Comparative Analysis of Crystal Violet-Binding Aptamers as Potential Cores for Binary Sensors
Abstract
1. Introduction
2. Results
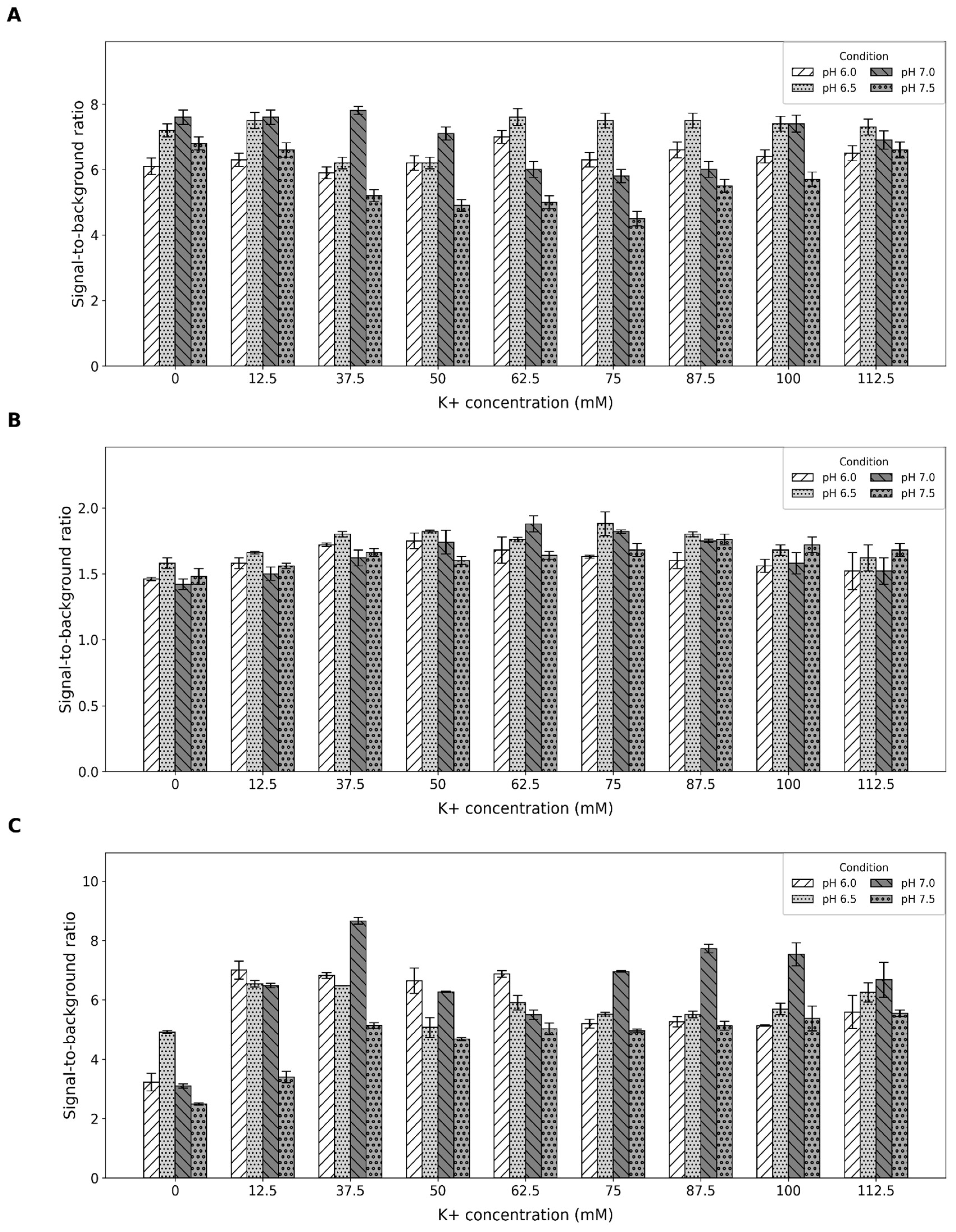
2.1. Buffer Optimization
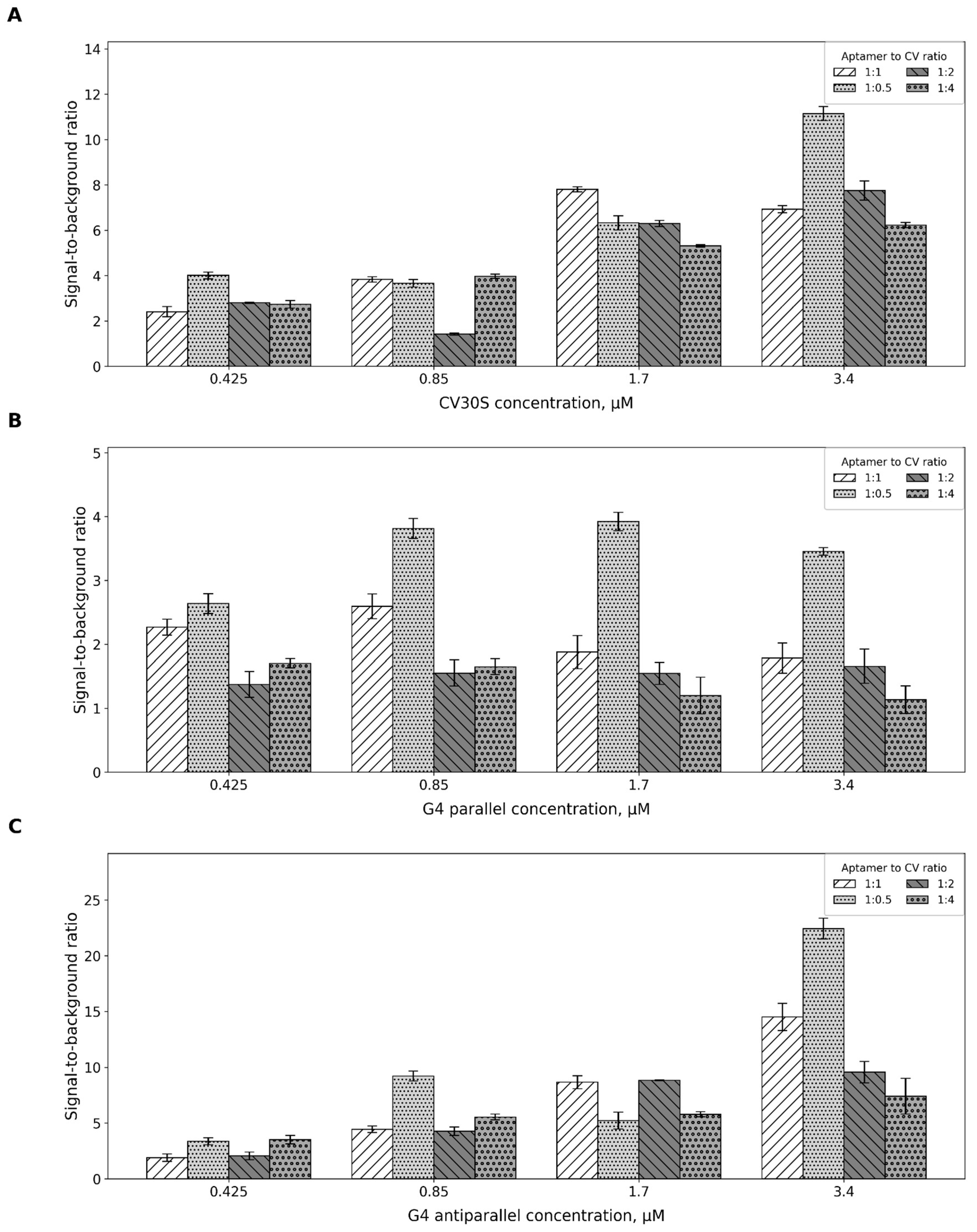
2.2. Aptamer-to-Fluorescent Ratio
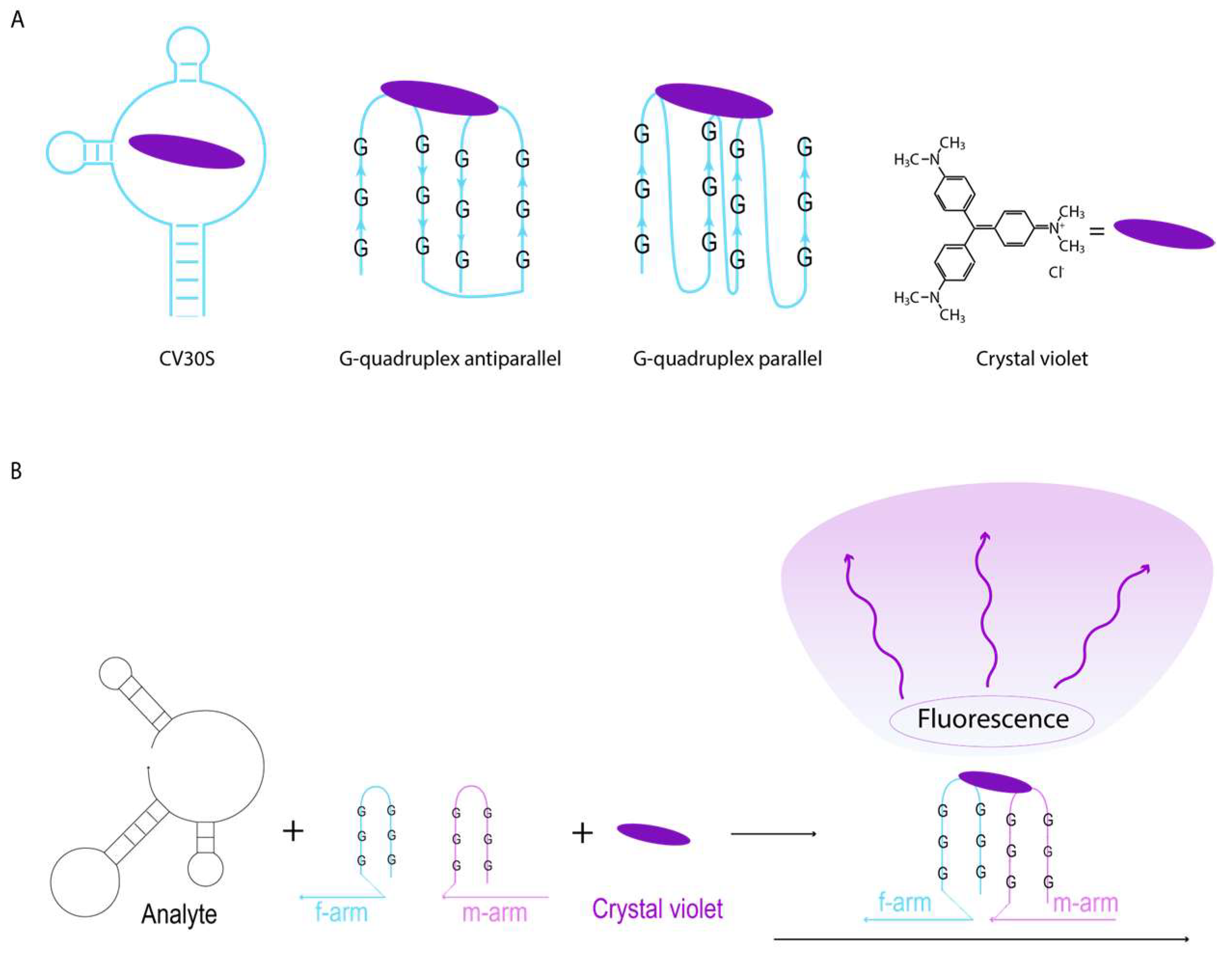
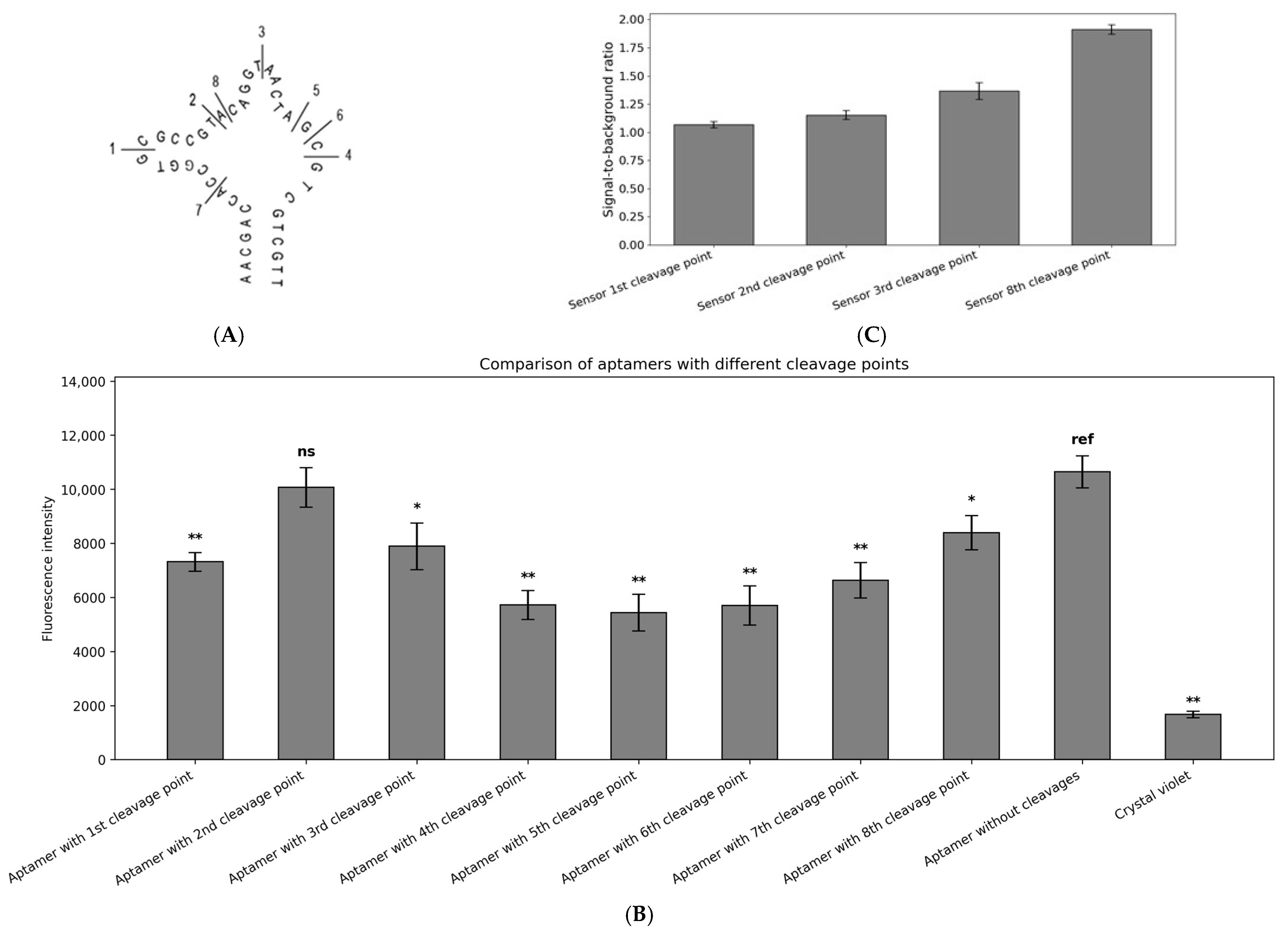
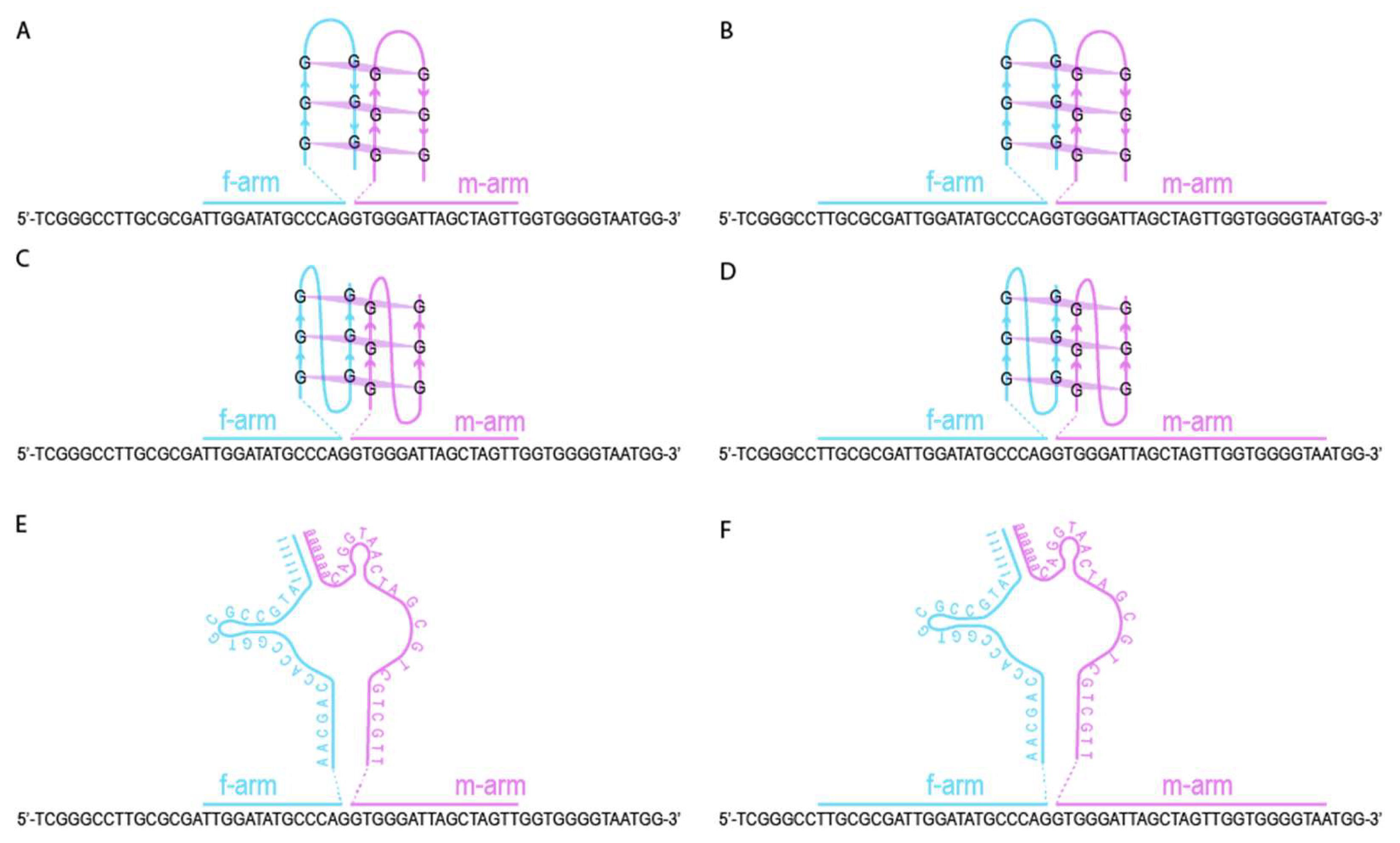
2.3. CV30S Aptamer Splitting
2.4. Design of DNA Nanosensors
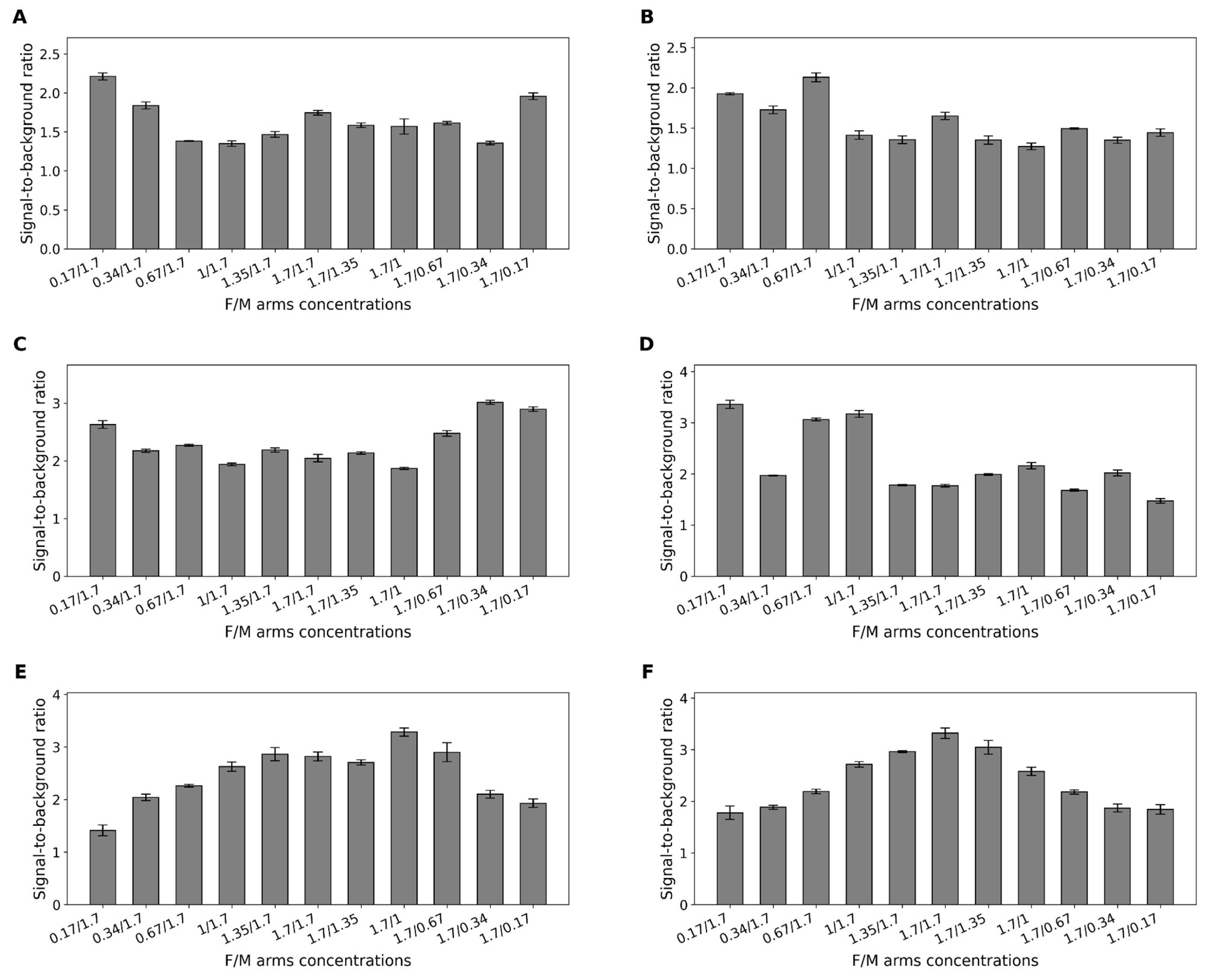
2.5. Optimization of Fragment Ratios in Binary Sensors
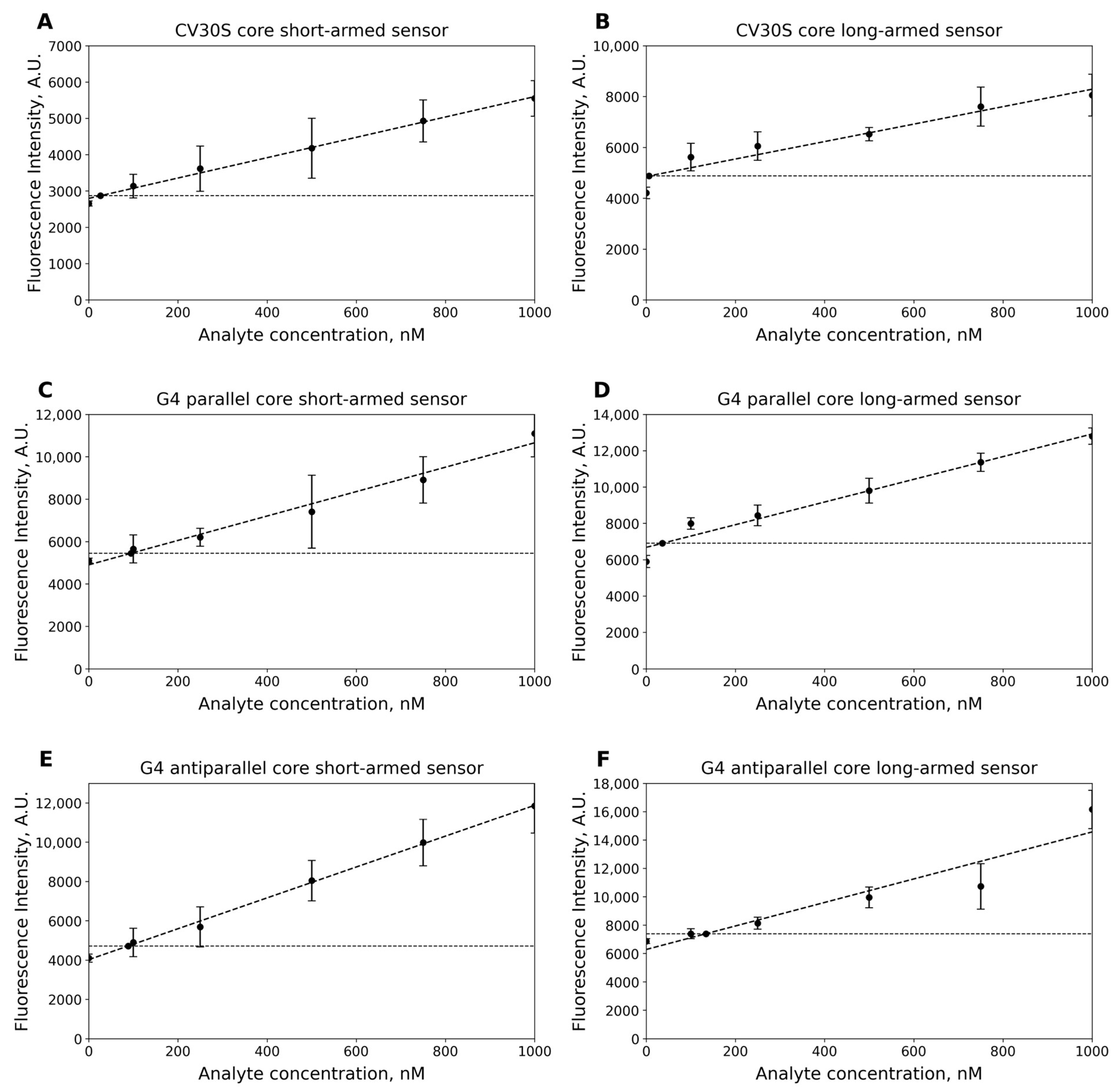
2.6. Limit of Detection (LOD)
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Buffer Optimization
4.3. Aptamer-to-CV Ratio
4.4. Aptamer Splitting
4.5. Binary Sensor Design
- While selecting the recognition site for the biosensor, the secondary structure of the target DNA region must be taken into account.
- If the target DNA sequence folds into a complex secondary structure forming a loop, the sensor arms should be placed on the same side of the loop.
- The melting temperature should be within the range of 35–40 °C for detection at 37 °C or 50–60 °C for detection at 55 °C.
- Each sequence should be linked to one of the aptamer sequences through a T-linker composed of two to six thymidine residues.
- Each sensor arm must be checked for secondary structure formation at 37 °C and 55 °C. They should not form loop structures, and the overall Gibbs free energy of the structure should not exceed -4 kcal/mol.
4.6. Binary Sensor Arm Ratio Optimization
4.7. Limits of Detection
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CV | Crystal Violet |
| G4 | G-Quadruplex |
| LOD | Limit of Detection |
| S/B | Signal-to-Background |
References
- Radom, F.; Jurek, P.M.; Mazurek, M.P.; Otlewski, J.; Jeleń, F. Aptamers: Molecules of great potential. Biotechnol. Adv. 2013, 31, 1260–1274. [Google Scholar] [CrossRef]
- Ruscito, A.; DeRosa, M.C. Small-molecule binding aptamers: Selection strategies, characterization, and applications. Front. Chem. 2016, 4, 14. [Google Scholar] [CrossRef]
- Yang, H.; Wang, W.; Zeng, Y.; Tang, R.; Yan, H.; Yang, Y.; Li, L. A novel composite of aptamer-based Ti3C2Tx and molecularly imprinted polymer with double recognition property for sensitive electrochemical detection of ofloxacin. J. Anal. Test. 2025, 9, 109–117. [Google Scholar] [CrossRef]
- Sequeira-Antunes, B.; Ferreira, H.A. Nucleic acid aptamer-based biosensors: A review. Biomedicines 2023, 11, 3201. [Google Scholar] [CrossRef]
- Ning, Y.; Hu, J.; Lu, F. Aptamers used for biosensors and targeted therapy. Biomed. Pharmacother. 2020, 132, 110902. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shen, C.; Wu, C.; Zhan, Z.; Qu, R.; Xie, Y.; Chen, P. Self-assembled DNA machine and selective complexation recognition enable rapid homogeneous portable quantification of lung cancer CTCs. Research 2024, 7, 0352. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Tai, X.; Ren, Q.; Ren, A. Structure-based insights into fluorogenic RNA aptamers. Acta Biochim. Biophys. Sin. 2024, 57, 108. [Google Scholar] [CrossRef]
- Ouellet, J. RNA fluorescence with light-up aptamers. Front. Chem. 2016, 4, 29. [Google Scholar] [CrossRef]
- Ma, Z.; Kulsoom; Wang, F. Lighting Up Endogenous RNA: Fluorescent Aptamer Sensors for Live-Cell Imaging. Chem. Biomed. Imaging 2025, 1–4. [Google Scholar] [CrossRef]
- Kolpashchikov, D.M.; Spelkov, A.A. Binary (Split) Light-up Aptameric Sensors. Angew. Chem. 2021, 133, 5040–5051. [Google Scholar] [CrossRef]
- Kikuchi, N.; Kolpashchikov, D.M. A universal split spinach aptamer (USSA) for nucleic acid analysis and DNA computation. Chem. Commun. 2017, 53, 4977–4980. [Google Scholar] [CrossRef]
- Gerasimova, Y.V.; Kolpashchikov, D.M. Detection of bacterial 16S rRNA using a molecular beacon-based X sensor. Biosens. Bioelectron. 2013, 41, 386–390. [Google Scholar] [CrossRef]
- Kikuchi, N.; Reed, A.; Gerasimova, Y.V.; Kolpashchikov, D.M. Split dapoxyl aptamer for sequence-selective analysis of nucleic acid sequence based amplification amplicons. Anal. Chem. 2019, 91, 2667–2671. [Google Scholar] [CrossRef]
- Pothoulakis, G.; Ceroni, F.; Reeve, B.; Ellis, T. The spinach RNA aptamer as a characterization tool for synthetic biology. ACS Synth. Biol. 2014, 3, 182–187. [Google Scholar] [CrossRef]
- Feoktistova, M.; Geserick, P.; Leverkus, M. Crystal violet assay for determining viability of cultured cells. Cold Spring Harb. Protoc. 2016, 2016, pdb.prot087379. [Google Scholar] [CrossRef]
- Borowicz, M.; Krzyżanowska, D.M.; Jafra, S. Crystal violet-based assay for the assessment of bacterial biofilm formation in medical tubing. J. Microbiol. Methods 2023, 204, 106656. [Google Scholar] [CrossRef]
- Lei, A.; Li, M.; Zhuo, S.; Li, Q.; Li, J. A label-free fluorescence and naked-eye detection for ochratoxin A based on aptamer antiparallel G-quadruplex/crystal violet. Food Biosci. 2024, 57, 103603. [Google Scholar] [CrossRef]
- Lu, P.; Aodeng, G.; Ai, J. Progress in ion detection and clinical diagnosis based on G-quadruplex combined with fluorescence properties. Adv. Sens. Energy Mat. 2024, 3, 100112. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, J.; Zhang, Y.; Xu, L.; Gao, T.; Wang, B.; Pei, R. Selection and characterization of a DNA aptamer to crystal violet. Photochem. Photobiol. Sci. 2018, 17, 800–806. [Google Scholar] [CrossRef]
- Zheng, B.; Cheng, S.; Dong, H.; Liang, H.; Liu, J.; Lam, M.H.W. Label free determination of potassium ions using crystal violet and thrombin-binding aptamer. Anal. Lett. 2014, 47, 1726–1736. [Google Scholar] [CrossRef]
- Di Leva, F.S.; Novellino, E.; Cavalli, A.; Parrinello, M.; Limongelli, V. Mechanistic insight into ligand binding to G-quadruplex DNA. Nucleic Acids Res. 2014, 42, 5447–5455. [Google Scholar] [CrossRef]
- Santos, T.; Salgado, G.F.; Cabrita, E.J.; Cruz, C. G-quadruplexes and their ligands: Biophysical methods to unravel G-quadruplex/ligand interactions. Pharmaceuticals 2021, 14, 769. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, Y.; Liu, Z.; Lin, B.; Yi, H.; Xu, F.; Yao, S. Insight into G-quadruplex-hemin DNAzyme/RNAzyme: Adjacent adenine as the intramolecular species for remarkable enhancement of enzymatic activity. Nucleic Acids Res. 2016, 44, 7373–7384. [Google Scholar] [CrossRef]
- Rubel, M.S.; Shkodenko, L.A.; Gorbenko, D.A.; Solyanikova, V.V.; Maltzeva, Y.I.; Rubel, A.A.; Kolpashchikov, D.M. Detection of Multiplex NASBA RNA Products Using Colorimetric Split G Quadruplex Probes. In RNA Nanostructures: Design, Characterization, and Applications; Springer: New York, NY, USA, 2023; pp. 287–298. [Google Scholar]
- Gorbenko, D.A.; Shkodenko, L.A.; Rubel, M.S.; Slita, A.V.; Nikitina, E.V.; Martens, E.A.; Kolpashchikov, D.M. DNA nanomachine for visual detection of structured RNA and double stranded DNA. Chem. Commun. 2022, 58, 5395–5398. [Google Scholar] [CrossRef]
- Maltzeva, Y.I.; Gorbenko, D.A.; Nikitina, E.V.; Rubel, M.S.; Kolpashchikov, D.M. Visual Detection of Stem-Loop Primer Amplification (SPA) Products without Denaturation Using Peroxidase-like DNA Machines (PxDM). Int. J. Mol. Sci. 2023, 24, 7812. [Google Scholar] [CrossRef]
- Tsukakoshi, K.; Yamagishi, Y.; Kanazashi, M.; Nakama, K.; Oshikawa, D.; Savory, N.; Ikebukuro, K. G-quadruplex-forming aptamer enhances the peroxidase activity of myoglobin against luminol. Nucleic Acids Res. 2021, 49, 6069–6081. [Google Scholar] [PubMed]
- Chiorcea-Paquim, A.M.; Oliveira-Brett, A.M. Guanine quadruplex electrochemical aptasensors. Chemosensors 2016, 4, 13. [Google Scholar] [CrossRef]
- Cao, Y.; Li, W.; Pei, R. Engineering smart thioflavin T binders with switchable DNA topologies for constructing multiple label-free biosensors. Chem. Eng. J. 2024, 493, 152655. [Google Scholar] [CrossRef]
- Yang, H.; Zhou, Y.; Liu, J. G-quadruplex DNA for construction of biosensors. TrAC Trends Anal. Chem. 2020, 132, 116060. [Google Scholar] [CrossRef]
- Kong, D.M.; Guo, J.H.; Yang, W.; Ma, Y.E.; Shen, H.X. Crystal violet–G-quadruplex complexes as fluorescent sensors for homogeneous detection of potassium ion. Biosens. Bioelectron. 2009, 25, 88–93. [Google Scholar] [CrossRef]
- Le, H.T.; Miller, M.C.; Buscaglia, R.; Dean, W.L.; Holt, P.A.; Chaires, J.B.; Trent, J.O. Not all G-quadruplexes are created equally: An investigation of the structural polymorphism of the c-Myc G-quadruplex-forming sequence and its interaction with the porphyrin TMPyP4. Org. Biomol. Chem. 2012, 10, 9393–9404. [Google Scholar] [CrossRef]
- Beaudoin, J.D.; Perreault, J.P. Potassium ions modulate a G-quadruplex-ribozyme’s activity. RNA 2008, 14, 1018–1025. [Google Scholar] [CrossRef]
- Benabou, S.; Mazzini, S.; Avino, A.; Eritja, R.; Gargallo, R. A pH-dependent bolt involving cytosine bases located in the lateral loops of antiparallel G-quadruplex structures within the SMARCA4 gene promotor. Sci. Rep. 2019, 9, 15807. [Google Scholar] [CrossRef]
- Vummidi, B.R.; Alzeer, J.; Luedtke, N.W. Fluorescent probes for G-quadruplex structures. ChemBioChem 2013, 14, 540–558. [Google Scholar] [CrossRef]
- Bhasikuttan, A.C.; Mohanty, J. Targeting G-quadruplex structures with extrinsic fluorogenic dyes: Promising fluorescence sensors. Chem. Commun. 2015, 51, 7581–7597. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, L.; Dong, S.; Wang, E. How to split a G-quadruplex for DNA detection: New insight into the formation of DNA split G-quadruplex. Chem. Sci. 2015, 6, 4822–4827. [Google Scholar] [CrossRef]
- MacDougall, D.; Crummett, W.B. Guidelines for data acquisition and data quality evaluation in environmental chemistry. Anal. Chem. 1980, 52, 2242–2249. [Google Scholar] [CrossRef]
- Kikuchi, N.; Kolpashchikov, D.M. Split spinach aptamer for highly selective recognition of DNA and RNA at ambient temperatures. ChemBioChem 2016, 17, 1589–1592. [Google Scholar] [CrossRef]
- Markham, N.R.; Zuker, M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005, 33, W577–W581. [Google Scholar] [CrossRef] [PubMed]
- Filatov, P.V.; Ateiah, M.; Berezovskaya, M.Y.; Rubel, M.S.; Kolpashchikov, D.M. DNAzyme 10-23-Based Nanomachines for Nucleic Acid Recognition. J. Vis. Exp. 2024, 204, e66461. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bobkov, G.A.; Yushkov, G.S.; Kuzmin, A.D.; Popysheva, T.D.; Stepchenkova, E.I.; Rubel, M.S. Comparative Analysis of Crystal Violet-Binding Aptamers as Potential Cores for Binary Sensors. Int. J. Mol. Sci. 2025, 26, 9833. https://doi.org/10.3390/ijms26199833
Bobkov GA, Yushkov GS, Kuzmin AD, Popysheva TD, Stepchenkova EI, Rubel MS. Comparative Analysis of Crystal Violet-Binding Aptamers as Potential Cores for Binary Sensors. International Journal of Molecular Sciences. 2025; 26(19):9833. https://doi.org/10.3390/ijms26199833
Chicago/Turabian StyleBobkov, Gleb A., Gleb S. Yushkov, Andrei D. Kuzmin, Tatiana D. Popysheva, Elena I. Stepchenkova, and Maria S. Rubel. 2025. "Comparative Analysis of Crystal Violet-Binding Aptamers as Potential Cores for Binary Sensors" International Journal of Molecular Sciences 26, no. 19: 9833. https://doi.org/10.3390/ijms26199833
APA StyleBobkov, G. A., Yushkov, G. S., Kuzmin, A. D., Popysheva, T. D., Stepchenkova, E. I., & Rubel, M. S. (2025). Comparative Analysis of Crystal Violet-Binding Aptamers as Potential Cores for Binary Sensors. International Journal of Molecular Sciences, 26(19), 9833. https://doi.org/10.3390/ijms26199833








