Urokinase Plasminogen Activation System Modulation in Transformed Cell Lines
Abstract
1. Introduction
2. Results
2.1. Cloning of Urokinase, PAI1 and uPAR Sequences and Production of Strains with Different Urokinase Activity
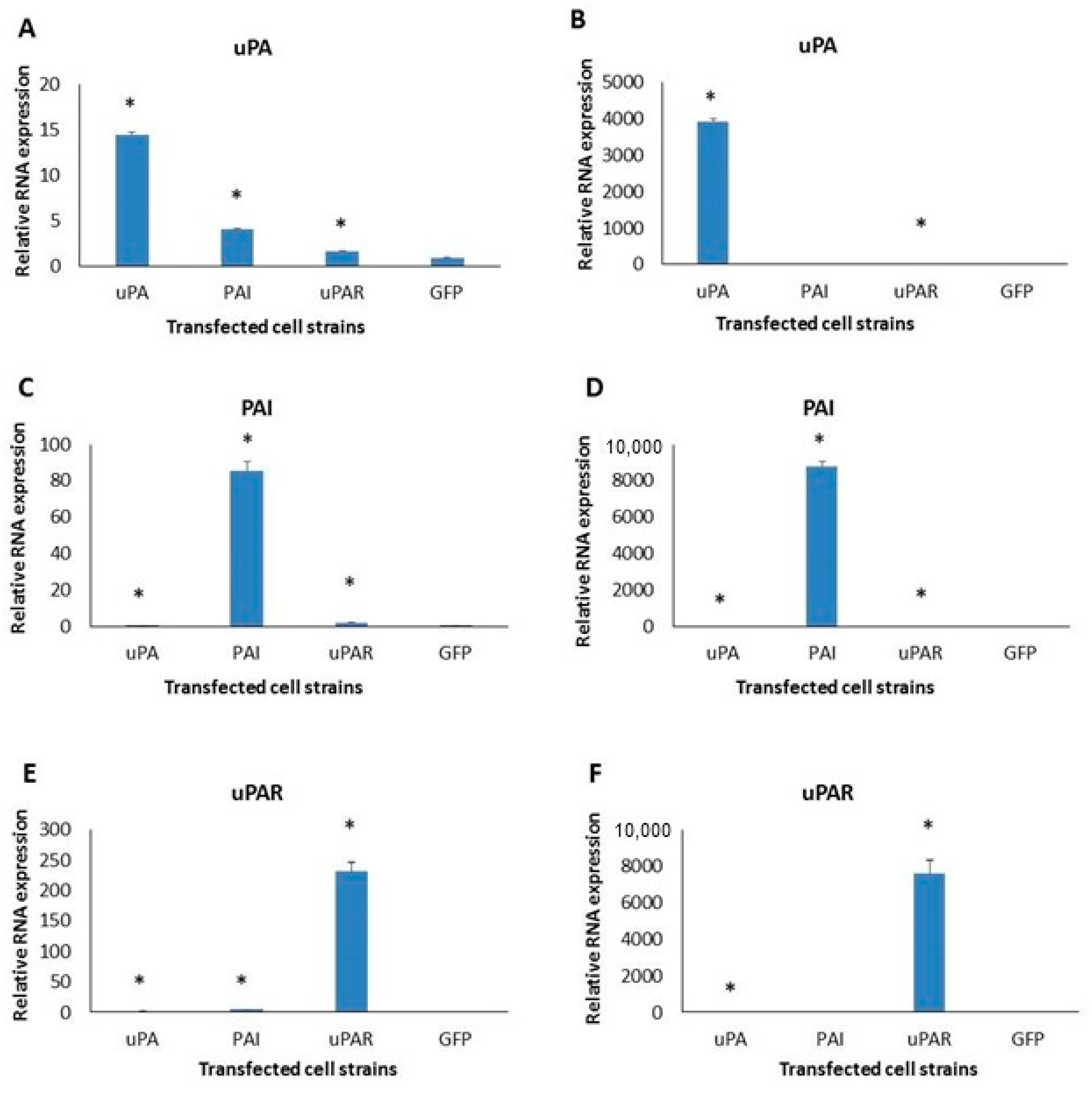
2.2. Expression of uPA System Molecules in Two Cell Lines
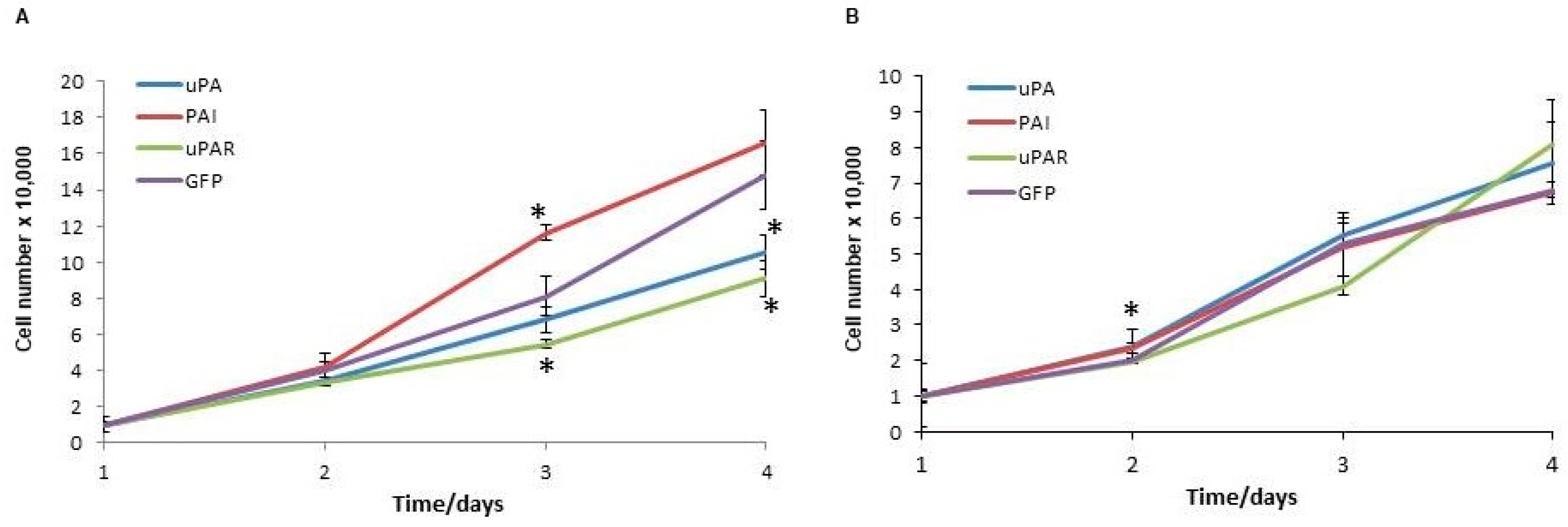
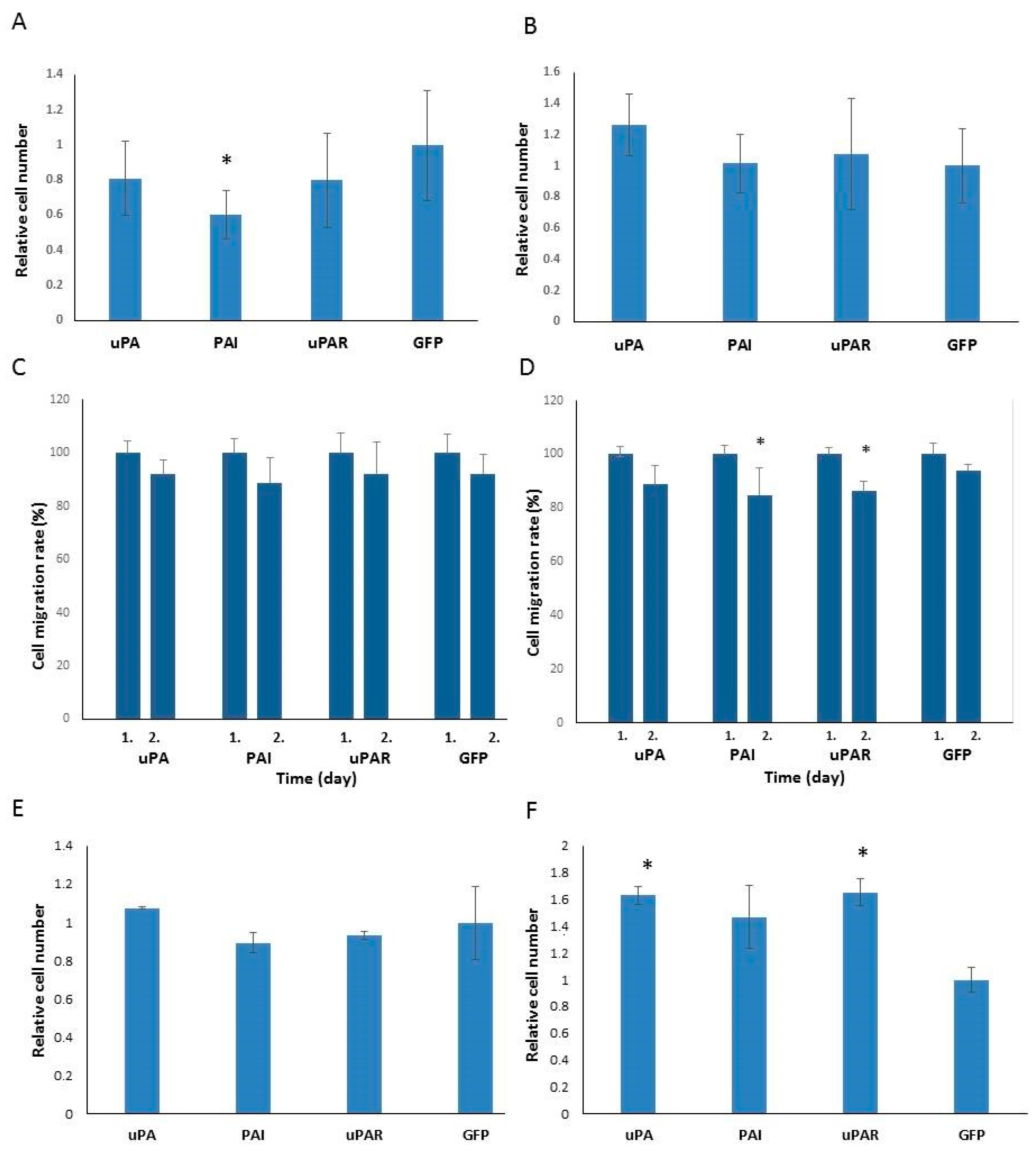
2.3. Analysis of Cell Proliferation and Migration
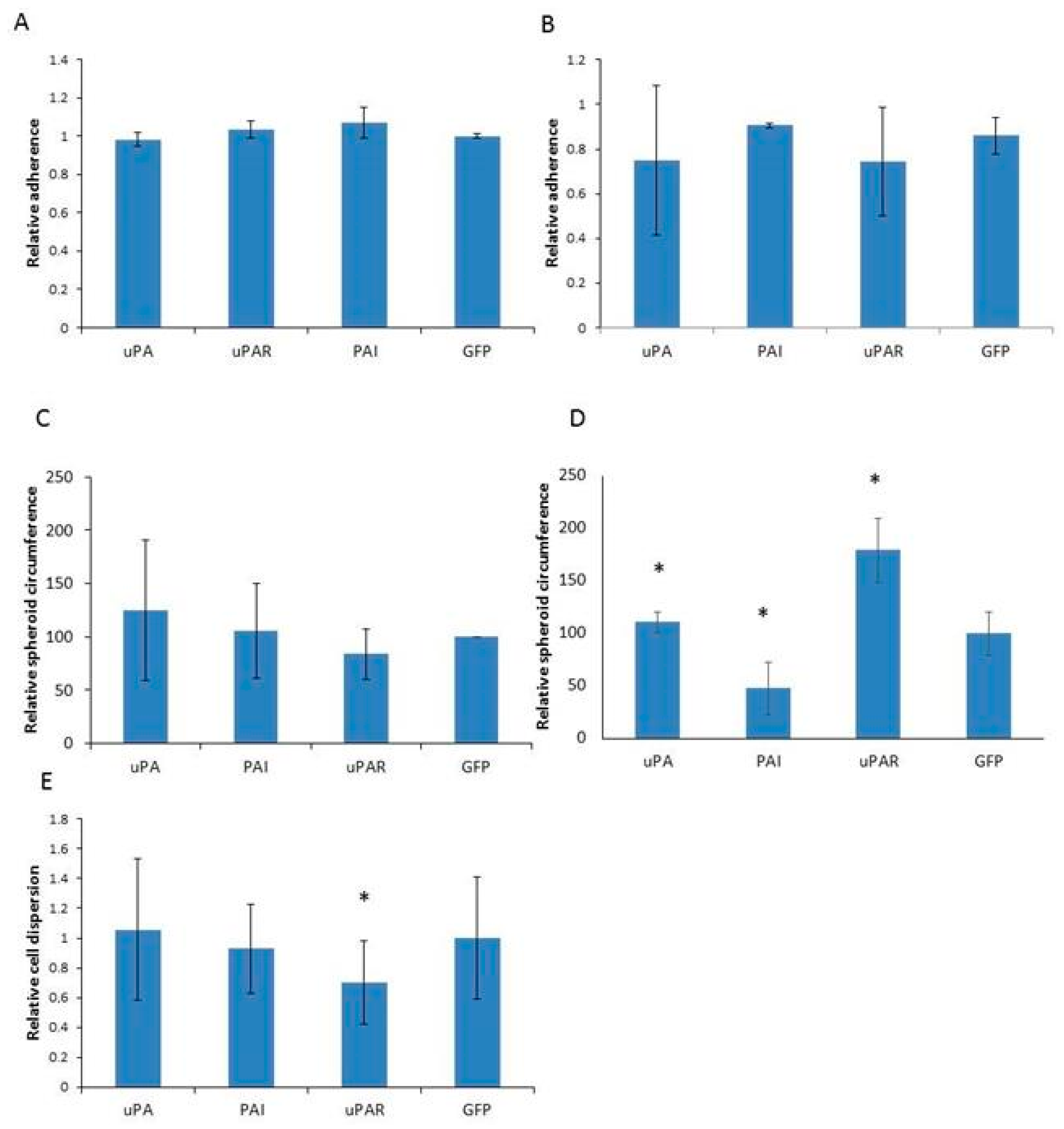
2.4. Analysis of Cell Adhesion
3. Discussion
4. Materials and Methods
4.1. Cloning of uPA, PAI1 and uPAR Sequences
4.2. Cell Culture, Growth Assessment and Transfection
4.3. Western Blot Analysis
4.4. Enzyme Activity Assays
4.5. Migration and Invasion Assays
4.6. Spheroid Formation and Cell Adhesion Assay
4.7. qRT-PCR Assay
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Danø, K.; Andreasen, P.A.; Grøndahl-Hansen, J.; Kristensen, P.; Nielsen, L.S.; Skriver, L. Plasminogen activators, tissue degradation, and cancer. Adv. Cancer Res. 1985, 44, 139–266. [Google Scholar] [CrossRef]
- Irigoyen, J.P.; Muñoz-Cánoves, P.; Montero, L.; Koziczak, M.; Nagamine, Y. The plasminogen activator system: Biology and regulation. Cell Mol. Life Sci. 1999, 56, 104–132. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Morgan, D.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 6th ed.; Garland Science, Taylor & Francis Group: New York, NY, USA, 2015. [Google Scholar]
- Odekon, L.E.; Blasi, F.; Rifkin, D.B. Requirement for receptor-bound urokinase in plasmin-dependent cellular conversion of latent TGF-beta to TGF-beta. J. Cell Physiol. 1994, 158, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Lyons, R.M.; Gentry, L.E.; Purchio, A.F.; Moses, H.L. Mechanism of activation of latent recombinant transforming growth factor beta 1 by plasmin. J. Cell Biol. 1990, 110, 1361–1367. [Google Scholar] [CrossRef]
- Tagirasa, R.; Yoo, E. Role of Serine Proteases at the Tumor-Stroma Interface. Front. Immunol. 2022, 13, 832418. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, V.; Jayaraman, P.S.; Zaitsev, S.V.; Lebedeva, T.; Bdeir, K.; Kershaw, R.; Holman, K.R.; Parfyonova, Y.V.; Semina, E.V.; Beloglazova, I.B.; et al. Urokinase-type Plasminogen Activator (uPA) Promotes Angiogenesis by Attenuating Proline-rich Homeodomain Protein (PRH) Transcription Factor Activity and De-repressing Vascular Endothelial Growth Factor (VEGF) Receptor Expression. J. Biol. Chem. 2016, 291, 15029–15045. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Chen, X.; Zhang, H.; Li, X.; Wang, J.; Han, W.; Zhang, L.Y.; Fu, L.; Li, Y.; Nie, C.; et al. Urokinase plasminogen activator secreted by cancer-associated fibroblasts induces tumor progression via PI3K/AKT and ERK signaling in esophageal squamous cell carcinoma. Oncotarget 2017, 8, 42300–42313. [Google Scholar] [CrossRef][Green Version]
- Merino, P.; Diaz, A.; Torre, E.R.; Yepes, M. Urokinase-type plasminogen activator (uPA) regulates the expression and function of growth-associated protein 43 (GAP-43) in the synapse. J. Biol. Chem. 2020, 295, 619–630. [Google Scholar] [CrossRef]
- Chen, G.; Sun, J.; Xie, M.; Yu, S.; Tang, Q.; Chen, L. PLAU Promotes Cell Proliferation and Epithelial-Mesenchymal Transition in Head and Neck Squamous Cell Carcinoma. Front. Genet. 2021, 12, 651882. [Google Scholar] [CrossRef]
- Semina, E.V.; Rubina, K.A.; Shmakova, A.A.; Rysenkova, K.D.; Klimovich, P.S.; Aleksanrushkina, N.A.; Sysoeva, V.Y.; Kara-gyaur, M.N.; Tkachuk, V.A. Downregulation of uPAR promotes urokinase translocation into the nucleus and epithelial to mesenchymal transition in neuroblastoma. J. Cell Physiol. 2020, 235, 6268–6286. [Google Scholar] [CrossRef]
- Stepanova, V.; Mukhina, S.; Köhler, E.; Resink, T.J.; Erne, P.; Tkachuk, V.A. Urokinase plasminogen activator induces human smooth muscle cell migration and proliferation via distinct receptor-dependent and proteolysis-dependent mechanisms. Mol. Cell Biochem. 1999, 195, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Maupas-Schwalm, F.; Bedel, A.; Augé, N.; Grazide, M.H.; Mucher, E.; Thiers, J.C.; Salvayre, R.; Nègre-Salvayre, A. Integrin alpha(v)beta(3), metalloproteinases, and sphingomyelinase-2 mediate urokinase mitogenic effect. Cell Signal 2009, 21, 1925–1934. [Google Scholar] [CrossRef] [PubMed]
- Cullen, J.P.; Nicholl, S.M.; Sayeed, S.; Sitzmann, J.V.; Okada, S.S.; Cahill, P.A.; Redmond, E.M. Plasminogen activator inhibitor-1 deficiency enhances flow-induced smooth muscle cell migration. Thromb. Res. 2004, 114, 57–65. [Google Scholar] [CrossRef]
- Blasi, F.; Sidenius, N. The urokinase receptor: Focused cell surface proteolysis, cell adhesion and signaling. FEBS Lett. 2010, 584, 1923–1930. [Google Scholar] [CrossRef] [PubMed]
- De Lorenzi, V.; Sarra Ferraris, G.M.; Madsen, J.B.; Lupia, M.; Andreasen, P.A.; Sidenius, N. Urokinase links plasminogen activation and cell adhesion by cleavage of the RGD motif in vitronectin. EMBO Rep. 2016, 17, 982–998. [Google Scholar] [CrossRef]
- Biagioni, A.; Laurenzana, A.; Chillà, A.; Del Rosso, M.; Andreucci, E.; Poteti, M.; Bani, D.; Guasti, D.; Fibbi, G.; Margheri, F. uPAR Knockout Results in a Deep Glycolytic and OXPHOS Reprogramming in Melanoma and Colon Carcinoma Cell Lines. Cells 2020, 9, 308. [Google Scholar] [CrossRef] [PubMed]
- Biagioni, A.; Laurenzana, A.; Menicacci, B.; Peppicelli, S.; Andreucci, E.; Bianchini, F.; Guasti, D.; Paoli, P.; Serratì, S.; Mocali, A.; et al. uPAR-expressing melanoma exosomes promote angiogenesis by VE-Cadherin, EGFR and uPAR overexpression and rise of ERK1,2 signaling in endothelial cells. Cell Mol. Life Sci. 2021, 78, 3057–3072. [Google Scholar] [CrossRef] [PubMed]
- Sarno, F.; Goubert, D.; Logie, E.; Rutten, M.G.S.; Koncz, M.; Deben, C.; Niemarkt, A.E.; Altucci, L.; Verschure, P.J.; Kiss, A.; et al. Functional Validation of the Putative Oncogenic Activity of PLAU. Biomedicines 2022, 11, 102. [Google Scholar] [CrossRef] [PubMed]
- Holst-Hansen, C.; Johannessen, B.; Høyer-Hansen, G.; Rømer, J.; Ellis, V.; Brünner, N. Urokinase-type plasminogen activation in three human breast cancer cell lines correlates with their in vitro invasiveness. Clin. Exp. Metastasis 1996, 14, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Gandhari, M.; Arens, N.; Majety, M.; Dorn-Beineke, A.; Hildenbrand, R. Urokinase-type plasminogen activator induces pro- liferation in breast cancer cells. Int. J. Oncol. 2006, 28, 1463–1470. [Google Scholar]
- Jo, M.; Thomas, K.S.; Marozkina, N.; Amin, T.J.; Silva, C.M.; Parsons, S.J.; Gonias, S.L. Dynamic assembly of the urokinase-type plasminogen activator signaling receptor complex determines the mitogenic activity of urokinase-type plasminogen activator. J. Biol. Chem. 2005, 280, 17449–17457. [Google Scholar] [CrossRef] [PubMed]
- Carlin, S.M.; Resink, T.J.; Tamm, M.; Roth, M. Urokinase signal transduction and its role in cell migration. FASEB J. 2005, 19, 195–202. [Google Scholar] [CrossRef]
- Kozlova, N.; Samoylenko, A.; Drobot, L.; Kietzmann, T. Urokinase is a negative modulator of Egf-dependent proliferation and motility in the two breast cancer cell lines MCF-7 and MDA-MB-231. Mol. Carcinog. 2016, 55, 170–181. [Google Scholar] [CrossRef]
- Krüger, A.; Soeltl, R.; Lutz, V.; Wilhelm, O.G.; Magdolen, V.; Rojo, E.E.; Hantzopoulos, P.A.; Graeff, H.; Gänsbacher, B.; Schmitt, M. Reduction of breast carcinoma tumor growth and lung colonization by overexpression of the soluble urokinase-type plasminogen activator receptor (CD87). Cancer Gene Ther. 2000, 7, 292–299. [Google Scholar] [CrossRef][Green Version]
- Furuya, H.; Sasaki, Y.; Chen, R.; Peres, R.; Hokutan, K.; Murakami, K.; Kim, N.; Chan, O.T.M.; Pagano, I.; Dyrskjøt, L.; et al. PAI-1 is a potential transcriptional silencer that supports bladder cancer cell activity. Sci. Rep. 2022, 12, 12186. [Google Scholar] [CrossRef] [PubMed]
- Gomes-Giacoia, E.; Miyake, M.; Goodison, S.; Rosser, C.J. Targeting plasminogen activator inhibitor-1 inhibits angiogenesis and tumor growth in a human cancer xenograft model. Mol. Cancer Ther. 2013, 12, 2697–2708. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Xing, Z.H.; Jiang, Q.W.; Yang, Y.; Huang, J.R.; Yuan, M.L.; Wei, M.N.; Li, Y.; Wang, S.T.; Liu, K.; et al. Targeting uPAR by CRISPR/Cas9 System Attenuates Cancer Malignancy and Multidrug Resistance. Front. Oncol. 2019, 9, 80. [Google Scholar] [CrossRef]
- Duffy, M.J.; Duggan, C.; Mulcahy, H.E.; McDermott, E.W.; O’Higgins, N.J. Urokinase plasminogen activator: A prognostic marker in breast cancer including patients with axillary node-negative disease. Clin. Chem. 1998, 44 Pt 1, 1177–1183. [Google Scholar] [CrossRef]
- Look, M.P.; van Putten, W.L.; Duffy, M.J.; Harbeck, N.; Christensen, I.J.; Thomssen, C.; Kates, R.; Spyratos, F.; Fernö, M.; Ep- penberger-Castori, S.; et al. Pooled analysis of prognostic impact of urokinase-type plasminogen activator and its inhibitor PAI-1 in 8377 breast cancer patients. J. Natl. Cancer Inst. 2002, 94, 116–128. [Google Scholar] [CrossRef]
- Ossowski, L.; Russo-Payne, H.; Wilson, E.L. Inhibition of urokinase-type plasminogen activator by antibodies: The effect on dissemination of a human tumor in the nude mouse. Cancer Res. 1991, 51, 274–281. [Google Scholar]
- Kobayashi, H.; Fujishiro, S.; Terao, T. Impact of urokinase-type plasminogen activator and its inhibitor type 1 on prognosis in cervical cancer of the uterus. Cancer Res. 1994, 54, 6539–6548. [Google Scholar]
- Conese, M.; Blasi, F. The urokinase/urokinase-receptor system and cancer invasion. Bailliere’s Clin. Haematol. 1995, 8, 365–389. [Google Scholar] [CrossRef]
- Jänicke, F.; Prechtl, A.; Thomssen, C.; Harbeck, N.; Meisner, C.; Untch, M.; Sweep, C.G.; Selbmann, H.K.; Graeff, H.; Schmitt, M. German N0 Study Group Randomized adjuvant chemotherapy trial in high-risk, lymph node-negative breast cancer patients identified by urokinase-type plasminogen activator and plasminogen activator inhibitor type 1. J. Natl. Cancer Inst. 2001, 93, 913–920. [Google Scholar] [CrossRef][Green Version]
- Gao, Y.; Ma, X.; Lu, H.; Xu, P.; Xu, C. PLAU is associated with cell migration and invasion and is regulated by transcription factor YY1 in cervical cancer. Oncol. Rep. 2023, 49, 25. [Google Scholar] [CrossRef] [PubMed]
- Minaei, E.; Mueller, S.A.; Ashford, B.; Thind, A.S.; Mitchell, J.; Perry, J.R.; Genenger, B.; Clark, J.R.; Gupta, R.; Ranson, M. Cancer Progression Gene Expression Profiling Identifies the Urokinase Plasminogen Activator Receptor as a Biomarker of Metastasis in Cutaneous Squamous Cell Carcinoma. Front. Oncol. 2022, 12, 835929. [Google Scholar] [CrossRef] [PubMed]
- Jo, M.; Lester, R.D.; Montel, V.; Eastman, B.; Takimoto, S.; Gonias, S.L. Reversibility of epithelial-mesenchymal transition (EMT) induced in breast cancer cells by activation of urokinase receptor-dependent cell signaling. J. Biol. Chem. 2009, 284, 22825–22833. [Google Scholar] [CrossRef] [PubMed]
- Seker, F.; Cingoz, A.; Sur-Erdem, I.; Erguder, N.; Erkent, A.; Uyulur, F.; Selvan, M.E.; Gumus, Z.H.; Gonen, M.; Byraktar, H.; et al. Identification of SERPINE1 as a regulator of glioblastoma cell dispersal with transcriptome profiling. Cancers 2019, 11, 1651. [Google Scholar] [CrossRef] [PubMed]
- Ferraris, G.M.; Schulte, C.; Buttiglione, V.; De Lorenzi, V.; Piontini, A.; Galluzzi, M.; Podestà, A.; Madsen, C.D.; Sidenius, N. The interaction between uPAR and vitronectin triggers ligand-independent adhesion signalling by integrins. EMBO J. 2014, 33, 2458–2472. [Google Scholar] [CrossRef]
- Gorrasi, A.; Li Santi, A.; Amodio, G.; Alfano, D.; Remondelli, P.; Montuori, N.; Ragno, P. The urokinase receptor takes control of cell migration by recruiting integrins and FPR1 on the cell surface. PLoS ONE 2014, 9, e86352. [Google Scholar] [CrossRef] [PubMed]
- Margheri, F.; Luciani, C.; Taddei, M.L.; Giannoni, E.; Laurenzana, A.; Biagioni, A.; Chillà, A.; Chiarugi, P.; Fibbi, G.; Del Rosso, M. The receptor for urokinase-plasminogen activator (uPAR) controls plasticity of cancer cell movement in mesenchymal and amoeboid migration style. Oncotarget 2014, 5, 1538–1553. [Google Scholar] [CrossRef]
- Expression Atlas. Available online: https://www.ebi.ac.uk/gxa/home (accessed on 17 November 2023).
- Sánchez-Tilló, E.; de Barrios, O.; Siles, L.; Amendola, P.G.; Darling, D.S.; Cuatrecasas, M.; Castells, A.; Postigo, A. ZEB1 Promotes invasiveness of colorectal carcinoma cells through the opposing regulation of uPA and PAI-1. Clin. Cancer Res. 2013, 19, 1071–1082. [Google Scholar] [CrossRef] [PubMed]
- Integrated DNA Technologies. Available online: https://www.idtdna.com/pages (accessed on 24 November 2017).
- Primer Designing Tool. Available online: https://www.ncbi.nlm.nih.gov/tools/primer-blast (accessed on 11 November 2020).
- Giard, D.J.; Aaronson, S.A.; Todaro, G.J.; Arnstein, P.; Kersey, J.H.; Dosik, H.; Parks, W.P. In vitro cultivation of human tumors: Establishment of cell lines derived from a series of solid tumors. J. Natl. Cancer Inst. 1973, 51, 1417–1423. [Google Scholar] [CrossRef]
- Sambrook, J.F.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Matulić, M.; Brdar, B. Urokinase-type Plasminogen Activator and Plasminogen Activator Inhibitor Induction by Etoposide in a Glioblastoma Cell Strain. Food Tech. Biotech. 2002, 40, 1–7. [Google Scholar]
- Horvat, L.; Madunić, J.; Grubar, M.; Antica, M.; Matulić, M. Induction of Urokinase Activity by Retinoic Acid in Two Cell Lines of Neuronal Origin. Biomedicines 2019, 7, 70. [Google Scholar] [CrossRef] [PubMed]
- Justus, C.R.; Leffler, N.; Ruiz-Echevarria, M.; Yang, L.V. In vitro cell migration and invasion assays. J. Vis. Exp. 2014, 88, 51046. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Culej Bošnjak, D.; Balent, T.; Korać, P.; Antica, M.; Matulić, M. Urokinase Plasminogen Activation System Modulation in Transformed Cell Lines. Int. J. Mol. Sci. 2025, 26, 675. https://doi.org/10.3390/ijms26020675
Culej Bošnjak D, Balent T, Korać P, Antica M, Matulić M. Urokinase Plasminogen Activation System Modulation in Transformed Cell Lines. International Journal of Molecular Sciences. 2025; 26(2):675. https://doi.org/10.3390/ijms26020675
Chicago/Turabian StyleCulej Bošnjak, Diana, Tihana Balent, Petra Korać, Mariastefania Antica, and Maja Matulić. 2025. "Urokinase Plasminogen Activation System Modulation in Transformed Cell Lines" International Journal of Molecular Sciences 26, no. 2: 675. https://doi.org/10.3390/ijms26020675
APA StyleCulej Bošnjak, D., Balent, T., Korać, P., Antica, M., & Matulić, M. (2025). Urokinase Plasminogen Activation System Modulation in Transformed Cell Lines. International Journal of Molecular Sciences, 26(2), 675. https://doi.org/10.3390/ijms26020675





