Long Non-Coding RNAs in Malignant Human Brain Tumors: Driving Forces Behind Progression and Therapy
Abstract
1. Introduction
2. Research Progress on the Function of lncRNA in Malignant Brain Tumors
2.1. The Biological Functions of lncRNA in Malignant Brain Tumors
2.2. LncRNAs Regulate the Progression of Malignant Brain Tumors and Relate to Prognosis
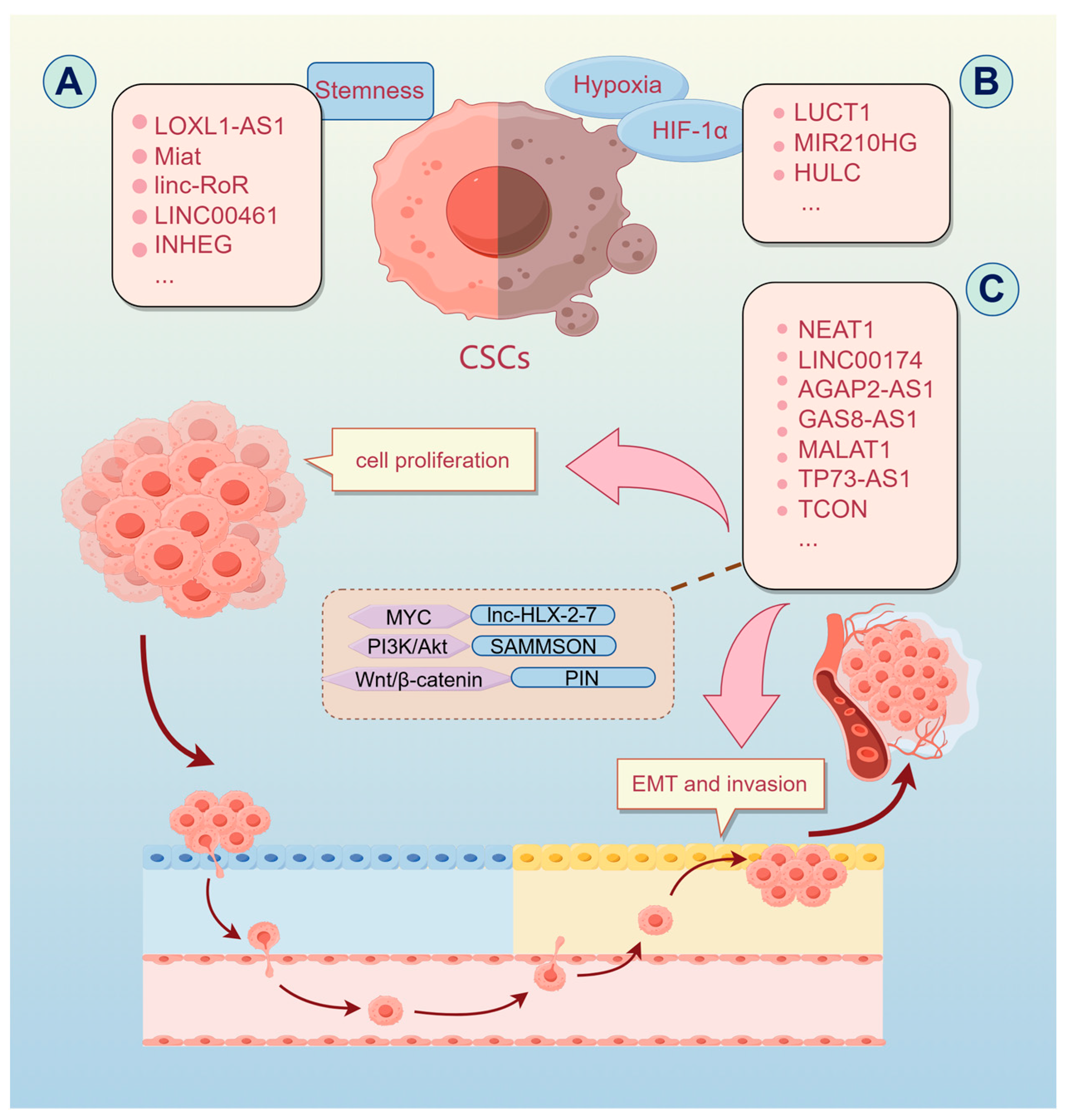
2.2.1. Tumor Stemness
2.2.2. Cell Proliferation and Metastasis
2.2.3. Multiple Feature-Related lncRNA Prognostic Models from Database Mining
2.3. lncRNA-miRNA Regulatory Network in Malignant Brain Tumors
3. Research Progress of lncRNAs in the Treatment of Malignant Brain Tumors
3.1. Chemosensitivity
3.1.1. TMZ Resistance
3.1.2. Overcoming the Blood–Brain Barrier
3.2. Radiosensitivity
3.3. Immunotherapy
4. Conclusions and Future Prospects
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nemeth, K.; Bayraktar, R.; Ferracin, M.; Calin, G.A. Non-coding RNAs in disease: From mechanisms to therapeutics. Nat. Rev. Genet. 2024, 25, 211–232. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhan, L.; Jiang, X.; Tang, X. Comprehensive review for non-coding RNAs: From mechanisms to therapeutic applications. Biochem. Pharmacol. 2024, 224, 116218. [Google Scholar] [CrossRef]
- Uppaluri, K.R.; Challa, H.J.; Gaur, A.; Jain, R.; Krishna Vardhani, K.; Geddam, A.; Natya, K.; Aswini, K.; Palasamudram, K.; Sri Manjari, K. Unlocking the potential of non-coding RNAs in cancer research and therapy. Transl. Oncol. 2023, 35, 101730. [Google Scholar] [CrossRef]
- Núñez-Martínez, H.N.; Recillas-Targa, F. Emerging Functions of lncRNA Loci beyond the Transcript Itself. Int. J. Mol. Sci. 2022, 23, 6258. [Google Scholar] [CrossRef]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.-T.; Lin, J.-F.; Li, T.; Li, J.-J.; Xu, R.-H.; Ju, H.-Q. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun. 2021, 41, 109–120. [Google Scholar] [CrossRef]
- McCabe, E.M.; Rasmussen, T.P. lncRNA involvement in cancer stem cell function and epithelial-mesenchymal transitions. Semin. Cancer Biol. 2021, 75, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. LncRNA-encoded peptides in cancer. J. Hematol. Oncol. 2024, 17, 66. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Gahan, P.B. Interplay between LncRNAs and microRNAs in Breast Cancer. Int. J. Mol. Sci. 2023, 24, 8095. [Google Scholar] [CrossRef] [PubMed]
- Braga, E.A.; Fridman, M.V.; Moscovtsev, A.A.; Filippova, E.A.; Dmitriev, A.A.; Kushlinskii, N.E. LncRNAs in Ovarian Cancer Progression, Metastasis, and Main Pathways: ceRNA and Alternative Mechanisms. Int. J. Mol. Sci. 2020, 21, 8855. [Google Scholar] [CrossRef]
- Ciafrè, S.A.; Russo, M.; Michienzi, A.; Galardi, S. Long Noncoding RNAs and Cancer Stem Cells: Dangerous Liaisons Managing Cancer. Int. J. Mol. Sci. 2023, 24, 1828. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Meng, Q.; Qian, J.; Li, M.; Gu, C.; Yang, Y. Review: RNA-based diagnostic markers discovery and therapeutic targets development in cancer. Pharmacol. Ther. 2022, 234, 108123. [Google Scholar] [CrossRef]
- Dai, J.; Su, Y.; Zhong, S.; Cong, L.; Liu, B.; Yang, J.; Tao, Y.; He, Z.; Chen, C.; Jiang, Y. Exosomes: Key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 2020, 5, 145. [Google Scholar] [CrossRef]
- Wu, P.; Mo, Y.; Peng, M.; Tang, T.; Zhong, Y.; Deng, X.; Xiong, F.; Guo, C.; Wu, X.; Li, Y.; et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol. Cancer 2020, 19, 22. [Google Scholar] [CrossRef] [PubMed]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef]
- Chen, B.; Dragomir, M.P.; Yang, C.; Li, Q.; Horst, D.; Calin, G.A. Targeting non-coding RNAs to overcome cancer therapy resistance. Signal Transduct. Target. Ther. 2022, 7, 121. [Google Scholar] [CrossRef]
- Park, E.-G.; Pyo, S.-J.; Cui, Y.; Yoon, S.-H.; Nam, J.-W. Tumor immune microenvironment lncRNAs. Brief. Bioinform. 2022, 23, bbab504. [Google Scholar] [CrossRef] [PubMed]
- Esposito, R.; Bosch, N.; Lanzós, A.; Polidori, T.; Pulido-Quetglas, C.; Johnson, R. Hacking the Cancer Genome: Profiling Therapeutically Actionable Long Non-coding RNAs Using CRISPR-Cas9 Screening. Cancer Cell 2019, 35, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.-H.; Yoon, G.; Ryu, Y.; Jeong, D.; Song, J.; Kim, Y.S.; Ahn, Y.; Kook, H.; Kim, Y.-K. Human lncRNA SUGCT-AS1 Regulates the Proinflammatory Response of Macrophage. Int. J. Mol. Sci. 2023, 24, 13315. [Google Scholar] [CrossRef]
- Schaff, L.R.; Mellinghoff, I.K. Glioblastoma and Other Primary Brain Malignancies in Adults: A Review. JAMA 2023, 329, 574–587. [Google Scholar] [CrossRef]
- Mellinghoff, I.K.; Cloughesy, T.F. Balancing Risk and Efficiency in Drug Development for Rare and Challenging Tumors: A New Paradigm for Glioma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2022, 40, 3510–3519. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Z.; Landry, A.P.; Raleigh, D.R.; Sahm, F.; Walsh, K.M.; Goldbrunner, R.; Yefet, L.S.; Tonn, J.C.; Gui, C.; Ostrom, Q.T.; et al. Meningioma: International Consortium on Meningiomas consensus review on scientific advances and treatment paradigms for clinicians, researchers, and patients. Neuro Oncol. 2024, 26, 1742–1780. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, J.; Li, Y.; Song, T.; Wu, Y.; Fang, S.; Bu, D.; Li, H.; Sun, L.; Pei, D.; et al. NONCODEV6: An updated database dedicated to long non-coding RNA annotation in both animals and plants. Nucleic Acids Res. 2021, 49, D165–D171. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Yildiz, C.B.; Kundu, T.; Gehrmann, J.; Koesling, J.; Ravaei, A.; Wolff, P.; Kraft, F.; Maié, T.; Jakovcevski, M.; Pensold, D.; et al. EphrinA5 regulates cell motility by modulating Snhg15/DNA triplex-dependent targeting of DNMT1 to the Ncam1 promoter. Epigenetics Chromatin 2023, 16, 42. [Google Scholar] [CrossRef]
- Bartl, J.; Zanini, M.; Bernardi, F.; Forget, A.; Blümel, L.; Talbot, J.; Picard, D.; Qin, N.; Cancila, G.; Gao, Q.; et al. The HHIP-AS1 lncRNA promotes tumorigenicity through stabilization of dynein complex 1 in human SHH-driven tumors. Nat. Commun. 2022, 13, 4061. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimpour, A.; Sarfi, M.; Rezatabar, S.; Tehrani, S.S. Novel insights into the interaction between long non-coding RNAs and microRNAs in glioma. Mol. Cell. Biochem. 2021, 476, 2317–2335. [Google Scholar] [CrossRef]
- Chaudhary, R. Potential of long non-coding RNAs as a therapeutic target and molecular markers in glioblastoma pathogenesis. Heliyon 2021, 7, e06502. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Yu, Y.; Ni, X.; Li, C.; Hu, Y.; Wang, J.; Chen, F.; Xi, S.; Chen, Z. LncRNA LINC00998 inhibits the malignant glioma phenotype via the CBX3-mediated c-Met/Akt/mTOR axis. Cell Death Dis. 2020, 11, 1032. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Xin, T.; Pang, B.; Sun, J.; Liu, Z.H.; Qin, Z.; Ji, X.S.; Yang, F.; Wei, Y.B.; Wang, Z.X.; et al. A novel lncRNA MDHDH suppresses glioblastoma multiforme by acting as a scaffold for MDH2 and PSMA1 to regulate NAD+ metabolism and autophagy. J. Exp. Clin. Cancer Res. 2022, 41, 349. [Google Scholar] [CrossRef]
- Cardon, T.; Fournier, I.; Salzet, M. Unveiling a Ghost Proteome in the Glioblastoma Non-Coding RNAs. Front. Cell Dev. Biol. 2021, 9, 703583. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef]
- Hofman, D.A.; Ruiz-Orera, J.; Yannuzzi, I.; Murugesan, R.; Brown, A.; Clauser, K.R.; Condurat, A.L.; van Dinter, J.T.; Engels, S.A.G.; Goodale, A.; et al. Translation of non-canonical open reading frames as a cancer cell survival mechanism in childhood medulloblastoma. Mol. Cell 2024, 84, 261–276. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Zhang, Z.; Jia, L.; Zhang, H.; Zhang, S.; Wang, H.; Cheng, Z. Micropeptide AF127577.4-ORF hidden in a lncRNA diminishes glioblastoma cell proliferation via the modulation of ERK2/METTL3 interaction. Sci. Rep. 2024, 14, 12090. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Beylerli, O.; Gareev, I.; Sufianov, A.; Ilyasova, T.; Guang, Y. Long noncoding RNAs as promising biomarkers in cancer. Non-Coding RNA Res. 2022, 7, 66–70. [Google Scholar] [CrossRef]
- Pokorná, M.; Černá, M.; Boussios, S.; Ovsepian, S.V.; O’Leary, V.B. lncRNA Biomarkers of Glioblastoma Multiforme. Biomedicines 2024, 12, 932. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Jiang, L.; Yang, H.; Chen, T.; Wu, X.; Lv, K. Analyzing the lncRNA, miRNA, and mRNA-associated ceRNA networks to reveal potential prognostic biomarkers for glioblastoma multiforme. Cancer Cell Int. 2020, 20, 393. [Google Scholar] [CrossRef]
- Do, A.D.; Wu, K.-S.; Chu, S.-S.; Giang, L.H.; Lin, Y.-L.; Chang, C.-C.; Wong, T.-T.; Hsieh, C.-L.; Sung, S.-Y. LOXL1-AS1 contributes to metastasis in sonic-hedgehog medulloblastoma by promoting cancer stem-like phenotypes. J. Exp. Clin. Cancer Res. 2024, 43, 130. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.-L.; Vasudevan, H.N.; Lockney, D.T.; Baum, R.; Hendrickson, R.C.; Raleigh, D.R.; Schmitt, A.M. Miat and interacting protein Metadherin maintain a stem-like niche to promote medulloblastoma tumorigenesis and treatment resistance. Proc. Natl. Acad. Sci. USA 2022, 119, e2203738119. [Google Scholar] [CrossRef]
- Kovalenko, T.F.; Yadav, B.; Anufrieva, K.S.; Rubtsov, Y.P.; Zatsepin, T.S.; Shcherbinina, E.Y.; Solyus, E.M.; Staroverov, D.B.; Larionova, T.D.; Latyshev, Y.A.; et al. Functions of long non-coding RNA ROR in patient-derived glioblastoma cells. Biochimie 2022, 200, 131–139. [Google Scholar] [CrossRef]
- Wu, A.-C.; Yang, W.-B.; Chang, K.-Y.; Lee, J.-S.; Liou, J.-P.; Su, R.-Y.; Cheng, S.M.; Hwang, D.-Y.; Kikkawa, U.; Hsu, T.-I.; et al. HDAC6 involves in regulating the lncRNA-microRNA-mRNA network to promote the proliferation of glioblastoma cells. J. Exp. Clin. Cancer Res. 2022, 41, 47. [Google Scholar] [CrossRef]
- Liu, L.; Liu, Z.; Liu, Q.; Wu, W.; Lin, P.; Liu, X.; Zhang, Y.; Wang, D.; Prager, B.C.; Gimple, R.C.; et al. LncRNA INHEG promotes glioma stem cell maintenance and tumorigenicity through regulating rRNA 2′-O-methylation. Nat. Commun. 2023, 14, 7526. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Shah, H.; Hao, J.; Lin, J.; Prayson, R.A.; Xie, L.; Bao, S.; Chakraborty, A.A.; Jankowsky, E.; Zhao, J.; et al. Long non-coding RNA lung cancer-associated transcript-1 promotes glioblastoma progression by enhancing Hypoxia-inducible factor 1 alpha activity. Neuro Oncol. 2024, 26, 1388–1401. [Google Scholar] [CrossRef]
- Ho, K.-H.; Shih, C.-M.; Liu, A.-J.; Chen, K.-C. Hypoxia-inducible lncRNA MIR210HG interacting with OCT1 is involved in glioblastoma multiforme malignancy. Cancer Sci. 2022, 113, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-P.; Liu, Y.; Xiao, L.-M.; Chen, L.-K.; Tao, E.-X.; Zeng, E.-M.; Xu, C.-H. Induction of cancer cell stemness in glioma through glycolysis and the long noncoding RNA HULC-activated FOXM1/AGR2/HIF-1α axis. Lab. Investig. 2022, 102, 691–701. [Google Scholar] [CrossRef]
- Katsushima, K.; Lee, B.; Kunhiraman, H.; Zhong, C.; Murad, R.; Yin, J.; Liu, B.; Garancher, A.; Gonzalez-Gomez, I.; Monforte, H.L.; et al. The long noncoding RNA lnc-HLX-2-7 is oncogenic in Group 3 medulloblastomas. Neuro Oncol. 2021, 23, 572–585. [Google Scholar] [CrossRef] [PubMed]
- Katsushima, K.; Joshi, K.; Yuan, M.; Romero, B.; Batish, M.; Stapleton, S.; Jallo, G.; Kolanthai, E.; Seal, S.; Saulnier, O.; et al. A therapeutically targetable positive feedback loop between lnc-HLX-2-7, HLX, and MYC that promotes group 3 medulloblastoma. Cell Rep. 2024, 43, 113938. [Google Scholar] [CrossRef]
- Liang, J.; Liu, C.; Xu, D.; Xie, K.; Li, A. LncRNA NEAT1 facilitates glioma progression via stabilizing PGK1. J. Transl. Med. 2022, 20, 80. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Q.; Bao, Z.; Guo, L.; Chen, H.; Lv, T.; Xu, T.; Zhang, X.; Zhou, C.; Sun, L. LINC00174 is a favorable prognostic biomarker in glioblastoma via promoting proliferative phenotype. Cancer Biomark. 2020, 28, 421–427. [Google Scholar] [CrossRef]
- Liu, S.J.; Dang, H.X.; Lim, D.A.; Feng, F.Y.; Maher, C.A. Long noncoding RNAs in cancer metastasis. Nat. Rev. Cancer 2021, 21, 446–460. [Google Scholar] [CrossRef]
- Tian, Y.; Zheng, Y.; Dong, X. AGAP2-AS1 serves as an oncogenic lncRNA and prognostic biomarker in glioblastoma multiforme. J. Cell Biochem. 2019, 120, 9056–9062. [Google Scholar] [CrossRef]
- Li, Q.; Cheng, Y. Long non-coding RNA LBX2-AS1 activates IL4R to promote glioblastoma metastasis and angiogenesis by binding to the transcription factor NFKB1. Folia Neuropathol. 2024, 62, 293–304. [Google Scholar] [CrossRef]
- Leung, D.H.L.; Phon, B.W.S.; Sivalingam, M.; Radhakrishnan, A.K.; Kamarudin, M.N.A. Regulation of EMT Markers, Extracellular Matrix, and Associated Signalling Pathways by Long Non-Coding RNAs in Glioblastoma Mesenchymal Transition: A Scoping Review. Biology 2023, 12, 818. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Jiang, T.; Huang, R.; Xiao, X. LncRNA GAS8-AS1 downregulates lncRNA NEAT1 to inhibit glioblastoma cell proliferation. Brain Behav. 2021, 11, e02128. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yuan, X.; Yan, D.; Li, D.; Guan, F.; Dong, Y.; Wang, H.; Liu, X.; Yang, B. Long Non-Coding RNA MALAT1 Decreases the Sensitivity of Resistant Glioblastoma Cell Lines to Temozolomide. Cell Physiol. Biochem. 2017, 42, 1192–1201. [Google Scholar] [CrossRef]
- Varon, M.; Levy, T.; Mazor, G.; Ben David, H.; Marciano, R.; Krelin, Y.; Prasad, M.; Elkabets, M.; Pauck, D.; Ahmadov, U.; et al. The long noncoding RNA TP73-AS1 promotes tumorigenicity of medulloblastoma cells. Int. J. Cancer 2019, 145, 3402–3413. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.; Wang, K.; Xie, P.; Zuo, J.; Liu, W.; Liu, C. LncRNA SAMMSON Knockdown Inhibits the Malignancy of Glioblastoma Cells by Inactivation of the PI3K/Akt Pathway. Cell. Mol. Neurobiol. 2021, 41, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Chen, Z.; Shen, L.; Tang, T.; Yang, M.; Zheng, X. Long Noncoding RNA LINC-PINT Suppresses Cell Proliferation, Invasion, and EMT by Blocking Wnt/β-Catenin Signaling in Glioblastoma. Front. Pharmacol. 2020, 11, 586653. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Wang, Y.; Zhang, L.; Wang, J.; Wang, W.; Han, X.; Mu, C.; Gao, D. Identification of novel LncRNA targeting Smad2/PKCα signal pathway to negatively regulate malignant progression of glioblastoma. J. Cell. Physiol. 2020, 235, 3835–3848. [Google Scholar] [CrossRef]
- Wedemeyer, M.A.; Muskens, I.; Strickland, B.A.; Aurelio, O.; Martirosian, V.; Wiemels, J.L.; Weisenberger, D.J.; Wang, K.; Mukerjee, D.; Rhie, S.K.; et al. Epigenetic dysregulation in meningiomas. Neuro-Oncol. Adv. 2022, 4, vdac084. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Liu, Z.; Gao, J.; Bing, X.; He, D.; Liu, W.; Wang, Y.; Wei, Y.; Yin, X.; Zhang, F.; et al. N6-Methyladenosine-modified lncRNA LINREP promotes Glioblastoma progression by recruiting the PTBP1/HuR complex. Cell Death Differ. 2023, 30, 54–68. [Google Scholar] [CrossRef]
- Li, L.; Zhou, A.; Wei, Y.; Liu, F.; Li, P.; Fang, R.; Ma, L.; Zhang, S.; Wang, L.; Liu, J.; et al. Critical role of lncEPAT in coupling dysregulated EGFR pathway and histone H2A deubiquitination during glioblastoma tumorigenesis. Sci. Adv. 2022, 8, eabn2571. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Peng, Z.; Li, P.; Fu, H.; Feng, J.; Zhang, Y.; Liu, T.; Liu, Y.; Liu, Q.; Liu, Q.; et al. lncRNA RMST Suppressed GBM Cell Mitophagy through Enhancing FUS SUMOylation. Mol. Ther. Nucleic Acids 2020, 19, 1198–1208. [Google Scholar] [CrossRef] [PubMed]
- Balandeh, E.; Mohammadshafie, K.; Mahmoudi, Y.; Hossein Pourhanifeh, M.; Rajabi, A.; Bahabadi, Z.R.; Mohammadi, A.H.; Rahimian, N.; Hamblin, M.R.; Mirzaei, H. Roles of Non-coding RNAs and Angiogenesis in Glioblastoma. Front. Cell Dev. Biol. 2021, 9, 716462. [Google Scholar] [CrossRef] [PubMed]
- Malgulwar, P.B.; Nambirajan, A.; Singh, M.; Suri, V.; Sarkar, C.; Sharma, M.C. Expression and Clinical Significance of Translation Regulatory Long Non-Coding RNA 1 (TRERNA1) in Ependymomas. Pathol. Oncol. Res. 2020, 26, 1975–1981. [Google Scholar] [CrossRef] [PubMed]
- Amer, R.G.; Ezz El Arab, L.R.; Abd El Ghany, D.; Saad, A.S.; Bahie-Eldin, N.; Swellam, M. Prognostic utility of lncRNAs (LINC00565 and LINC00641) as molecular markers in glioblastoma multiforme (GBM). J. Neurooncol. 2022, 158, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yang, S.; Lv, C.; Liu, Y. Cancer-associated fibroblasts suppressed ferroptosis in glioblastoma via upregulating lncRNA DLEU1. Am. J. Physiol. Cell Physiol. 2023, 324, C1039–C1052. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Zhong, W.; Liu, D.; Sun, X.; Hao, S.; Yang, Y.; Ao, L.; Zhou, J.; Xia, Y.; Zhou, Y.; et al. Computational identification of immune-related lncRNA signature for predicting the prognosis and immune landscape of human glioblastoma multiforme. Front. Immunol. 2022, 13, 932938. [Google Scholar] [CrossRef]
- Yu, W.; Ma, Y.; Hou, W.; Wang, F.; Cheng, W.; Qiu, F.; Wu, P.; Zhang, G. Identification of Immune-Related lncRNA Prognostic Signature and Molecular Subtypes for Glioblastoma. Front. Immunol. 2021, 12, 706936. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, N.; Wu, W.; Zhou, R.; Li, S.; Wang, Z.; Dai, Z.; Zhang, L.; Liu, Z.; Zhang, J.; et al. Machine learning-based tumor-infiltrating immune cell-associated lncRNAs for predicting prognosis and immunotherapy response in patients with glioblastoma. Brief. Bioinform. 2022, 23, bbac386. [Google Scholar] [CrossRef]
- Liu, D.-H.; Yang, X.; Guo, J.-F.; Meng, H.; Shen, S.-H. Immune-related lncRNAs, LINC01268 and CTB-31O20.2, as favorable prognostic markers for glioma inhibition. Transl. Cancer Res. 2022, 11, 823–834. [Google Scholar] [CrossRef]
- Cao, Y.; Zhu, H.; Tan, J.; Yin, W.; Zhou, Q.; Xin, Z.; Wu, Z.; Jiang, Z.; Guo, Y.; Kuang, Y.; et al. Development of an Immune-Related LncRNA Prognostic Signature for Glioma. Front. Genet. 2021, 12, 678436. [Google Scholar] [CrossRef]
- Cheng, M.; Sun, L.; Huang, K.; Yue, X.; Chen, J.; Zhang, Z.; Zhao, B.; Bian, E. A Signature of Nine lncRNA Methylated Genes Predicts Survival in Patients With Glioma. Front. Oncol. 2021, 11, 646409. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Wang, X.; Zhong, J.; Chen, H.; Song, D.; Xu, T.; Tian, K.; Sun, P.; Sun, N.; Qin, J.; et al. Using integrated analysis from multicentre studies to identify RNA methylation-related lncRNA risk stratification systems for glioma. Cancer Cell Int. 2023, 23, 156. [Google Scholar] [CrossRef] [PubMed]
- Tu, Z.; Wu, L.; Wang, P.; Hu, Q.; Tao, C.; Li, K.; Huang, K.; Zhu, X. N6-Methylandenosine-Related lncRNAs Are Potential Biomarkers for Predicting the Overall Survival of Lower-Grade Glioma Patients. Front. Cell Dev. Biol. 2020, 8, 642. [Google Scholar] [CrossRef] [PubMed]
- Shao, D.; Li, Y.; Wu, J.; Zhang, B.; Xie, S.; Zheng, X.; Jiang, Z. An m6A/m5C/m1A/m7G-Related Long Non-coding RNA Signature to Predict Prognosis and Immune Features of Glioma. Front. Genet. 2022, 13, 903117. [Google Scholar] [CrossRef]
- Luo, Q.; Yang, Z.; Deng, R.; Pang, X.; Han, X.; Liu, X.; Du, J.; Tian, Y.; Wu, J.; Tang, C. Comprehensive analysis of prognosis of patients with GBM based on 4 m6A-related lncRNAs and immune cell infiltration. Heliyon 2023, 9, e12838. [Google Scholar] [CrossRef]
- Li, B.; Zhao, R.; Qiu, W.; Pan, Z.; Zhao, S.; Qi, Y.; Qiu, J.; Zhang, S.; Guo, Q.; Fan, Y.; et al. The N6-methyladenosine-mediated lncRNA WEE2-AS1 promotes glioblastoma progression by stabilizing RPN2. Theranostics 2022, 12, 6363–6379. [Google Scholar] [CrossRef] [PubMed]
- Joshi, K.; Yuan, M.; Katsushima, K.; Saulnier, O.; Ray, A.; Amankwah, E.; Stapleton, S.; Jallo, G.; Taylor, M.D.; Eberhart, C.G.; et al. Systematic transcriptomic analysis of childhood medulloblastoma identifies N6-methyladenosine-dependent lncRNA signatures associated with molecular subtype, immune cell infiltration, and prognosis. Acta Neuropathol. Commun. 2024, 12, 138. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, W.; Ma, S.; Guan, X.; Zhang, D.; Peng, J.; Wang, X.; Yuan, L.; Li, P.; Mao, B.; et al. Identification of a Glycolysis-Related LncRNA Signature to Predict Survival in Diffuse Glioma Patients. Front. Oncol. 2020, 10, 597877. [Google Scholar] [CrossRef] [PubMed]
- Lei, Q.; Yuan, B.; Liu, K.; Peng, L.; Xia, Z. A novel prognostic related lncRNA signature associated with amino acid metabolism in glioma. Front. Immunol. 2023, 14, 1014378. [Google Scholar] [CrossRef]
- Wang, Q.; Zheng, C.; Hou, H.; Bao, X.; Tai, H.; Huang, X.; Li, Z.; Li, Z.; Wang, Q.; Pan, Q.; et al. Interplay of Sphingolipid Metabolism in Predicting Prognosis of GBM Patients: Towards Precision Immunotherapy. J. Cancer 2024, 15, 275–292. [Google Scholar] [CrossRef] [PubMed]
- Xing, Z.; Liu, Z.; Fu, X.; Zhou, S.; Liu, L.; Dang, Q.; Guo, C.; Ge, X.; Lu, T.; Zheng, Y.; et al. Clinical Significance and Immune Landscape of a Pyroptosis-Derived LncRNA Signature for Glioblastoma. Front. Cell Dev. Biol. 2022, 10, 805291. [Google Scholar] [CrossRef]
- Tanzhu, G.; Li, N.; Li, Z.; Zhou, R.; Shen, L. Molecular Subtypes and Prognostic Signature of Pyroptosis-Related lncRNAs in Glioma Patients. Front. Oncol. 2022, 12, 779168. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Zhao, H.; Wang, F.; Peng, L.; Zhang, H.; Wang, Z.; Jiang, F.; Zhang, D.; Yin, M.; Li, S.; et al. Integrative analysis to screen novel pyroptosis-related LncRNAs for predicting clinical outcome of glioma and validation in tumor tissue. Aging 2023, 15, 1628–1651. [Google Scholar] [CrossRef]
- Mamivand, A.; Bayat, S.; Maghrouni, A.; Shabani, S.; Khoshnevisan, A.; Saffar, H.; Tabrizi, M. Data mining of bulk and single-cell RNA sequencing introduces OBI1-AS1 as an astrocyte marker with possible role in glioma recurrence and progression. Clin. Epigenetics 2022, 14, 35. [Google Scholar] [CrossRef] [PubMed]
- Stackhouse, C.T.; Anderson, J.C.; Yue, Z.; Nguyen, T.; Eustace, N.J.; Langford, C.P.; Wang, J.; Rowland, J.R.; Xing, C.; Mikhail, F.M.; et al. An in vivo model of glioblastoma radiation resistance identifies long noncoding RNAs and targetable kinases. JCI Insight 2022, 7, e148717. [Google Scholar] [CrossRef] [PubMed]
- Slavik, H.; Balik, V.; Kokas, F.Z.; Slavkovsky, R.; Vrbkova, J.; Rehulkova, A.; Lausova, T.; Ehrmann, J.; Gurska, S.; Uberall, I.; et al. Transcriptomic Profiling Revealed Lnc-GOLGA6A-1 as a Novel Prognostic Biomarker of Meningioma Recurrence. Neurosurgery 2022, 91, 360–369. [Google Scholar] [CrossRef]
- Xu, C.; Zhao, J.; Song, J.; Xiao, M.; Cui, X.; Xin, L.; Xu, J.; Zhang, Y.; Yi, K.; Hong, B.; et al. lncRNA PRADX is a Mesenchymal Glioblastoma Biomarker for Cellular Metabolism Targeted Therapy. Front. Oncol. 2022, 12, 888922. [Google Scholar] [CrossRef] [PubMed]
- Tao, C.; Luo, H.; Chen, L.; Li, J.; Zhu, X.; Huang, K. Identification of an epithelial-mesenchymal transition related long non-coding RNA (LncRNA) signature in Glioma. Bioengineered 2021, 12, 4016–4031. [Google Scholar] [CrossRef]
- Yang, X.; Niu, S.; Liu, J.; Fang, J.; Wu, Z.; Ling, S.; Di, G.; Jiang, X. Identification of an epithelial-mesenchymal transition-related lncRNA prognostic signature for patients with glioblastoma. Sci. Rep. 2021, 11, 23694. [Google Scholar] [CrossRef]
- Huang, K.; Yue, X.; Zheng, Y.; Zhang, Z.; Cheng, M.; Li, L.; Chen, Z.; Yang, Z.; Bian, E.; Zhao, B. Development and Validation of an Mesenchymal-Related Long Non-Coding RNA Prognostic Model in Glioma. Front. Oncol. 2021, 11, 726745. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Zhang, Z.; Li, D.; Lv, Q.; Chen, S.; Zhang, Z.; Wu, M. LncRNA PELATON, a Ferroptosis Suppressor and Prognositic Signature for GBM. Front. Oncol. 2022, 12, 817737. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Ye, Y.; Tian, W.; Qiu, H. A Novel lncRNA Panel Related to Ferroptosis, Tumor Progression, and Microenvironment is a Robust Prognostic Indicator for Glioma Patients. Front. Cell Dev. Biol. 2021, 9, 788451. [Google Scholar] [CrossRef]
- Zheng, J.; Zhou, Z.; Qiu, Y.; Wang, M.; Yu, H.; Wu, Z.; Wang, X.; Jiang, X. A Prognostic Ferroptosis-Related lncRNAs Signature Associated With Immune Landscape and Radiotherapy Response in Glioma. Front. Cell Dev. Biol. 2021, 9, 675555. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Gu, Y.; Hong, S.; Yu, B.; Zhang, J.-H.; Liu, J.-N. Comprehensive analysis of lncRNA-TF crosstalks and identification of prognostic regulatory feedback loops of glioblastoma using lncRNA/TF-mediated ceRNA network. J. Cell Biochem. 2020, 121, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Long, S.; Wu, B.; Yang, L.; Wang, L.; Wang, B.; Yan, Y.; Jiang, J.; Yang, B.; Zhou, Q.; Shi, M.; et al. Novel tumor necrosis factor-related long non-coding RNAs signature for risk stratification and prognosis in glioblastoma. Front. Neurol. 2023, 14, 1054686. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.-F.; Wang, Y.-Z.; Wen, G.-B.; Jiang, J.-J. Prognostic model of kidney renal clear cell carcinoma using aging-related long noncoding RNA signatures identifies THBS1-IT1 as a potential prognostic biomarker for multiple cancers. Aging 2023, 15, 8630–8663. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Cheng, Y.; Li, R.; Lian, M.; Guo, S.; Liang, C. Development of a novel angiogenesis-related lncRNA signature to predict the prognosis and immunotherapy of glioblastoma multiforme. Transl. Cancer Res. 2023, 12, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Wu, Y.; Zhuo, Q.; Zuo, Y.; Lin, J.; Shi, H.; Zhou, H.; Xu, Z. Comprehensive analysis of oxidative stress-related lncRNA signatures in glioma reveals the discrepancy of prognostic and immune infiltration. Sci. Rep. 2023, 13, 7731. [Google Scholar] [CrossRef]
- Hao, S.; Gao, M.; Li, Q.; Shu, L.; Wang, P.; Hao, G. Machine learning predicts cuproptosis-related lncRNAs and survival in glioma patients. Sci. Rep. 2024, 14, 22323. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Zhu, L.; Gu, J.; Wang, T.; Shi, L.; Li, C.; Chen, L.; Xie, S.; Lu, Y. A necroptosis-related lncRNA signature was identified to predict the prognosis and immune microenvironment of IDH-wild-type GBM. Front. Oncol. 2022, 12, 1024208. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zhang, Y.; He, Y.; Liu, Y.; Qi, P. Screening and Bioinformatics Analysis of Competitive Endogenous RNA Regulatory Network --Related to Circular RNA in Breast Cancer. Biomed. Res. Int. 2021, 2021, 5575286. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Yang, G.; Kang, Y.; Li, S.; Duan, R.; Shen, L.; Jiang, W.; Qian, B.; Yin, Z.; Liang, T. Construction and Analysis of a ceRNA Network Reveals Potential Prognostic Markers in Colorectal Cancer. Front. Genet. 2020, 11, 418. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Li, M.; Tayier, T.; Zhang, M.; Chen, L.; Feng, S. Bioinformatics analysis of lncRNA-associated ceRNA network in melanoma. J. Cancer 2021, 12, 2921–2932. [Google Scholar] [CrossRef]
- Zeng, Z.-M.; Chen, Y.-Y.; Wen, X.-C.; Geng, X.-C.; Zhu, Y.-X.; Hao, L.-C.; Dong, Z.-S.; Yang, J.-F.; Wang, T.-T.; Zhang, R.-B.; et al. Whole-transcriptome sequencing with ceRNA regulation network construction and verification in glioblastoma. Am. J. Transl. Res. 2023, 15, 4291–4313. [Google Scholar]
- Bazrgar, M.; Mirmotalebisohi, S.A.; Ahmadi, M.; Azimi, P.; Dargahi, L.; Zali, H.; Ahmadiani, A. Comprehensive analysis of lncRNA-associated ceRNA network reveals novel potential prognostic regulatory axes in glioblastoma multiforme. J. Cell Mol. Med. 2024, 28, e18392. [Google Scholar] [CrossRef]
- Wang, L.; Cho, K.B.; Li, Y.; Tao, G.; Xie, Z.; Guo, B. Long Noncoding RNA (lncRNA)-Mediated Competing Endogenous RNA Networks Provide Novel Potential Biomarkers and Therapeutic Targets for Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 5758. [Google Scholar] [CrossRef]
- Li, B.; Shen, M.; Yao, H.; Chen, X.; Xiao, Z. Long Noncoding RNA TP73-AS1 Modulates Medulloblastoma Progression In Vitro And In Vivo By Sponging miR-494-3p And Targeting EIF5A2. Onco Targets Ther. 2019, 12, 9873–9885. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, N.; Fu, J.; Zhou, W. Long noncoding RNA HOTAIR promotes medulloblastoma growth, migration and invasion by sponging miR-1/miR-206 and targeting YY1. Biomed. Pharmacother. 2020, 124, 109887. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.-H.; Fan, W.-J.; An, Z.-J.; Sun, Y. Inhibition of Long Noncoding RNA CRNDE Increases Chemosensitivity of Medulloblastoma Cells by Targeting miR-29c-3p. Oncol. Res. 2020, 28, 95. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Ge, Y.; Wang, D.; Liu, Q.; Sun, S.; Hua, L.; Deng, J.; Luan, S.; Cheng, H.; Xie, Q.; et al. LncRNA-IMAT1 Promotes Invasion of Meningiomas by Suppressing KLF4/hsa-miR22-3p/Snai1 Pathway. Mol. Cells 2022, 45, 388–402. [Google Scholar] [CrossRef]
- Zheng, J.; Pang, C.-H.; Du, W.; Wang, L.; Sun, L.-G.; Xing, Z.-Y. An allele of rs619586 polymorphism in MALAT1 alters the invasiveness of meningioma via modulating the expression of collagen type V alpha (COL5A1). J. Cell Mol. Med. 2020, 24, 10223–10232. [Google Scholar] [CrossRef] [PubMed]
- Liao, K.; Lin, Y.; Gao, W.; Xiao, Z.; Medina, R.; Dmitriev, P.; Cui, J.; Zhuang, Z.; Zhao, X.; Qiu, Y.; et al. Blocking lncRNA MALAT1/miR-199a/ZHX1 Axis Inhibits Glioblastoma Proliferation and Progression. Mol. Ther. Nucleic Acids 2019, 18, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Yi, X.; Xu, J.; Huang, Z.; Bu, X.; Wang, D.; Ge, H.; Zhang, G.; Gu, J.; Kang, D.; et al. Long Non-Coding RNA MEG3 Modifies Cell-Cycle, Migration, Invasion, and Proliferation Through AKAP12 by Sponging miR-29c in Meningioma Cells. Front. Oncol. 2020, 10, 537763. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Huang, M.-Y. Tumor-Suppressive Function of lncRNA-MEG3 in Glioma Cells by Regulating miR-6088/SMARCB1 Axis. Biomed. Res. Int. 2020, 2020, 4309161. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, R.; Li, Q.; Li, Y.; Xuan, T.; Cao, S.; Zheng, J. SNHG1/miR-556-5p/TCF12 feedback loop enhances the tumorigenesis of meningioma through Wnt signaling pathway. J. Cell Biochem. 2020, 121, 1880–1889. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Du, J.; Liu, J.; Hou, K.; Ji, H.; Ma, S.; Ba, Y.; Chen, L.; Xie, R.; Hu, S. FtMt promotes glioma tumorigenesis and angiogenesis via lncRNA SNHG1/miR-9-5p axis. Cell Signal 2020, 75, 109749. [Google Scholar] [CrossRef]
- Wang, X.; Tian, W.; Wu, L.; Wei, Z.; Li, W.; Xu, Y.; Li, Y. LncRNA SNHG4 regulates miR-138/c-Met axis to promote the proliferation of glioblastoma cells. Neuroreport 2020, 31, 657–662. [Google Scholar] [CrossRef]
- Chen, Y.; Yuan, S.; Ning, T.; Xu, H.; Guan, B. SNHG7 Facilitates Glioblastoma Progression by Functioning as a Molecular Sponge for MicroRNA-449b-5p and Thereby Increasing MYCN Expression. Technol. Cancer Res. Treat. 2020, 19, 1533033820945802. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Zhao, Z.; Shang, J.; Li, G.; Zhang, R. LncRNA NEAT1 promotes glioma cancer progression via regulation of miR-98-5p/BZW1. Biosci. Rep. 2021, 41, BSR20200767. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, H.; Wang, J.; Niu, W.; Deng, C.; Zhou, M. LncRNA NEAT1 Enhances Glioma Progression via Regulating the miR-128-3p/ITGA5 Axis. Mol. Neurobiol. 2021, 58, 5163–5177. [Google Scholar] [CrossRef] [PubMed]
- Shou, J.; Gao, H.; Cheng, S.; Wang, B.; Guan, H. LncRNA HOXA-AS2 promotes glioblastoma carcinogenesis by targeting miR-885-5p/RBBP4 axis. Cancer Cell Int. 2021, 21, 39. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wang, L. HOXA-AS2 enhances GBM cell malignancy by suppressing miR-2116-3p thereby upregulating SERPINA3. BMC Cancer 2022, 22, 366. [Google Scholar]
- Chen, W.; Li, Q.; Zhang, G.; Wang, H.; Zhu, Z.; Chen, L. LncRNA HOXA-AS3 promotes the malignancy of glioblastoma through regulating miR-455-5p/USP3 axis. J. Cell Mol. Med. 2020, 24, 11755–11767. [Google Scholar] [CrossRef]
- Li, H.L.; Han, P.H.; Pan, D.Q.; Chen, G.; Lu, X.H.; Li, J. LncRNA XIST regulates cell proliferation, migration and invasion of glioblastoma via regulating miR-448 and ROCK1. J. Biol. Regul. Homeost. Agents 2020, 34, 2049–2058. [Google Scholar] [PubMed]
- Cheng, Z.; Luo, C.; Guo, Z. LncRNA-XIST/microRNA-126 sponge mediates cell proliferation and glucose metabolism through the IRS1/PI3K/Akt pathway in glioma. J. Cell Biochem. 2020, 121, 2170–2183. [Google Scholar] [CrossRef]
- Luo, C.; Quan, Z.; Zhong, B.; Zhang, M.; Zhou, B.; Wang, S.; Luo, X.; Tang, C. lncRNA XIST promotes glioma proliferation and metastasis through miR-133a/SOX4. Exp. Ther. Med. 2020, 19, 1641–1648. [Google Scholar] [CrossRef]
- Gong, M.; Wang, X.; Mu, L.; Wang, Y.; Pan, J.; Yuan, X.; Zhou, H.; Xing, J.; Wang, R.; Sun, J.; et al. Steroid receptor coactivator-1 enhances the stemness of glioblastoma by activating long noncoding RNA XIST/miR-152/KLF4 pathway. Cancer Sci. 2021, 112, 604–618. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Yu, Q.; Peng, Y.; Zhou, S.; Liu, Z.; Deng, Y.; Guo, L.; Zhao, S.; Chen, G. Novel LncRNA OXCT1-AS1 indicates poor prognosis and contributes to tumorigenesis by regulating miR-195/CDC25A axis in glioblastoma. J. Exp. Clin. Cancer Res. 2021, 40, 123. [Google Scholar] [CrossRef]
- Liu, K.; Deng, Y.; Yang, Y.; Wang, H.; Zhou, P. MicorRNA-195 links long non-coding RNA SEMA3B antisense RNA 1 (head to head) and cyclin D1 to regulate the proliferation of glioblastoma cells. Bioengineered 2022, 13, 8798–8805. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Wang, S.; Li, Q.; Ma, Y.; Sun, P. Long noncoding RNA LINC00460 targets miR-539/MMP-9 to promote meningioma progression and metastasis. Biomed. Pharmacother. 2018, 105, 677–682. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Ren, J.; Ma, J.; Wu, J.; Zhang, R.; Yuan, H.; Han, X. LINC00702/miR-4652-3p/ZEB1 axis promotes the progression of malignant meningioma through activating Wnt/β-catenin pathway. Biomed. Pharmacother. 2019, 113, 108718. [Google Scholar] [CrossRef]
- Cao, J.; Tang, Z.; Su, Z. Long non-coding RNA LINC01426 facilitates glioblastoma progression via sponging miR-345-3p and upregulation of VAMP8. Cancer Cell Int. 2020, 20, 327. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Lv, L.; Zhang, Z.; Wei, S.; Zheng, T. LINC00294 negatively modulates cell proliferation in glioma through a neurofilament medium-mediated pathway via interacting with miR-1278. J. Gene Med. 2020, 22, e3235. [Google Scholar] [CrossRef]
- Yang, J.; Yu, D.; Liu, X.; Changyong, E.; Yu, S. LINC00641/miR-4262/NRGN axis confines cell proliferation in glioma. Cancer Biol. Ther. 2020, 21, 758–766. [Google Scholar] [CrossRef]
- Du, X.; Tu, Y.; Liu, S.; Zhao, P.; Bao, Z.; Li, C.; Li, J.; Pan, M.; Ji, J. LINC00511 contributes to glioblastoma tumorigenesis and epithelial-mesenchymal transition via LINC00511/miR-524-5p/YB1/ZEB1 positive feedback loop. J. Cell Mol. Med. 2020, 24, 1474–1487. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Cheng, Y. LINC01198 facilitates gliomagenesis through activating PI3K/AKT pathway. RNA Biol. 2020, 17, 1040–1052. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.; Xie, M. LINC01579 promotes cell proliferation by acting as a ceRNA of miR-139-5p to upregulate EIF4G2 expression in glioblastoma. J. Cell. Physiol. 2019, 234, 23658–23666. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Yu, B.; Li, Y.; Zhang, W.; Alvarez, A.A.; Hu, B.; Cheng, S.-Y.; Feng, H. TGF-β-activated lncRNA LINC00115 is a critical regulator of glioma stem-like cell tumorigenicity. EMBO Rep. 2019, 20, e48170. [Google Scholar] [CrossRef]
- Dong, N.; Qi, W.; Wu, L.; Li, J.; Zhang, X.; Wu, H.; Zhang, W.; Jiang, J.; Zhang, S.; Fu, W.; et al. LINC00606 promotes glioblastoma progression through sponge miR-486-3p and interaction with ATP11B. J. Exp. Clin. Cancer Res. 2024, 43, 139. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, L.; Liang, Z.; Xi, Y. Long Non-Coding RNA BCAR4 Promotes Growth, Invasion and Tumorigenicity by Targeting miR-2276 to Upregulate MMP7 Expression in Glioma. Onco Targets Ther. 2019, 12, 10963–10973. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Chen, Q.; Liu, J.; Liao, Y.; Jiang, Q. Silencing of lncRNA CHRM3-AS2 Expression Exerts Anti-Tumour Effects Against Glioma via Targeting microRNA-370-5p/KLF4. Front. Oncol. 2022, 12, 856381. [Google Scholar] [CrossRef] [PubMed]
- Liao, K.; Qian, Z.; Zhang, S.; Chen, B.; Li, Z.; Huang, R.; Cheng, L.; Wang, T.; Yang, R.; Lan, J.; et al. The LGMN pseudogene promotes tumor progression by acting as a miR-495-3p sponge in glioblastoma. Cancer Lett. 2020, 490, 111–123. [Google Scholar] [CrossRef]
- Xie, P.; Li, X.; Chen, R.; Liu, Y.; Liu, D.; Liu, W.; Cui, G.; Xu, J. Upregulation of HOTAIRM1 increases migration and invasion by glioblastoma cells. Aging (Albany NY) 2020, 13, 2348–2364. [Google Scholar] [CrossRef]
- Li, D.; Hu, J.; Li, S.; Zhou, C.; Feng, M.; Li, L.; Gao, Y.; Chen, X.; Wu, X.; Cao, Y.; et al. LINC01393, a Novel Long Non-Coding RNA, Promotes the Cell Proliferation, Migration and Invasion through MiR-128-3p/NUSAP1 Axis in Glioblastoma. Int. J. Mol. Sci. 2023, 24, 5878. [Google Scholar] [CrossRef] [PubMed]
- Li, X.X.; Yu, Q. Linc01094 Accelerates the Growth and Metastatic-Related Traits of Glioblastoma by Sponging miR-126-5p. Onco Targets Ther. 2020, 13, 9917–9928. [Google Scholar] [CrossRef]
- Hu, S.; Yao, Y.; Hu, X.; Zhu, Y. LncRNA DCST1-AS1 downregulates miR-29b through methylation in glioblastoma (GBM) to promote cancer cell proliferation. Clin. Transl. Oncol. 2020, 22, 2230–2235. [Google Scholar] [CrossRef]
- Gu, J.; Ye, Y.; Sunil, R.; Zhan, W.; Yu, R. Downregulation of lncRNA SATB2-AS1 facilitates glioma cell proliferation by sponging miR-671-5p. Exp. Ther. Med. 2023, 26, 503. [Google Scholar] [CrossRef]
- Zhou, C.; Jiang, X.; Liang, A.; Zhu, R.; Yang, Y.; Zhong, L.; Wan, D. LncRNA FEZF1-AS1 aggravates cell proliferation and migration in glioblastoma. Neurosci. Lett. 2021, 764, 136245. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Chen, Y.; Zeng, Y.; Xu, C.; Huang, J.; Hu, W.; Chen, X.; Fu, H. Long non-coding RNA PSMA3-AS1 promotes glioma progression through modulating the miR-411-3p/HOXA10 pathway. BMC Cancer 2021, 21, 844. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Niu, W.; Mu, M.; Hu, S.; Niu, C. Long non-coding RNA LPP-AS2 promotes glioma tumorigenesis via miR-7-5p/EGFR/PI3K/AKT/c-MYC feedback loop. J. Exp. Clin. Cancer Res. 2020, 39, 196. [Google Scholar] [CrossRef]
- Wu, J.; Li, R.; Li, L.; Gu, Y.; Zhan, H.; Zhou, C.; Zhong, C. MYC-activated lncRNA HNF1A-AS1 overexpression facilitates glioma progression via cooperating with miR-32-5p/SOX4 axis. Cancer Med. 2020, 9, 6387–6398. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, H.; Xu, M.; Chen, F.; Li, W.; Hu, H.; Yuan, Q.; Su, Y.; Liu, X.; Wuri, J.; et al. Long noncoding RNA HAS2-AS1 promotes tumor progression in glioblastoma via functioning as a competing endogenous RNA. J. Cell Biochem. 2020, 121, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Tang, Q.; Feng, H.; Zhu, L.; Wang, Y. lncRNA KTN1-AS1 promotes glioma cell proliferation and invasion by negatively regulating miR-505-3p. Oncol. Rep. 2020, 44, 2645–2655. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Cai, Y. Long non-coding RNA (lncRNA) HOXD-AS2 promotes glioblastoma cell proliferation, migration and invasion by regulating the miR-3681-5p/MALT1 signaling pathway. Bioengineered 2021, 12, 9113–9127. [Google Scholar] [CrossRef]
- Ma, C.; Wang, H.; Zong, G.; He, J.; Wang, Y.; Yang, F.; Yang, Z.; Bian, E.; Zhao, B. EGR1 modulated LncRNA HNF1A-AS1 drives glioblastoma progression via miR-22-3p/ENO1 axis. Cell Death Discov. 2021, 7, 350. [Google Scholar] [CrossRef]
- Wang, Z.; Han, Y.; Li, Q.; Wang, B.; Ma, J. LncRNA DLGAP1-AS1 accelerates glioblastoma cell proliferation through targeting miR-515-5p/ROCK1/NFE2L1 axis and activating Wnt signaling pathway. Brain Behav. 2021, 11, e2321. [Google Scholar] [CrossRef]
- Li, H.; Wang, D.; Yi, B.; Cai, H.; Wang, Y.; Lou, X.; Xi, Z.; Li, Z. SUMOylation of IGF2BP2 promotes vasculogenic mimicry of glioma via regulating OIP5-AS1/miR-495-3p axis. Int. J. Biol. Sci. 2021, 17, 2912–2930. [Google Scholar] [CrossRef]
- Gao, W.; Li, H.; Liu, Y.; Zhang, Y.; Zhao, H.; Liu, F. Long non-coding RNA FLVCR1-AS1 promotes glioma cell proliferation and invasion by negatively regulating miR-30b-3p. Mol. Med. Rep. 2020, 22, 723–732. [Google Scholar] [CrossRef]
- Zeng, S.; Zhou, C.; Yang, D.-H.; Xu, L.-S.; Yang, H.-J.; Xu, M.-H.; Wang, H. LEF1-AS1 is implicated in the malignant development of glioblastoma via sponging miR-543 to upregulate EN2. Brain Res. 2020, 1736, 146781. [Google Scholar] [CrossRef]
- Lu, Y.; Guo, G.; Hong, R.; Chen, X.; Sun, Y.; Liu, F.; Zhang, Z.; Jin, X.; Dong, J.; Yu, K.; et al. LncRNA HAS2-AS1 Promotes Glioblastoma Proliferation by Sponging miR-137. Front. Oncol. 2021, 11, 634893. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cui, Y.; Ma, W.; Wang, M.; Cai, Y.; Jiang, Y. LncRNA RBPMS-AS1 promotes NRGN transcription to enhance the radiosensitivity of glioblastoma through the microRNA-301a-3p/CAMTA1 axis. Transl. Oncol. 2022, 15, 101282. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Zhang, Y.; He, H.; Chen, C.; Zhang, B.; Cai, M. LncRNA FEZF1-AS1 Sponges miR-34a to Upregulate Notch-1 in Glioblastoma. Cancer Manag. Res. 2020, 12, 1827–1833. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Wang, F.; Han, G. LncRNA PSMB8-AS1 acts as ceRNA of miR-22-3p to regulate DDIT4 expression in glioblastoma. Neurosci. Lett. 2020, 728, 134896. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhu, X.; Song, Z.; Guo, M.; Liang, J.; Yan, D. FGD5-AS1 facilitates glioblastoma progression by activation of Wnt/β-catenin signaling via regulating miR-129-5p/HNRNPK axis. Life Sci. 2020, 256, 117998. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Ren, K.; Zhao, J.; Li, J.; Jia, B.; Wu, X.; Dou, Y.; Fei, X.; Huan, Y.; He, X.; et al. LncRNA GAS5 represses stemness and malignancy of gliomas via elevating the SPACA6-miR-125a/let-7e Axis. Front. Oncol. 2022, 12, 803652. [Google Scholar] [CrossRef]
- Shree, B.; Tripathi, S.; Sharma, V. Transforming Growth Factor-Beta-Regulated LncRNA-MUF Promotes Invasion by Modulating the miR-34a Snail1 Axis in Glioblastoma Multiforme. Front. Oncol. 2021, 11, 788755. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wang, W.; Wu, Z.; Chen, S.; Chen, X.; Zhuang, S.; Song, G.; Lv, Y.; Lin, Y. Over-expression of lncRNA TMEM161B-AS1 promotes the malignant biological behavior of glioma cells and the resistance to temozolomide via up-regulating the expression of multiple ferroptosis-related genes by sponging hsa-miR-27a-3p. Cell Death Discov. 2021, 7, 311. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Zhang, J.; Zhao, L.; Fu, X.; Zhao, Q.; Chao, M.; Cao, H.; Jiao, Y.; Hu, Y.; Chen, C.; et al. Long Noncoding RNA NONHSAT079852.2 Contributes to GBM Recurrence by Functioning as a ceRNA for has-mir-10401-3p to Facilitate HSPA1A Upregulation. Front. Oncol. 2021, 11, 636632. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Dong, H.; Dong, Y.; Yang, X. Long non-coding RNA Unigene56159 promotes glioblastoma multiforme cell proliferation and invasion through negatively regulating microRNA-194-5p. Mol. Med. Rep. 2020, 21, 768–776. [Google Scholar] [CrossRef]
- Ma, Q.; Wang, X.; Li, J. LncRNA RP1-86C11.7 exacerbates the glioma progression and oncogenicity by hsa-miR-144-3p/TFRC signaling. Transl. Oncol. 2021, 14, 101215. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Li, T.; Wang, Y.; Wang, Y.; Gu, Q.; Li, Z. LncRNA MYCNOS promotes glioblastoma cell proliferation by regulating miR-216b/FOXM1 axis. Metab. Brain Dis. 2021, 36, 1185–1189. [Google Scholar] [CrossRef] [PubMed]
- Geng, S.; Tu, S.; Fu, W.; Wang, J.; Bai, Z. LncRNA PITPNA-AS1 stimulates cell proliferation and suppresses cell apoptosis in glioblastoma via targeting miR-223-3p/EGFR axis and activating PI3K/AKT signaling pathway. Cell Cycle 2021, 20, 1988–1998. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Tao, X.; Ji, B.-W.; Gong, J. Long Non-coding RNA COX10-AS1 Promotes Glioma Progression by Competitively Binding miR-1-3p to Regulate ORC6 Expression. Neuroscience 2024, 540, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Pan, L.; Yu, Z.; Ni, W. The lncRNA RP3-439F8.1 promotes GBM cell proliferation and progression by sponging miR-139-5p to upregulate NR5A2. Pathol.-Res. Pract. 2021, 223, 153319. [Google Scholar] [CrossRef]
- Zhao, H.; Li, J.; Yan, X.; Bian, X. LncRNA MAFG-AS1 Suppresses the Maturation of miR-34a to Promote Glioblastoma Cell Proliferation. Cancer Manag. Res. 2021, 13, 3493–3501. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Chen, B.; Jiao, H.; Yi, L. Long noncoding RNA UNC5B-AS1 suppresses cell proliferation by sponging miR-24-3p in glioblastoma multiforme. BMC Med. Genom. 2024, 17, 83. [Google Scholar] [CrossRef]
- Tian, Y.; Gao, X.; Yang, X.; Chen, S.; Ren, Y. Glioma-derived exosome Lncrna Agap2-As1 promotes glioma proliferation and metastasis by mediating Tgf-β1 secretion of myeloid-derived suppressor cells. Heliyon 2024, 10, e29949. [Google Scholar] [CrossRef]
- Rezaee, A.; Tehrany, P.M.; Tirabadi, F.J.; Sanadgol, N.; Karimi, A.S.; Ajdari, A.; Eydivandi, S.; Etemad, S.; Rajabi, R.; Rahmanian, P.; et al. Epigenetic regulation of temozolomide resistance in human cancers with an emphasis on brain tumors: Function of non-coding RNAs. Biomed. Pharmacother. 2023, 165, 115187. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Wang, B.; Zhao, S.; Xu, J. Inhibition of lncRNA NEAT1 sensitizes medulloblastoma cells to cisplatin through modulating the miR-23a-3p-glutaminase (GLS) axis. Bioengineered 2022, 13, 7670–7682. [Google Scholar] [CrossRef]
- Han, J.; Yu, X.; Wang, S.; Wang, Y.; Liu, Q.; Xu, H.; Wang, X. IGF2BP2 Induces U251 Glioblastoma Cell Chemoresistance by Inhibiting FOXO1-Mediated PID1 Expression Through Stabilizing lncRNA DANCR. Front. Cell Dev. Biol. 2021, 9, 659228. [Google Scholar] [CrossRef] [PubMed]
- Lang, F.; Liu, Y.; Chou, F.-J.; Yang, C. Genotoxic therapy and resistance mechanism in gliomas. Pharmacol. Ther. 2021, 228, 107922. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Wang, X.; Wang, Y.; Wang, K. Functions and underlying mechanisms of lncRNA HOTAIR in cancer chemotherapy resistance. Cell Death Discov. 2022, 8, 383. [Google Scholar] [CrossRef]
- Yang, E.; Hong, B.; Wang, Y.; Wang, Q.; Zhao, J.; Cui, X.; Wu, Y.; Yang, S.; Su, D.; Liu, X.; et al. EPIC-0628 abrogates HOTAIR/EZH2 interaction and enhances the temozolomide efficacy via promoting ATF3 expression and inhibiting DNA damage repair in glioblastoma. Cancer Lett. 2024, 588, 216812. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yang, S.; Cui, X.; Wang, Q.; Yang, E.; Tong, F.; Hong, B.; Xiao, M.; Xin, L.; Xu, C.; et al. A novel compound EPIC-0412 reverses temozolomide resistance via inhibiting DNA repair/MGMT in glioblastoma. Neuro Oncol. 2023, 25, 857–870. [Google Scholar] [CrossRef] [PubMed]
- Xin, L.; Tan, Y.; Zhu, Y.; Cui, X.; Wang, Q.; Zhao, J.; Tian, S.; Xu, C.; Xiao, M.; Hong, B.; et al. EPIC-0307-mediated selective disruption of PRADX-EZH2 interaction and enhancement of temozolomide sensitivity to glioblastoma via inhibiting DNA repair and MGMT. Neuro Oncol. 2023, 25, 1976–1988. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Quan, W.; Yu, D.-H.; Chen, X.; Wang, Z.-F.; Li, Z.-Q. High expression of LncRNA HOTAIR is a risk factor for temozolomide resistance in glioblastoma via activation of the miR-214/β-catenin/MGMT pathway. Sci. Rep. 2024, 14, 26224. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Hu, J.; Han, B.; Tan, S.; Jia, W.; Xin, Y. A positive feedback loop of lncRNA-RMRP/ZNRF3 axis and Wnt/β-catenin signaling regulates the progression and temozolomide resistance in glioma. Cell Death Dis. 2021, 12, 952. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-X.; Ren, P.; Cao, Y.-Y.; Wang, T.-T.; Huang, G.-H.; Li, Y.; Zhou, S.; Yang, W.; Yang, L.; Liu, G.-L.; et al. HOXD-AS2-STAT3 feedback loop attenuates sensitivity to temozolomide in glioblastoma. CNS Neurosci. Ther. 2023, 29, 3430–3445. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Xu, J.; Fan, Y.; Qi, Y.; Wang, S.; Zhao, S.; Guo, X.; Xue, H.; Deng, L.; Zhao, R.; et al. PDIA3P1 promotes Temozolomide resistance in glioblastoma by inhibiting C/EBPβ degradation to facilitate proneural-to-mesenchymal transition. J. Exp. Clin. Cancer Res. 2022, 41, 223. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Zhang, S.; Li, X.; Qian, W.; Li, M.; Chen, S.; Wu, X.; Yu, X.; Li, Z.; Tang, M.; et al. Dynamic structural remodeling of LINC01956 enhances temozolomide resistance in MGMT-methylated glioblastoma. Sci. Transl. Med. 2024, 16, eado1573. [Google Scholar] [CrossRef]
- Liu, B.; Zhou, J.; Wang, C.; Chi, Y.; Wei, Q.; Fu, Z.; Lian, C.; Huang, Q.; Liao, C.; Yang, Z.; et al. LncRNA SOX2OT promotes temozolomide resistance by elevating SOX2 expression via ALKBH5-mediated epigenetic regulation in glioblastoma. Cell Death Dis. 2020, 11, 384. [Google Scholar] [CrossRef]
- Yan, Y.; Xu, Z.; Chen, X.; Wang, X.; Zeng, S.; Zhao, Z.; Qian, L.; Li, Z.; Wei, J.; Huo, L.; et al. Novel Function of lncRNA ADAMTS9-AS2 in Promoting Temozolomide Resistance in Glioblastoma via Upregulating the FUS/MDM2 Ubiquitination Axis. Front. Cell Dev. Biol. 2019, 7, 217. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Huang, X.; Zhang, Y.; Wang, J.; Li, H.; Huang, H. PSMG3-AS1 enhances glioma resistance to temozolomide via stabilizing c-Myc in the nucleus. Brain Behav. 2022, 12, e2531. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, Z.; Tian, K.; Meng, X.; Wang, X.; Song, D.; Wang, X.; Xu, T.; Sun, P.; Zhong, J.; et al. LncRNA-Mediated TPI1 and PKM2 Promote Self-Renewal and Chemoresistance in GBM. Adv. Sci. 2024, 11, e2402600. [Google Scholar] [CrossRef]
- Liang, J.; Xie, J.-X.; He, J.; Li, Y.; Wei, D.; Zhou, R.; Wei, G.; Liu, X.; Chen, Q.; Li, D. Inhibiting lncRNA NEAT1 Increases Glioblastoma Response to TMZ by Reducing Connexin 43 Expression. Cancer Rep. 2024, 7, e70031. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Liu, M.; Long, W.; Yuan, J.; Li, H.; Zhang, C.; Tang, G.; Jiang, W.; Yuan, X.; Wu, M.; et al. Knockdown lncRNA CRNDE enhances temozolomide chemosensitivity by regulating autophagy in glioblastoma. Cancer Cell Int. 2021, 21, 456. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Xu, N.; Liu, B.; Wang, C.; He, Z.; Lenahan, C.; Tang, W.; Zeng, H.; Guo, H. lncRNA XLOC013218 promotes cell proliferation and TMZ resistance by targeting the PIK3R2-mediated PI3K/AKT pathway in glioma. Cancer Sci. 2022, 113, 2681–2692. [Google Scholar] [CrossRef]
- Dong, J.; Peng, Y.; Zhong, M.; Xie, Z.; Jiang, Z.; Wang, K.; Wu, Y. Implication of lncRNA ZBED3-AS1 downregulation in acquired resistance to Temozolomide and glycolysis in glioblastoma. Eur. J. Pharmacol. 2023, 938, 175444. [Google Scholar] [CrossRef]
- Yuan, S.; Yan, Q.; Zhao, Z.-Y.; Zhang, J.-L.; Zhang, H.; Yin, H.; Yuan, Z. STAT3-mediated upregulation of LINC00520 contributed to temozolomide chemoresistance in glioblastoma by interacting with RNA-binding protein LIN28B. Cancer Cell Int. 2022, 22, 248. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ma, F.; Zhang, Z.; Guo, Y.; Shen, H.; Chen, H. LncRNA-PVT1 was identified as a key regulator for TMZ resistance and STAT-related pathway in glioma. BMC Cancer 2023, 23, 455. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhou, J.; Liu, Y.; Ni, H.; Zhou, B. Targeting MAGI2-AS3-modulated Akt-dependent ATP-binding cassette transporters as a possible strategy to reverse temozolomide resistance in temozolomide-resistant glioblastoma cells. Drug Dev. Res. 2023, 84, 1482–1495. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhao, H.; Song, J.; Wang, F.; Chen, M. LINC00174 down-regulation decreases chemoresistance to temozolomide in human glioma cells by regulating miR-138-5p/SOX9 axis. Hum. Cell 2020, 33, 159–174. [Google Scholar] [CrossRef]
- Wang, X.; Yu, X.; Xu, H.; Wei, K.; Wang, S.; Wang, Y.; Han, J. Serum-derived extracellular vesicles facilitate temozolomide resistance in glioblastoma through a HOTAIR-dependent mechanism. Cell Death Dis. 2022, 13, 344. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, G.; Gao, Y.; Liang, H. HOTAIR/miR-125 axis-mediated Hexokinase 2 expression promotes chemoresistance in human glioblastoma. J. Cell Mol. Med. 2020, 24, 5707–5717. [Google Scholar] [CrossRef]
- Zhou, L.; Ma, J. MIR99AHG/miR-204-5p/TXNIP/Nrf2/ARE Signaling Pathway Decreases Glioblastoma Temozolomide Sensitivity. Neurotox. Res. 2022, 40, 1152–1162. [Google Scholar] [CrossRef]
- Shang, C.; Tang, W.; Pan, C.; Hu, X.; Hong, Y. Long non-coding RNA TUSC7 inhibits temozolomide resistance by targeting miR-10a in glioblastoma. Cancer Chemother. Pharmacol. 2018, 81, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.; Yang, Y.; Mu, Z.; Sun, R.; Li, X.; Dong, J. Silencing lncRNA LINC01410 suppresses cell viability yet promotes apoptosis and sensitivity to temozolomide in glioblastoma cells by inactivating PTEN/AKT pathway via targeting miR-370-3p. Immunopharmacol. Immunotoxicol. 2021, 43, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Wang, Q.; Chen, L.; Zhao, D.; Tang, J.; Xu, J.; He, Z. LncRNA UCA1/miR-182-5p/MGMT axis modulates glioma cell sensitivity to temozolomide through MGMT-related DNA damage pathways. Hum. Pathol. 2022, 123, 59–73. [Google Scholar] [CrossRef]
- Lu, Y.; Tian, M.; Liu, J.; Wang, K. LINC00511 facilitates Temozolomide resistance of glioblastoma cells via sponging miR-126-5p and activating Wnt/β-catenin signaling. J. Biochem. Mol. Toxicol. 2021, 35, e22848. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, X.; Zhou, Y.; Huang, X.; Jiang, X. Long non-coding RNA OIP5-AS1 inhibition upregulates microRNA-129-5p to repress resistance to temozolomide in glioblastoma cells via downregulating IGF2BP2. Cell Biol. Toxicol. 2022, 38, 963–977. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Wei, Y.; Wang, X.; Zhang, Z.; Yin, J.; Li, W.; Chen, L.; Lyu, X.; Shi, Z.; Yan, W.; et al. DNA-methylation-mediated activating of lncRNA SNHG12 promotes temozolomide resistance in glioblastoma. Mol. Cancer 2020, 19, 28. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Lin, D.; Jin, L.; Wang, J.; Lin, Z.; Zhang, S.; Lin, G. LncRNA HOXA-AS2 Promotes Temozolomide Resistance in Glioblastoma by Regulated miR-302a-3p/IGF1 Axis. Genet. Res. 2022, 2022, 3941952. [Google Scholar] [CrossRef]
- Wei, C.; Jiang, W.; Wang, R.; Zhong, H.; He, H.; Gao, X.; Zhong, S.; Yu, F.; Guo, Q.; Zhang, L.; et al. Brain endothelial GSDMD activation mediates inflammatory BBB breakdown. Nature 2024, 629, 893–900. [Google Scholar] [CrossRef]
- Upton, D.H.; Ung, C.; George, S.M.; Tsoli, M.; Kavallaris, M.; Ziegler, D.S. Challenges and opportunities to penetrate the blood-brain barrier for brain cancer therapy. Theranostics 2022, 12, 4734–4752. [Google Scholar] [CrossRef]
- Singh, P.; Singh, A.; Shah, S.; Vataliya, J.; Mittal, A.; Chitkara, D. RNA Interference Nanotherapeutics for Treatment of Glioblastoma Multiforme. Mol. Pharm. 2020, 17, 4040–4066. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Piao, L.; Sun, G.; Lv, C.; Jing, Y.; Jin, R. Dual functional nanoparticles efficiently across the blood-brain barrier to combat glioblastoma via simultaneously inhibit the PI3K pathway and NKG2A axis. J. Drug Target. 2021, 29, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sui, L.; Huang, J.; Miao, L.; Nie, Y.; Wang, K.; Yang, Z.; Huang, Q.; Gong, X.; Nan, Y.; et al. MoS2-based nanocomposites for cancer diagnosis and therapy. Bioact. Mater. 2021, 6, 4209–4242. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Wan, Z.; Wang, C.; Lu, F.; Wei, M.; Wang, D.; Hao, Q. Designer exosomes for targeted and efficient ferroptosis induction in cancer via chemo-photodynamic therapy. Theranostics 2021, 11, 8185–8196. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, P.; Tan, H.; Chen, X.; Wang, Q.; Chen, T. Exosomes as Smart Nanoplatforms for Diagnosis and Therapy of Cancer. Front. Oncol. 2021, 11, 743189. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef]
- Chen, R.; Xu, X.; Tao, Y.; Qian, Z.; Yu, Y. Exosomes in hepatocellular carcinoma: A new horizon. Cell Commun. Signal 2019, 17, 1. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.-X.; Sun, C.; Wang, L.; Guo, X.-L. New insight into isolation, identification techniques and medical applications of exosomes. J. Control Release 2019, 308, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, D.; Ho, E.A. Challenges in the development and establishment of exosome-based drug delivery systems. J. Control Release 2021, 329, 894–906. [Google Scholar] [CrossRef]
- Gaurav, I.; Thakur, A.; Iyaswamy, A.; Wang, X.; Chen, X.; Yang, Z. Factors Affecting Extracellular Vesicles Based Drug Delivery Systems. Molecules 2021, 26, 1544. [Google Scholar] [CrossRef]
- Tang, Y.; Zhang, P.; Wang, Y.; Wang, J.; Su, M.; Wang, Y.; Zhou, L.; Zhou, J.; Xiong, W.; Zeng, Z.; et al. The Biogenesis, Biology, and Clinical Significance of Exosomal PD-L1 in Cancer. Front. Immunol. 2020, 11, 604. [Google Scholar] [CrossRef]
- Li, Z.; Meng, X.; Wu, P.; Zha, C.; Han, B.; Li, L.; Sun, N.; Qi, T.; Qin, J.; Zhang, Y.; et al. Glioblastoma Cell-Derived lncRNA-Containing Exosomes Induce Microglia to Produce Complement C5, Promoting Chemotherapy Resistance. Cancer Immunol. Res. 2021, 9, 1383–1399. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.-Y.; Wang, G.-H.; Xu, J.-J.; Li, X.-L.; Lin, X.-Y.; Fang, X.; Zhang, H.-X.; Feng, M.; Jiang, C.-M. CREB-induced LINC00473 promotes chemoresistance to TMZ in glioblastoma by regulating O6-methylguanine-DNA-methyltransferase expression via CEBPα binding. Neuropharmacology 2024, 243, 109790. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yin, J.; Lu, C.; Wei, Y.; Zeng, A.; You, Y. Exosomal transfer of long non-coding RNA SBF2-AS1 enhances chemoresistance to temozolomide in glioblastoma. J. Exp. Clin. Cancer Res. 2019, 38, 166. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Zhang, S.; He, J. The Mechanism of Long Non-coding RNA in Cancer Radioresistance/Radiosensitivity: A Systematic Review. Front. Pharmacol. 2022, 13, 879704. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Luo, L.; Zhang, J.; Zhai, D.; Huang, D.; Yin, J.; Zhou, Q.; Zhang, Q.; Zheng, G. lncRNA LINC01057 promotes mesenchymal differentiation by activating NF-κB signaling in glioblastoma. Cancer Lett. 2021, 498, 152–164. [Google Scholar] [CrossRef]
- Zhang, X.; Li, R. lncRNA MAFG-AS1 enhances radioresistance of glioblastoma cells via miR-642a-5p/Notch1 axis. Acta Neurobiol. Exp. 2022, 82, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Wei, Y.; Zhang, Q.; Sun, M.; Wang, Y.; Hou, J.; Zhang, P.; Lv, X.; Su, D.; Jiang, Y.; et al. Multiomics analyses reveal DARS1-AS1/YBX1-controlled posttranscriptional circuits promoting glioblastoma tumorigenesis/radioresistance. Sci. Adv. 2023, 9, eadf3984. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Ding, F.; Cheng, Z.; Ge, X.; Li, Y.; Zeng, A.; Zhang, J.; Yan, W.; Shi, Z.; Qian, X.; et al. METTL3-mediated m6A modification of LINC00839 maintains glioma stem cells and radiation resistance by activating Wnt/β-catenin signaling. Cell Death Dis. 2023, 14, 417. [Google Scholar] [CrossRef]
- Fan, Y.; Gao, Z.; Xu, J.; Wang, H.; Guo, Q.; Li, B.; Li, M.; Xu, H.; Qi, Y.; Zhao, S.; et al. SPI1-mediated MIR222HG transcription promotes proneural-to-mesenchymal transition of glioma stem cells and immunosuppressive polarization of macrophages. Theranostics 2023, 13, 3310–3329. [Google Scholar] [CrossRef]
- Ahmadov, U.; Picard, D.; Bartl, J.; Silginer, M.; Trajkovic-Arsic, M.; Qin, N.; Blümel, L.; Wolter, M.; Lim, J.K.M.; Pauck, D.; et al. The long non-coding RNA HOTAIRM1 promotes tumor aggressiveness and radiotherapy resistance in glioblastoma. Cell Death Dis. 2021, 12, 885. [Google Scholar] [CrossRef]
- Xu, S.; Luo, C.; Chen, D.; Tang, L.; Chen, L.; Liu, Z. Whole transcriptome and proteome analyses identify potential targets and mechanisms underlying tumor treating fields against glioblastoma. Cell Death Dis. 2022, 13, 721. [Google Scholar] [CrossRef]
- Toker, J.; Iorgulescu, J.B.; Ling, A.L.; Villa, G.R.; Gadet, J.A.M.A.; Parida, L.; Getz, G.; Wu, C.J.; Reardon, D.A.; Chiocca, E.A.; et al. Clinical Importance of the lncRNA NEAT1 in Cancer Patients Treated with Immune Checkpoint Inhibitors. Clin. Cancer Res. 2023, 29, 2226–2238. [Google Scholar] [CrossRef] [PubMed]
- Yi, K.; Cui, X.; Liu, X.; Wang, Y.; Zhao, J.; Yang, S.; Xu, C.; Yang, E.; Xiao, M.; Hong, B.; et al. PTRF/Cavin-1 as a Novel RNA-Binding Protein Expedites the NF-κB/PD-L1 Axis by Stabilizing lncRNA NEAT1, Contributing to Tumorigenesis and Immune Evasion in Glioblastoma. Front. Immunol. 2021, 12, 802795. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.; Xie, D.-K.; Li, J.; Qiang, Y.-J.; Fan, S.-Y.; Wang, T.-T.; Han, Y.-Y.; Zang, J.; Yang, Y.; Zhao, J.-L.; et al. Glioma-Stem-Cell-Derived Exosomes Remodeled Glioma-Associated Macrophage via NEAT1/miR-125a/STAT3 Pathway. Cancers 2024, 16, 2500. [Google Scholar] [CrossRef]
- Dong, F.; Qin, X.; Wang, B.; Li, Q.; Hu, J.; Cheng, X.; Guo, D.; Cheng, F.; Fang, C.; Tan, Y.; et al. ALKBH5 Facilitates Hypoxia-Induced Paraspeckle Assembly and IL8 Secretion to Generate an Immunosuppressive Tumor Microenvironment. Cancer Res. 2021, 81, 5876–5888. [Google Scholar] [CrossRef]
- Huang, L.; Wang, Z.; Liao, C.; Zhao, Z.; Gao, H.; Huang, R.; Chen, J.; Wu, F.; Zeng, F.; Zhang, Y.; et al. PVT1 promotes proliferation and macrophage immunosuppressive polarization through STAT1 and CX3CL1 regulation in glioblastoma multiforme. CNS Neurosci. Ther. 2024, 30, e14566. [Google Scholar] [CrossRef]
- Zheng, P.; Zhang, X.; Ren, D.; Zhang, Y. RP11-552D4.1: A novel m6a-related LncRNA associated with immune status in glioblastoma. Aging 2022, 14, 7348–7363. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Gao, Y.; Zhong, J.; Wu, X.; Leng, Z.; Liu, M.; Wang, Y.; Wang, Y.; Yang, X.; Huang, N.; et al. Lnc-H19-derived protein shapes the immunosuppressive microenvironment of glioblastoma. Cell Rep. Med. 2024, 5, 101806. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Cheng, L.; Hu, L.; Lou, D.; Zhang, T.; Li, J.; Zhu, Q.; Liu, F. An integrative microfluidic device for isolation and ultrasensitive detection of lung cancer-specific exosomes from patient urine. Biosens. Bioelectron. 2020, 163, 112290. [Google Scholar] [CrossRef]
- Kashefi-Kheyrabadi, L.; Kim, J.; Chakravarty, S.; Park, S.; Gwak, H.; Kim, S.-I.; Mohammadniaei, M.; Lee, M.-H.; Hyun, K.-A.; Jung, H.-I. Detachable microfluidic device implemented with electrochemical aptasensor (DeMEA) for sequential analysis of cancerous exosomes. Biosens. Bioelectron. 2020, 169, 112622. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Park, U.; Koo, H.-J.; Park, J.-S.; Lee, D.H.; Kim, K.; Choi, J. Exosome-mediated diagnosis of pancreatic cancer using lectin-conjugated nanoparticles bound to selective glycans. Biosens. Bioelectron. 2021, 177, 112980. [Google Scholar] [CrossRef] [PubMed]
- Hajikarimloo, B.; Habibi, M.A.; Alvani, M.S.; Meinagh, S.O.; Kooshki, A.; Afkhami-Ardakani, O.; Rasouli, F.; Tos, S.M.; Tavanaei, R.; Akhlaghpasand, M.; et al. Machine learning-based models for prediction of survival in medulloblastoma: A systematic review and meta-analysis. Neurol. Sci. 2024. [Google Scholar] [CrossRef]
- Huang, Q.; Wu, W.; Ai, K.; Liu, J. Highly Sensitive Polydiacetylene Ensembles for Biosensing and Bioimaging. Front. Chem. 2020, 8, 565782. [Google Scholar] [CrossRef]
- Schönholzer, M.T.; Migliavacca, J.; Alvarez, E.; Santhana Kumar, K.; Neve, A.; Gries, A.; Ma, M.; Grotzer, M.A.; Baumgartner, M. Real-time sensing of MAPK signaling in medulloblastoma cells reveals cellular evasion mechanism counteracting dasatinib blockade of ERK activation during invasion. Neoplasia 2020, 22, 470–483. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xiao, X.; Han, X.; Yao, L.; Lan, W. Natural flavonoids alleviate glioblastoma multiforme by regulating long non-coding RNA. Biomed. Pharmacother. 2023, 161, 114477. [Google Scholar] [CrossRef] [PubMed]
- Stevanovic, M.; Kovacevic-Grujicic, N.; Petrovic, I.; Drakulic, D.; Milivojevic, M.; Mojsin, M. Crosstalk between SOX Genes and Long Non-Coding RNAs in Glioblastoma. Int. J. Mol. Sci. 2023, 24, 6392. [Google Scholar] [CrossRef]
- Garcia-Padilla, C.; Lozano-Velasco, E.; Muñoz-Gallardo, M.D.M.; Castillo-Casas, J.M.; Caño-Carrillo, S.; Martínez-Amaro, F.J.; García-López, V.; Aránega, A.; Franco, D.; García-Martínez, V.; et al. LncRNA H19 Impairs Chemo and Radiotherapy in Tumorigenesis. Int. J. Mol. Sci. 2022, 23, 8309. [Google Scholar] [CrossRef] [PubMed]
- Botti, G.; Scognamiglio, G.; Aquino, G.; Liguori, G.; Cantile, M. LncRNA HOTAIR in Tumor Microenvironment: What Role? Int. J. Mol. Sci. 2019, 20, 2279. [Google Scholar] [CrossRef]
- Goyal, B.; Yadav, S.R.M.; Awasthee, N.; Gupta, S.; Kunnumakkara, A.B.; Gupta, S.C. Diagnostic, prognostic, and therapeutic significance of long non-coding RNA MALAT1 in cancer. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2021, 1875, 188502. [Google Scholar] [CrossRef]
- Li, K.; Yao, T.; Zhang, Y.; Li, W.; Wang, Z. NEAT1 as a competing endogenous RNA in tumorigenesis of various cancers: Role, mechanism and therapeutic potential. Int. J. Biol. Sci. 2021, 17, 3428–3440. [Google Scholar] [CrossRef] [PubMed]
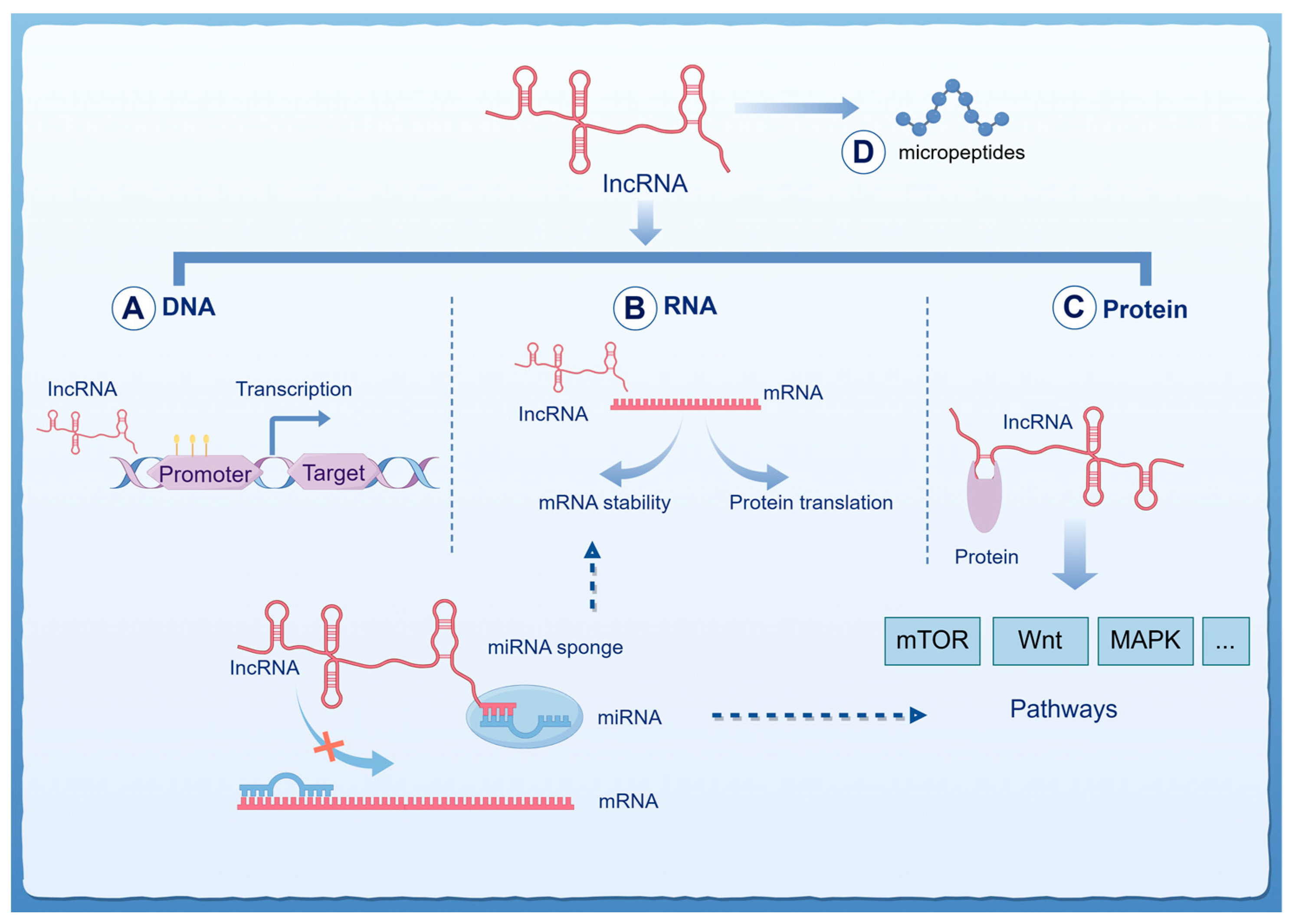
| lncRNA | miRNA | Progression | Cancer |
|---|---|---|---|
| TP73-AS1 | miR-494-3p | Promotes tumor progression [110] | Medulloblastoma |
| HOTAIR | miR-1/miR-206 | Tumor growth, migration, invasion, and apoptosis [111] | Medulloblastoma |
| CRNDE | miR-29c-3p | Proliferation, migration, invasion, apoptosis, and chemosensitivity [112] | Medulloblastoma |
| IMAT1 | hsa-miR22-3p | Invasiveness [113] | Meningioma |
| MALAT1 | miR-145 | Invasiveness [114] | Meningioma |
| MALAT1 | miR-199a | Cell proliferation and apoptosis (associated with prognosis) [115] | GB |
| MEG3 * | miR-29c | Cell cycle, migration, invasion, and proliferation [116] | Meningioma |
| MEG3 * | miR-6088 | Proliferation and EMT [117] | Glioma |
| SNHG1 | miR-556-5p | Tumorigenesis [118] | Meningioma |
| SNHG1 | miR-9-5p | Tumorigenesis and angiogenesis [119] | GB |
| SNHG4 | miR-138 | Proliferation [120] | GB |
| SNHG7 | MicroRNA-449b-5p | Motility, migration, and invasion [121] | GB |
| NEAT1 | miR-98-5p | Cancer progression [122] | GB |
| NEAT1 | miR-128-3p | Occurrence and progression [123] | GB |
| HOXA-AS2 | miR-885-5p | Tumorigenesis [124] | GB |
| HOXA-AS2 | miR-2116-3p | Growth and proliferation [125] | GB |
| HOXA-AS3 | miR-455-5p | Proliferation, migration, invasion, and prognosis [126] | GB |
| XIST | miR-448 | Tumorigenesis [127] | GB |
| XIST | miR-126 | Cell viability, migration, invasion, apoptosis resistance, and glucose metabolism [128] | GB |
| XIST | miR-133a | Proliferation and metastasis [129] | GB |
| XIST | miR-152 | Proliferation and stemness [130] | GB |
| OXCT1-AS1 | miR-195 | Tumor progression [131] | GB |
| SEMA3B-AS1 * | miR-195 | Tumor progression [132] | GB |
| LINC00460 | miR-539 | Proliferation, metastasis, and malignant transformation [133] | Malignant meningioma |
| LINC00702 | miR-4652-3p | Tumor progression [134] | Malignant meningioma |
| LINC01426 | miR-345-3p | Cell proliferation and prognosis [135] | GB |
| LINC00294 * | miR-1278 | Cell proliferation [136] | Glioma |
| LINC00641 * | miR-4262 | Cell proliferation and apoptosis [137] | Glioma |
| LINC00511 | miR-524-5p | Cell proliferation, EMT, and prognosis [138] | GB |
| LINC01198 | miR-129-5p | Cell proliferation and apoptosis [139] | GB |
| LINC01579 | miR-139-5p | Cell proliferation and apoptosis [140] | GB |
| LINC00115 | miR-200 | Key regulators of self-renewal and tumorigenicity of GSCS [141] | GB |
| LINC00606 | miR-486-3p | Proliferation, migration, and apoptosis [142] | GB |
| BCAR4 | miR-2276 | Invasion, tumorigenesis, and prognosis [143] | GB |
| CHRM3-AS2 | miRNA-370-5P | Proliferation, invasion, and migration [144] | GB |
| LGMNP1 | miR-495-3p | Proliferation and invasion [145] | GB |
| HOTAIRM1 | miR-153-5p | Migration and invasion and potential molecular markers [146] | GB |
| LINC01393 | miR-128-3p | Cell proliferation, migration, and invasion [147] | GB |
| Linc01094 | miR-126-5p | Growth and invasion [148] | GB |
| DCST1-AS1 | miR-29b | Cell proliferation [149] | GB |
| SATB2-AS1 * | miR-671-5p | Cell proliferation and apoptosis and glycolysis metabolism [150] | Low-grade glioma and GB |
| FEZF1-AS1 | miR-363-3p | Cell proliferation and apoptosis and glycolysis metabolism [151] | GB |
| PSMA3-AS1 | miR-411-3p | Proliferation and apoptosis [152] | GB |
| LPP-AS2 | miR-7-5p | Tumorigenesis [153] | Glioma |
| HNF1A-AS1 | miR-32-5p | Tumor progression [154] | Glioma |
| HAS2-AS1 | miR-608 | Migration, invasion, and prognosis [155] | GB |
| KTN1-AS1 | miR-505-3p | Proliferation and invasion [156] | GB |
| HOXD-AS2 | miR-3681-5p | Proliferation, migration, and invasion [157] | GB |
| HNF1A-AS1 | miR-22-3p | Malignant behavior of tumors [158] | GB |
| DLGAP1-AS1 | miRNA-515-5p | Tumor progression [159] | GB |
| OIP5-AS1 | miR-495-3p | Angiogenesis [160] | GB |
| FLVCR1-AS1 | miR-30b-3p | Proliferation, invasion, novel therapeutic targets, and diagnostic biomarkers [161] | GB |
| LEF1-AS1 | miR-543 | Colony formation, invasion, and migration [162] | GB |
| HAS2-AS1 | miR-137 | Cell proliferation and tumorigenicity [163] | GB |
| RBPMS-AS1 * | miR-301a-3p | Cell proliferation, apoptosis, and radiosensitivity [164] | GB |
| FEZF1-AS1 | miR-34a | Invasion and migration [165] | GB |
| PSMB8-AS1 | miR-22-3p | Cell proliferation, apoptosis, and radiation resistance [166] | GB |
| FGD5-AS1 | miR-129-5p | Proliferation, migration, and EMT progression [167] | GB |
| GAS5 * | miR-let-7e and miR-125a | Cell migration and invasion, stemness and proliferation of GSCs, and prognostic biomarkers [168] | GB |
| MUF | miR-34a | Proliferation, migration, and invasion; TMZ sensitivity; therapeutic target [169] | GB |
| TMEM161B-AS1 | hsa-miR-27a-3p | Proliferation, migration, invasion, and apoptosis; ferroptosis; TMZ sensitivity [170] | GB |
| NONHSAT079852.2 | hsa-miR-10401-3p | Cell proliferation and invasion, neoplasm progression and recurrence, diagnostic biomarkers, and therapeutic targets [171] | GB |
| Unigene56159 | miR-194-5p | Proliferation and invasion and potential therapeutic targets [172] | GB |
| RP1-86C11.7 | hsa-miR-144-3p | Cell proliferation and intracellular iron levels [173] | GB |
| MYCNOS | miR-216b | Cell proliferation [174] | GB |
| PITPNA-AS1 | miR-223-3p | Cell proliferation and apoptosis and potential biomarkers for diagnosis and treatment [175] | GB |
| COX10-AS1 | miR-1-3p | Cell proliferation, migration, and invasion and tumorigenesis [176] | GB |
| RP3-439F8.1 | miR-139-5p | Cell proliferation, colony formation, invasion, and cell cycle [177] | GB |
| MAFG-AS1 | miR-34a | Cell proliferation [178] | GB |
| UNC5B-AS1 * | miR-24-3p | Cell proliferation [179] | GB |
| AGAP2-AS1 | miR-486-3p | Growth and metastasis [180] | GB |
| LncRNA | Signal Axis/miRNA |
|---|---|
| Linc00942 | TPI1/SOX9 [197] |
| NEAT1 | Connexin 43 [198] |
| CRNDE | LC3 II/I, Beclin-1, ATG-5, p62 [199] |
| XLOC013218 | PI3K/AKT [200] |
| ZBED3-AS1 * | THBD [201] |
| LINC00520 | RNA-binding protein LIN28B [202] |
| PVT1 | JAK/STAT [203] |
| MAGI2-AS3 * | Akt pathway [204] |
| LINC00174 | miR-138-5p/SOX9 [205] |
| HOTAIR | miR-526b-3p [206] |
| HOTAIR | mir-125 [207] |
| MIR99AHG | miR-204-5p [208] |
| TUSC7 * | miR-10a [209] |
| LINC01410 | miR-370-3p [210] |
| UCA1 | miR-182-5p [211] |
| LINC00511 | miR-126-5p [212] |
| OIP5-AS1 | miRNA-129-5p [213] |
| SNHG12 | miR-129-5p [214] |
| HOXA-AS2 | miR-302a-3p/IGF1 [215] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pei, D.; Zhang, D.; Guo, Y.; Chang, H.; Cui, H. Long Non-Coding RNAs in Malignant Human Brain Tumors: Driving Forces Behind Progression and Therapy. Int. J. Mol. Sci. 2025, 26, 694. https://doi.org/10.3390/ijms26020694
Pei D, Zhang D, Guo Y, Chang H, Cui H. Long Non-Coding RNAs in Malignant Human Brain Tumors: Driving Forces Behind Progression and Therapy. International Journal of Molecular Sciences. 2025; 26(2):694. https://doi.org/10.3390/ijms26020694
Chicago/Turabian StylePei, Dakun, Dandan Zhang, Yan Guo, Hongbo Chang, and Hongjuan Cui. 2025. "Long Non-Coding RNAs in Malignant Human Brain Tumors: Driving Forces Behind Progression and Therapy" International Journal of Molecular Sciences 26, no. 2: 694. https://doi.org/10.3390/ijms26020694
APA StylePei, D., Zhang, D., Guo, Y., Chang, H., & Cui, H. (2025). Long Non-Coding RNAs in Malignant Human Brain Tumors: Driving Forces Behind Progression and Therapy. International Journal of Molecular Sciences, 26(2), 694. https://doi.org/10.3390/ijms26020694





