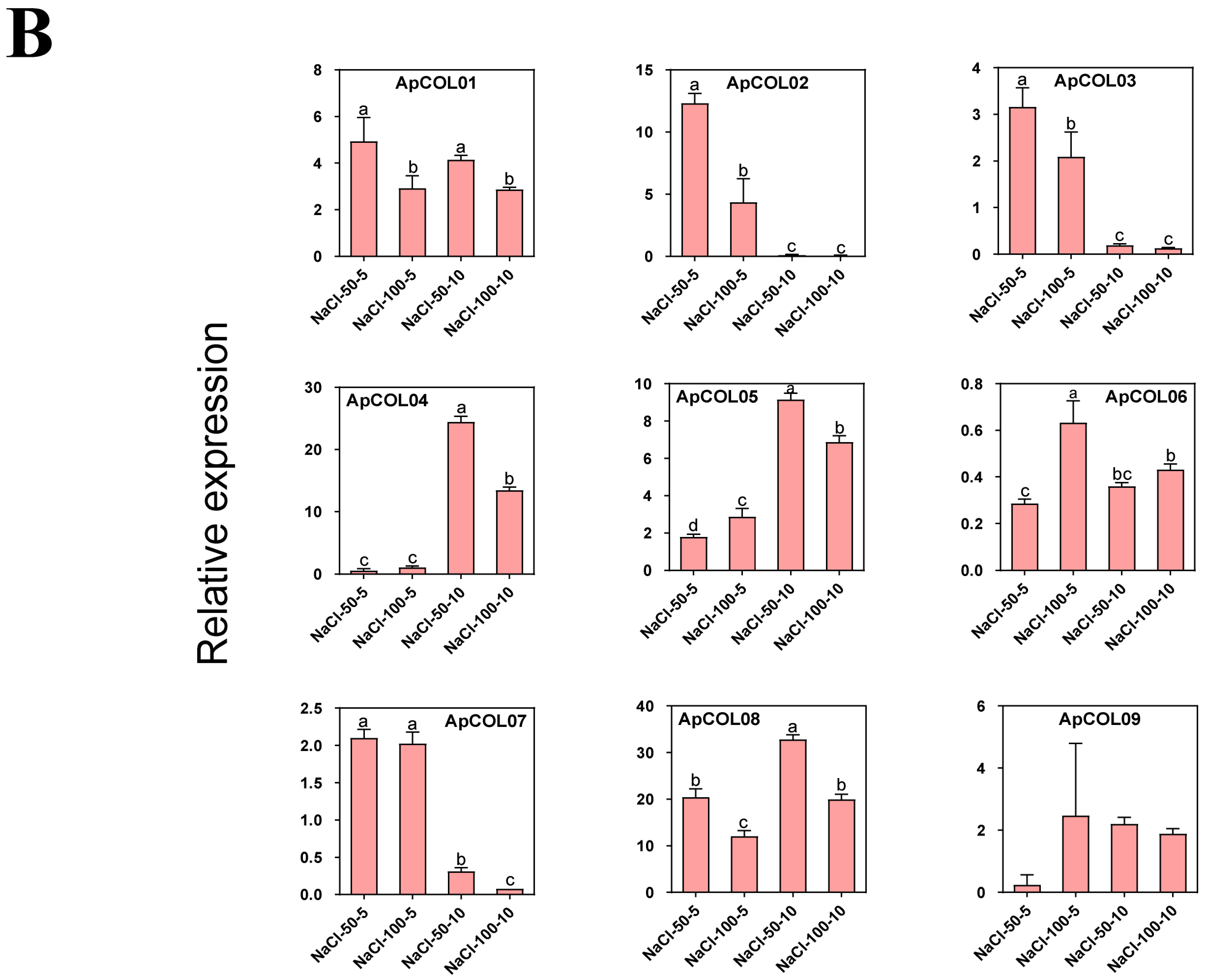
2.8. Expression Profiles of ApCOL Genes Under Salt Stresses
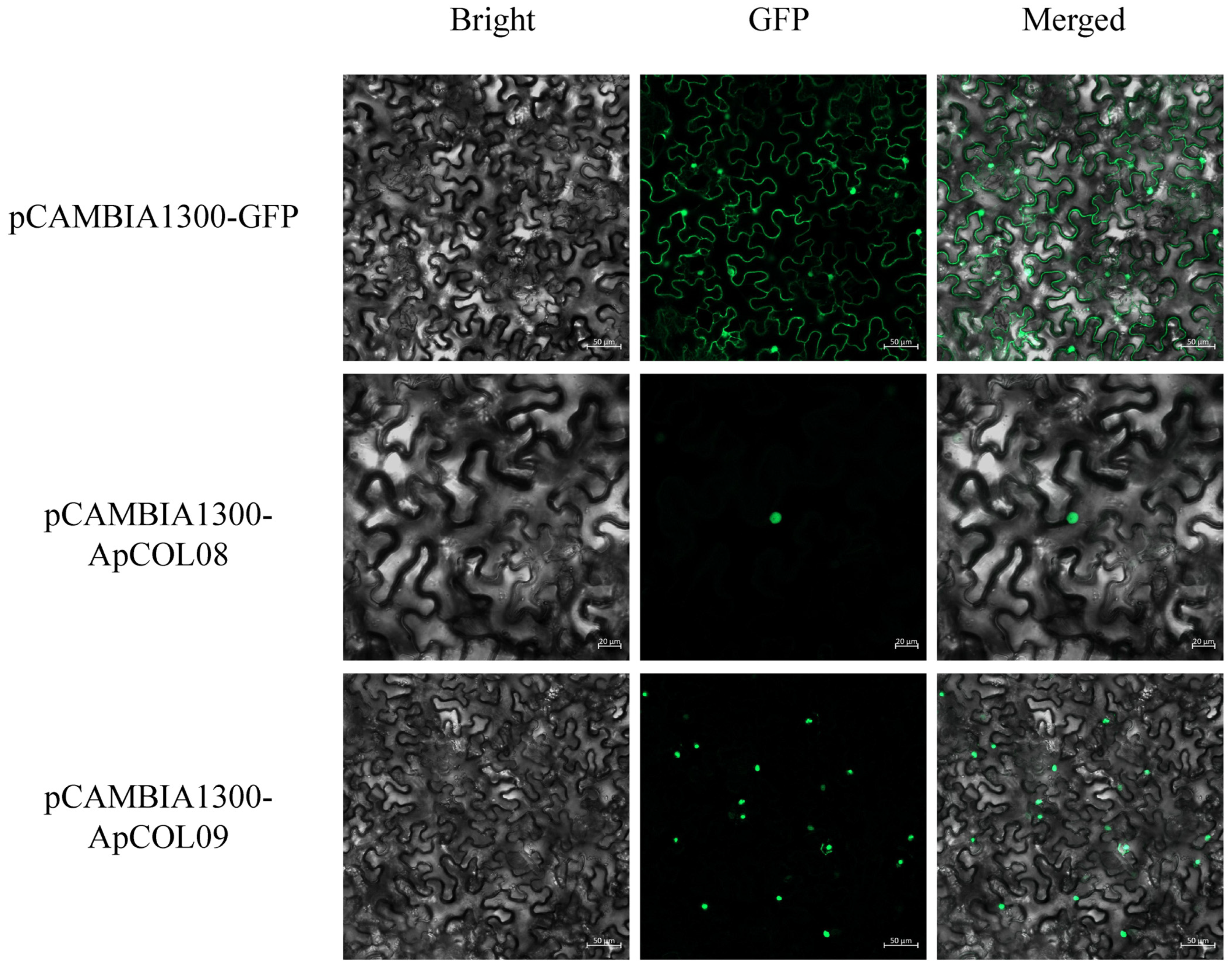
2.9. Subcellular Localization of Key ApCOL Proteins
2.10. Heterologous Expression Verification of ApCOL08 Salt Tolerance Function in Yeast
3. Discussion
4. Materials and Methods
4.1. Identification and Update of the COL Genes
4.2. Chromosomal Location and Syntenic Analysis of ApCOLs
4.3. Phylogenetic Analysis and Subfamily Classification
4.4. Sequence and Structural Analysis of COL Proteins
4.5. Analysis of Cis-Regulatory Elements and Transcription Factor Binding Sites
4.6. Plant Materials and Treatment Methods
4.7. RNA Extraction and Construction of Gene Expression Profiles
4.8. Subcellular Localization Analysis
4.9. Heterologous Expression of Genes in Yeast
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fitter, A.H.; Fitter, R.S.R. Rapid Changes in Flowering Time in British Plants. Science 2002, 296, 1689–1691. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, A.; Schmid, M. Regulation of flowering time: All roads lead to Rome. Cell Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef]
- Song, Y.H.; Ito, S.; Imaizumi, T. Flowering time regulation: Photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 2013, 18, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Blümel, M.; Dally, N.; Jung, C. Flowering time regulation in crops—What did we learn from Arabidopsis? Curr. Opin. Biotechnol. 2015, 32, 121–129. [Google Scholar] [CrossRef]
- Shim, J.S.; Kubota, A.; Imaizumi, T. Circadian Clock and Photoperiodic Flowering in Arabidopsis: CONSTANS Is a Hub for Signal Integration. Plant Physiol. 2017, 173, 5–15. [Google Scholar] [CrossRef]
- Kinmonth-Schultz, H.A.; Tong, X.; Lee, J.; Song, Y.H.; Ito, S.; Kim, S.H.; Imaizumi, T. Cool night-time temperatures induce the expression of CONSTANS and FLOWERING LOCUS T to regulate flowering in Arabidopsis. New Phytol. 2016, 211, 208–224. [Google Scholar] [CrossRef] [PubMed]
- Putterill, J.; Varkonyi-Gasic, E. FT and florigen long-distance flowering control in plants. Curr. Opin. Plant Biol. 2016, 33, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Robson, F.; Costa, M.M.R.; Hepworth, S.R.; Vizir, I.; Piñeiro, M.; Reeves, P.H.; Putterill, J.; Coupland, G. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2002, 28, 619–631. [Google Scholar] [CrossRef]
- Khanna, R.; Kronmiller, B.; Maszle, D.R.; Coupland, G.; Holm, M.; Mizuno, T.; Wu, S.-H. The Arabidopsis B-Box Zinc Finger Family. Plant Cell 2009, 21, 3416–3420. [Google Scholar] [CrossRef]
- Wenkel, S.; Turck, F.; Singer, K.; Gissot, L.; Le Gourrierec, J.; Samach, A.; Coupland, G. CONSTANS and the CCAAT Box Binding Complex Share a Functionally Important Domain and Interact to Regulate Flowering of Arabidopsis. Plant Cell 2006, 18, 2971–2984. [Google Scholar] [CrossRef]
- Lagercrantz, U.; Axelsson, T. Rapid Evolution of the Family of CONSTANS LIKE Genes in Plants. Mol. Biol. Evol. 2000, 17, 1499–1507. [Google Scholar] [CrossRef] [PubMed]
- Hassidim, M.; Harir, Y.; Yakir, E.; Kron, I.; Green, R.M. Over-expression of CONSTANS-LIKE 5 can induce flowering in short-day grown Arabidopsis. Planta 2009, 230, 481–491. [Google Scholar] [CrossRef]
- Griffiths, S.; Dunford, R.P.; Coupland, G.; Laurie, D.A. The Evolution of CONSTANS-LIKE Gene Families in Barley, Rice, and Arabidopsis. Plant Physiol. 2003, 131, 1855–1867. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, R.; Hu, Q.; Wei, W.; Liu, J. Conservation and Divergence of the CONSTANS-LIKE (COL) Genes Related to Flowering and Circadian Rhythm in Brassica napus. Front. Plant Sci. 2021, 12, 760379. [Google Scholar] [CrossRef] [PubMed]
- Niu, T.; Wang, X.; Abbas, M.; Shen, J.; Liu, R.; Wang, Z.; Liu, A. Expansion of CONSTANS-LIKE genes in sunflower confers putative neofunctionalization in the adaptation to abiotic stresses. Ind. Crops Prod. 2022, 176, 114400. [Google Scholar] [CrossRef]
- Wong, A.C.S.; Hecht, V.r.F.G.; Picard, K.; Diwadkar, P.; Laurie, R.E.; Wen, J.; Mysore, K.; Macknight, R.C.; Weller, J.L. Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Front. Plant Sci. 2014, 5, 486. [Google Scholar] [CrossRef] [PubMed]
- Somers, D.E.; Wu, F.; Price, B.W.; Haider, W.; Seufferheld, G.; Nelson, R.; Hanzawa, Y. Functional and Evolutionary Characterization of the CONSTANS Gene Family in Short-Day Photoperiodic Flowering in Soybean. PLoS ONE 2014, 9, e85754. [Google Scholar]
- Wu, K.; Cai, D.; Liu, H.; Sang, N.; Huang, X. Identification and characterization of CONSTANS-LIKE (COL) gene family in upland cotton (Gossypium hirsutum L.). PLoS ONE 2017, 12, e0179038. [Google Scholar]
- Chaurasia, A.K.; Patil, H.B.; Azeez, A.; Subramaniam, V.R.; Krishna, B.; Sane, A.P.; Sane, P.V. Molecular characterization of CONSTANS-LIKE (COL) genes in banana (Musa acuminata L. AAA Group, cv. Grand Nain). Physiol. Mol. Biol. Plants 2016, 22, 1–15. [Google Scholar] [CrossRef]
- Datta, S.; Hettiarachchi, G.H.C.M.; Deng, X.-W.; Holm, M. Arabidopsis CONSTANS-LIKE 3 Is a Positive Regulator of Red Light Signaling and Root Growth. Plant Cell 2006, 18, 70–84. [Google Scholar] [CrossRef]
- Cheng, X.F.; Wang, Z.Y. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 2005, 43, 758–768. [Google Scholar] [CrossRef]
- Takase, T.; Kakikubo, Y.; Nakasone, A.; Nishiyama, Y.; Yasuhara, M.; Tokioka-Ono, Y.; Kiyosue, T. Characterization and transgenic study of CONSTANS-LIKE8 (COL8) gene in Arabidopsis thaliana: Expression of 35S:COL8 delays flowering under long-day conditions. Plant Biotechnol. 2011, 28, 439–446. [Google Scholar] [CrossRef]
- Yano, M.; Katayose, Y.; Ashikari, M.; Yamanouchi, U.; Monna, L.; Fuse, T.; Baba, T.; Yamamoto, K.; Umehara, Y.; Nagamura, Y.; et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 2000, 12, 2473–2483. [Google Scholar] [CrossRef] [PubMed]
- Hayama, R.; Yokoi, S.; Tamaki, S.; Yano, M.; Shimamoto, K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 2003, 422, 719–722. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Gu, F.; Dong, S.; Liu, W.; Wang, H.; Chen, Z.; Wang, J. CONSTANS-LIKE 9 (COL9) delays the flowering time in Oryza sativa by repressing the Ehd1 pathway. Biochem. Biophys. Res. Commun. 2016, 479, 173–178. [Google Scholar] [CrossRef]
- Min, J.H.; Chung, J.S.; Lee, K.H.; Kim, C.S. The CONSTANS-LIKE 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis. J. Integr. Plant Biol. 2014, 57, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Shen, J.; Xu, Y.; Li, X.; Xiao, J.; Xiong, L. Ghd2, a CONSTANS-LIKE gene, confers drought sensitivity through regulation of senescence in rice. J. Exp. Bot. 2016, 67, 5785–5798. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Shan, J.; Liu, T.; Wang, Q.; Ji, Y.; Zhang, Y.; Wang, M.; Xia, N.; Zhao, L. CONSTANS-LIKE 1a positively regulates salt and drought tolerance in soybean. Plant Physiol. 2023, 191, 2427–2446. [Google Scholar] [CrossRef]
- Chen, J.; Chen, J.-y.; Wang, J.-n.; Kuang, J.-f.; Shan, W.; Lu, W.-j. Molecular characterization and expression profiles of MaCOL1, a CONSTANS-LIKE gene in banana fruit. Gene 2012, 496, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Xia, P.; Li, Q.; Liang, Z.; Zhang, X.; Yan, K. Spaceflight breeding could improve the volatile constituents of Andrographis paniculata. Ind. Crops Prod. 2021, 171, 113967. [Google Scholar] [CrossRef]
- Samy, R.P.; Thwin, M.M.; Gopalakrishnakone, P.; Ignacimuthu, S. Ethnobotanical survey of folk plants for the treatment of snakebites in Southern part of Tamilnadu, India. J. Ethnopharmacol. 2008, 115, 302–312. [Google Scholar] [CrossRef]
- Lim, J.C.W.; Chan, T.K.; Ng, D.S.W.; Sagineedu, S.R.; Stanslas, J.; Wong, W.S.F. Andrographolide and its analogues: Versatile bioactive molecules for combating inflammation and cancer. Clin. Exp. Pharmacol. Physiol. 2012, 39, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Mishra, K.; Dash, A.P.; Swain, B.K.; Dey, N. Anti-malarial activities of Andrographis paniculata and Hedyotis corymbosa extracts and their combination with curcumin. Malar. J. 2009, 8, 26. [Google Scholar] [CrossRef]
- Yu, B.; Dai, C.-Q.; Jiang, Z.-Y.; Li, E.-Q.; Chen, C.; Wu, X.-L.; Chen, J.; Liu, Q.; Zhao, C.-L.; He, J.-X.; et al. Andrographolide as an Anti-H1N1 drug and the mechanism related to retinoic acid-inducible gene-I-like receptors signaling pathway. Chin. J. Integr. Med. 2014, 20, 540–545. [Google Scholar] [CrossRef]
- Zeng, W.J.; Xu, L.; He, Q.L.; Zhang, L.L.; Xu, H.L.; Liang, Z.S. Agronomic traits of Andrographis paniculata and their correlation with diterpene lactones. Chin. J. Chin. Mater. Med. 2019, 44, 3233–3238. [Google Scholar]
- Xie, R.; Lin, Z.; Zhong, C.; Li, S.; Chen, B.; Wu, Y.; Huang, L.; Yao, H.; Shi, P.; Huang, J. Deciphering the potential anti-COVID-19 active ingredients inAndrographis paniculata(Burm. F.) Nees by combination of network pharmacology, molecular docking, and molecular dynamics. RSC Adv. 2021, 11, 36511–36517. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xu, L.; Wang, F.; Chai, W.; Xu, B.; Xu, X.; Zhang, X.; Liang, Z. Preliminary analysis on the formation of Andrographis paniculata quality between the two main producing areas. J. Zhejiang Sci-Tech Univ. 2021, 45, 416–422. [Google Scholar]
- Hossain, M.S. The effect of salinity stress on the morpho-physiology and protein profile of Andrographis paniculata. Master’s Thesis, Kulliyyah of Science, International Islamic University Malaysia, Jalan Gombak, Malaysia, 2016. [Google Scholar]
- Wang, Q.; Xu, L.; Zhang, M.; Chai, W.; Zhang, X.; Xu, X.; Liang, Z. Mechanism of salicylic acid ameliorates salt-induced changes in Andrographis paniculata. China J. Chin. Mater. Medica 2020, 45, 5465–5471. [Google Scholar]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Shen, X.; Sun, Y.; Liu, Q.; Ma, N.; Zhang, X.; Jia, Q.; Liang, Z.; Wang, D. Genome-wide investigation of ARF transcription factor gene family and its responses to abiotic stress in Coix (Coix lacryma-jobi L.). Protoplasma 2023, 260, 1389–1405. [Google Scholar] [CrossRef] [PubMed]
- Anuradha, V.E.; Jaleel, C.A.; Salem, M.A.; Gomathinayagam, M.; Panneerselvam, R. Plant growth regulators induced changes in antioxidant potential and andrographolide content in Andrographis paniculata Wall.ex Nees. Pestic. Biochem. Physiol. 2010, 98, 312–316. [Google Scholar] [CrossRef]
- Sharma, S.N.; Jha, Z.; Sinha, R.K.; Geda, A.K. Jasmonate-induced biosynthesis of andrographolide in Andrographis paniculata. Physiol. Plant 2014, 153, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Srinath, M.; Shailaja, A.; Bindu, B.B.V.; Giri, C.C. Molecular Cloning and Differential Gene Expression Analysis of 1-Deoxy-D-xylulose 5-Phosphate Synthase (DXS) in Andrographis paniculata (Burm. f) Nees. Mol. Biotechnol. 2020, 63, 109–124. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Fang, Y.; Yang, D. Biosynthesis, Regulation and Metabolic Engineering of Terpenoids in Plants. J. Zhejiang Sci-Tech Univ. 2017, 37, 255–264. [Google Scholar]
- Sun, W.; Leng, L.; Yin, Q.; Xu, M.; Huang, M.; Xu, Z.; Zhang, Y.; Yao, H.; Wang, C.; Xiong, C.; et al. The genome of the medicinal plant Andrographis paniculata provides insight into the biosynthesis of the bioactive diterpenoid neoandrographolide. Plant J. 2019, 97, 841–857. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-Z.; Ren, Y.-J.; Miao, Y. Roles of PPR Proteins in Plant Growth and Development. J. Trop. Subtrop. Bot. 2019, 27, 225–234. [Google Scholar]
- Pan, G.; Li, Z.; Yin, M.; Huang, S.; Tao, J.; Chen, A.; Li, J.; Tang, H.; Chang, L.; Deng, Y.; et al. Genome-wide identification, expression, and sequence analysis of CONSTANS-LIKE gene family in cannabis reveals a potential role in plant flowering time regulation. BMC Plant Biol. 2021, 21, 142. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Jin, M.; Wang, J.; Wu, F.; Sheng, P.; Cheng, Z.; Wang, J.; Zheng, X.; Chen, L.; Wang, M.; et al. OsCOL10, a CONSTANS-LIKE Gene, Functions as a Flowering Time Repressor Downstream of Ghd7 in Rice. Plant Cell Physiol. 2016, 57, 798–812. [Google Scholar] [CrossRef] [PubMed]
- Li, X.L.; Ljungdahl, L.G. Expression of Aureobasidium pullulans xynA in, and secretion of the xylanase from, Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1996, 62, 209–213. [Google Scholar] [CrossRef]
- Esquivel, B.D.; Rybak, J.M.; Barker, K.S.; Fortwendel, J.R.; Rogers, P.D.; White, T.C.; Idnurm, A. Characterization of the Efflux Capability and Substrate Specificity of Aspergillus fumigatus PDR5-like ABC Transporters Expressed in Saccharomyces cerevisiae. MBio 2020, 11, e00338-20. [Google Scholar] [CrossRef]
- Devarenne, T.P.; Liu, H.; Dong, S.; Sun, D.; Liu, W.; Gu, F.; Liu, Y.; Guo, T.; Wang, H.; Wang, J.; et al. CONSTANS-Like 9 (OsCOL9) Interacts with Receptor for Activated C-Kinase 1(OsRACK1) to Regulate Blast Resistance through Salicylic Acid and Ethylene Signaling Pathways. PLoS ONE 2016, 11, e0166249. [Google Scholar]
- Wang, H.; Zhang, Z.; Li, H.; Zhao, X.; Liu, X.; Ortiz, M.; Lin, C.; Liu, B. CONSTANS-LIKE 7 regulates branching and shade avoidance response in Arabidopsis. J. Exp. Bot. 2013, 64, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ji, R.; Li, H.; Zhao, T.; Liu, J.; Lin, C.; Liu, B. CONSTANS-LIKE 7 (COL7) Is Involved in Phytochrome B (phyB)-Mediated Light-Quality Regulation of Auxin Homeostasis. Mol. Plant 2014, 7, 1429–1440. [Google Scholar] [CrossRef]
- Casal, J. Signalling for developmental plasticity. Trends Plant Sci. 2004, 9, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; He, Y.; Niu, S.; Yan, S.; Zhang, Y. Identification and characterization of the CONSTANS (CO)/CONSTANS-LIKE (COL) genes related to photoperiodic signaling and flowering in tomato. Plant Sci. 2020, 301, 110653. [Google Scholar] [CrossRef]
- López, M.; Larrea, H.; Alvarenga, N.; González, D.; Iehisa, J.C.M. CONSTANS-LIKE genes are associated with flowering time in sesame. Theor. Exp. Plant Physiol. 2023, 35, 341–353. [Google Scholar] [CrossRef]
- Wang, L.; Xue, J.; Dai, W.; Tang, Y.; Gong, P.; Wang, Y.; Zhang, C. Genome-wide Identification, Phylogenetic Analysis, and Expression Profiling of CONSTANS-LIKE (COL) Genes in Vitis vinifera. J. Plant Growth Regul. 2018, 38, 631–643. [Google Scholar] [CrossRef]
- Song, N.; Xu, Z.; Wang, J.; Qin, Q.; Jiang, H.; Si, W.; Li, X. Genome-wide analysis of maize CONSTANS-LIKE gene family and expression profiling under light/dark and abscisic acid treatment. Gene 2018, 673, 1–11. [Google Scholar] [CrossRef]
- Ke, Y.-Z.; Wu, Y.-W.; Zhou, H.-J.; Chen, P.; Wang, M.-M.; Liu, M.-M.; Li, P.-F.; Yang, J.; Li, J.-N.; Du, H. Genome-wide survey of the bHLH super gene family in Brassica napus. BMC Plant Biol. 2020, 20, 115. [Google Scholar] [CrossRef] [PubMed]
- Valcarcel, J.; Koralewski, T.E.; Krutovsky, K.V. Evolution of Exon-Intron Structure and Alternative Splicing. PLoS ONE 2011, 6, e18055. [Google Scholar]
- Han, Y.; Hou, Z.; He, Q.; Zhang, X.; Yan, K.; Han, R.; Liang, Z. Genome-Wide Characterization and Expression Analysis of bZIP Gene Family Under Abiotic Stress in Glycyrrhiza uralensis. Front. Genet. 2021, 12, 754237. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Cong, Q.; Cheng, L. BBX transcriptional factors family in plants–a review. Chin. J. Biotechnol. 2020, 36, 666–677. [Google Scholar]
- He, F.; Shi, Y.-J.; Li, J.-L.; Lin, T.-T.; Zhao, K.-J.; Chen, L.-H.; Mi, J.-X.; Zhang, F.; Zhong, Y.; Lu, M.-M.; et al. Genome-wide analysis and expression profiling of Cation/H+ exchanger (CAX) family genes reveal likely functions in cadmium stress responses in poplar. Int. J. Biol. Macromol. 2022, 204, 76–88. [Google Scholar] [CrossRef]
- He, F.; Shi, Y.-J.; Mi, J.-X.; Zhao, K.-J.; Cui, X.-L.; Chen, L.-H.; Yang, H.-B.; Zhang, F.; Zhao, Q.; Huang, J.-L.; et al. Genome-Wide Investigation of the NF-X1 Gene Family in Populus trichocarpa Expression Profiles during Development and Stress. Int. J. Mol. Sci. 2021, 22, 4664. [Google Scholar] [CrossRef]
- He, F.; Shi, Y.-J.; Zhao, Q.; Zhao, K.-J.; Cui, X.-L.; Chen, L.-H.; Yang, H.-B.; Zhang, F.; Mi, J.-X.; Huang, J.-L.; et al. Genome-wide investigation and expression profiling of polyphenol oxidase (PPO) family genes uncover likely functions in organ development and stress responses in Populus trichocarpa. BMC Genom. 2021, 22, 731. [Google Scholar] [CrossRef]
- Zhao, P.; Wang, D.; Wang, R.; Kong, N.; Zhang, C.; Yang, C.; Wu, W.; Ma, H.; Chen, Q. Genome-wide analysis of the potato Hsp20 gene family: Identification, genomic organization and expression profiles in response to heat stress. BMC Genom. 2018, 19, 61. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, T.; Wu, X.; Yao, X.; Ai, H.; Zhang, Y.; Gan, Z.; Huang, X. Genome-Wide Identification, Characterization and Expression Profiling of the CONSTANS-LIKE Genes in Potato (Solanum tuberosum L.). Genes 2023, 14, 1174. [Google Scholar] [CrossRef]
- Turck, F.; Fornara, F.; Coupland, G. Regulation and Identity of Florigen: FLOWERING LOCUS T Moves Center Stage. Annu. Rev. Plant Biol. 2008, 59, 573–594. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, L.; Zou, H.; Qiu, L.; Zheng, Y.; Yang, D.; Wang, Y. Effects of Light on Secondary Metabolite Biosynthesis in Medicinal Plants. Front. Plant Sci. 2021, 12, 781236. [Google Scholar] [CrossRef]
- Zemlyanskaya, E.V.; Dolgikh, V.A.; Levitsky, V.G.; Mironova, V. Transcriptional regulation in plants: Using omics data to crack the cis-regulatory code. Curr. Opin. Plant Biol. 2021, 63, 102058. [Google Scholar] [CrossRef]
- Hu, W.; Zheng, Y.; Zheng, J.; Yan, K.; Liang, Z.; Xia, P. Binding proteins PnCOX11 and PnDCD strongly respond to GA and ABA in Panax notoginseng. Int. J. Biol. Macromol. 2022, 212, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Zheng, X.-M.; Chen, D.; Zhang, Y.; Ma, W.; Zhang, H.; Sun, L.; Yang, Z.; Zhao, C.; Zhan, X.; et al. OsCOL16, encoding a CONSTANS-LIKE protein, represses flowering by up-regulating Ghd7 expression in rice. Plant Sci. 2017, 260, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Irfanqureshi, M.; Israr, M.; Abdin, M.; Iqbal, M. Responses of Artemisia annua L. to lead and salt-induced oxidative stress. Environ. Exp. Bot. 2005, 53, 185–193. [Google Scholar] [CrossRef]
- Wang, Q.; Zeng, W.; Ali, B.; Zhang, X.; Xu, L.; Liang, Z. Genome-wide identification of WRKY gene family and expression analysis under abiotic stresses in Andrographis paniculata. Biocell 2021, 45, 1107–1119. [Google Scholar] [CrossRef]
- Li, J.-L.; Li, H.; Zhao, J.-J.; Yang, P.; Xiang, X.; Wei, S.-Y.; Wang, T.; Shi, Y.-J.; Huang, J.; He, F. Genome-wide identification and characterization of the RZFP gene family and analysis of its expression pattern under stress in Populus trichocarpa. Int. J. Biol. Macromol. 2024, 255, 128108. [Google Scholar] [CrossRef]
- Xu, F.-C.; Wang, M.-J.; Guo, Y.-W.; Song, J.; Gao, W.; Long, L. The Na+/H+ antiporter GbSOS1 interacts with SIP5 and regulates salt tolerance in Gossypium barbadense. Plant Sci. 2023, 330, 111658. [Google Scholar] [CrossRef] [PubMed]
- Jia, T.; Hou, J.; Iqbal, M.Z.; Zhang, Y.; Cheng, B.; Feng, H.; Li, Z.; Liu, L.; Zhou, J.; Feng, G.; et al. Overexpression of the white clover TrSAMDC1 gene enhanced salt and drought resistance in Arabidopsis thaliana. Plant Physiol. Biochem. 2021, 165, 147–160. [Google Scholar] [CrossRef]
- Baghban, R.; Farajnia, S.; Rajabibazl, M.; Ghasemi, Y.; Mafi, A.; Hoseinpoor, R.; Rahbarnia, L.; Aria, M. Yeast Expression Systems: Overview and Recent Advances. Mol. Biotechnol. 2019, 61, 365–384. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar Gustavo, A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.h.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Hwang, J.K. Prediction of protein subcellular localization. Proteins 2006, 64, 643–651. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Peer, Y.V.d.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Ismail, N.A.; Hossain, M.S.; Husna, N.; Mustafa, M.; Phang, I.C. Morpho-physiological characteristics, selected macronutrient uptake, and oxidative stress level of Andrographis paniculata under saline condition. J. Teknol. 2015, 77, 135–140. [Google Scholar] [CrossRef][Green Version]
- Zaheer, M.; Giri, C.C. Multiple shoot induction and jasmonic versus salicylic acid driven elicitation for enhanced andrographolide production in Andrographis paniculata. Plant Cell Tissue Organ. Cult. 2015, 122, 553–563. [Google Scholar] [CrossRef]
- Xu, J.; Xu, H.; Zhao, H.; Liu, H.; Xu, L.; Liang, Z. Genome-wide investigation of bHLH genes and expression analysis under salt and hormonal treatments in Andrographis paniculata. Ind. Crops Prod. 2022, 183, 114928. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Ibrahim, H.R.; Matsuzaki, T.; Aoki, T. Genetic evidence that antibacterial activity of lysozyme is independent of its catalytic function. FEBS Lett. 2001, 506, 27–32. [Google Scholar] [CrossRef]
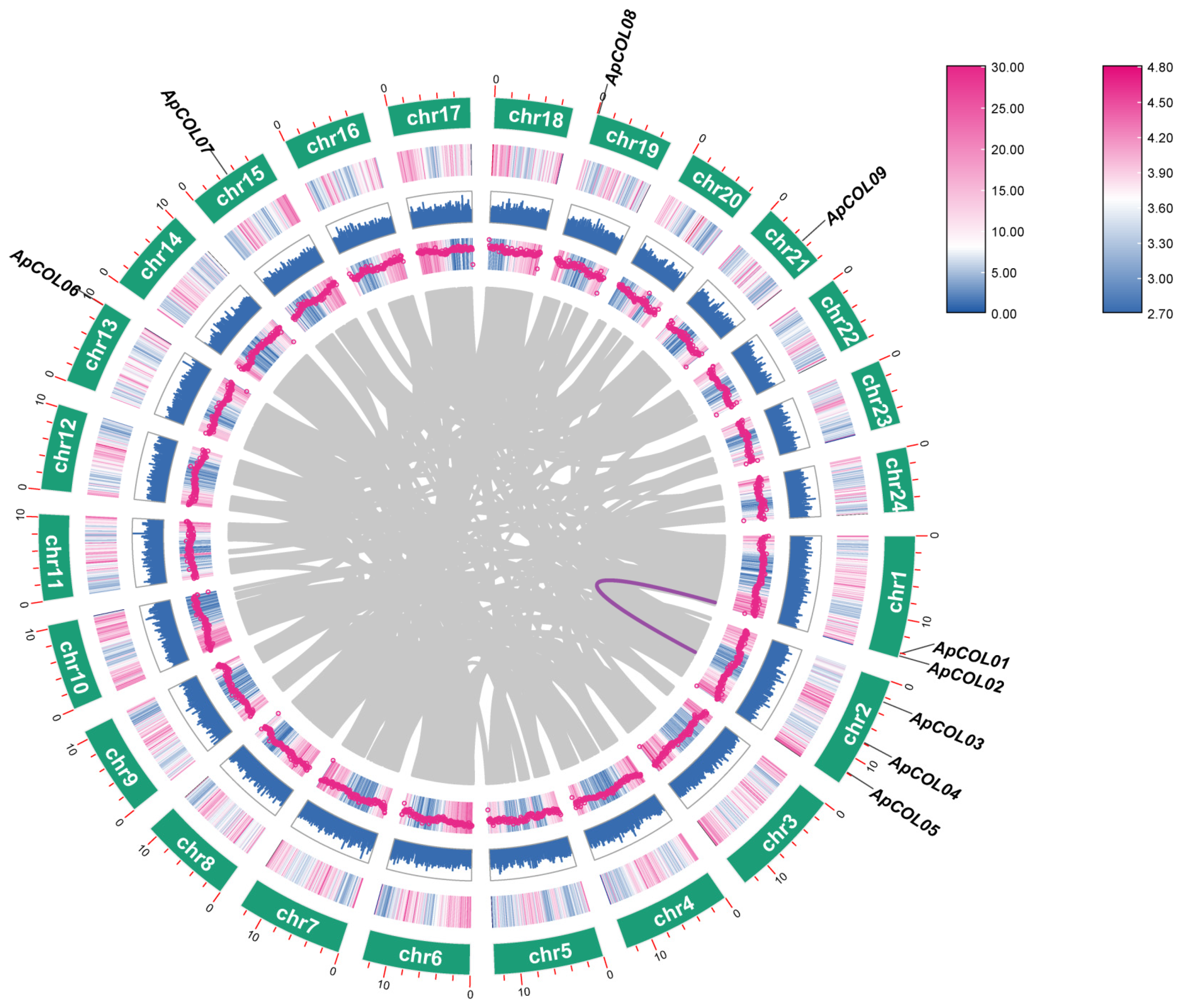
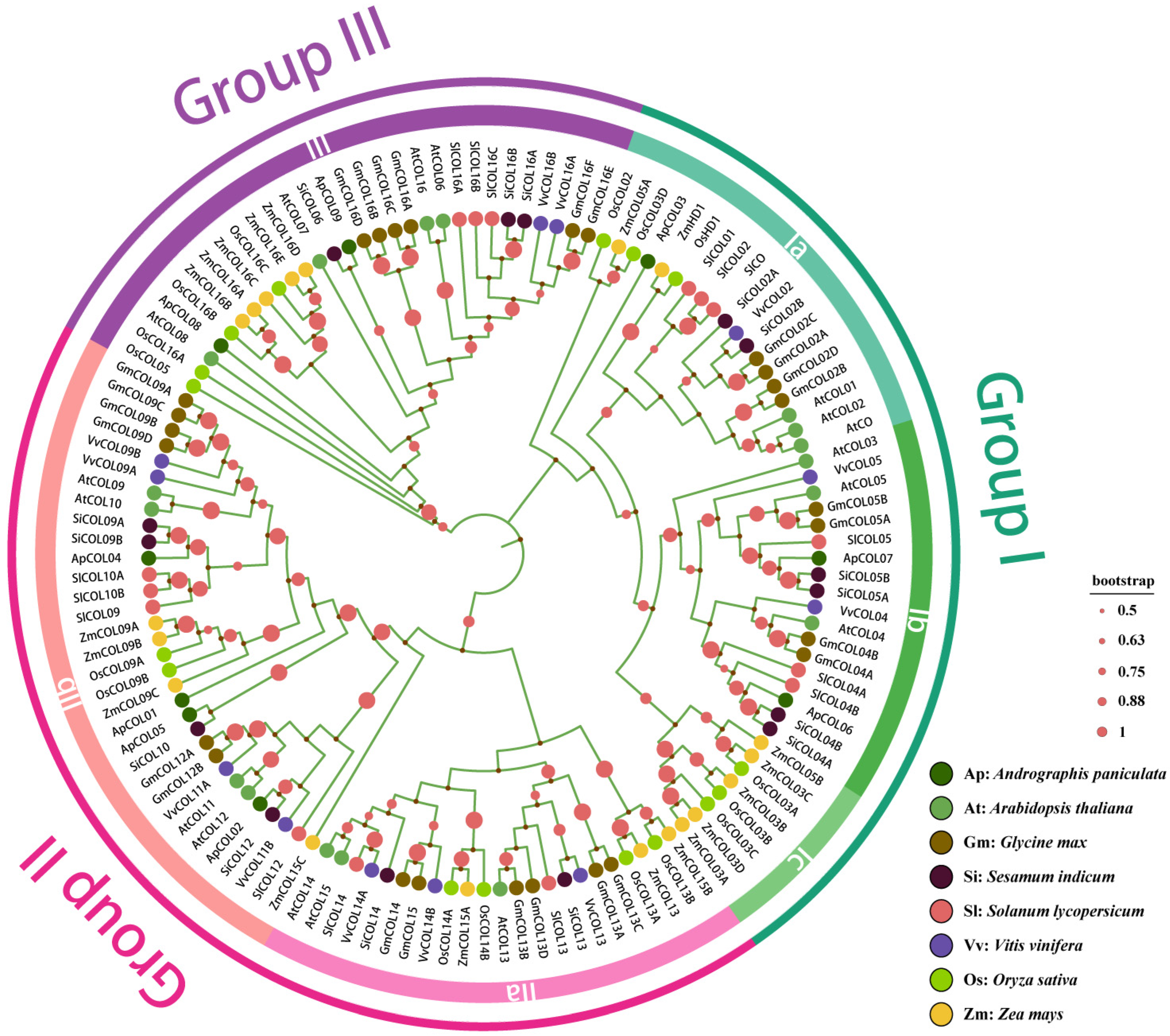
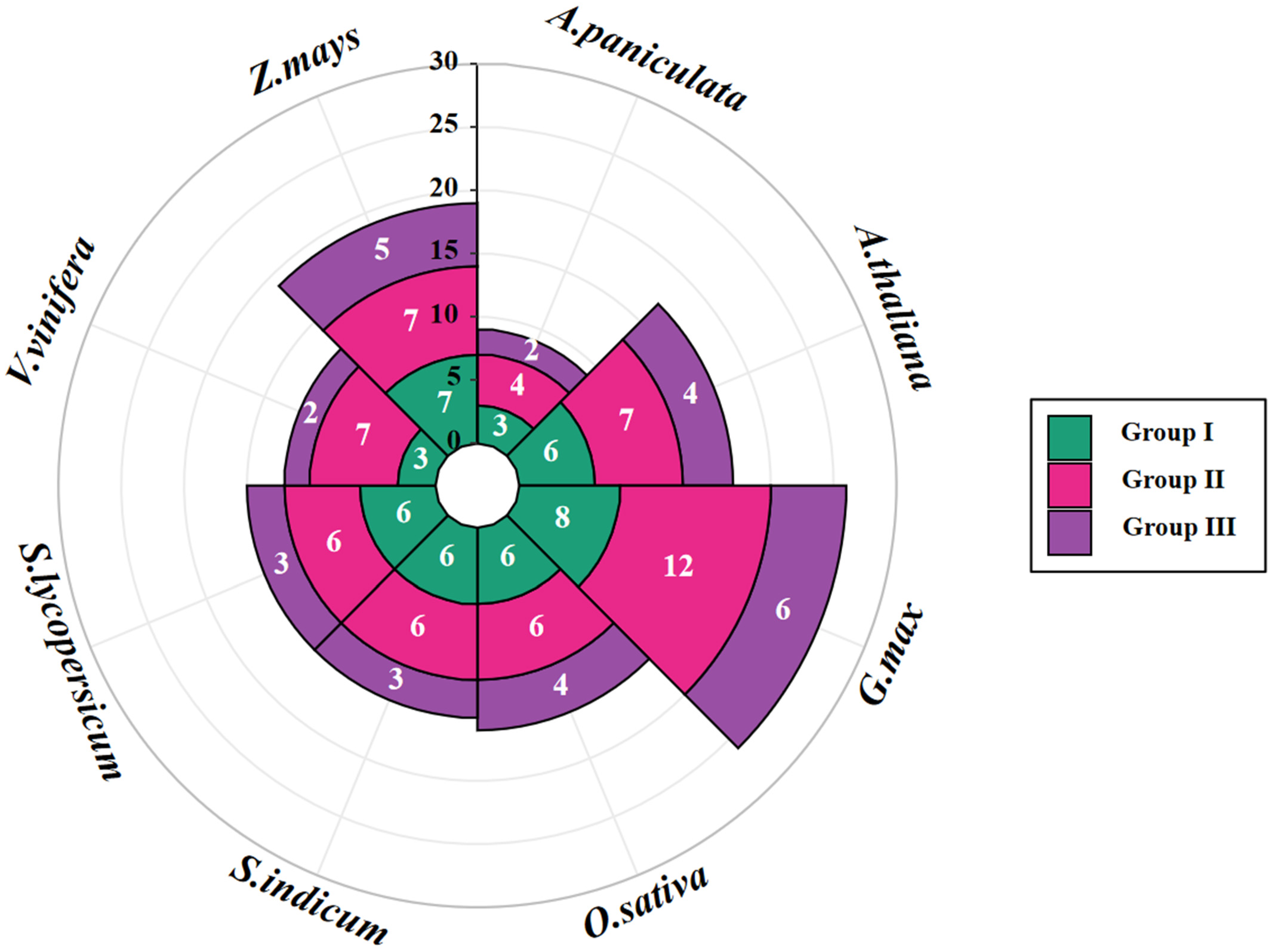
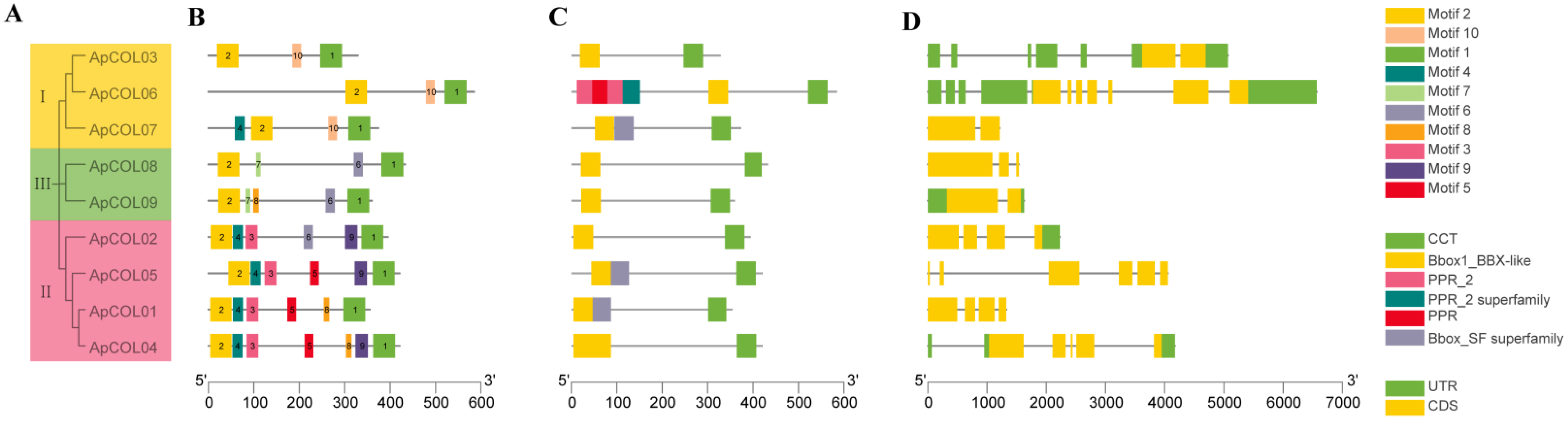

| Name | Gene ID | Chromosome | Strand | Gene Length (bp) | Protein | Predicted Subcellular Localization | ||
|---|---|---|---|---|---|---|---|---|
| Length (aa) | MW (kDa) | pI | ||||||
| ApCOL01 | CXN00019358 | Chr1 | + | 1327 | 264 | 39.13744 | 5.5 | Nucleus |
| ApCOL02 | CXN00016323 | Chr1 | + | 2228 | 247 | 43.25575 | 5.1 | Chloroplast |
| ApCOL03 | CXN00002171 | Chr2 | + | 5067 | 326 | 35.31866 | 5.84 | Nucleus |
| ApCOL04 | CXN00012269 | Chr2 | − | 4173 | 260 | 46.36184 | 6.24 | Nucleus |
| ApCOL05 | CXN00007803 | Chr2 | + | 4053 | 204 | 47.51274 | 5.1 | Nucleus |
| ApCOL06 | CXN00005866 | Chr13 | − | 1355 | 352 | 36.45549 | 5.2 | Nucleus |
| ApCOL07 | CXN00017383 | Chr15 | − | 1212 | 207 | 41.87428 | 6.81 | Chloroplast |
| ApCOL08 | CXN00004299 | Chr19 | − | 1536 | 211 | 48.41006 | 5.42 | Nucleus |
| ApCOL09 | CXN00005183 | Chr21 | + | 1625 | 238 | 40.98486 | 5.95 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Xu, J.; Xu, X.; Liu, H.; Chang, Q.; Xu, L.; Liang, Z. Genome-Wide Identification of CONSTANS-like (COL) Gene Family and the Potential Function of ApCOL08 Under Salt Stress in Andrographis paniculata. Int. J. Mol. Sci. 2025, 26, 724. https://doi.org/10.3390/ijms26020724
Zhao Y, Xu J, Xu X, Liu H, Chang Q, Xu L, Liang Z. Genome-Wide Identification of CONSTANS-like (COL) Gene Family and the Potential Function of ApCOL08 Under Salt Stress in Andrographis paniculata. International Journal of Molecular Sciences. 2025; 26(2):724. https://doi.org/10.3390/ijms26020724
Chicago/Turabian StyleZhao, Yizhu, Jiahao Xu, Xinyi Xu, Hui Liu, Qinxiang Chang, Ling Xu, and Zongsuo Liang. 2025. "Genome-Wide Identification of CONSTANS-like (COL) Gene Family and the Potential Function of ApCOL08 Under Salt Stress in Andrographis paniculata" International Journal of Molecular Sciences 26, no. 2: 724. https://doi.org/10.3390/ijms26020724
APA StyleZhao, Y., Xu, J., Xu, X., Liu, H., Chang, Q., Xu, L., & Liang, Z. (2025). Genome-Wide Identification of CONSTANS-like (COL) Gene Family and the Potential Function of ApCOL08 Under Salt Stress in Andrographis paniculata. International Journal of Molecular Sciences, 26(2), 724. https://doi.org/10.3390/ijms26020724






