A Systematic Review of Microbiota in Cirrhosis: A Change Towards a More Pathogenic Predisposition
Abstract
1. Introduction
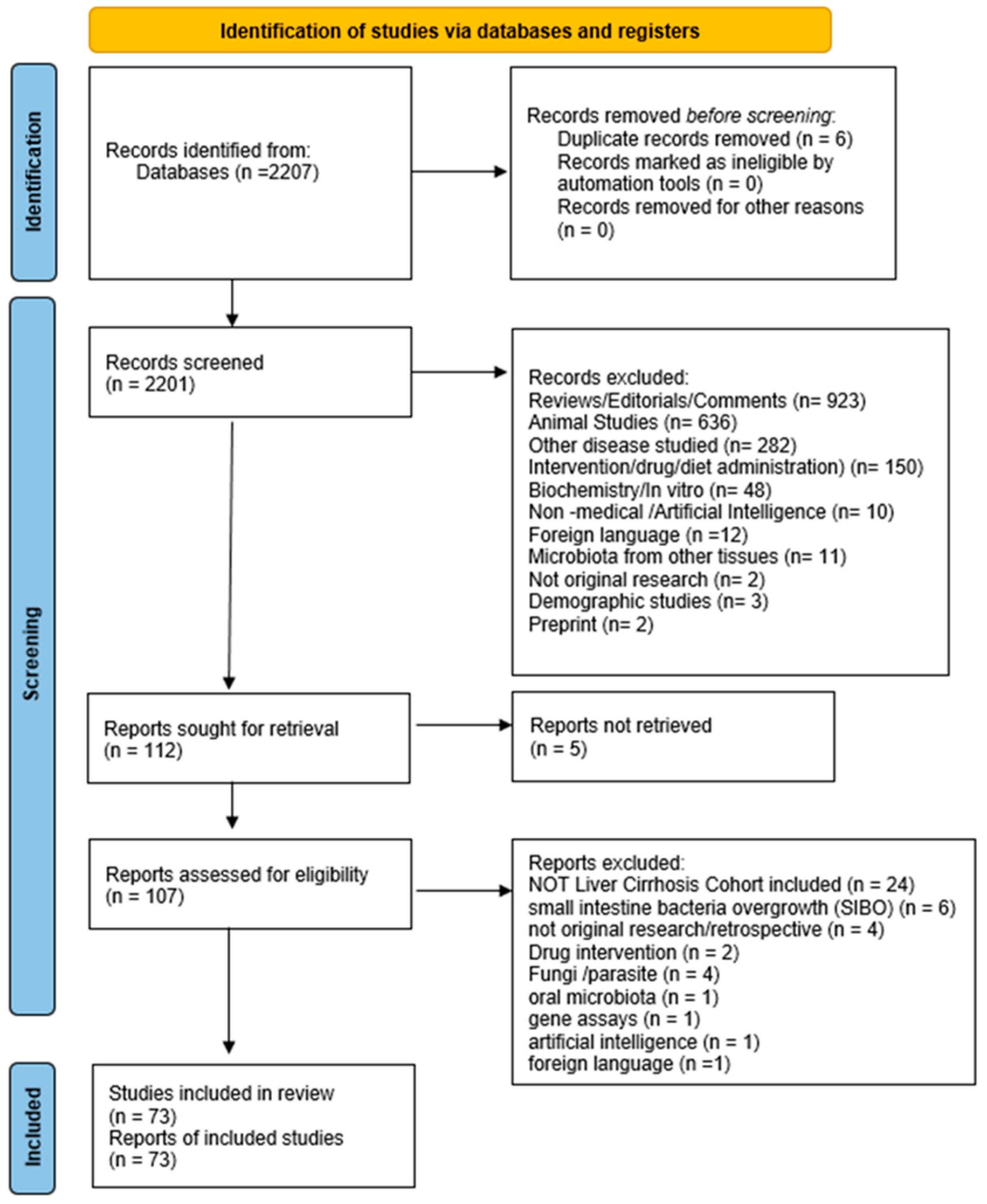
2. Methods
2.1. Data Identification
2.2. Inclusion and Exclusion Criteria
2.3. Data Extraction and Methodological Assessment
2.4. Endpoints
3. Results and Discussion
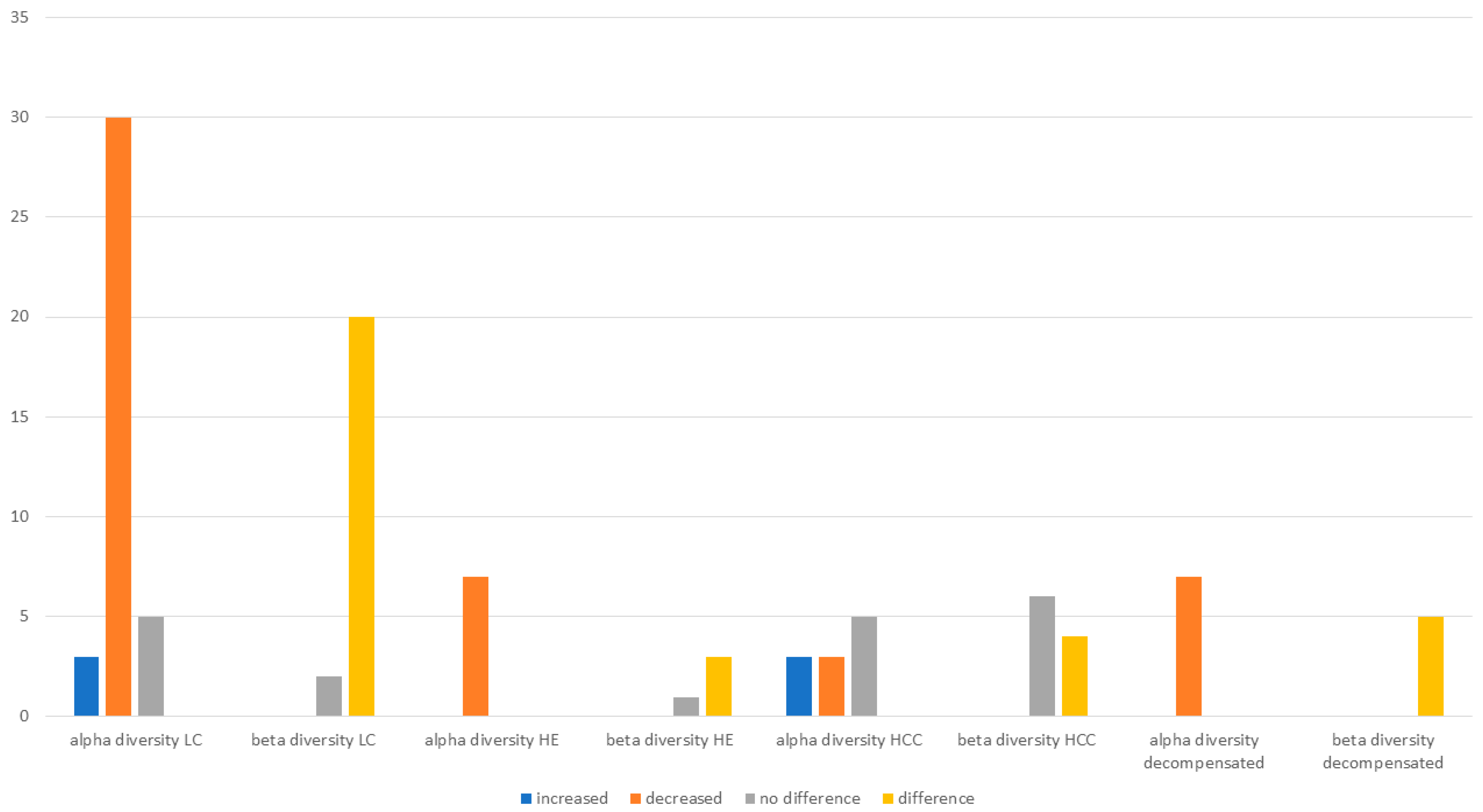
3.1. Microbiota Diversity in Cirrhosis
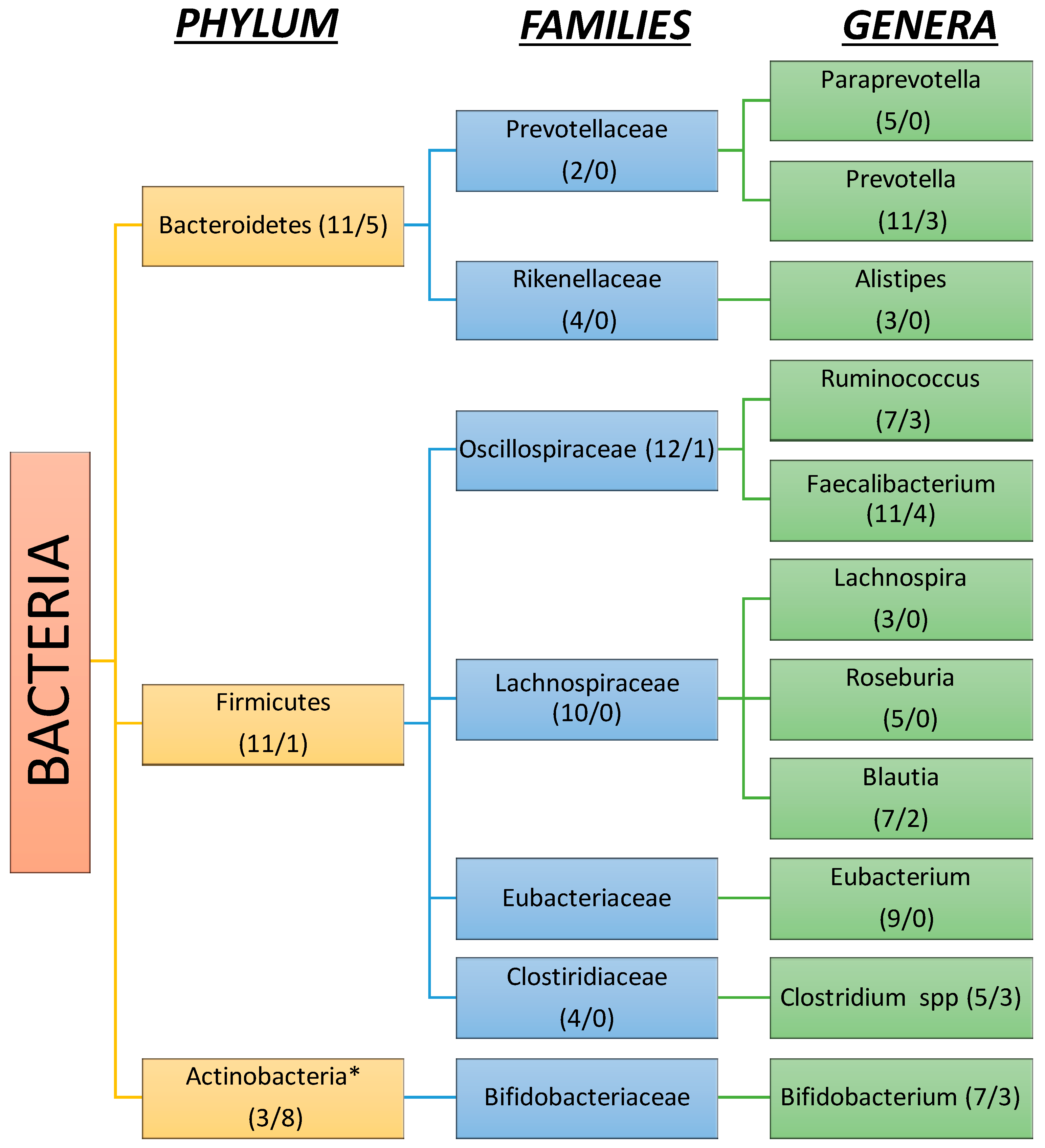
3.2. Gut Microbiota Composition in Liver Cirrhosis
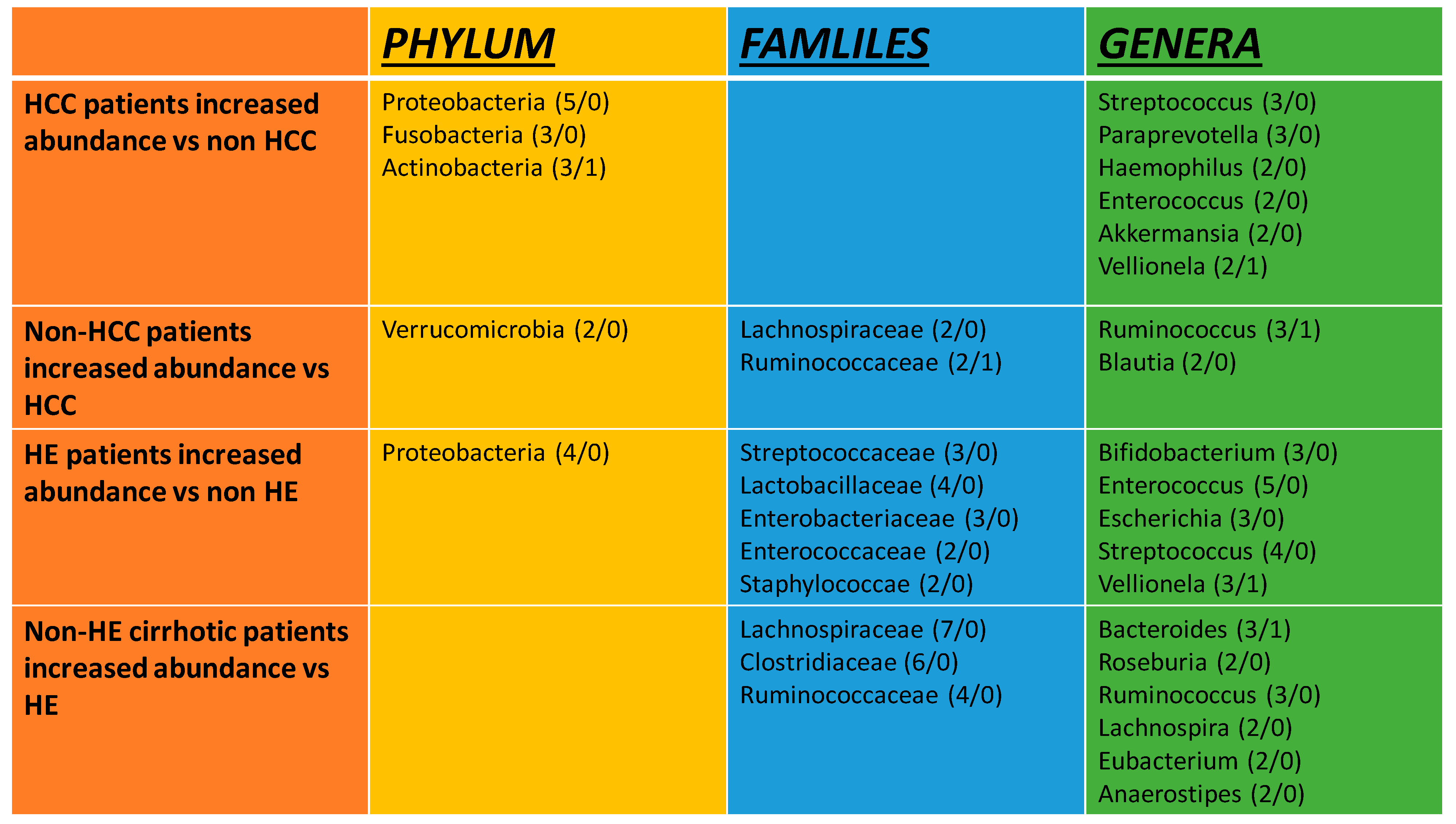
3.3. Gut Microbiota Composition in Hepatic Encephalopathy
3.4. Gut Microbiota Composition in Hepatocellular Carcinoma
3.5. Gut Microbiota Composition in Other Complications
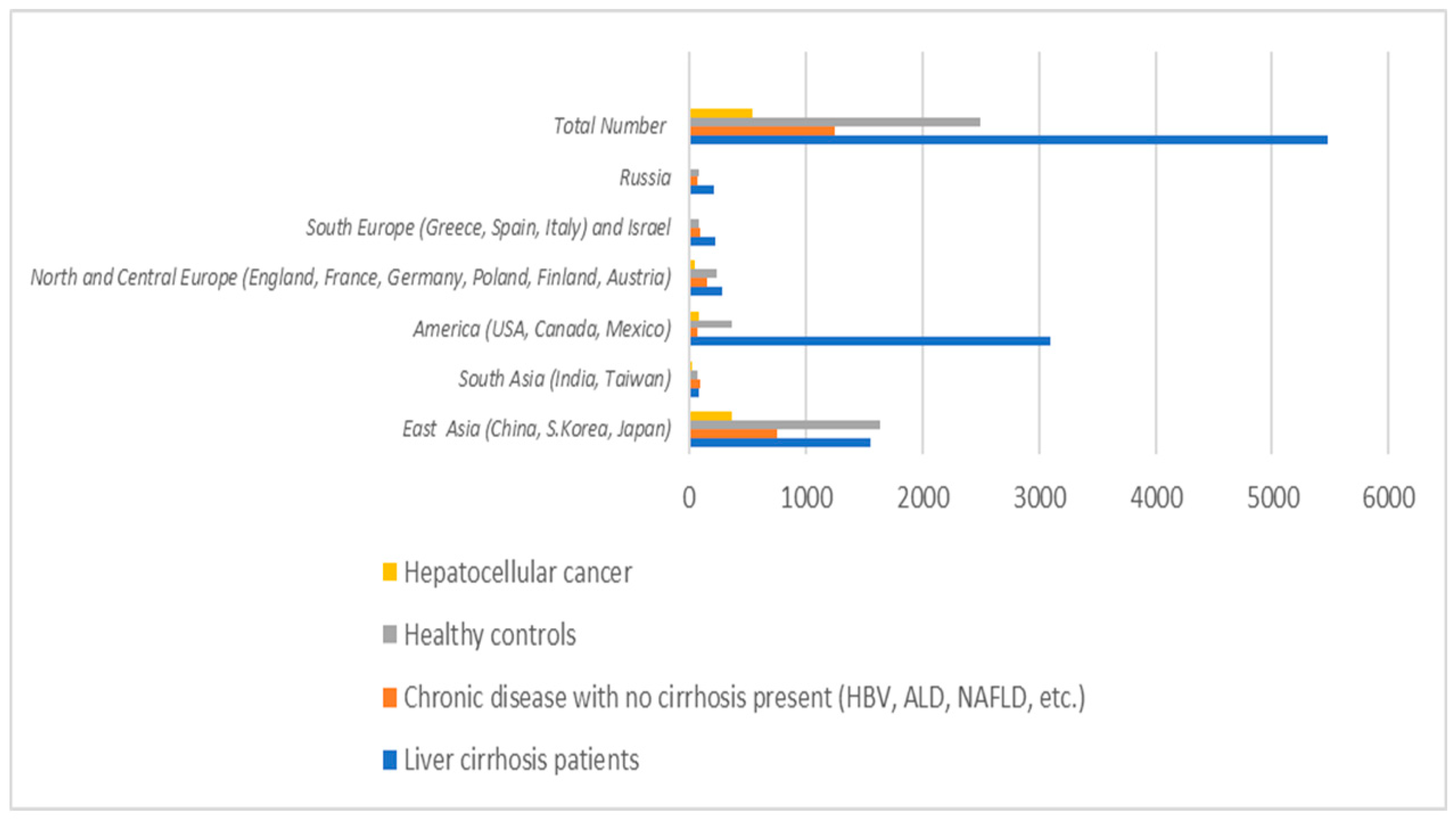
3.6. Gut Microbiota Assessment in Liver Cirrhosis Various Etiologies
4. Conclusions
Supplementary Materials
Funding
Data Availability Statement
Conflicts of Interest
References
- Albillos, A.; de Gottardi, A.; Rescigno, M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J. Hepatol. 2020, 72, 558–577. [Google Scholar] [CrossRef]
- Arab, J.P.; Martin-Mateos, R.M.; Shah, V.H. Gut-liver axis, cirrhosis and portal hypertension: The chicken and the egg. Hepatol. Int. 2018, 12, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Xirouchakis, E.; Manousou, P.; Tsartsali, L.; Georgopoulos, S.D.; Burroughs, A.K. Insights into the pathogenesis of NAFLD: The role of metabolic and pro-inflammatory mediators. Ann. Gastroenterol. 2009, 22, 24–33. [Google Scholar]
- Collins, S.L.; Stine, J.G.; Bisanz, J.E.; Okafor, C.D.; Patterson, A.D. Bile acids and the gut microbiota: Metabolic interactions and impacts on disease. Nat. Rev. Microbiol. 2023, 21, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Meroni, M.; Longo, M.; Dongiovanni, P. Alcohol or Gut Microbiota: Who Is the Guilty? Int. J. Mol. Sci. 2019, 20, 4568. [Google Scholar] [CrossRef]
- Park, J.W.; Kim, S.E.; Lee, N.Y.; Kim, J.H.; Jung, J.H.; Jang, M.K.; Park, S.H.; Lee, M.S.; Kim, D.J.; Kim, H.S.; et al. Role of Microbiota-Derived Metabolites in Alcoholic and Non-Alcoholic Fatty Liver Diseases. Int. J. Mol. Sci. 2021, 23, 426. [Google Scholar] [CrossRef]
- Jenne, C.N.; Kubes, P. Immune surveillance by the liver. Nat. Immunol. 2013, 14, 996–1006. [Google Scholar] [CrossRef]
- Forlano, R.; Martinez-Gili, L.; Takis, P.; Miguens-Blanco, J.; Liu, T.; Triantafyllou, E.; Skinner, C.; Loomba, R.; Thursz, M.; Marchesi, J.R.; et al. Disruption of gut barrier integrity and host-microbiome interactions underlie MASLD severity in patients with type-2 diabetes mellitus. Gut Microbes 2024, 16, 2304157. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, J.R.; Adams, D.H.; Fava, F.; Hermes, G.D.; Hirschfield, G.M.; Hold, G.; Quraishi, M.N.; Kinross, J.; Smidt, H.; Tuohy, K.M.; et al. The gut microbiota and host health: A new clinical frontier. Gut 2016, 65, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Acharya, C.; Bajaj, J.S. Altered Microbiome in Patients with Cirrhosis and Complications. Clin. Gastroenterol. Hepatol. 2019, 17, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Maslennikov, R.; Ivashkin, V.; Efremova, I.; Poluektova, E.; Kudryavtseva, A.; Krasnov, G. Gut dysbiosis and small intestinal bacterial overgrowth as independent forms of gut microbiota disorders in cirrhosis. World J. Gastroenterol. 2022, 28, 1067–1077. [Google Scholar] [CrossRef] [PubMed]
- Gkolfakis, P.; Tziatzios, G.; Leite, G.; Papanikolaou, I.S.; Xirouchakis, E.; Panayiotides, I.G.; Karageorgos, A.; Millan, M.J.; Mathur, R.; Weitsman, S.; et al. Prevalence of Small Intestinal Bacterial Overgrowth Syndrome in Patients with Non-Alcoholic Fatty Liver Disease/Non-Alcoholic Steatohepatitis: A Cross-Sectional Study. Microorganisms 2023, 11, 723. [Google Scholar] [CrossRef] [PubMed]
- Efremova, I.; Maslennikov, R.; Alieva, A.; Poluektova, E.; Ivashkin, V. Small Intestinal Bacterial Overgrowth Is Associated with Poor Prognosis in Cirrhosis. Microorganisms 2023, 11, 1017. [Google Scholar] [CrossRef]
- Fitriakusumah, Y.; Lesmana, C.R.A.; Bastian, W.P.; Jasirwan, C.O.M.; Hasan, I.; Simadibrata, M.; Kurniawan, J.; Sulaiman, A.S.; Gani, R.A. The role of Small Intestinal Bacterial Overgrowth (SIBO) in Non-alcoholic Fatty Liver Disease (NAFLD) patients evaluated using Controlled Attenuation Parameter (CAP) Transient Elastography (TE): A tertiary referral center experience. BMC Gastroenterol. 2019, 19, 43. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jin, Y.; Li, J.; Zhao, L.; Li, Z.; Xu, J.; Zhao, F.; Feng, J.; Chen, H.; Fang, C.; et al. Small Bowel Transit and Altered Gut Microbiota in Patients with Liver Cirrhosis. Front. Physiol. 2018, 9, 470. [Google Scholar] [CrossRef]
- Xirouchakis, E.; Kranidioti, H.; Hadziyanni, E.; Kourikou, A.; Reppas, C.; Vertzoni, M.; Papadopoulos, N.; Deutsch, M.; Papatheodoridis, G.; Manolakopoulos, S. The effect of propranolol on gastrointestinal motility and permeability in patients with cirrhosis and significant portal hypertension. BMC Gastroenterol. 2024, 24, 420. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Group, P. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. Ann. Intern. Med. 2009, 151, 264–269. [Google Scholar] [CrossRef]
- Shea, B.J.; Reeves, B.C.; Wells, G.; Thuku, M.; Hamel, C.; Moran, J.; Moher, D.; Tugwell, P.; Welch, V.; Kristjansson, E.; et al. AMSTAR 2: A critical appraisal tool for systematic reviews that include randomised or non-randomised studies of healthcare interventions, or both. BMJ 2017, 358, j4008. [Google Scholar] [CrossRef]
- Yang, J.; He, Q.; Lu, F.; Chen, K.; Ni, Z.; Wang, H.; Zhou, C.; Zhang, Y.; Chen, B.; Bo, Z.; et al. A distinct microbiota signature precedes the clinical diagnosis of hepatocellular carcinoma. Gut Microbes 2023, 15, 2201159. [Google Scholar] [CrossRef] [PubMed]
- Jinato, T.; Sikaroodi, M.; Fagan, A.; Sterling, R.K.; Lee, H.; Puri, P.; Davis, B.C.; Fuchs, M.; Gavis, E.; Gillevet, P.M.; et al. Alterations in gut virome are associated with cognitive function and minimal hepatic encephalopathy cross-sectionally and longitudinally in cirrhosis. Gut Microbes 2023, 15, 2288168. [Google Scholar] [CrossRef]
- Wang, Q.; Tang, X.; Qiao, W.; Sun, L.; Shi, H.; Chen, D.; Xu, B.; Liu, Y.; Zhao, J.; Huang, C.; et al. Machine learning-based characterization of the gut microbiome associated with the progression of primary biliary cholangitis to cirrhosis. Microbes Infect. 2024, 26, 105368. [Google Scholar] [CrossRef]
- Efremova, I.; Maslennikov, R.; Poluektova, E.; Zharkova, M.; Kudryavtseva, A.; Krasnov, G.; Fedorova, M.; Shirokova, E.; Kozlov, E.; Levshina, A.; et al. Gut Dysbiosis and Hemodynamic Changes as Links of the Pathogenesis of Complications of Cirrhosis. Microorganisms 2023, 11, 2202. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, S.; Xu, C.; Yu, L.; Chu, S.; Bao, J.; Wang, J.; Wang, J. Identification of Diagnostic Biomarkers for Compensatory Liver Cirrhosis Based on Gut Microbiota and Urine Metabolomics Analyses. Mol. Biotechnol. 2023, 66, 3164–3181. [Google Scholar] [CrossRef] [PubMed]
- Aliwa, B.; Horvath, A.; Traub, J.; Feldbacher, N.; Habisch, H.; Fauler, G.; Madl, T.; Stadlbauer, V. Altered gut microbiome, bile acid composition and metabolome in sarcopenia in liver cirrhosis. J. Cachexia Sarcopenia Muscle 2023, 14, 2676–2691. [Google Scholar] [CrossRef]
- Lin, M.J.; Su, T.H.; Chen, C.C.; Wu, W.K.; Hsu, S.J.; Tseng, T.C.; Liao, S.H.; Hong, C.M.; Yang, H.C.; Liu, C.J.; et al. Diversity and composition of gut microbiota in healthy individuals and patients at different stages of hepatitis B virus-related liver disease. Gut Pathog. 2023, 15, 24. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Chen, C.; Zuo, S.; Cao, K.; Li, H. Integrative analysis of the gut microbiota and faecal and serum short-chain fatty acids and tryptophan metabolites in patients with cirrhosis and hepatic encephalopathy. J. Transl. Med. 2023, 21, 395. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Zhang, Q.; Shi, K.; Zhang, Y.; Zhu, B.; Bi, Y.; Wang, X. Gut microbiota dysbiosis with hepatitis B virus liver disease and association with immune response. Front. Cell. Infect. Microbiol. 2023, 13, 1152987. [Google Scholar] [CrossRef]
- Lu, G.; Zhang, Y.; Ren, Y.; Shi, J.S.; Xu, Z.H.; Geng, Y. Diversity and Comparison of Intestinal Desulfovibrio in Patients with Liver Cirrhosis and Healthy People. Microorganisms 2023, 11, 276. [Google Scholar] [CrossRef]
- Shen, Y.; Wu, S.D.; Chen, Y.; Li, X.Y.; Zhu, Q.; Nakayama, K.; Zhang, W.Q.; Weng, C.Z.; Zhang, J.; Wang, H.K.; et al. Alterations in gut microbiome and metabolomics in chronic hepatitis B infection-associated liver disease and their impact on peripheral immune response. Gut Microbes 2023, 15, 2155018. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.W.; Chu, Y.D.; Hsu, C.W.; Chen, Y.C.; Liang, K.H.; Yeh, C.T. Multi-Omics Analyses Identify Signatures in Patients with Liver Cirrhosis and Hepatocellular Carcinoma. Cancers 2022, 15, 210. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Lv, H.; Lv, J.; Shi, Y.; Huang, H.; Chen, L.; Shi, D. Alterations of gut microbiota in cirrhotic patients with spontaneous bacterial peritonitis: A distinctive diagnostic feature. Front. Cell. Infect. Microbiol. 2022, 12, 999418. [Google Scholar] [CrossRef]
- Maslennikov, R.; Ivashkin, V.; Alieva, A.; Poluektova, E.; Kudryavtseva, A.; Krasnov, G.; Zharkova, M.; Zharikov, Y. Gut dysbiosis and body composition in cirrhosis. World J. Hepatol. 2022, 14, 1210–1225. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.; Feng, H. Changes in intestinal microbiota of HBV-associated liver cirrhosis with/without hepatic encephalopathy. Medicine 2022, 101, e29935. [Google Scholar] [CrossRef] [PubMed]
- Philips, C.A.; Ahamed, R.; Abduljaleel, J.K.P.; Rajesh, S.; Augustine, P. Identification and Analysis of Gut Microbiota and Functional Metabolism in Decompensated Cirrhosis with Infection. J. Clin. Transl. Hepatol. 2023, 11, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, J.S.; Pena-Rodriguez, M.; La Reau, A.; Phillips, W.; Fuchs, M.; Davis, B.C.; Sterling, R.K.; Sikaroodi, M.; Fagan, A.; Shamsaddini, A.; et al. Longitudinal transkingdom gut microbial approach towards decompensation in outpatients with cirrhosis. Gut 2023, 72, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Chi, X.; Zhao, Y.; Liu, S.; Xing, H. Characteristics and Clinical Significance of Intestinal Microbiota in Patients with Chronic Hepatitis B Cirrhosis and Type 2 Diabetes Mellitus. J. Diabetes Res. 2022, 2022, 1826181. [Google Scholar] [CrossRef]
- Ullah, N.; Kakakhel, M.A.; Khan, I.; Gul Hilal, M.; Lajia, Z.; Bai, Y.; Sajjad, W.; Yuxi, L.; Ullah, H.; Almohaimeed, H.M.; et al. Structural and compositional segregation of the gut microbiota in HCV and liver cirrhotic patients: A clinical pilot study. Microb. Pathog. 2022, 171, 105739. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Reddy, K.R.; Tandon, P.; Garcia-Tsao, G.; Kamath, P.S.; O’Leary, J.G.; Wong, F.; Lai, J.; Vargas, H.; Thuluvath, P.J.; et al. Association of serum metabolites and gut microbiota at hospital admission with nosocomial infection development in patients with cirrhosis. Liver Transpl. 2022, 28, 1831–1840. [Google Scholar] [CrossRef]
- Baltazar-Diaz, T.A.; Gonzalez-Hernandez, L.A.; Aldana-Ledesma, J.M.; Pena-Rodriguez, M.; Vega-Magana, A.N.; Zepeda-Morales, A.S.M.; Lopez-Roa, R.I.; Del Toro-Arreola, S.; Martinez-Lopez, E.; Salazar-Montes, A.M.; et al. Escherichia/Shigella, SCFAs, and Metabolic Pathways-The Triad That Orchestrates Intestinal Dysbiosis in Patients with Decompensated Alcoholic Cirrhosis from Western Mexico. Microorganisms 2022, 10, 1231. [Google Scholar] [CrossRef]
- Zheng, R.; Wang, G.; Pang, Z.; Ran, N.; Gu, Y.; Guan, X.; Yuan, Y.; Zuo, X.; Pan, H.; Zheng, J.; et al. Liver cirrhosis contributes to the disorder of gut microbiota in patients with hepatocellular carcinoma. Cancer Med. 2020, 9, 4232–4250. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, J.S.; Fagan, A.; McGeorge, S.; Sterling, R.K.; Rogal, S.; Sikaroodi, M.; Gillevet, P.M. Area Deprivation Index and Gut-Brain Axis in Cirrhosis. Clin. Transl. Gastroenterol. 2022, 13, e00495. [Google Scholar] [CrossRef] [PubMed]
- Shu, W.; Shanjian, C.; Jinpiao, L.; Qishui, O. Gut microbiota dysbiosis in patients with hepatitis B virus-related cirrhosis. Ann. Hepatol. 2022, 27, 100676. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Yan, G.; Feng, F.; Wang, M.; Long, F. Characterization of intestinal microbiota and serum metabolites in patients with mild hepatic encephalopathy. Open Life Sci. 2022, 17, 139–154. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.S.; Jacobs, J.P.; Agopian, V.; Pisegna, J.R.; Ayoub, W.; Durazo, F.; Enayati, P.; Sundaram, V.; Benhammou, J.N.; Noureddin, M.; et al. Duodenal Microbiome and Serum Metabolites Predict Hepatocellular Carcinoma in a Multicenter Cohort of Patients with Cirrhosis. Dig. Dis. Sci. 2022, 67, 3831–3841. [Google Scholar] [CrossRef]
- Shen, T.D.; Daniel, S.G.; Patel, S.; Kaplan, E.; Phung, L.; Lemelle-Thomas, K.; Chau, L.; Herman, L.; Trisolini, C.; Stonelake, A.; et al. The Mucosally-Adherent Rectal Microbiota Contains Features Unique to Alcohol-Related Cirrhosis. Gut Microbes 2021, 13, 1987781. [Google Scholar] [CrossRef]
- Chen, T.; Ding, R.; Chen, X.; Lu, Y.; Shi, J.; Lu, Y.; Tang, B.; Zhang, W.; Ye, C.; Yuan, M.; et al. Firmicutes and Blautia in gut microbiota lessened in chronic liver diseases and hepatocellular carcinoma patients: A pilot study. Bioengineered 2021, 12, 8233–8246. [Google Scholar] [CrossRef]
- Maslennikov, R.; Ivashkin, V.; Efremova, I.; Alieva, A.; Kashuh, E.; Tsvetaeva, E.; Poluektova, E.; Shirokova, E.; Ivashkin, K. Gut dysbiosis is associated with poorer long-term prognosis in cirrhosis. World J. Hepatol. 2021, 13, 557–570. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, Z.; Mo, Z.S.; Yang, X.H.; Lin, B.L.; Peng, L.; Xu, Y.; Lei, C.Y.; Zhuang, X.D.; Lu, L.; et al. Gut microbiota as prognosis markers for patients with HBV-related acute-on-chronic liver failure. Gut Microbes 2021, 13, 1921925. [Google Scholar] [CrossRef]
- Zhong, X.; Cui, P.; Jiang, J.; Ning, C.; Liang, B.; Zhou, J.; Tian, L.; Zhang, Y.; Lei, T.; Zuo, T.; et al. Streptococcus, the Predominant Bacterium to Predict the Severity of Liver Injury in Alcoholic Liver Disease. Front. Cell. Infect. Microbiol. 2021, 11, 649060. [Google Scholar] [CrossRef] [PubMed]
- Gui, Q.F.; Jin, H.L.; Zhu, F.; Lu, H.F.; Zhang, Q.; Xu, J.; Yang, Y.M.; Xiao, C. Gut microbiota signatures in Schistosoma japonicum infection-induced liver cirrhosis patients: A case-control study. Infect. Dis. Poverty 2021, 10, 43. [Google Scholar] [CrossRef]
- Bloom, P.P.; Luevano, J.M., Jr.; Miller, K.J.; Chung, R.T. Deep stool microbiome analysis in cirrhosis reveals an association between short-chain fatty acids and hepatic encephalopathy. Ann. Hepatol. 2021, 25, 100333. [Google Scholar] [CrossRef]
- Tang, Y.; Zhou, H.; Xiang, Y.; Cui, F. The diagnostic potential of gut microbiome for early hepatitis B virus-related hepatocellular carcinoma. Eur. J. Gastroenterol. Hepatol. 2021, 33, e167–e175. [Google Scholar] [CrossRef]
- Yokoyama, K.; Tsuchiya, N.; Yamauchi, R.; Miyayama, T.; Uchida, Y.; Shibata, K.; Fukuda, H.; Umeda, K.; Takata, K.; Tanaka, T.; et al. Exploratory Research on the Relationship between Human Gut Microbiota and Portal Hypertension. Intern. Med. 2020, 59, 2089–2094. [Google Scholar] [CrossRef] [PubMed]
- Zha, H.; Chen, Y.; Wu, J.; Chang, K.; Lu, Y.; Zhang, H.; Xie, J.; Wang, Q.; Tang, R.; Li, L. Characteristics of three microbial colonization states in the duodenum of the cirrhotic patients. Future Microbiol. 2020, 15, 855–868. [Google Scholar] [CrossRef] [PubMed]
- Lapidot, Y.; Amir, A.; Nosenko, R.; Uzan-Yulzari, A.; Veitsman, E.; Cohen-Ezra, O.; Davidov, Y.; Weiss, P.; Bradichevski, T.; Segev, S.; et al. Alterations in the Gut Microbiome in the Progression of Cirrhosis to Hepatocellular Carcinoma. mSystems 2020, 5, e00153-20. [Google Scholar] [CrossRef]
- Chen, Z.; Xie, Y.; Zhou, F.; Zhang, B.; Wu, J.; Yang, L.; Xu, S.; Stedtfeld, R.; Chen, Q.; Liu, J.; et al. Featured Gut Microbiomes Associated with the Progression of Chronic Hepatitis B Disease. Front. Microbiol. 2020, 11, 383. [Google Scholar] [CrossRef] [PubMed]
- Sole, C.; Guilly, S.; Da Silva, K.; Llopis, M.; Le-Chatelier, E.; Huelin, P.; Carol, M.; Moreira, R.; Fabrellas, N.; De Prada, G.; et al. Alterations in Gut Microbiome in Cirrhosis as Assessed by Quantitative Metagenomics: Relationship with Acute-on-Chronic Liver Failure and Prognosis. Gastroenterology 2021, 160, 206–218.e13. [Google Scholar] [CrossRef] [PubMed]
- Sydor, S.; Best, J.; Messerschmidt, I.; Manka, P.; Vilchez-Vargas, R.; Brodesser, S.; Lucas, C.; Wegehaupt, A.; Wenning, C.; Assmuth, S.; et al. Altered Microbiota Diversity and Bile Acid Signaling in Cirrhotic and Noncirrhotic NASH-HCC. Clin. Transl. Gastroenterol. 2020, 11, e00131. [Google Scholar] [CrossRef] [PubMed]
- Seok, J.; Suk, K.T. Gut-microbiome Taxonomic Profiling as Non-invasive Biomarkers for the Early Detection of Alcoholic Hepatocellular Carcinoma. J. Liver Cancer 2020, 20, 32–40. [Google Scholar] [CrossRef]
- Haraguchi, M.; Miuma, S.; Masumoto, H.; Ichikawa, T.; Kanda, Y.; Sasaki, R.; Fukushima, M.; Miyaaki, H.; Taura, N.; Nakao, K. Bacteroides in colonic mucosa-associated microbiota affects the development of minimal hepatic encephalopathy in patients with cirrhosis. Hepatol. Int. 2019, 13, 482–489. [Google Scholar] [CrossRef]
- Addolorato, G.; Ponziani, F.R.; Dionisi, T.; Mosoni, C.; Vassallo, G.A.; Sestito, L.; Petito, V.; Picca, A.; Marzetti, E.; Tarli, C.; et al. Gut microbiota compositional and functional fingerprint in patients with alcohol use disorder and alcohol-associated liver disease. Liver Int. 2020, 40, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.D.; Peng, X.B.; Zhao, R.R.; Ma, C.Q.; Li, J.N.; Yao, L.Q. The intestinal microbial community dissimilarity in hepatitis B virus-related liver cirrhosis patients with and without at alcohol consumption. Gut Pathog. 2019, 11, 58. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, F.; Lu, H.; Wang, B.; Chen, Y.; Lei, D.; Wang, Y.; Zhu, B.; Li, L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology 2011, 54, 562–572. [Google Scholar] [CrossRef]
- Wei, X.; Jiang, S.; Zhao, X.; Li, H.; Lin, W.; Li, B.; Lu, J.; Sun, Y.; Yuan, J. Community-Metabolome Correlations of Gut Microbiota from Child-Turcotte-Pugh of A and B Patients. Front. Microbiol. 2016, 7, 1856. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, J.S.; Ridlon, J.M.; Hylemon, P.B.; Thacker, L.R.; Heuman, D.M.; Smith, S.; Sikaroodi, M.; Gillevet, P.M. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G168–G175. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wu, D.; Ahmed, A.; Li, X.; Ma, Y.; Tang, L.; Mo, D.; Ma, Y.; Xin, Y. Comparison of the gut microbe profiles and numbers between patients with liver cirrhosis and healthy individuals. Curr. Microbiol. 2012, 65, 7–13. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, Y.N.; Chen, T.; Ren, C.H.; Li, X.; Liu, G.X. Relationship between intestinal microbial dysbiosis and primary liver cancer. Hepatobiliary Pancreat. Dis. Int. 2019, 18, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Grat, M.; Wronka, K.M.; Krasnodebski, M.; Masior, L.; Lewandowski, Z.; Kosinska, I.; Grat, K.; Stypulkowski, J.; Rejowski, S.; Wasilewicz, M.; et al. Profile of Gut Microbiota Associated with the Presence of Hepatocellular Cancer in Patients with Liver Cirrhosis. Transplant. Proc. 2016, 48, 1687–1691. [Google Scholar] [CrossRef]
- Astbury, S.; Atallah, E.; Vijay, A.; Aithal, G.P.; Grove, J.I.; Valdes, A.M. Lower gut microbiome diversity and higher abundance of proinflammatory genus Collinsella are associated with biopsy-proven nonalcoholic steatohepatitis. Gut Microbes 2020, 11, 569–580. [Google Scholar] [CrossRef]
- Jin, M.; Kalainy, S.; Baskota, N.; Chiang, D.; Deehan, E.C.; McDougall, C.; Tandon, P.; Martinez, I.; Cervera, C.; Walter, J.; et al. Faecal microbiota from patients with cirrhosis has a low capacity to ferment non-digestible carbohydrates into short-chain fatty acids. Liver Int. 2019, 39, 1437–1447. [Google Scholar] [CrossRef]
- Ponziani, F.R.; Bhoori, S.; Castelli, C.; Putignani, L.; Rivoltini, L.; Del Chierico, F.; Sanguinetti, M.; Morelli, D.; Paroni Sterbini, F.; Petito, V.; et al. Hepatocellular Carcinoma Is Associated with Gut Microbiota Profile and Inflammation in Nonalcoholic Fatty Liver Disease. Hepatology 2019, 69, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, J.S.; Fagan, A.; White, M.B.; Wade, J.B.; Hylemon, P.B.; Heuman, D.M.; Fuchs, M.; John, B.V.; Acharya, C.; Sikaroodi, M.; et al. Specific Gut and Salivary Microbiota Patterns Are Linked with Different Cognitive Testing Strategies in Minimal Hepatic Encephalopathy. Am. J. Gastroenterol. 2019, 114, 1080–1090. [Google Scholar] [CrossRef]
- Ciocan, D.; Voican, C.S.; Wrzosek, L.; Hugot, C.; Rainteau, D.; Humbert, L.; Cassard, A.M.; Perlemuter, G. Bile acid homeostasis and intestinal dysbiosis in alcoholic hepatitis. Aliment. Pharmacol. Ther. 2018, 48, 961–974. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Li, A.; Jiang, J.; Zhou, L.; Yu, Z.; Lu, H.; Xie, H.; Chen, X.; Shao, L.; Zhang, R.; et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 2019, 68, 1014–1023. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, J.S.; Hylemon, P.B.; Ridlon, J.M.; Heuman, D.M.; Daita, K.; White, M.B.; Monteith, P.; Noble, N.A.; Sikaroodi, M.; Gillevet, P.M. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G675–G685. [Google Scholar] [CrossRef] [PubMed]
- Qin, N.; Yang, F.; Li, A.; Prifti, E.; Chen, Y.; Shao, L.; Guo, J.; Le Chatelier, E.; Yao, J.; Wu, L.; et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014, 513, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhai, H.; Geng, J.; Yu, R.; Ren, H.; Fan, H.; Shi, P. Large-scale survey of gut microbiota associated with MHE Via 16S rRNA-based pyrosequencing. Am. J. Gastroenterol. 2013, 108, 1601–1611. [Google Scholar] [CrossRef] [PubMed]
- Grat, M.; Holowko, W.; Wronka, K.M.; Grat, K.; Lewandowski, Z.; Kosinska, I.; Krasnodebski, M.; Wasilewicz, M.; Galecka, M.; Szachta, P.; et al. The relevance of intestinal dysbiosis in liver transplant candidates. Transpl. Infect. Dis. 2015, 17, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Tuomisto, S.; Pessi, T.; Collin, P.; Vuento, R.; Aittoniemi, J.; Karhunen, P.J. Changes in gut bacterial populations and their translocation into liver and ascites in alcoholic liver cirrhotics. BMC Gastroenterol. 2014, 14, 40. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, J.S.; Heuman, D.M.; Hylemon, P.B.; Sanyal, A.J.; White, M.B.; Monteith, P.; Noble, N.A.; Unser, A.B.; Daita, K.; Fisher, A.R.; et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J. Hepatol. 2014, 60, 940–947. [Google Scholar] [CrossRef]
- Ahluwalia, V.; Betrapally, N.S.; Hylemon, P.B.; White, M.B.; Gillevet, P.M.; Unser, A.B.; Fagan, A.; Daita, K.; Heuman, D.M.; Zhou, H.; et al. Impaired Gut-Liver-Brain Axis in Patients with Cirrhosis. Sci. Rep. 2016, 6, 26800. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, J.S.; Vargas, H.E.; Reddy, K.R.; Lai, J.C.; O’Leary, J.G.; Tandon, P.; Wong, F.; Mitrani, R.; White, M.B.; Kelly, M.; et al. Association Between Intestinal Microbiota Collected at Hospital Admission and Outcomes of Patients with Cirrhosis. Clin. Gastroenterol. Hepatol. 2019, 17, 756–765.e753. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Betrapally, N.S.; Hylemon, P.B.; Thacker, L.R.; Daita, K.; Kang, D.J.; White, M.B.; Unser, A.B.; Fagan, A.; Gavis, E.A.; et al. Gut Microbiota Alterations can predict Hospitalizations in Cirrhosis Independent of Diabetes Mellitus. Sci. Rep. 2015, 5, 18559. [Google Scholar] [CrossRef] [PubMed]
- Albhaisi, S.; Shamsaddini, A.; Fagan, A.; McGeorge, S.; Sikaroodi, M.; Gavis, E.; Patel, S.; Davis, B.C.; Acharya, C.; Sterling, R.K.; et al. Gut Microbial Signature of Hepatocellular Cancer in Men with Cirrhosis. Liver Transpl. 2021, 27, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Oh, T.G.; Kim, S.M.; Caussy, C.; Fu, T.; Guo, J.; Bassirian, S.; Singh, S.; Madamba, E.V.; Bettencourt, R.; Richards, L.; et al. A Universal Gut-Microbiome-Derived Signature Predicts Cirrhosis. Cell Metab. 2020, 32, 878–888.e876. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Thacker, L.R.; Fagan, A.; White, M.B.; Gavis, E.A.; Hylemon, P.B.; Brown, R.; Acharya, C.; Heuman, D.M.; Fuchs, M.; et al. Gut microbial RNA and DNA analysis predicts hospitalizations in cirrhosis. JCI Insight 2018, 3, e98019. [Google Scholar] [CrossRef] [PubMed]
- Dubinkina, V.B.; Tyakht, A.V.; Odintsova, V.Y.; Yarygin, K.S.; Kovarsky, B.A.; Pavlenko, A.V.; Ischenko, D.S.; Popenko, A.S.; Alexeev, D.G.; Taraskina, A.Y.; et al. Links of gut microbiota composition with alcohol dependence syndrome and alcoholic liver disease. Microbiome 2017, 5, 141. [Google Scholar] [CrossRef]
- Caussy, C.; Tripathi, A.; Humphrey, G.; Bassirian, S.; Singh, S.; Faulkner, C.; Bettencourt, R.; Rizo, E.; Richards, L.; Xu, Z.Z.; et al. A gut microbiome signature for cirrhosis due to nonalcoholic fatty liver disease. Nat. Commun. 2019, 10, 1406. [Google Scholar] [CrossRef]
- Vilstrup, H.; Amodio, P.; Bajaj, J.; Cordoba, J.; Ferenci, P.; Mullen, K.D.; Weissenborn, K.; Wong, P. Hepatic encephalopathy in chronic liver disease: 2014 Practice Guideline by the American Association for the Study of Liver Diseases and the European Association for the Study of the Liver. Hepatology 2014, 60, 715–735. [Google Scholar] [CrossRef] [PubMed]
- Patidar, K.R.; Bajaj, J.S. Covert and Overt Hepatic Encephalopathy: Diagnosis and Management. Clin. Gastroenterol. Hepatol. 2015, 13, 2048–2061. [Google Scholar] [CrossRef]
- Llovet, J.M.; Zucman-Rossi, J.; Pikarsky, E.; Sangro, B.; Schwartz, M.; Sherman, M.; Gores, G. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2016, 2, 16018. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Chen, S.; Fu, Y.; Wu, W.; Chen, T.; Chen, J.; Yang, B.; Ou, Q. Gut microbiota dysbiosis in patients with hepatitis B virus-induced chronic liver disease covering chronic hepatitis, liver cirrhosis and hepatocellular carcinoma. J. Viral Hepat. 2020, 27, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Gerard, P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens 2013, 3, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Hollister, E.B.; Gao, C.; Versalovic, J. Compositional and functional features of the gastrointestinal microbiome and their effects on human health. Gastroenterology 2014, 146, 1449–1458. [Google Scholar] [CrossRef] [PubMed]
- El Aidy, S.; van den Bogert, B.; Kleerebezem, M. The small intestine microbiota, nutritional modulation and relevance for health. Curr. Opin. Biotechnol. 2015, 32, 14–20. [Google Scholar] [CrossRef]
- Luo, W.; Guo, S.; Zhou, Y.; Zhao, J.; Wang, M.; Sang, L.; Chang, B.; Wang, B. Hepatocellular Carcinoma: How the Gut Microbiota Contributes to Pathogenesis, Diagnosis, and Therapy. Front. Microbiol. 2022, 13, 873160. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, J.S.; Fagan, A.; Gavis, E.A.; Mousel, T.; Gallagher, M.L.; Puri, P.; Fuchs, M.; Davis, B.C.; Hylemon, P.B.; Zhou, H.; et al. The RIVET RCT: Rifamycin SV MMX improves muscle mass, physical function, and ammonia in cirrhosis and minimal encephalopathy. Hepatol. Commun. 2024, 8, e0384. [Google Scholar] [CrossRef] [PubMed]
- Scarpellini, E.; Abenavoli, L.; Cassano, V.; Rinninella, E.; Sorge, M.; Capretti, F.; Rasetti, C.; Svegliati Baroni, G.; Luzza, F.; Santori, P.; et al. The Apparent Asymmetrical Relationship Between Small Bowel Bacterial Overgrowth, Endotoxemia, and Liver Steatosis and Fibrosis in Cirrhotic and Non-Cirrhotic Patients: A Single-Center Pilot Study. Front. Med. 2022, 9, 872428. [Google Scholar] [CrossRef]
- Yokoyama, K.; Sakamaki, A.; Takahashi, K.; Naruse, T.; Sato, C.; Kawata, Y.; Tominaga, K.; Abe, H.; Sato, H.; Tsuchiya, A.; et al. Hydrogen-producing small intestinal bacterial overgrowth is associated with hepatic encephalopathy and liver function. PLoS ONE 2022, 17, e0264459. [Google Scholar] [CrossRef] [PubMed]
- Leitner, U.; Brits, A.; Xu, D.; Patil, S.; Sun, J. Efficacy of probiotics on improvement of health outcomes in cirrhotic liver disease patients: A systematic review and meta-analysis of randomised controlled trials. Eur. J. Pharmacol. 2024, 981, 176874. [Google Scholar] [CrossRef]
- Guo, Y.; Zhang, Y.; Gerhard, M.; Gao, J.J.; Mejias-Luque, R.; Zhang, L.; Vieth, M.; Ma, J.L.; Bajbouj, M.; Suchanek, S.; et al. Effect of Helicobacter pylori on gastrointestinal microbiota: A population-based study in Linqu, a high-risk area of gastric cancer. Gut 2020, 69, 1598–1607. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xirouchakis, E.; Pelekanos, A.; Xirouchakis, S.; Kranidioti, H.; Manolakopoulos, S. A Systematic Review of Microbiota in Cirrhosis: A Change Towards a More Pathogenic Predisposition. Int. J. Mol. Sci. 2025, 26, 527. https://doi.org/10.3390/ijms26020527
Xirouchakis E, Pelekanos A, Xirouchakis S, Kranidioti H, Manolakopoulos S. A Systematic Review of Microbiota in Cirrhosis: A Change Towards a More Pathogenic Predisposition. International Journal of Molecular Sciences. 2025; 26(2):527. https://doi.org/10.3390/ijms26020527
Chicago/Turabian StyleXirouchakis, Elias, Alexandros Pelekanos, Spyridon Xirouchakis, Hariklia Kranidioti, and Spilios Manolakopoulos. 2025. "A Systematic Review of Microbiota in Cirrhosis: A Change Towards a More Pathogenic Predisposition" International Journal of Molecular Sciences 26, no. 2: 527. https://doi.org/10.3390/ijms26020527
APA StyleXirouchakis, E., Pelekanos, A., Xirouchakis, S., Kranidioti, H., & Manolakopoulos, S. (2025). A Systematic Review of Microbiota in Cirrhosis: A Change Towards a More Pathogenic Predisposition. International Journal of Molecular Sciences, 26(2), 527. https://doi.org/10.3390/ijms26020527





