Viral Metagenomic Next-Generation Sequencing for One Health Discovery and Surveillance of (Re)Emerging Viruses: A Deep Review
Abstract
1. Introduction
1.1. Metagenomic Next-Generation Sequencing: Transforming Virus Discovery and Surveillance
1.2. Pathogen Discovery and Surveillance: One Health Imperative
2. Viral Metagenomic Next-Generation Sequencing Workflows
2.1. Strategic Sample Selection and Quality Control in vmNGS
2.1.1. Environmental Settings
2.1.2. Clinical and Postmortem Context
2.2. Increasing Sensitivity with Host Depletion and Virus Enrichment
2.2.1. Depletion of Non-Viral Material
2.2.2. Virus Enrichment
2.3. Next-Generation Sequencing Technologies
2.4. Bioinformatic Analysis for Virus Discovery, Characterisation, and Molecular Epidemiology
2.5. Maintaining Quality Control and Standardisation of vmNGS
3. Emerging Virus Discovery with vmNGS
3.1. The Central Role of vmNGS in COVID-19 Discovery and Response
3.2. A Henipavirus Emerging in the Shadow of COVID-19
4. Passive Surveillance of Reemerging Viruses Using vmNGS
4.1. vmNGS to Monitor Mpox Epidemics and Global Spread
4.2. vmNGS in the Response and Understanding of Zika Virus Reemergence
4.3. vmNGS to Monitor the Spread and Reemergence of Arboviruses
5. Enhancing (Re)Emerging Virus Surveillance with vmNGS
5.1. Active and Passive (Re)Emerging Virus Surveillance in Animals
5.1.1. Farm Animals: The Need for vmNGS Surveillance
| Livestock | Zoonotic Virus | Location | Year | References |
|---|---|---|---|---|
| Cattle | Bovine coronavirus | Russia | Pre-1889 | [199,200,209] |
| Swine | Influenza virus H1N1 | Mexico | 2009 | [210] |
| Hepatitis E Virus | Multiple (e.g., USA) | Multiple (e.g., 1998) | [211] | |
| Nipah virus | Malaysia and Singapore | 1998 | [212] | |
| Japanese encephalitis virus 1 | Unknown | Unknown | [213] | |
| Poultry | Avian influenza A virus | USA or France | Pre-1918 | [210,214] |
| Newcastle Disease virus | USA | 1965 | [215] | |
| West Nile virus | Israel | 1998 | [216] | |
| Horses | Hendra virus | Australia | 1994 | [217] |
5.1.2. Wildlife Reservoirs and Metagenomic Surveillance: Preventing Zoonotic Spillover
5.1.3. Companion Animals: A Critical Interface for One Health Surveillance
5.1.4. Expanding the Scope of Arbovirus Surveillance
5.2. Influenza: A Recurring Issue
5.3. Rooting Through the Rubbish: Wastewater Surveillance
6. Discussion
7. Conclusions
8. Future Directions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACE2 | Angiotensin-converting enzyme 2 |
| APOBEC3 | Apolipoprotein B mRNA editing enzyme, catalytic subunit 3 |
| DRC | Democratic Republic of Congo |
| ds-cDNA | double stranded complementary dideoxynucleic acid |
| HC | Hybrid Capture |
| HIV | Human Immunodeficiency Virus |
| HPAIV | Highly Pathogenic Influenza A virus |
| IAV | Influenza A virus |
| LMIC | Low- and Middle-Income Countries |
| LOD | Limit of Detection |
| MERS-CoV | Middle East Respiratory Syndrome Coronavirus |
| ONT | Oxford Nanopore Technologies |
| OROV | Oropouche Virus |
| PCR | Polymerase Chain Reaction |
| PEG | Polyethylene Glycol |
| QC | Quality Control |
| rRNA | Ribosomal Ribonucleic acid |
| SARS-CoV-2 | Severe Acute Respiratory Syndrome Coronavirus 2 |
| SBV | Schmallenberg Virus |
| SISPA | Sequence-independent single primer amplification |
| SNP | Single nucleotide polymorphism |
| TMPRSS2 | Transmembrane Protease, Serine 2 |
| TNA | Total nucleic acid |
| vmNGS | Viral Metagenomic Next-Generation Sequencing |
| WGS | Whole genome sequencing |
| WHO | World Health Organisation |
References
- Shanmugaraj, B.; Kothalam, R.; Tharik, M.S.; Azeeze, A. A Brief Overview on the Threat of Zoonotic Viruses. Microbes Infect. Dis. 2024, 6, 2034–2041. [Google Scholar] [CrossRef]
- Burrell, C.J.; Mackay, P.; Greenaway, P.J.; Hofschneider, P.H.; Murray, K. Expression in Escherichia Coli of Hepatitis B Virus DNA Sequences Cloned in Plasmid PBR322. Nature 1979, 279, 43–47. [Google Scholar] [CrossRef]
- Wang, K.S.; Choo, Q.L.; Weiner, A.J.; Ou, J.H.; Najarian, R.C.; Thayer, R.M.; Mullenbach, G.T.; Denniston, K.J.; Gerin, J.L.; Houghton, M. Structure, Sequence and Expression of the Hepatitis Delta (δ) Viral Genome. Nature 1986, 323, 508–514. [Google Scholar] [CrossRef]
- Desai, S.M.; Kalyanaraman, V.S.; Casey, J.M.; Srinivasan, A.; Andersen, P.R.; Devare, S.G. Molecular Cloning and Primary Nucleotide Sequence Analysis of a Distinct Human Immunodeficiency Virus Isolate Reveal Significant Divergence in Its Genomic Sequences. Proc. Natl. Acad. Sci. USA 1986, 83, 8380–8384. [Google Scholar] [CrossRef] [PubMed]
- Choo, Q.L.; Richman, K.H.; Han, J.H.; Berger, K.; Lee, C.; Dong, C.; Gallegos, C.; Coit, D.; Medina-Selby, A.; Barr, P.J.; et al. Genetic Organization and Diversity of the Hepatitis C Virus. Proc. Natl. Acad. Sci. USA 1991, 88, 2451. [Google Scholar] [CrossRef] [PubMed]
- Kato, N.; Hijikata, M.; Ootsuyama, Y.; Nakagawa, M.; Ohkoshi, S.; Sugimura, T.; Shimotohno, K. Molecular Cloning of the Human Hepatitis C Virus Genome from Japanese Patients with Non-A, Non-B Hepatitis. Proc. Natl. Acad. Sci. USA 1990, 87, 9524. [Google Scholar] [CrossRef] [PubMed]
- Henderson, W.W.; Monroe, M.C.; St. Jeor, S.C.; Thayer, W.P.; Rowe, J.E.; Peters, C.J.; Nichol, S.T. Naturally Occurring Sin Nombre Virus Genetic Reassortants. Virology 1995, 214, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Chizhikov, V.E.; Spiropoulou, C.F.; Morzunov, S.P.; Monroe, M.C.; Peters, C.J.; Nichol, S.T. Complete Genetic Characterization and Analysis of Isolation of Sin Nombre Virus. J. Virol. 1995, 69, 8132–8136. [Google Scholar] [CrossRef]
- Chew, M.H.L.; Arguin, P.M.; Shay, D.K.; Goh, K.T.; Rollin, P.E.; Shieh, W.J.; Zaki, S.R.; Rota, P.A.; Ling, A.E.; Ksiazek, T.G.; et al. Risk Factors for Nipah Virus Infection among Abattoir Workers in Singapore. J. Infect. Dis. 2000, 181, 1760–1763. [Google Scholar] [CrossRef]
- Drosten, C.; Günther, S.; Preiser, W.; van der Werf, S.; Brodt, H.-R.; Becker, S.; Rabenau, H.; Panning, M.; Kolesnikova, L.; Fouchier, R.A.M.; et al. Identification of a Novel Coronavirus in Patients with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003, 348, 1967–1976. [Google Scholar] [CrossRef]
- Peiris, J.S.M.; Lai, S.T.; Poon, L.L.M.; Guan, Y.; Yam, L.Y.C.; Lim, W.; Nicholls, J.; Yee, W.K.S.; Yan, W.W.; Cheung, M.T.; et al. Coronavirus as a Possible Cause of Severe Acute Respiratory Syndrome. Lancet 2003, 361, 1319–1325. [Google Scholar] [CrossRef] [PubMed]
- Van Der Hoek, L.; Pyrc, K.; Jebbink, M.F.; Vermeulen-Oost, W.; Berkhout, R.J.M.; Wolthers, K.C.; Wertheim-Van Dillen, P.M.E.; Kaandorp, J.; Spaargaren, J.; Berkhout, B. Identification of a New Human Coronavirus. Nat. Med. 2004, 10, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Shinde, V.; Bridges, C.B.; Uyeki, T.M.; Shu, B.; Balish, A.; Xu, X.; Lindstrom, S.; Gubareva, L.V.; Deyde, V.; Garten, R.J.; et al. Triple-Reassortant Swine Influenza A (H1) in Humans in the United States, 2005–2009. N. Engl. J. Med. 2009, 360, 2616–2625. [Google Scholar] [CrossRef] [PubMed]
- Greninger, A.L.; Chen, E.C.; Sittler, T.; Scheinerman, A.; Roubinian, N.; Yu, G.; Kim, E.; Pillai, D.R.; Guyard, C.; Mazzulli, T.; et al. A Metagenomic Analysis of Pandemic Influenza A (2009 H1N1) Infection in Patients from North America. PLoS ONE 2010, 5, e13381. [Google Scholar] [CrossRef]
- Bhatt, D.L.; Va Boston, M.P.H.; Harrington, R.A. Brief Report: Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2013, 369, 394. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579, 270. [Google Scholar] [CrossRef]
- Makino, S.; Chang, M.F.; Shieh, C.K.; Kamahora, T.; Vannier, D.M.; Govindarajan, S.; Lai, M.M.C. Molecular Cloning and Sequencing of a Human Hepatitis Delta (δ) Virus RNA. Nature 1987, 329, 343–346. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA Sequencing with Chain-Terminating Inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of Age: Ten Years of next-Generation Sequencing Technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef]
- Thermes, C. Ten Years of Next-Generation Sequencing Technology. Trends Genet. 2014, 30, 418–426. [Google Scholar] [CrossRef]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G.; et al. Correction: Satam et al. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology 2024, 13, 286. [Google Scholar] [CrossRef] [PubMed]
- Ksiazek, T.G.; Erdman, D.; Goldsmith, C.S.; Zaki, S.R.; Peret, T.; Emery, S.; Tong, S.; Urbani, C.; Comer, J.A.; Lim, W.; et al. A Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003, 348, 1953–1966. [Google Scholar] [CrossRef] [PubMed]
- Morse, S.S.; Mazet, J.A.K.; Woolhouse, M.; Parrish, C.R.; Carroll, D.; Karesh, W.B.; Zambrana-Torrelio, C.; Lipkin, W.I.; Daszak, P. Prediction and Prevention of the next Pandemic Zoonosis. Lancet 2012, 380, 1956. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global Trends in Emerging Infectious Diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef]
- Wolfe, N.D.; Dunavan, C.P.; Diamond, J. Origins of Major Human Infectious Diseases. Nature 2007, 447, 279–283. [Google Scholar] [CrossRef]
- Jones, B.A.; Grace, D.; Kock, R.; Alonso, S.; Rushton, J.; Said, M.Y.; McKeever, D.; Mutua, F.; Young, J.; McDermott, J.; et al. Zoonosis Emergence Linked to Agricultural Intensification and Environmental Change. Proc. Natl. Acad. Sci. USA 2013, 110, 8399–8404. [Google Scholar] [CrossRef]
- Singh, B.B.; Ward, M.P.; Kostoulas, P.; Dhand, N.K. Zoonosis–Why We Should Reconsider “What’s in a Name?”. Front. Public Health 2023, 11, 1133330. [Google Scholar] [CrossRef]
- Figueiredo, L.T.M. Human Urban Arboviruses Can Infect Wild Animals and Jump to Sylvatic Maintenance Cycles in South America. Front. Cell. Infect. Microbiol. 2019, 9, 453237. [Google Scholar] [CrossRef]
- Guan, Y.; Zheng, B.J.; He, Y.Q.; Liu, X.L.; Zhuang, Z.X.; Cheung, C.L.; Luo, S.W.; Li, P.H.; Zhang, L.J.; Guan, Y.J.; et al. Isolation and Characterization of Viruses Related to the SARS Coronavirus from Animals in Southern China. Science 2003, 302, 276–278. [Google Scholar] [CrossRef]
- Neumann, G.; Kawaoka, Y. Highly Pathogenic H5N1 Avian Influenza Virus Outbreak in Cattle: The Knowns and Unknowns. Nat. Rev. Microbiol. 2024, 22, 525–526. [Google Scholar] [CrossRef]
- Caserta, L.C.; Frye, E.A.; Butt, S.L.; Laverack, M.; Nooruzzaman, M.; Covaleda, L.M.; Thompson, A.C.; Koscielny, M.P.; Cronk, B.; Johnson, A.; et al. Spillover of Highly Pathogenic Avian Influenza H5N1 Virus to Dairy Cattle. Nature 2024, 634, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Thomson, T.N.; Marsland, M.J.; Minko, C.; Snow, K.J.; Friedman, N.D. Japanese Encephalitis: A Rapid Review of Reported Prevalence of Infection, Clinical Disease and Sequelae in Immunologically Naive Populations to Inform Australia’s Response. Aust. N. Z. J. Public Health 2023, 47, 100041. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Smith, K.; Sarker, S.; Peters, A.; Adriaanse, K.; Eden, P.; Ghorashi, S.A.; Forwood, J.K.; Raidal, S.R. Repeat Spillover of beak and feather disease virus into an endangered parrot highlights the risk associated with endemic pathogen loss in endangered species. J. Wildl. Dis. 2020, 56, 896–906. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, J.H.O.; Eldholm, V.; Seligman, S.J.; Lundkvist, Å.; Falconar, A.K.; Gaunt, M.W.; Musso, D.; Nougairède, A.; Charrel, R.; Gould, E.A.; et al. Erratum for Pettersson et al., “How Did Zika Virus Emerge in the Pacific Islands and Latin America?”. mBio 2018, 9, e00386-18, Erratum in mBio 2016, 7, e01239-16. https://doi.org/10.1128/mBio.01239-16.. [Google Scholar] [CrossRef]
- Gibbs, E.P.J. The Evolution of One Health: A Decade of Progress and Challenges for the Future. Vet. Rec. 2014, 174, 85–91. [Google Scholar] [CrossRef]
- Finch, A.; Vora, N.M.; Hassan, L.; Walzer, C.; Plowright, R.K.; Alders, R.; Suit-B, C.Y.; Amuasi, J.H.; Mulumba, M.; Loch-Temzelides, T.; et al. The Promise and Compromise of the WHO Pandemic Agreement for Spillover Prevention and One Health. Lancet 2025, 405, 1800–1802. [Google Scholar] [CrossRef]
- Samarasekera, U. New EU Health Programme Comes into Force. Lancet 2021, 397, 1252–1253. [Google Scholar] [CrossRef]
- Hamisu, A.W.; Blake, I.M.; Sume, G.; Braka, F.; Jimoh, A.; Dahiru, H.; Bonos, M.; Dankoli, R.; Bello, A.M.; Yusuf, K.M.; et al. Characterizing Environmental Surveillance Sites in Nigeria and Their Sensitivity to Detect Poliovirus and Other Enteroviruses. J. Infect. Dis. 2020, 225, 1377. [Google Scholar] [CrossRef]
- Minor, N.R.; Ramuta, M.D.; Stauss, M.R.; Harwood, O.E.; Brakefield, S.F.; Alberts, A.; Vuyk, W.C.; Bobholz, M.J.; Rosinski, J.R.; Wolf, S.; et al. Author Correction: Metagenomic Sequencing Detects Human Respiratory and Enteric Viruses in Air Samples Collected from Congregate Settings. Sci. Rep. 2024, 14, 1316, Erratum in Sci. Rep. 2023, 1. https://doi.org/10.1038/s41598-023-48352-6.. [Google Scholar] [CrossRef]
- Fourgeaud, J.; Regnault, B.; Ok, V.; Da Rocha, N.; Sitterlé, É.; Mekouar, M.; Faury, H.; Milliancourt-Seels, C.; Jagorel, F.; Chrétien, D.; et al. Performance of Clinical Metagenomics in France: A Prospective Observational Study. Lancet Microbe 2024, 5, e52–e61. [Google Scholar] [CrossRef]
- Yandle, Z.; Gonzalez, G.; Carr, M.; Matthijnssens, J.; De Gascun, C. A Viral Metagenomic Protocol for Nanopore Sequencing of Group A Rotavirus. J. Virol. Methods 2023, 312, 114664. [Google Scholar] [CrossRef]
- Cebriá-Mendoza, M.; Arbona, C.; Larrea, L.; Díaz, W.; Arnau, V.; Peña, C.; Bou, J.V.; Sanjuán, R.; Cuevas, J.M. Deep Viral Blood Metagenomics Reveals Extensive Anellovirus Diversity in Healthy Humans. Sci. Rep. 2021, 11, 6921. [Google Scholar] [CrossRef]
- Ogunbayo, A.E.; Sabiu, S.; Nyaga, M.M. Evaluation of Extraction and Enrichment Methods for Recovery of Respiratory RNA Viruses in a Metagenomics Approach. J. Virol. Methods 2023, 314, 114677. [Google Scholar] [CrossRef]
- Edridge, A.W.D.; Deijs, M.; Van Zeggeren, I.E.; Kinsella, C.M.; Jebbink, M.F.; Bakker, M.; Van de Beek, D.; Brouwer, M.C.; Van der Hoek, L. Viral Metagenomics on Cerebrospinal Fluid. Genes 2019, 10, 332. [Google Scholar] [CrossRef]
- Benoit, P.; Brazer, N.; de Lorenzi-Tognon, M.; Kelly, E.; Servellita, V.; Oseguera, M.; Nguyen, J.; Tang, J.; Omura, C.; Streithorst, J.; et al. Seven-Year Performance of a Clinical Metagenomic next-Generation Sequencing Test for Diagnosis of Central Nervous System Infections. Nat. Med. 2024, 30, 3522–3533. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.; Naccache, S.N.; Samayoa, E.; Messacar, K.; Arevalo, S.; Federman, S.; Stryke, D.; Pham, E.; Fung, B.; Bolosky, W.J.; et al. Laboratory Validation of a Clinical Metagenomic Sequencing Assay for Pathogen Detection in Cerebrospinal Fluid. Genome Res. 2019, 29, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.R.; Sample, H.A.; Zorn, K.C.; Arevalo, S.; Yu, G.; Neuhaus, J.; Federman, S.; Stryke, D.; Briggs, B.; Langelier, C.; et al. Clinical Metagenomic Sequencing for Diagnosis of Meningitis and Encephalitis. N. Engl. J. Med. 2019, 380, 2327–2340. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wan, W.; Yu, K.; Lemey, P.; Pettersson, J.H.O.; Bi, Y.; Lu, M.; Li, X.; Chen, Z.; Zheng, M.; et al. Farmed Fur Animals Harbour Viruses with Zoonotic Spillover Potential. Nature 2024, 634, 228. [Google Scholar] [CrossRef]
- He, X.; Wang, X.; Fan, G.; Li, F.; Wu, W.; Wang, Z.; Fu, M.; Wei, X.; Ma, S.; Ma, X. Metagenomic Analysis of Viromes in Tissues of Wild Qinghai Vole from the Eastern Tibetan Plateau. Sci. Rep. 2022, 12, 17239. [Google Scholar] [CrossRef]
- Tan, J.K.; Servellita, V.; Stryke, D.; Kelly, E.; Streithorst, J.; Sumimoto, N.; Foresythe, A.; Huh, H.J.; Nguyen, J.; Oseguera, M.; et al. Laboratory Validation of a Clinical Metagenomic Next-Generation Sequencing Assay for Respiratory Virus Detection and Discovery. Nat. Commun. 2024, 15, 9016. [Google Scholar] [CrossRef]
- Parris, D.J.; Kariithi, H.; Suarez, D.L. Non-Target RNA Depletion Strategy to Improve Sensitivity of next-Generation Sequencing for the Detection of RNA Viruses in Poultry. J. Vet. Diagn. Investig. 2022, 34, 638–645. [Google Scholar] [CrossRef]
- Zhang, X.L.; Li, W.F.; Yuan, S.; Guo, J.Y.; Li, Z.L.; Chi, S.H.; Huang, W.J.; Li, X.W.; Huang, S.J.; Shao, J.W. Meta-Transcriptomic Analysis Reveals a New Subtype of Genotype 3 Avian Hepatitis e Virus in Chicken Flocks with High Mortality in Guangdong, China. BMC Vet. Res. 2019, 15, 131. [Google Scholar] [CrossRef] [PubMed]
- Prakoso, D.; Dark, M.J.; Barbet, A.F.; Salemi, M.; Barr, K.L.; Liu, J.J.; Wenzlow, N.; Waltzek, T.B.; Long, M.T. Viral Enrichment Methods Affect the Detection but Not Sequence Variation of West Nile Virus in Equine Brain Tissue. Front. Vet. Sci. 2018, 5, 418004. [Google Scholar] [CrossRef] [PubMed]
- Reyes, G.R.; Kim, J.P. Sequence-Independent, Single-Primer Amplification (SISPA) of Complex DNA Populations. Mol. Cell Probes 1991, 5, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Chrzastek, K.; Lee, D.H.; Smith, D.; Sharma, P.; Suarez, D.L.; Pantin-Jackwood, M.; Kapczynski, D.R. Use of Sequence-Independent, Single-Primer-Amplification (SISPA) for Rapid Detection, Identification, and Characterization of Avian RNA Viruses. Virology 2017, 509, 159. [Google Scholar] [CrossRef]
- Electron Microscopy and a Sequence-Independent, Single-Primer Amplification (SISPA) Viromics Approach for Monkeypox Virus Genome Determination—MPXV/Genome Reports—Virological. Available online: https://virological.org/t/electron-microscopy-and-a-sequence-independent-single-primer-amplification-sispa-viromics-approach-for-monkeypox-virus-genome-determination/899 (accessed on 3 April 2025).
- Peserico, A.; Marcacci, M.; Malatesta, D.; Di Domenico, M.; Pratelli, A.; Mangone, I.; D’Alterio, N.; Pizzurro, F.; Cirone, F.; Zaccaria, G.; et al. Diagnosis and Characterization of Canine Distemper Virus through Sequencing by MinION Nanopore Technology. Sci. Rep. 2019, 9, 1714. [Google Scholar] [CrossRef]
- Jothikumar, N.; Cromeans, T.; Shivajothi, J.; Vinjé, J.; Murphy, J. Development and Evaluation of a Ligation-Free Sequence-Independent, Single-Primer Amplification (LF-SISPA) Assay for Whole Genome Characterization of Viruses. J. Virol. Methods 2021, 299, 114346. [Google Scholar] [CrossRef]
- Piantadosi, A.; Mukerji, S.S.; Ye, S.; Leone, M.J.; Freimark, L.M.; Park, D.; Adams, G.; Lemieux, J.; Kanjilal, S.; Solomon, I.H.; et al. Enhanced Virus Detection and Metagenomic Sequencing in Patients with Meningitis and Encephalitis. mBio 2021, 12, e01143-21. [Google Scholar] [CrossRef]
- Mourik, K.; Sidorov, I.; Carbo, E.C.; van der Meer, D.; Boot, A.; Kroes, A.C.M.; Claas, E.C.J.; Boers, S.A.; de Vries, J.J.C. Comparison of the Performance of Two Targeted Metagenomic Virus Capture Probe-Based Methods Using Reference Control Materials and Clinical Samples. J. Clin. Microbiol. 2024, 62, e00345-24. [Google Scholar] [CrossRef]
- Mao, W.; Wang, J.; Li, T.; Wu, J.; Wang, J.; Wen, S.; Huang, J.; Shi, Y.; Zheng, K.; Zhai, Y.; et al. Hybrid Capture-Based Sequencing Enables Highly Sensitive Zoonotic Virus Detection Within the One Health Framework. Pathogens 2025, 14, 264. [Google Scholar] [CrossRef]
- Carbo, E.C.; Buddingh, E.P.; Karelioti, E.; Sidorov, I.A.; Feltkamp, M.C.W.; von dem Borne, P.A.; Verschuuren, J.J.G.M.; Kroes, A.C.M.; Claas, E.C.J.; de Vries, J.J.C. Improved Diagnosis of Viral Encephalitis in Adult and Pediatric Hematological Patients Using Viral Metagenomics. J. Clin. Virol. 2020, 130, 104566. [Google Scholar] [CrossRef]
- Castellot, A.; Camacho, J.; Fernández-García, M.D.; Tarragó, D. Shotgun Metagenomics to Investigate Unknown Viral Etiologies of Pediatric Meningoencephalitis. PLoS ONE 2023, 18, e0296036. [Google Scholar] [CrossRef] [PubMed]
- Child, H.T.; Airey, G.; Maloney, D.M.; Parker, A.; Wild, J.; McGinley, S.; Evens, N.; Porter, J.; Templeton, K.; Paterson, S.; et al. Erratum for Child et al., “Comparison of Metagenomic and Targeted Methods for Sequencing Human Pathogenic Viruses from Wastewater”. mBio 2024, 15, e0255824, Erratum in mBio 2023, 6, e0146823. https://doi.org/10.1128/mbio.02558-24.. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, W.; Bovinder Ylitalo, E.; Castel, G.; Sjödin, A.; Larsson, P.; Wigren Byström, J.; Forsell, M.N.E.; Ahlm, C.; Pettersson, L.; Tuiskunen Bäck, A. Hybrid Capture-Based next-Generation Sequencing of New and Old World Orthohantavirus Strains and Wild-Type Puumala Isolates from Humans and Bank Voles. J. Clin. Virol. 2024, 172, 105672. [Google Scholar] [CrossRef] [PubMed]
- Kuchinski, K.S.; Loos, K.D.; Suchan, D.M.; Russell, J.N.; Sies, A.N.; Kumakamba, C.; Muyembe, F.; Kingebeni, P.M.; Lukusa, I.N.; N’Kawa, F.; et al. Targeted Genomic Sequencing with Probe Capture for Discovery and Surveillance of Coronaviruses in Bats. Elife 2022, 11, e79777. [Google Scholar] [CrossRef]
- Nabel, C.S.; Sameroff, S.; Shilling, D.; Alapat, D.; Ruth, J.R.; Kawano, M.; Sato, Y.; Stone, K.; Spetalen, S.; Valdivieso, F.; et al. Virome Capture Sequencing Does Not Identify Active Viral Infection in Unicentric and Idiopathic Multicentric Castleman Disease. PLoS ONE 2019, 14, e0218660. [Google Scholar] [CrossRef]
- Shafer, M.M.; Bobholz, M.J.; Vuyk, W.C.; Gregory, D.A.; Roguet, A.; Haddock Soto, L.A.; Rushford, C.; Janssen, K.H.; Emmen, I.E.; Ries, H.J.; et al. Tracing the Origin of SARS-CoV-2 Omicron-like Spike Sequences Detected in an Urban Sewershed: A Targeted, Longitudinal Surveillance Study of a Cryptic Wastewater Lineage. Lancet Microbe 2024, 5, e335. [Google Scholar] [CrossRef]
- Hassard, F.; Vu, M.; Rahimzadeh, S.; Castro-Gutierrez, V.; Stanton, I.; Burczynska, B.; Wildeboer, D.; Baio, G.; Brown, M.R.; Garelick, H.; et al. Wastewater Monitoring for Detection of Public Health Markers during the COVID-19 Pandemic: Near-Source Monitoring of Schools in England over an Academic Year. PLoS ONE 2023, 18, e0286259. [Google Scholar] [CrossRef]
- Stockdale, S.R.; Blanchard, A.A.; Nayak, A.; Husain, A.; Nashine, R.; Dudani, H.; McClure, C.P.; Tarr, A.W.; Nag, A.; Meena, E.; et al. RNA-Seq of Untreated Wastewater to Assess COVID-19 and Emerging and Endemic Viruses for Public Health Surveillance. Lancet Reg. Health Southeast Asia 2023, 14, 100205. [Google Scholar] [CrossRef]
- Miyani, B.; Li, Y.; Guzman, H.P.; Briceno, R.K.; Vieyra, S.; Hinojosa, R.; Xagoraraki, I. Bioinformatics-Based Screening Tool Identifies a Wide Variety of Human and Zoonotic Viruses in Trujillo-Peru Wastewater. One Health 2024, 18, 100756. [Google Scholar] [CrossRef]
- Whitehouse, E.R.; Gerloff, N.; English, R.; Reckling, S.K.; Alazawi, M.A.; Fuschino, M.; St George, K.; Lang, D.; Rosenberg, E.S.; Omoregie, E.; et al. Wastewater Surveillance for Poliovirus in Selected Jurisdictions, United States, 2022–2023. Emerg. Infect. Dis. 2024, 30, 2279. [Google Scholar] [CrossRef]
- Zhang, T.; Li, H.; Jiang, M.; Hou, H.; Gao, Y.; Li, Y.; Wang, F.; Wang, J.; Peng, K.; Liu, Y.X. Nanopore Sequencing: Flourishing in Its Teenage Years. J. Genet. Genom. 2024, 51, 1361–1374. [Google Scholar] [CrossRef]
- Vandenbogaert, M.; Kwasiborski, A.; Gonofio, E.; Descorps-Declère, S.; Selekon, B.; Nkili Meyong, A.A.; Ouilibona, R.S.; Gessain, A.; Manuguerra, J.C.; Caro, V.; et al. Nanopore Sequencing of a Monkeypox Virus Strain Isolated from a Pustular Lesion in the Central African Republic. Sci. Rep. 2022, 12, 10768. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gao, L.; Zhu, C.; Jin, J.; Song, C.; Dong, H.; Li, Z.; Wang, Z.; Chen, Y.; Yang, Z.; et al. Clinical Value of Metagenomic Next-Generation Sequencing by Illumina and Nanopore for the Detection of Pathogens in Bronchoalveolar Lavage Fluid in Suspected Community-Acquired Pneumonia Patients. Front. Cell. Infect. Microbiol. 2022, 12, 1021320. [Google Scholar] [CrossRef] [PubMed]
- Hong, N.T.T.; Anh, N.T.; Mai, N.T.H.; Nghia, H.D.T.; Nhu, L.N.T.; Thanh, T.T.; Phu, N.H.; Deng, X.; van Doorn, H.R.; van Vinh Chau, N.; et al. Performance of Metagenomic Next-Generation Sequencing for the Diagnosis of Viral Meningoencephalitis in a Resource-Limited Setting. Open Forum Infect. Dis. 2020, 7, ofaa046. [Google Scholar] [CrossRef] [PubMed]
- Horiba, K.; Torii, Y.; Aizawa, Y.; Yamaguchi, M.; Haruta, K.; Okumura, T.; Suzuki, T.; Kawano, Y.; Kawada, J.I.; Hara, S.; et al. Performance of Nanopore and Illumina Metagenomic Sequencing for Pathogen Detection and Transcriptome Analysis in Infantile Central Nervous System Infections. Open Forum Infect. Dis. 2022, 9, ofac504. [Google Scholar] [CrossRef]
- Kalantar, K.L.; Carvalho, T.; De Bourcy, C.F.A.; Dimitrov, B.; Dingle, G.; Egger, R.; Han, J.; Holmes, O.B.; Juan, Y.F.; King, R.; et al. IDseq—An Open Source Cloud-Based Pipeline and Analysis Service for Metagenomic Pathogen Detection and Monitoring. Gigascience 2020, 9, giaa111. [Google Scholar] [CrossRef]
- Pérot, P.; Bigot, T.; Temmam, S.; Regnault, B.; Eloit, M. Microseek: A Protein-Based Metagenomic Pipeline for Virus Diagnostic and Discovery. Viruses 2022, 14, 1990. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. MetaSPAdes: A New Versatile Metagenomic Assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Antipov, D.; Raiko, M.; Lapidus, A.; Pevzner, P.A. MetaviralSPAdes: Assembly of Viruses from Metagenomic Data. Bioinformatics 2020, 36, 4126–4129. [Google Scholar] [CrossRef]
- Ibañez-Lligoña, M.; Colomer-Castell, S.; González-Sánchez, A.; Gregori, J.; Campos, C.; Garcia-Cehic, D.; Andrés, C.; Piñana, M.; Pumarola, T.; Rodríguez-Frias, F.; et al. Bioinformatic Tools for NGS-Based Metagenomics to Improve the Clinical Diagnosis of Emerging, Re-Emerging and New Viruses. Viruses 2023, 15, 587. [Google Scholar] [CrossRef] [PubMed]
- Elrashedy, A.; Mousa, W.; Nayel, M.; Salama, A.; Zaghawa, A.; Elsify, A.; Hasan, M.E. Advances in Bioinformatics and Multi-Omics Integration: Transforming Viral Infectious Disease Research in Veterinary Medicine. Virol. J. 2025, 22, 22. [Google Scholar] [CrossRef]
- Qayyum, H.; Ishaq, Z.; Ali, A.; Kayani, M.U.R.; Huang, L. Genome-Resolved Metagenomics from Short-Read Sequencing Data in the Era of Artificial Intelligence. Funct. Integr. Genom. 2025, 25, 124. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; He, Y.; Fang, P.; Mei, S.Q.; Xu, Z.; Wu, W.C.; Tian, J.H.; Zhang, S.; Zeng, Z.Y.; Gou, Q.Y.; et al. Using Artificial Intelligence to Document the Hidden RNA Virosphere. Cell 2024, 187, 6929–6942.e16. [Google Scholar] [CrossRef] [PubMed]
- Wadas, I.; Domingues, I. Systematic Review of Phylogenetic Analysis Techniques for RNA Viruses Using Bioinformatics. Int. J. Mol. Sci. 2025, 26, 2180. [Google Scholar] [CrossRef]
- Wee, J.J.; Wei, G.W. Rapid Response to Fast Viral Evolution Using AlphaFold 3-Assisted Topological Deep Learning. Virus Evol. 2025, 11, 26. [Google Scholar] [CrossRef]
- Yu, D.; Chojnowski, G.; Rosenthal, M.; Kosinski, J. AlphaPulldown—A Python Package for Protein–Protein Interaction Screens Using AlphaFold-Multimer. Bioinformatics 2023, 39, btac749. [Google Scholar] [CrossRef]
- Jurasz, H.; Pawłowski, T.; Perlejewski, K. Contamination Issue in Viral Metagenomics: Problems, Solutions, and Clinical Perspectives. Front. Microbiol. 2021, 12, 745076. [Google Scholar] [CrossRef]
- Dsa, O.C.; Kadni, T.S.; Sudheesh, N. From Cold Chain to Ambient Temperature: Transport of Viral Specimens—A Review. Ann. Med. 2023, 55, 2257711. [Google Scholar] [CrossRef]
- Chheda, U.; Pradeepan, S.; Esposito, E.; Strezsak, S.; Fernandez-Delgado, O.; Kranz, J. Factors Affecting Stability of RNA—Temperature, Length, Concentration, PH, and Buffering Species. J. Pharm. Sci. 2024, 113, 377–385. [Google Scholar] [CrossRef]
- Parmar, S.; Sridhar, S.; Forrest, S.; Kean, I.; Young, J.; Bartholdson Scott, J.; Maes, M.; Pereira-Dias, J.; Routledge, M.; Sparkes, D.; et al. A Blueprint for the Implementation of a Validated Approach for the Detection of SARS-Cov2 in Clinical Samples in Academic Facilities. Wellcome Open Res. 2020, 5, 110. [Google Scholar] [CrossRef]
- Lam, C.; Gray, K.; Gall, M.; Sadsad, R.; Arnott, A.; Johnson-Mackinnon, J.; Fong, W.; Basile, K.; Kok, J.; Dwyer, D.E.; et al. SARS-CoV-2 Genome Sequencing Methods Differ in Their Abilities to Detect Variants from Low-Viral-Load Samples. J. Clin. Microbiol. 2021, 59. [Google Scholar] [CrossRef]
- Liu, T.; Chen, Z.; Chen, W.; Chen, X.; Hosseini, M.; Yang, Z.; Li, J.; Ho, D.; Turay, D.; Gheorghe, C.P.; et al. A Benchmarking Study of SARS-CoV-2 Whole-Genome Sequencing Protocols Using COVID-19 Patient Samples. iScience 2021, 24, 102892. [Google Scholar] [CrossRef]
- Buddle, S.; Forrest, L.; Akinsuyi, N.; Martin Bernal, L.M.; Brooks, T.; Venturini, C.; Miller, C.; Brown, J.R.; Storey, N.; Atkinson, L.; et al. Evaluating Metagenomics and Targeted Approaches for Diagnosis and Surveillance of Viruses. Genome Med. 2024, 16, 111. [Google Scholar] [CrossRef] [PubMed]
- Schönegger, D.; Moubset, O.; Margaria, P.; Menzel, W.; Winter, S.; Roumagnac, P.; Marais, A.; Candresse, T. Benchmarking of Virome Metagenomic Analysis Approaches Using a Large, 60+ Members, Viral Synthetic Community. J. Virol. 2023, 97, e01300-23. [Google Scholar] [CrossRef] [PubMed]
- Endrullat, C.; Glökler, J.; Franke, P.; Frohme, M. Standardization and Quality Management in Next-Generation Sequencing. Appl. Transl. Genom. 2016, 10, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Temeeyasen, G.; Sharafeldin, T.A.; Lin, C.M.; Hause, B.M. Spillover of Canine Parvovirus Type 2 to Pigs, South Dakota, USA, 2020. Emerg. Infect. Dis. 2022, 28, 471. [Google Scholar] [CrossRef]
- Zhang, X.-A.; Li, H.; Jiang, F.-C.; Zhu, F.; Zhang, Y.-F.; Chen, J.-J.; Tan, C.-W.; Anderson, D.E.; Fan, H.; Dong, L.-Y.; et al. A Zoonotic Henipavirus in Febrile Patients in China. N. Engl. J. Med. 2022, 387, 470–472. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A New Coronavirus Associated with Human Respiratory Disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904.e9. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasllieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greeneugh, T.C.; et al. Angiotensin-Converting Enzyme 2 Is a Functional Receptor for the SARS Coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef]
- Samavati, L.; Uhal, B.D. ACE2, Much More Than Just a Receptor for SARS-COV-2. Front. Cell. Infect. Microbiol. 2020, 10, 554397. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Bogner, P.; Capua, I.; Cox, N.J.; Lipman, D.J. A Global Initiative on Sharing Avian Flu Data. Nature 2006, 442, 981. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. NextStrain: Real-Time Tracking of Pathogen Evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 Novel Coronavirus (2019-NCoV) by Real-Time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef]
- Eurosurveillance Editorial Team. Correction for Euro Surveill. 2020;25(23). Eurosurveillance 2021, 26, 210325c. [Google Scholar] [CrossRef]
- Sharma, O.; Sultan, A.A.; Ding, H.; Triggle, C.R. A Review of the Progress and Challenges of Developing a Vaccine for COVID-19. Front. Immunol. 2020, 11, 585354. [Google Scholar] [CrossRef]
- Thanh Le, T.; Andreadakis, Z.; Kumar, A.; Gómez Román, R.; Tollefsen, S.; Saville, M.; Mayhew, S. The COVID-19 Vaccine Development Landscape. Nat. Rev. Drug Discov. 2020, 19, 305–306. [Google Scholar] [CrossRef]
- Zhu, F.C.; Li, Y.H.; Guan, X.H.; Hou, L.H.; Wang, W.J.; Li, J.X.; Wu, S.P.; Wang, B.; Wang, Z.; Wang, L.; et al. Safety, Tolerability, and Immunogenicity of a Recombinant Adenovirus Type-5 Vectored COVID-19 Vaccine: A Dose-Escalation, Open-Label, Non-Randomised, First-in-Human Trial. Lancet 2020, 395, 1845. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.C.; Guan, X.H.; Li, Y.H.; Huang, J.Y.; Jiang, T.; Hou, L.H.; Li, J.X.; Yang, B.F.; Wang, L.; Wang, W.J.; et al. Immunogenicity and Safety of a Recombinant Adenovirus Type-5-Vectored COVID-19 Vaccine in Healthy Adults Aged 18 Years or Older: A Randomised, Double-Blind, Placebo-Controlled, Phase 2 Trial. Lancet 2020, 396, 479. [Google Scholar] [CrossRef] [PubMed]
- Mahase, E. Covid-19: Moderna Applies for US and EU Approval as Vaccine Trial Reports 94.1% Efficacy. BMJ 2020, 371, m4709. [Google Scholar] [CrossRef] [PubMed]
- Mahase, E. Covid-19: Vaccine Candidate May Be More than 90% Effective, Interim Results Indicate. BMJ 2020, 371, m4347. [Google Scholar] [CrossRef]
- Kirste, I.; Hortsch, S.; Grunert, V.P.; Legault, H.; Maglinao, M.; Eichenlaub, U.; Kashlan, B.; Pajon, R.; Jochum, S. Quantifying the Vaccine-Induced Humoral Immune Response to Spike-Receptor Binding Domain as a Surrogate for Neutralization Testing Following MRNA-1273 (Spikevax) Vaccination Against COVID-19. Infect. Dis. Ther. 2023, 12, 177–191. [Google Scholar] [CrossRef]
- Lucas, C.; Vogels, C.B.F.; Yildirim, I.; Rothman, J.E.; Lu, P.; Monteiro, V.; Gehlhausen, J.R.; Campbell, M.; Silva, J.; Tabachnikova, A.; et al. Impact of Circulating SARS-CoV-2 Variants on MRNA Vaccine-Induced Immunity. Nature 2021, 600, 523–529. [Google Scholar] [CrossRef]
- Singh, P.; Sharma, K.; Bhargava, A.; Negi, S.S. Genomic Characterization of Influenza A (H1N1)Pdm09 and SARS-CoV-2 from Influenza Like Illness (ILI) and Severe Acute Respiratory Illness (SARI) Cases Reported between July–December, 2022. Sci. Rep. 2024, 14, 10660. [Google Scholar] [CrossRef]
- Cardona-Ospina, J.A.; Rojas-Gallardo, D.M.; Garzón-Castaño, S.C.; Jiménez-Posada, E.V.; Rodríguez-Morales, A.J. Phylodynamic Analysis in the Understanding of the Current COVID-19 Pandemic and Its Utility in Vaccine and Antiviral Design and Assessment. Hum. Vaccin. Immunother. 2021, 17, 2437. [Google Scholar] [CrossRef]
- Sah, R.; Mohanty, A.; Chakraborty, S.; Dhama, K. Langya Virus: A Newly Identified Zoonotic Henipavirus. J. Med. Virol. 2022, 94, 5621–5622. [Google Scholar] [CrossRef]
- Olamilekan Adesola, R.; Viola Miranda, A.; Shun Joshua Tran, Y.; Idris, I.; Lin, X.; Kouwenhoven, M.B.N.; Eliseo Lucero-Prisno, D., III. Langya Virus Outbreak: Current Challenges and Lesson Learned from Previous Henipavirus Outbreaks in China, Australia, and Southeast Asia. Bull. Natl. Res. Cent. 2023, 47, 87. [Google Scholar] [CrossRef]
- Wise, E.L.; Márquez, S.; Mellors, J.; Paz, V.; Atkinson, B.; Gutierrez, B.; Zapata, S.; Coloma, J.; Pybus, O.G.; Jackson, S.K.; et al. Oropouche Virus Cases Identified in Ecuador Using an Optimised QRT-PCR Informed by Metagenomic Sequencing. PLoS Negl. Trop. Dis. 2020, 14, e0007897. [Google Scholar] [CrossRef] [PubMed]
- Wise, E.L.; Pullan, S.T.; Márquez, S.; Paz, V.; Mosquera, J.D.; Zapata, S.; Jackson, S.K.; Fejer, G.; Trueba, G.; Logue, C.H. Isolation of Oropouche Virus from Febrile Patient, Ecuador. Emerg. Infect. Dis. 2018, 24, 935–937. [Google Scholar] [CrossRef] [PubMed]
- Isidro, J.; Borges, V.; Pinto, M.; Sobral, D.; Santos, J.D.; Nunes, A.; Mixão, V.; Ferreira, R.; Santos, D.; Duarte, S.; et al. Phylogenomic Characterization and Signs of Microevolution in the 2022 Multi-Country Outbreak of Monkeypox Virus. Nat. Med. 2022, 28, 1569. [Google Scholar] [CrossRef] [PubMed]
- Treutiger, C.J.; Filén, F.; Rehn, M.; Aarum, J.; Jacks, A.; Gisslén, M.; Sturegård, E.; Karlberg, M.L.; Lindsjö, O.K.; Sondén, K. First Case of Mpox with Monkeypox Virus Clade Ib Outside Africa in a Returning Traveller, Sweden, August 2024: Public Health Measures. Eurosurveillance 2024, 29, 2400740. [Google Scholar] [CrossRef]
- Calvet, G.; Aguiar, R.S.; Melo, A.S.O.; Sampaio, S.A.; de Filippis, I.; Fabri, A.; Araujo, E.S.M.; de Sequeira, P.C.; de Mendonça, M.C.L.; de Oliveira, L.; et al. Detection and Sequencing of Zika Virus from Amniotic Fluid of Fetuses with Microcephaly in Brazil: A Case Study. Lancet Infect. Dis. 2016, 16, 653–660. [Google Scholar] [CrossRef]
- Chiu, C.Y.; Coffey, L.L.; Murkey, J.; Symmes, K.; Sample, H.A.; Wilson, M.R.; Naccache, S.N.; Arevalo, S.; Somasekar, S.; Federman, S.; et al. Diagnosis of Fatal Human Case of St. Louis Encephalitis Virus Infection by Metagenomic Sequencing, California, 2016. Emerg. Infect. Dis. 2017, 23, 1964. [Google Scholar] [CrossRef]
- Li, T.; Mbala-Kingebeni, P.; Naccache, S.N.; Thézé, J.; Bouquet, J.; Federman, S.; Somasekar, S.; Yu, G.; Martin, C.S.S.; Achari, A.; et al. Metagenomic Next-Generation Sequencing of the 2014 Ebola Virus Disease Outbreak in the Democratic Republic of the Congo. J. Clin. Microbiol. 2019, 57, e00827-19. [Google Scholar] [CrossRef]
- Akingbola, A.; Abiodun, A.; Idahor, C.; Peters, F.; Ojo, O.; Jessica, O.U.; Alao, U.H.; Adewole, O.; Owolabi, A.; Chuku, J. Genomic Evolution and Epidemiological Impact of Ongoing Clade Ib MPox Disease: A Narrative Review. Glob. Health Epidemiol. Genom. 2025, 2025, 8845911. [Google Scholar] [CrossRef]
- Americo, J.L.; Earl, P.L.; Moss, B. Virulence Differences of Mpox (Monkeypox) Virus Clades I, IIa, and IIb.1 in a Small Animal Model. Proc. Natl. Acad. Sci. USA 2023, 120, e2220415120. [Google Scholar] [CrossRef]
- Akingbola, A.; Adegbesan, C.A.; Adewole, O.; Idahor, C.; Odukoya, T.; Nwaeze, E.; Mayowa, S.; Abdullahi, O.; Mariaria, P.K. Understanding the Resurgence of Mpox: Key Drivers and Lessons from Recent Outbreaks in Africa. Trop. Med. Health 2025, 53, 47. [Google Scholar] [CrossRef]
- Sharif, N.; Sharif, N.; Alzahrani, K.J.; Halawani, I.F.; Alzahrani, F.M.; Díez, I.D.l.T.; Lipari, V.; Flores, M.A.L.; Parvez, A.K.; Dey, S.K. Molecular Epidemiology, Transmission and Clinical Features of 2022-Mpox Outbreak: A Systematic Review. Health Sci. Rep. 2023, 6, e1603. [Google Scholar] [CrossRef]
- Van Dijck, C.; Hoff, N.A.; Mbala-Kingebeni, P.; Low, N.; Cevik, M.; Rimoin, A.W.; Kindrachuk, J.; Liesenborghs, L. Emergence of Mpox in the Post-Smallpox Era—A Narrative Review on Mpox Epidemiology. Clin. Microbiol. Infect. 2023, 29, 1487–1492. [Google Scholar] [CrossRef]
- Yinka-Ogunleye, A.; Aruna, O.; Dalhat, M.; Ogoina, D.; McCollum, A.; Disu, Y.; Mamadu, I.; Akinpelu, A.; Ahmad, A.; Burga, J.; et al. Outbreak of Human Monkeypox in Nigeria in 2017–2018: A Clinical and Epidemiological Report. Lancet Infect. Dis. 2019, 19, 872. [Google Scholar] [CrossRef]
- Vaughan, A.; Aarons, E.; Astbury, J.; Balasegaram, S.; Beadsworth, M.; Beck, C.R.; Chand, M.; O’connor, C.; Dunning, J.; Ghebrehewet, S.; et al. Two Cases of Monkeypox Imported to the United Kingdom, September 2018. Eurosurveillance 2018, 23, 1800509. [Google Scholar] [CrossRef]
- Erez, N.; Achdout, H.; Milrot, E.; Schwartz, Y.; Wiener-Well, Y.; Paran, N.; Politi, B.; Tamir, H.; Israely, T.; Weiss, S.; et al. Diagnosis of Imported Monkeypox, Israel, 2018. Emerg. Infect. Dis. 2019, 25, 980. [Google Scholar] [CrossRef]
- Ng, O.T.; Lee, V.; Marimuthu, K.; Vasoo, S.; Chan, G.; Lin, R.T.P.; Leo, Y.S. A Case of Imported Monkeypox in Singapore. Lancet Infect. Dis. 2019, 19, 1166. [Google Scholar] [CrossRef] [PubMed]
- Mauldin, M.R.; McCollum, A.M.; Nakazawa, Y.J.; Mandra, A.; Whitehouse, E.R.; Davidson, W.; Zhao, H.; Gao, J.; Li, Y.; Doty, J.; et al. Exportation of Monkeypox Virus From the African Continent. J. Infect. Dis. 2022, 225, 1367–1376. [Google Scholar] [CrossRef] [PubMed]
- First Draft Genome Sequence of Monkeypox Virus Associated with the Suspected Multi-Country Outbreak, May 2022 (Confirmed Case in Portugal)—MPXV / Genome Reports—Virological. Available online: https://virological.org/t/first-draft-genome-sequence-of-monkeypox-virus-associated-with-the-suspected-multi-country-outbreak-may-2022-confirmed-case-in-portugal/799 (accessed on 29 July 2025).
- Ylaya, E.M.; Grande, P.G.; Dancel, L.L.; Nicolasora, A.D.; Polotan, F.G.; Pantoni, R.A.; Melo, E.; Ortia, S.P.; Manalo, J.I.; Abulencia, M.F.; et al. Case Report: A Comprehensive Report on the First Confirmed Mpox Case in the Philippines during the 2022 Mpox Global Outbreak: From Clinical Presentation to Shotgun Metagenomic Sequencing Analysis. Front. Med. 2024, 11, 1387407. [Google Scholar] [CrossRef] [PubMed]
- Claro, I.M.; Romano, C.M.; Candido, D.d.S.; de Lima, E.L.; Lindoso, J.A.L.; Ramundo, M.S.; Moreira, F.R.R.; Barra, L.A.C.; Borges, L.M.S.; Medeiros, L.A.; et al. Shotgun Metagenomic Sequencing of the First Case of Monkeypox Virus in Brazil, 2022. Rev. Inst. Med. Trop. Sao Paulo 2022, 64, e48. [Google Scholar] [CrossRef]
- Tiwari, A.; Kalonji, T.; Miller, T.; Van Den Bossche, T.; Krolicka, A.; Muhindo-Mavoko, H.; Mitashi, P.; Tahita, M.C.; Lood, R.; Pitkänen, T.; et al. Emergence and Global Spread of Mpox Clade Ib: Challenges and the Role of Wastewater and Environmental Surveillance. J. Infect. Dis. 2025, 231, e825–e829. [Google Scholar] [CrossRef]
- Srivastava, S.; Laxmi; Sharma, K.; Sridhar, S.B.; Talath, S.; Shareef, J.; Mehta, R.; Satapathy, P.; Sah, R. Clade Ib: A New Emerging Threat in the Mpox Outbreak. Front. Pharmacol. 2024, 15, 1504154. [Google Scholar] [CrossRef]
- Vakaniaki, E.H.; Kacita, C.; Kinganda-Lusamaki, E.; O’Toole, Á.; Wawina-Bokalanga, T.; Mukadi-Bamuleka, D.; Amuri-Aziza, A.; Malyamungu-Bubala, N.; Mweshi-Kumbana, F.; Mutimbwa-Mambo, L.; et al. Sustained Human Outbreak of a New MPXV Clade I Lineage in Eastern Democratic Republic of the Congo. Nat. Med. 2024, 30, 2791–2795. [Google Scholar] [CrossRef]
- Brosius, I.; Vakaniaki, E.H.; Mukari, G.; Munganga, P.; Tshomba, J.C.; De Vos, E.; Bangwen, E.; Mujula, Y.; Tsoumanis, A.; Van Dijck, C.; et al. Epidemiological and Clinical Features of Mpox during the Clade Ib Outbreak in South Kivu, Democratic Republic of the Congo: A Prospective Cohort Study. Lancet 2025, 405, 547–559. [Google Scholar] [CrossRef]
- Masirika, L.M.; Udahemuka, J.C.; Schuele, L.; Nieuwenhuijse, D.F.; Ndishimye, P.; Boter, M.; Mbiribindi, J.B.; Kacita, C.; Lang, T.; Gortázar, C.; et al. Epidemiological and Genomic Evolution of the Ongoing Outbreak of Clade Ib Mpox Virus in the Eastern Democratic Republic of the Congo. Nat. Med. 2025, 31, 1459–1463. [Google Scholar] [CrossRef]
- Pan, D.; Nazareth, J.; Sze, S.; Martin, C.A.; Decker, J.; Fletcher, E.; Déirdre Hollingsworth, T.; Barer, M.R.; Pareek, M.; Tang, J.W. Transmission of Monkeypox/Mpox Virus: A Narrative Review of Environmental, Viral, Host, and Population Factors in Relation to the 2022 International Outbreak. J. Med. Virol. 2023, 95, e28534. [Google Scholar] [CrossRef]
- Marty, A.M. Why Is There Expanding Community Transmission of Monkeypox in 2022? Lancet Microbe 2022, 3, e810–e811. [Google Scholar] [CrossRef]
- Schuele, L.; Masirika, L.M.; Udahemuka, J.C.; Siangoli, F.B.; Mbiribindi, J.B.; Ndishimye, P.; Aarestrup, F.M.; Koopmans, M.; Oude Munnink, B.B.; Molenkamp, R.; et al. Real-Time PCR Assay to Detect the Novel Clade Ib Monkeypox Virus, September 2023 to May 2024. Eurosurveillance 2024, 29, 2400486. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, R.P.; Fogel, R.; Limson, J. Overview of Diagnostic Methods, Disease Prevalence and Transmission of Mpox (Formerly Monkeypox) in Humans and Animal Reservoirs. Microorganisms 2023, 11, 1186. [Google Scholar] [CrossRef] [PubMed]
- Ortins-Pina, A.; Hegemann, B.; Saggini, A.; Deml, K.F.; Wallerius, K.; Hörster, S.; Kraft, S.; Weyers, W. Histopathological Features of Human Mpox: Report of Two Cases and Review of the Literature. J. Cutan. Pathol. 2023, 50, 706–710. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.W.; Choi, G.K.Y.; Yip, C.C.Y.; Cheng, V.C.C.; Yuen, K.Y. Zika Fever and Congenital Zika Syndrome: An Unexpected Emerging Arboviral Disease. J. Infect. 2016, 72, 507–524. [Google Scholar] [CrossRef]
- Singh, R.K.; Dhama, K.; Karthik, K.; Tiwari, R.; Khandia, R.; Munjal, A.; Iqbal, H.M.N.; Malik, Y.S.; Bueno-Marí, R. Advances in Diagnosis, Surveillance, and Monitoring of Zika Virus: An Update. Front. Microbiol. 2018, 8, 304633. [Google Scholar] [CrossRef] [PubMed]
- Landry, M.L.; St George, K. Laboratory Diagnosis of Zika Virus Infection. Arch. Pathol. Lab. Med. 2017, 141, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Gourinat, A.C.; O’Connor, O.; Calvez, E.; Goarant, C.; Dupont-Rouzeyrol, M. Detection of Zika Virus in Urine. Emerg. Infect. Dis. 2015, 21, 84–86. [Google Scholar] [CrossRef] [PubMed]
- Rossini, G.; Gaibani, P.; Vocale, C.; Cagarelli, R.; Landini, M.P. Comparison of Zika Virus (ZIKV) RNA Detection in Plasma, Whole Blood and Urine—Case Series of Travel-Associated ZIKV Infection Imported to Italy, 2016. J. Infect. 2017, 75, 242–245. [Google Scholar] [CrossRef]
- Quick, J.; Grubaugh, N.D.; Pullan, S.T.; Claro, I.M.; Smith, A.D.; Gangavarapu, K.; Oliveira, G.; Robles-Sikisaka, R.; Rogers, T.F.; Beutler, N.A.; et al. Multiplex PCR Method for MinION and Illumina Sequencing of Zika and Other Virus Genomes Directly from Clinical Samples. Nat. Protoc. 2017, 12, 1261–1276. [Google Scholar] [CrossRef]
- Metsky, H.C.; Matranga, C.B.; Wohl, S.; Schaffner, S.F.; Freije, C.A.; Winnicki, S.M.; West, K.; Qu, J.; Baniecki, M.L.; Gladden-Young, A.; et al. Zika Virus Evolution and Spread in the Americas. Nature 2017, 546, 411–415. [Google Scholar] [CrossRef]
- Gu, S.H.; Song, D.H.; Lee, D.; Jang, J.; Kim, M.Y.; Jung, J.; Woo, K.I.; Kim, M.; Seog, W.; Oh, H.S.; et al. Whole-Genome Sequence Analysis of Zika Virus, Amplified from Urine of Traveler from the Philippines. Virus Genes. 2017, 53, 918–921. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, H.N.; Pareek, V.; Raza, K.; Dantham, S.; Kumar, P.; Mochan, S.; Faiq, M.A. A Possible Mechanism of Zika Virus Associated Microcephaly: Imperative Role of Retinoic Acid Response Element (RARE) Consensus Sequence Repeats in the Viral Genome. Front. Hum. Neurosci. 2016, 10, 403. [Google Scholar] [CrossRef]
- Jun, S.R.; Wassenaar, T.M.; Wanchai, V.; Patumcharoenpol, P.; Nookaew, I.; Ussery, D.W. Suggested Mechanisms for Zika Virus Causing Microcephaly: What Do the Genomes Tell Us? BMC Bioinform. 2017, 18, 471. [Google Scholar] [CrossRef]
- Faizan, M.I.; Abdullah, M.; Ali, S.; Naqvi, I.H.; Ahmed, A.; Parveen, S. Zika Virus-Induced Microcephaly and Its Possible Molecular Mechanism. Intervirology 2017, 59, 152–158. [Google Scholar] [CrossRef]
- Liang, Q.; Luo, Z.; Zeng, J.; Chen, W.; Foo, S.S.; Lee, S.A.; Ge, J.; Wang, S.; Goldman, S.A.; Zlokovic, B.V.; et al. Zika Virus NS4A and NS4B Proteins Deregulate Akt-MTOR Signaling in Human Fetal Neural Stem Cells to Inhibit Neurogenesis and Induce Autophagy. Cell Stem Cell 2016, 19, 663–671. [Google Scholar] [CrossRef]
- Tang, H.; Hammack, C.; Ogden, S.C.; Wen, Z.; Qian, X.; Li, Y.; Yao, B.; Shin, J.; Zhang, F.; Lee, E.M.; et al. Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell Stem Cell 2016, 18, 587–590. [Google Scholar] [CrossRef]
- Ayllón, T.; Campos, R.d.M.; Brasil, P.; Morone, F.C.; Câmara, D.C.P.; Meira, G.L.S.; Tannich, E.; Yamamoto, K.A.; Carvalho, M.S.; Pedro, R.S.; et al. Early Evidence for Zika Virus Circulation among Aedes Aegypti Mosquitoes, Rio de Janeiro, Brazil. Emerg. Infect. Dis. 2017, 23, 1411–1412. [Google Scholar] [CrossRef] [PubMed]
- Naccache, S.N.; Thézé, J.; Sardi, S.I.; Somasekar, S.; Greninger, A.L.; Bandeira, A.C.; Campos, G.S.; Tauro, L.B.; Faria, N.R.; Pybus, O.G.; et al. Distinct Zika Virus Lineage in Salvador, Bahia, Brazil. Emerg. Infect. Dis. 2016, 22, 1788–1792. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.K.; Bhattacharjee, S. Dengue Virus: Epidemiology, Biology, and Disease Aetiology. Can. J. Microbiol. 2021, 67, 687–702. [Google Scholar] [CrossRef]
- Hales, S.; De Wet, N.; Maindonald, J.; Woodward, A. Potential Effect of Population and Climate Changes on Global Distribution of Dengue Fever: An Empirical Model. Lancet 2002, 360, 830–834. [Google Scholar] [CrossRef]
- Chen, L.H.; Wilson, M.E. The Role of the Traveler in Emerging Infections and Magnitude of Travel. Med. Clin. N. Am. 2008, 92, 1409–1432. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Fu, J.; Jiang, D.; Hao, M.; Lin, G. Mapping the Spatial Distribution of Aedes Aegypti and Aedes Albopictus. Acta Trop. 2018, 178, 155–162. [Google Scholar] [CrossRef]
- Abbasi, E. Global Expansion of Aedes Mosquitoes and Their Role in the Transboundary Spread of Emerging Arboviral Diseases: A Comprehensive Review. IJID One Health 2025, 6, 100058. [Google Scholar] [CrossRef]
- Halstead, S.B.; O’Rourke, E.J. Dengue Viruses and Mononuclear Phagocytes. I. Infection Enhancement by Non-Neutralizing Antibody. J. Exp. Med. 1977, 146, 201–217. [Google Scholar] [CrossRef]
- Grange, L.; Simon-Loriere, E.; Sakuntabhai, A.; Gresh, L.; Paul, R.; Harris, E. Epidemiological Risk Factors Associated with High Global Frequency of Inapparent Dengue Virus Infections. Front. Immunol. 2014, 5, 92839. [Google Scholar] [CrossRef]
- Silva, J.P.; Fernandez-Sesma, A. Challenges on the Development of a Dengue Vaccine: A Comprehensive Review of the State of the Art. J. Gen. Virol. 2023, 104, 001831. [Google Scholar] [CrossRef]
- Vasconcelos, P.F.; Travassos da Rosa, A.P.; Rodrigues, S.G.; Travassos da Rosa, E.S.; Dégallier, N.; Travassos da Rosa, J.F. Inadequate Management of Natural Ecosystem in the Brazilian Amazon Region Results in the Emergence and Reemergence of Arboviruses. Cad. Saude Publica 2001, 17, S155–S164. [Google Scholar] [CrossRef] [PubMed]
- Moreira, F.R.R.; Dutra, J.V.R.; de Carvalho, A.H.B.; Reis, C.R.; Rios, J.S.H.; Ribeiro, M.d.O.; Arruda, M.B.; Alvarez, P.; Souza, R.P.; Voloch, C.; et al. Oropouche Virus Genomic Surveillance in Brazil. Lancet Infect. Dis. 2024, 24, e664–e666. [Google Scholar] [CrossRef] [PubMed]
- Tilston-Lunel, N.L. Oropouche Virus: An Emerging Orthobunyavirus. J. Gen. Virol. 2024, 105, 002027. [Google Scholar] [CrossRef] [PubMed]
- Naveca, F.G.; de Almeida, T.A.P.; Souza, V.; Nascimento, V.; Silva, D.; Nascimento, F.; Mejía, M.; de Oliveira, Y.S.; Rocha, L.; Xavier, N.; et al. Human Outbreaks of a Novel Reassortant Oropouche Virus in the Brazilian Amazon Region. Nat. Med. 2024, 30, 3509–3521. [Google Scholar] [CrossRef]
- Scachetti, G.C.; Forato, J.; Claro, I.M.; Hua, X.; Salgado, B.B.; Vieira, A.; Simeoni, C.L.; Barbosa, A.R.C.; Rosa, I.L.; de Souza, G.F.; et al. Re-Emergence of Oropouche Virus between 2023 and 2024 in Brazil: An Observational Epidemiological Study. Lancet Infect. Dis. 2025, 25, 166–175. [Google Scholar] [CrossRef]
- Pialoux, G.; Gaüzère, B.A.; Jauréguiberry, S.; Strobel, M. Chikungunya, an Epidemic Arbovirosis. Lancet Infect. Dis. 2007, 7, 319–327. [Google Scholar] [CrossRef]
- Tsetsarkin, K.A.; Vanlandingham, D.L.; McGee, C.E.; Higgs, S. A Single Mutation in Chikungunya Virus Affects Vector Specificity and Epidemic Potential. PLoS Pathog. 2007, 3, e201. [Google Scholar] [CrossRef]
- Vazeille, M.; Moutailler, S.; Coudrier, D.; Rousseaux, C.; Khun, H.; Huerre, M.; Thiria, J.; Dehecq, J.S.; Fontenille, D.; Schuffenecker, I.; et al. Two Chikungunya Isolates from the Outbreak of La Reunion (Indian Ocean) Exhibit Different Patterns of Infection in the Mosquito, Aedes Albopictus. PLoS ONE 2007, 2, e1168. [Google Scholar] [CrossRef]
- Tsetsarkin, K.A.; McGee, C.E.; Volk, S.M.; Vanlandingham, D.L.; Weaver, S.C.; Higgs, S. Epistatic Roles of E2 Glycoprotein Mutations in Adaption of Chikungunya Virus to Aedes Albopictus and Ae. Aegypti Mosquitoes. PLoS ONE 2009, 4, e6835. [Google Scholar] [CrossRef]
- Bonilauri, P.; Bellini, R.; Calzolari, M.; Angelini, R.; Venturi, L.; Fallacara, F.; Cordioli, P.; Angelini, P.; Venturelli, C.; Merialdi, G.; et al. Chikungunya Virus in Aedes Albopictus, Italy. Emerg. Infect. Dis. 2008, 14, 852. [Google Scholar] [CrossRef] [PubMed]
- Farooq, Z.; Segelmark, L.; Rocklöv, J.; Lillepold, K.; Sewe, M.O.; Briet, O.J.T.; Semenza, J.C. Impact of Climate and Aedes Albopictus Establishment on Dengue and Chikungunya Outbreaks in Europe: A Time-to-Event Analysis. Lancet Planet Health 2025, 9, e374–e383. [Google Scholar] [CrossRef] [PubMed]
- Adikari, T.N.; Riaz, N.; Sigera, C.; Leung, P.; Valencia, B.M.; Barton, K.; Smith, M.A.; Bull, R.A.; Li, H.; Luciani, F.; et al. Single Molecule, near Full-Length Genome Sequencing of Dengue Virus. Sci. Rep. 2020, 10, 18196. [Google Scholar] [CrossRef] [PubMed]
- Frumence, E.; Piorkowski, G.; Traversier, N.; Amaral, R.; Vincent, M.; Mercier, A.; Ayhan, N.; Souply, L.; Pezzi, L.; Lier, C.; et al. Genomic Insights into the Re-Emergence of Chikungunya Virus on Réunion Island, France, 2024 to 2025. Eurosurveillance 2025, 30, 2500344. [Google Scholar] [CrossRef]
- Deiana, M.; Malagò, S.; Mori, A.; Accordini, S.; Matucci, A.; Passarelli Mantovani, R.; Gianesini, N.; Huits, R.; Piubelli, C.; Gobbi, F.G.; et al. Full Genome Characterization of the First Oropouche Virus Isolate Imported in Europe from Cuba. Viruses 2024, 16, 1586. [Google Scholar] [CrossRef]
- Vogels, C.B.F.; Hill, V.; Breban, M.I.; Chaguza, C.; Paul, L.M.; Sodeinde, A.; Taylor-Salmon, E.; Ott, I.M.; Petrone, M.E.; Dijk, D.; et al. DengueSeq: A Pan-Serotype Whole Genome Amplicon Sequencing Protocol for Dengue Virus. BMC Genom. 2024, 25, 433. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, F.J.; Laubscher, F.; Chudzinski, V.; Kaiser, L.; Cordey, S. Direct Dengue Virus Genome Sequencing from Antigen NS1 Rapid Diagnostic Tests: A Proof-of-Concept with the Standard Q Dengue Duo Assay. Viruses 2023, 15, 2167. [Google Scholar] [CrossRef]
- Kabir, K.M.A.; Sigera, C.; Maduranga, S.; Weeratunga, P.; Rajapakse, S.; Fernando, D.; Lloyd, A.R.; Bull, R.A.; Rodrigo, C. Chikungunya Masquerading as Dengue Infection in Sri Lanka Uncovered by Metagenomics. PLoS ONE 2025, 20, e0326995. [Google Scholar] [CrossRef]
- Fourgeaud, J.; Regnault, B.; Faury, H.; Da Rocha, N.; Jamet, A.; Stirnemann, J.; Eloit, M.; Perot, P.; Leruez-Ville, M.; Driessen, M.; et al. Fetal Zika Virus Infection Diagnosed by Metagenomic Next-Generation Sequencing of Amniotic Fluid. Ultrasound Obstet. Gynecol. 2023, 61, 116–117. [Google Scholar] [CrossRef]
- Ashraf, S.; Jerome, H.; Bugembe, D.L.; Ssemwanga, D.; Byaruhanga, T.; Kayiwa, J.T.; Downing, R.; Salazar-Gonzalez, J.F.; Salazar, M.G.; Shepherd, J.G.; et al. Uncovering the Viral Aetiology of Undiagnosed Acute Febrile Illness in Uganda Using Metagenomic Sequencing. Nat. Commun. 2025, 16, 2844. [Google Scholar] [CrossRef] [PubMed]
- Levine, Z.C.; Sene, A.; Mkandawire, W.; Deme, A.B.; Ndiaye, T.; Sy, M.; Gaye, A.; Diedhiou, Y.; Mbaye, A.M.; Ndiaye, I.M.; et al. Investigating the Etiologies of Non-Malarial Febrile Illness in Senegal Using Metagenomic Sequencing. Nat. Commun. 2024, 15, 747. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, A.; Nakielny, S.; Hsu, J.; Kyohere, M.; Byaruhanga, O.; de Bourcy, C.; Egger, R.; Dimitrov, B.; Juan, Y.F.; Sheu, J.; et al. Metagenomic Next-Generation Sequencing of Samples from Pediatric Febrile Illness in Tororo, Uganda. PLoS ONE 2019, 14, e0218318. [Google Scholar] [CrossRef] [PubMed]
- Sardi, S.I.; Somasekar, S.; Naccache, S.N.; Bandeira, A.C.; Tauro, L.B.; Campos, G.S.; Chiub, C.Y. Coinfections of Zika and Chikungunya Viruses in Bahia, Brazil, Identified by Metagenomic next-Generation Sequencing. J. Clin. Microbiol. 2016, 54, 2348–2353. [Google Scholar] [CrossRef]
- Lipkin, W.I. The Changing Face of Pathogen Discovery and Surveillance. Nat. Rev. Microbiol. 2013, 11, 133–141. [Google Scholar] [CrossRef]
- Zhao, J.; Cui, W.; Tian, B.P. The Potential Intermediate Hosts for SARS-CoV-2. Front. Microbiol. 2020, 11, 580137. [Google Scholar] [CrossRef]
- Vijgen, L.; Keyaerts, E.; Moës, E.; Thoelen, I.; Wollants, E.; Lemey, P.; Vandamme, A.-M.; Van Ranst, M. Complete Genomic Sequence of Human Coronavirus OC43: Molecular Clock Analysis Suggests a Relatively Recent Zoonotic Coronavirus Transmission Event. J. Virol. 2005, 79, 1595. [Google Scholar] [CrossRef]
- Shaw, B.; Gatherer, D. Candidate Historical Events for the Emergence of Human Coronavirus OC43: A Critical Reassessment of the Molecular Evidence. PLoS ONE 2023, 18, e0285481. [Google Scholar] [CrossRef]
- Hoffmann, B.; Scheuch, M.; Höper, D.; Jungblut, R.; Holsteg, M.; Schirrmeier, H.; Eschbaumer, M.; Goller, K.V.; Wernike, K.; Fischer, M.; et al. Novel Orthobunyavirus in Cattle, Europe, 2011. Emerg. Infect. Dis. 2012, 18, 469. [Google Scholar] [CrossRef]
- Sedda, L.; Rogers, D.J. The Influence of the Wind in the Schmallenberg Virus Outbreak in Europe. Sci. Rep. 2013, 3, 3361. [Google Scholar] [CrossRef]
- Reusken, C.; van den Wijngaard, C.; van Beek, P.; Beer, M.; Bouwstra, R.; Godeke, G.J.; Isken, L.; van den Kerkhof, H.; van Pelt, W.; van der Poel, W.; et al. Lack of Evidence for Zoonotic Transmission of Schmallenberg Virus. Emerg. Infect. Dis. 2012, 18, 1746–1754. [Google Scholar] [CrossRef]
- Ducomble, T.; Wilking, H.; Stark, K.; Takla, A.; Askar, M.; Schaade, L.; Nitsche, A.; Kurth, A. Lack of Evidence for Schmallenberg Virus Infection in Highly Exposed Persons, Germany. Emerg. Infect. Dis. 2012, 18, 1333–1335. [Google Scholar] [CrossRef] [PubMed]
- Yanase, T.; Kato, T.; Aizawa, M.; Shuto, Y.; Shirafuji, H.; Yamakawa, M.; Tsuda, T. Genetic Reassortment between Sathuperi and Shamonda Viruses of the Genus Orthobunyavirus in Nature: Implications for Their Genetic Relationship to Schmallenberg Virus. Arch. Virol. 2012, 157, 1611–1616. [Google Scholar] [CrossRef] [PubMed]
- Collins, Á.B.; Doherty, M.L.; Barrett, D.J.; Mee, J.F. Schmallenberg Virus: A Systematic International Literature Review (2011-2019) from an Irish Perspective. Ir. Vet. J. 2019, 72, 9. [Google Scholar] [CrossRef] [PubMed]
- Goller, K.V.; Höper, D.; Schirrmeier, H.; Mettenleiter, T.C.; Beer, M. Schmallenberg Virus as Possible Ancestor of Shamonda Virus. Emerg. Infect. Dis. 2012, 18, 1644. [Google Scholar] [CrossRef]
- Tomochi, H.; Mekaru, Y.; Murota, K.; Konishi, M.; Ikeda, R.; Yanase, T. Emergence of a Natural Reassortant between Shamonda and Sathuperi Viruses of the Species Orthobunyavirus Schmallenbergense in Japan. Arch. Virol. 2025, 170, 44. [Google Scholar] [CrossRef]
- Zhang, X.M.; Herbst, W.; Kousoulas, K.G.; Storz, J. Biological and Genetic Characterization of a Hemagglutinating Coronavirus Isolated from a Diarrhoeic Child. J. Med. Virol. 2005, 44, 152. [Google Scholar] [CrossRef]
- Reid, A.H.; Fanning, T.G.; Hultin, J.V.; Taubenberger, J.K. Origin and Evolution of the 1918 “Spanish” Influenza Virus Hemagglutinin Gene. Proc. Natl. Acad. Sci. USA 1999, 96, 1651. [Google Scholar] [CrossRef]
- Meng, X.-J.; Halbur, P.G.; Shapiro, M.S.; Govindarajan, S.; Bruna, J.D.; Mushahwar, I.K.; Purcell, R.H.; Emerson, S.U. Genetic and Experimental Evidence for Cross-Species Infection by Swine Hepatitis E Virus. J. Virol. 1998, 72, 9714. [Google Scholar] [CrossRef]
- Chua, K.B. Nipah Virus: A Recently Emergent Deadly Paramyxovirus. Science 2000, 288, 1432–1435. [Google Scholar] [CrossRef]
- Ricklin, M.E.; García-Nicolás, O.; Brechbühl, D.; Python, S.; Zumkehr, B.; Nougairede, A.; Charrel, R.N.; Posthaus, H.; Oevermann, A.; Summerfield, A. Vector-Free Transmission and Persistence of Japanese Encephalitis Virus in Pigs. Nat. Commun. 2016, 7, 10832. [Google Scholar] [CrossRef]
- Agunos, A.; Pierson, F.W.; Lungu, B.; Dunn, P.A.; Tablante, N. Review of Nonfoodborne Zoonotic and Potentially Zoonotic Poultry Diseases. Avian Dis. 2016, 60, 553–575. [Google Scholar] [CrossRef]
- Hales, R.H.; Ostler, H.B. Newcastle Disease Conjunctivitis with Subepithelial Infiltrates. Br. J. Ophthalmol. 1973, 57, 694. [Google Scholar] [CrossRef] [PubMed]
- Bin, H.; Grossman, Z.; Pokamunski, S.; Malkinson, M.; Weiss, L.; Duvdevani, P.; Banet, C.; Weisman, Y.; Annis, E.; Gandaku, D.; et al. West Nile Fever in Israel 1999–2000. Ann. N. Y. Acad. Sci. 2001, 951, 127–142. [Google Scholar] [CrossRef] [PubMed]
- Field, H.E. Hendra Virus Ecology and Transmission. Curr. Opin. Virol. 2016, 16, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Oreshkova, N.; Molenaar, R.J.; Vreman, S.; Harders, F.; Oude Munnink, B.B.; Van Der Honing, R.W.H.; Gerhards, N.; Tolsma, P.; Bouwstra, R.; Sikkema, R.S.; et al. SARS-CoV-2 Infection in Farmed Minks, the Netherlands, April and May 2020. Eurosurveillance 2020, 25, 2001005, Erratum in Eurosurveillance 2021, 12. https://doi.org/10.2807/1560-7917.ES.2021.26.12.210325c.. [Google Scholar] [CrossRef]
- Krog, J.S.; Breum, S.; Jensen, T.H.; Larsen, L.E. Hepatitis E Virus Variant in Farmed Mink, Denmark. Emerg. Infect. Dis. 2013, 19, 2028. [Google Scholar] [CrossRef]
- Lindh, E.; Lounela, H.; Ikonen, N.; Kantala, T.; Savolainen-Kopra, C.; Kauppinen, A.; Österlund, P.; Kareinen, L.; Katz, A.; Nokireki, T.; et al. Highly Pathogenic Avian Influenza A(H5N1) Virus Infection on Multiple Fur Farms in the South and Central Ostrobothnia Regions of Finland, July 2023. Eurosurveillance 2023, 28, 2300400. [Google Scholar] [CrossRef]
- Wu, Z.; Lu, L.; Du, J.; Yang, L.; Ren, X.; Liu, B.; Jiang, J.; Yang, J.; Dong, J.; Sun, L.; et al. Comparative Analysis of Rodent and Small Mammal Viromes to Better Understand the Wildlife Origin of Emerging Infectious Diseases. Microbiome 2018, 6, 178. [Google Scholar] [CrossRef]
- Cui, X.; Fan, K.; Liang, X.; Gong, W.; Chen, W.; He, B.; Chen, X.; Wang, H.; Wang, X.; Zhang, P.; et al. Virus Diversity, Wildlife-Domestic Animal Circulation and Potential Zoonotic Viruses of Small Mammals, Pangolins and Zoo Animals. Nat. Commun. 2023, 14, 2488. [Google Scholar] [CrossRef]
- Zhang, N.; Hu, B.; Zhang, L.; Gan, M.; Ding, Q.; Pan, K.; Wei, J.; Xu, W.; Chen, D.; Zheng, S.; et al. Virome Landscape of Wild Rodents and Shrews in Central China. Microbiome 2025, 13, 63. [Google Scholar] [CrossRef] [PubMed]
- Bodewes, R.; Ruiz-Gonzalez, A.; Schapendonk, C.M.E.; Van Den Brand, J.M.A.; Osterhaus, A.D.M.E.; Smits, S.L. Viral Metagenomic Analysis of Feces of Wild Small Carnivores. Virol. J. 2014, 11, 89. [Google Scholar] [CrossRef] [PubMed]
- Leroy, E.M.; Kumulungui, B.; Pourrut, X.; Rouquet, P.; Hassanin, A.; Yaba, P.; Délicat, A.; Paweska, J.T.; Gonzalez, J.P.; Swanepoel, R. Fruit Bats as Reservoirs of Ebola Virus. Nature 2005, 438, 575–576. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Aravena, M.; McKee, C.; Gamble, A.; Lunn, T.; Morris, A.; Snedden, C.E.; Yinda, C.K.; Port, J.R.; Buchholz, D.W.; Yeo, Y.Y.; et al. Author Correction: Ecology, Evolution and Spillover of Coronaviruses from Bats. Nat. Rev. Microbiol. 2022, 20, 315, Correction in Nat. Rev. Microbiol. 2022, 20, 299 –314.https://doi.org/10.2807/1560-7917.ES.2021.26.12.210325c.. [Google Scholar] [CrossRef]
- Geldenhuys, M.; Mortlock, M.; Weyer, J.; Bezuidt, O.; Seamark, E.C.J.; Kearney, T.; Gleasner, C.; Erkkila, T.H.; Cui, H.; Markotter, W. A Metagenomic Viral Discovery Approach Identifies Potential Zoonotic and Novel Mammalian Viruses in Neoromicia Bats within South Africa. PLoS ONE 2018, 13, e0194527. [Google Scholar] [CrossRef]
- de Souza, P.J.; Fernandes, J.; Coelho, T.A.; Cosentino, M.; D’arc, M.; Alves, P.D.G.; Guterres, A.; Vilar, E.M.; de Lemos, E.R.S.; Cordeiro-Estrela, P.; et al. A Newly Bat-Borne Hantavirus Detected in Seba’s Short-Tailed Bats (Carollia perspicillata) in the Brazilian Atlantic Rainforest. Mem. Inst. Oswaldo Cruz 2024, 119, e240132. [Google Scholar] [CrossRef]
- Crook, J.M.; Murphy, I.; Carter, D.P.; Pullan, S.T.; Carroll, M.; Vipond, R.; Cunningham, A.A.; Bell, D. Metagenomic Identification of a New Sarbecovirus from Horseshoe Bats in Europe. Sci. Rep. 2021, 11, 14723. [Google Scholar] [CrossRef]
- Li, Y.; Altan, E.; Reyes, G.; Halstead, B.; Deng, X.; Delwart, E. Virome of Bat Guano from Nine Northern California Roosts. J. Virol. 2021, 95. [Google Scholar] [CrossRef]
- Hardmeier, I.; Aeberhard, N.; Qi, W.; Schoenbaechler, K.; Kraettli, H.; Hatt, J.M.; Fraefel, C.; Kubacki, J. Metagenomic Analysis of Fecal and Tissue Samples from 18 Endemic Bat Species in Switzerland Revealed a Diverse Virus Composition Including Potentially Zoonotic Viruses. PLoS ONE 2021, 16, e0252534. [Google Scholar] [CrossRef]
- Allen, T.; Murray, K.A.; Zambrana-Torrelio, C.; Morse, S.S.; Rondinini, C.; Di Marco, M.; Breit, N.; Olival, K.J.; Daszak, P. Global Hotspots and Correlates of Emerging Zoonotic Diseases. Nat. Commun. 2017, 8, 1124. [Google Scholar] [CrossRef]
- Meisner, J.; Baines, A.; Ngere, I.; Garcia, P.J.; Sa-Nguansilp, C.; Nguyen, N.; Niang, C.; Bardosh, K.; Nguyen, T.; Fenelon, H.; et al. Mapping Hotspots of Zoonotic Pathogen Emergence: An Integrated Model-Based and Participatory-Based Approach. Lancet Planet Health 2025, 9, e14–e22. [Google Scholar] [CrossRef]
- Desvars-Larrive, A.; Vogl, A.E.; Puspitarani, G.A.; Yang, L.; Joachim, A.; Käsbohrer, A. Author Correction: A One Health Framework for Exploring Zoonotic Interactions Demonstrated through a Case Study. Nat. Commun. 2024, 15, 6719, Erratum in Nat. Commun. 2024, 1. https://doi.org/10.1038/s41467-024-49967-7.. [Google Scholar] [CrossRef]
- Brunker, K.; Jaswant, G.; Thumbi, S.M.; Lushasi, K.; Lugelo, A.; Czupryna, A.M.; Ade, F.; Wambura, G.; Chuchu, V.; Steenson, R.; et al. Rapid In-Country Sequencing of Whole Virus Genomes to Inform Rabies Elimination Programmes. Wellcome Open Res. 2020, 5, 3. [Google Scholar] [CrossRef]
- Huaman, C.; Paskey, A.C.; Clouse, C.; Feasley, A.; Rader, M.; Rice, G.K.; Luquette, A.E.; Fitzpatrick, M.C.; Drumm, H.M.; Yan, L.; et al. Genomic Surveillance of Rabies Virus in Georgian Canines. Viruses 2023, 15, 1797. [Google Scholar] [CrossRef]
- Peterson, B.; Barnes, A.N. Feline-Human Zoonosis Transmission in North Africa: A Systematic Review. Vector-Borne Zoonotic Dis. 2020, 20, 731–744. [Google Scholar] [CrossRef]
- Bonilla-Aldana, D.K.; Bonilla-Aldana, J.L.; Acosta-España, J.D.; Rodriguez-Morales, A.J. Highly Pathogenic Avian Influenza H5N1 in Cats (Felis catus): A Systematic Review and Meta-Analysis. Animals 2025, 15, 1441. [Google Scholar] [CrossRef] [PubMed]
- Doliff, R.; Martens, P. Cats and SARS-CoV-2: A Scoping Review. Animals 2022, 12, 1413. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.-C.; Pan, Y.-F.; Zhou, W.-D.; Liao, Y.-Q.; Peng, M.-W.; Luo, G.-Y.; Xin, G.-Y.; Peng, Y.-N.; An, T.; Li, B.; et al. Meta-Transcriptomic Analysis of Companion Animal Infectomes Reveals Their Diversity and Potential Roles in Animal and Human Disease. mSphere 2024, 9, e00439-24. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Tao, J.; Li, B.; Shen, X.; Cheng, J.; Liu, H. The Gut Viral Metagenome Analysis of Domestic Dogs Captures Snapshot of Viral Diversity and Potential Risk of Coronavirus. Front. Vet. Sci. 2021, 8, 695088. [Google Scholar] [CrossRef]
- Patiño, L.; Benítez, A.D.; Carrazco-Montalvo, A.; Regato-Arrata, M. Genomics for Arbovirus Surveillance: Considerations for Routine Use in Public Health Laboratories. Viruses 2024, 16, 1242. [Google Scholar] [CrossRef]
- Chen, S.; Fang, Y.; Fujita, R.; Khater, E.I.M.; Li, Y.; Wang, W.; Qian, P.; Huang, L.; Guo, Z.; Zhang, Y.; et al. An Exploration of the Viral Coverage of Mosquito Viromes Using Meta-Viromic Sequencing: A Systematic Review and Meta-Analysis. Microorganisms 2024, 12, 1899. [Google Scholar] [CrossRef]
- Laiton-Donato, K.; Guzmán-Cardozo, C.; Peláez-Carvajal, D.; Ajami, N.J.; Navas, M.C.; Parra-Henao, G.; Usme-Ciro, J.A. Evolution and Emergence of Mosquito-Borne Viruses of Medical Importance: Towards a Routine Metagenomic Surveillance Approach. J. Trop. Ecol. 2023, 39, e13. [Google Scholar] [CrossRef]
- Taylor-Robinson, A.; Liang, Y.; Cintra, A.M.; Mayumi Noda-Nicolau, N.; Leite De Oliveira Soman, M.; Henrique De Andrade Affonso, P.; Valente, G.T.; Tommasini Grotto, R.M. The Main Arboviruses and Virus Detection Methods in Vectors: Current Approaches and Future Perspectives. Pathogens 2025, 14, 416. [Google Scholar] [CrossRef] [PubMed]
- Castilho de Arruda, L.D.; Giovanetti, M.; Fonseca, V.; Zardin, M.C.S.U.; Lichs, G.G.d.C.; Asato, S.; Esposito, A.O.P.; Tokeshi Müller, M.; Xavier, J.; Fritsch, H.; et al. Dengue Fever Surveillance in Mato Grosso Do Sul: Insights from Genomic Analysis and Implications for Public Health Strategies. Viruses 2023, 15, 1790. [Google Scholar] [CrossRef] [PubMed]
- Carrazco-Montalvo, A.; Gutiérrez-Pallo, D.; Arévalo, V.; Ponce, P.; Rodríguez-Polit, C.; Echeverría-Garcés, G.; Coloma, J.; Nipaz, V.; Cevallos, V. Entomo-Virological Surveillance and Genomic Insights into DENV-2 Genotype III Circulation in Rural Esmeraldas, Ecuador. Pathogens 2025, 14, 541. [Google Scholar] [CrossRef] [PubMed]
- Neto, Z.; Martinez, P.A.; Hill, S.C.; Jandondo, D.; Thézé, J.; Mirandela, M.; Aguiar, R.S.; Xavier, J.; Sebastião, C.D.S.; Cândido, A.L.M.; et al. Molecular and Genomic Investigation of an Urban Outbreak of Dengue Virus Serotype 2 in Angola, 2017–2019. PLoS Negl. Trop. Dis. 2022, 16, e0010255. [Google Scholar] [CrossRef]
- de Jesus, J.G.; Dutra, K.R.; Sales, F.C.d.S.; Claro, I.M.; Terzian, A.C.; Candido, D.d.S.; Hill, S.C.; Thézé, J.; Torres, C.; D’agostini, T.L.; et al. Genomic Detection of a Virus Lineage Replacement Event of Dengue Virus Serotype 2 in Brazil, 2019. Mem. Inst. Oswaldo Cruz 2020, 115, e190423. [Google Scholar] [CrossRef]
- Wang, Y.; Lin, X.; Li, C.; Liu, G.; Wang, S.; Chen, M.; Wei, X.; Wen, H.; Tao, Z.; Xu, Y. Metagenomic Sequencing Reveals Viral Diversity of Mosquitoes from Shandong Province, China. Microbiol. Spectr. 2024, 12, e03932-23. [Google Scholar] [CrossRef]
- Thannesberger, J.; Rascovan, N.; Eisenmann, A.; Klymiuk, I.; Zittra, C.; Fuehrer, H.P.; Scantlebury-Manning, T.; Gittens-St.Hilaire, M.; Austin, S.; Landis, R.C.; et al. Viral Metagenomics Reveals the Presence of Novel Zika Virus Variants in Aedes Mosquitoes from Barbados. Parasit. Vectors 2021, 14, 343. [Google Scholar] [CrossRef]
- Batovska, J.; Mee, P.T.; Sawbridge, T.I.; Rodoni, B.C.; Lynch, S.E. Enhanced Arbovirus Surveillance with High-Throughput Metatranscriptomic Processing of Field-Collected Mosquitoes. Viruses 2022, 14, 2759. [Google Scholar] [CrossRef]
- Mirza, J.D.; Guimarães, L.d.O.; Wilkinson, S.; Rocha, E.C.; Bertanhe, M.; Helfstein, V.C.; De-Deus, J.T.; Claro, I.M.; Cumley, N.; Quick, J.; et al. Tracking Arboviruses, Their Transmission Vectors and Potential Hosts by Nanopore Sequencing of Mosquitoes. Microb. Genom. 2024, 10, 001184. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.F.; Zhao, H.; Gou, Q.Y.; Shi, P.B.; Tian, J.H.; Feng, Y.; Li, K.; Yang, W.H.; Wu, D.; Tang, G.; et al. Metagenomic Analysis of Individual Mosquito Viromes Reveals the Geographical Patterns and Drivers of Viral Diversity. Nat. Ecol. Evol. 2024, 8, 947–959. [Google Scholar] [CrossRef] [PubMed]
- Harrington, W.N.; Kackos, C.M.; Webby, R.J. The Evolution and Future of Influenza Pandemic Preparedness. Exp. Mol. Med. 2021, 53, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Brüssow, H. The Beginning and Ending of a Respiratory Viral Pandemic-Lessons from the Spanish Flu. Microb. Biotechnol. 2022, 15, 1301–1317. [Google Scholar] [CrossRef]
- Neumann, G.; Noda, T.; Kawaoka, Y. Emergence and Pandemic Potential of Swine-Origin H1N1 Influenza Virus. Nature 2009, 459, 931. [Google Scholar] [CrossRef]
- Hall, V.; Cardona, C.; Mendoza, K.; Torchetti, M.; Lantz, K.; Bueno, I.; Franzen-Klein, D. Surveillance for Highly Pathogenic Avian Influenza A (H5N1) in a Raptor Rehabilitation Center—2022. PLoS ONE 2024, 19, e0299330. [Google Scholar] [CrossRef]
- Stachler, E.; Gnirke, A.; McMahon, K.; Gomez, M.; Stenson, L.; Guevara-Reyes, C.; Knoll, H.; Hill, T.; Hill, S.; Messer, K.S.; et al. Establishing Methods to Monitor Influenza (A)H5N1 Virus in Dairy Cattle Milk, Massachusetts, USA. Emerg. Infect. Dis. 2025, 31, S70. [Google Scholar] [CrossRef]
- Trogu, T.; Bellini, S.; Canziani, S.; Carrera, M.; Chiapponi, C.; Chiari, M.; Farioli, M.; Fusaro, A.; Savegnago, E.; Nucci, A.; et al. Surveillance for Avian Influenza in Wild Birds in the Lombardy Region (Italy) in the Period 2022–2024. Viruses 2024, 16, 1668. [Google Scholar] [CrossRef]
- Tiwari, A.; Meriläinen, P.; Lindh, E.; Kitajima, M.; Österlund, P.; Ikonen, N.; Savolainen-Kopra, C.; Pitkänen, T. Avian Influenza Outbreaks: Human Infection Risks for Beach Users—One Health Concern and Environmental Surveillance Implications. Sci. Total Environ. 2024, 943, 173692. [Google Scholar] [CrossRef]
- Niu, Q.; Jiang, Z.; Wang, L.; Ji, X.; Baele, G.; Qin, Y.; Lin, L.; Lai, A.; Chen, Y.; Veit, M.; et al. Prevention and Control of Avian Influenza Virus: Recent Advances in Diagnostic Technologies and Surveillance Strategies. Nat. Commun. 2025, 16, 3558. [Google Scholar] [CrossRef]
- Louis, S.; Mark-Carew, M.; Biggerstaff, M.; Yoder, J.; Boehm, A.B.; Wolfe, M.K.; Flood, M.; Peters, S.; Stobierski, M.G.; Coyle, J.; et al. Wastewater Surveillance for Influenza A Virus and H5 Subtype Concurrent with the Highly Pathogenic Avian Influenza A(H5N1) Virus Outbreak in Cattle and Poultry and Associated Human Cases—United States, May 12–July 13, 2024. MMWR Morb. Mortal. Wkly. Rep. 2024, 73, 804–809. [Google Scholar] [CrossRef]
- Hay, A.J.; McCauley, J.W. The WHO Global Influenza Surveillance and Response System (GISRS)—A Future Perspective. Influenza Other Respir. Viruses 2018, 12, 551–557. [Google Scholar] [CrossRef]
- Dormitzer, P.R.; Suphaphiphat, P.; Gibson, D.G.; Wentworth, D.E.; Stockwell, T.B.; Algire, M.A.; Alperovich, N.; Barro, M.; Brown, D.M.; Craig, S.; et al. Synthetic Generation of Influenza Vaccine Viruses for Rapid Response to Pandemics. Sci. Transl. Med. 2013, 5, 185ra68. [Google Scholar] [CrossRef]
- Lewandowski, K.; Xu, Y.; Pullan, S.T.; Lumley, S.F.; Foster, D.; Sanderson, N.; Vaughan, A.; Morgan, M.; Bright, N.; Kavanagh, J.; et al. Metagenomic Nanopore Sequencing of Influenza Virus Direct from Clinical Respiratory Samples. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef] [PubMed]
- Leguia, M.; Garcia-Glaessner, A.; Muñoz-Saavedra, B.; Juarez, D.; Barrera, P.; Calvo-Mac, C.; Jara, J.; Silva, W.; Ploog, K.; Amaro, L.; et al. Highly Pathogenic Avian Influenza A (H5N1) in Marine Mammals and Seabirds in Peru. Nat. Commun. 2023, 14, 5489. [Google Scholar] [CrossRef] [PubMed]
- Koopmans, M.P.G.; Barton Behravesh, C.; Cunningham, A.A.; Adisasmito, W.B.; Almuhairi, S.; Bilivogui, P.; Bukachi, S.A.; Casas, N.; Cediel Becerra, N.; Charron, D.F.; et al. The Panzootic Spread of Highly Pathogenic Avian Influenza H5N1 Sublineage 2.3.4.4b: A Critical Appraisal of One Health Preparedness and Prevention. Lancet Infect. Dis. 2024, 24, e774–e781. [Google Scholar] [CrossRef] [PubMed]
- Crits-Christoph, A.; Kantor, R.S.; Olm, M.R.; Whitney, O.N.; Al-Shayeb, B.; Lou, Y.C.; Flamholz, A.; Kennedy, L.C.; Greenwald, H.; Hinkle, A.; et al. Genome Sequencing of Sewage Detects Regionally Prevalent SARS-CoV-2 Variants. mBio 2021, 12, e02703-20. [Google Scholar] [CrossRef]
- Tisza, M.; Javornik Cregeen, S.; Avadhanula, V.; Zhang, P.; Ayvaz, T.; Feliz, K.; Hoffman, K.L.; Clark, J.R.; Terwilliger, A.; Ross, M.C.; et al. Wastewater Sequencing Reveals Community and Variant Dynamics of the Collective Human Virome. Nat. Commun. 2023, 14, 6878. [Google Scholar] [CrossRef]
- Karatas, M.; Bloemen, M.; Swinnen, J.; Roukaerts, I.; Van Gucht, S.; Van Ranst, M.; Wollants, E.; Matthijnssens, J. Untapped Potential of Wastewater for Animal and Potentially Zoonotic Virus Surveillance: Pilot Study to Detect Non-Human Animal Viruses in Urban Settings. Environ. Int. 2025, 199, 109500. [Google Scholar] [CrossRef]
- Zuckerman, N.S.; Bucris, E.; Morad-Eliyahu, H.; Weiss, L.; Vasserman, R.; Fratty, I.S.; Aguvaev, I.; Cohen-Said, Z.; Matar, R.; Erster, O.; et al. Environmental Surveillance of a Circulating Vaccine-Derived Poliovirus Type 2 Outbreak in Israel between 2022 and 2023: A Genomic Epidemiology Study. Lancet Microbe 2024, 5, 100893. [Google Scholar] [CrossRef]
- Hill, M.; Pollard, A.J. Detection of Poliovirus in London Highlights the Value of Sewage Surveillance. Lancet 2022, 400, 1491–1492. [Google Scholar] [CrossRef]
- Yek, C.; Pacheco, A.R.; Vanaerschot, M.; Bohl, J.A.; Fahsbender, E.; Aranda-Díaz, A.; Lay, S.; Chea, S.; Oum, M.H.; Lon, C.; et al. Metagenomic Pathogen Sequencing in Resource-Scarce Settings: Lessons Learned and the Road Ahead. Front. Epidemiol. 2022, 2, 926695. [Google Scholar] [CrossRef]
- Pronyk, P.M.; de Alwis, R.; Rockett, R.; Basile, K.; Boucher, Y.F.; Pang, V.; Sessions, O.; Getchell, M.; Golubchik, T.; Lam, C.; et al. Advancing Pathogen Genomics in Resource-Limited Settings. Cell Genom. 2023, 3, 100443. [Google Scholar] [CrossRef]
- Marais, G.; Hardie, D.; Brink, A. A Case for Investment in Clinical Metagenomics in Low-Income and Middle-Income Countries. Lancet Microbe 2023, 4, e192–e199. [Google Scholar] [CrossRef]
- Slavov, S.N. Routine Detection of Viruses Through Metagenomics: Where Do We Stand? Am. J. Trop. Med. Hyg. 2025, 112, 479–480. [Google Scholar] [CrossRef]
- Parties to the Nagoya Protocol. Available online: https://www.cbd.int/abs/nagoya-protocol/signatories (accessed on 24 September 2025).
- Directive-92/43-EN-Habitats Directive-EUR-Lex. Available online: https://eur-lex.europa.eu/eli/dir/1992/43/oj/eng (accessed on 24 September 2025).
- Mammola, S.; Riccardi, N.; Prié, V.; Correia, R.; Cardoso, P.; Lopes-Lima, M.; Sousa, R. Towards a Taxonomically Unbiased European Union Biodiversity Strategy for 2030. Proc. R. Soc. B 2020, 287, 20202166. [Google Scholar] [CrossRef]
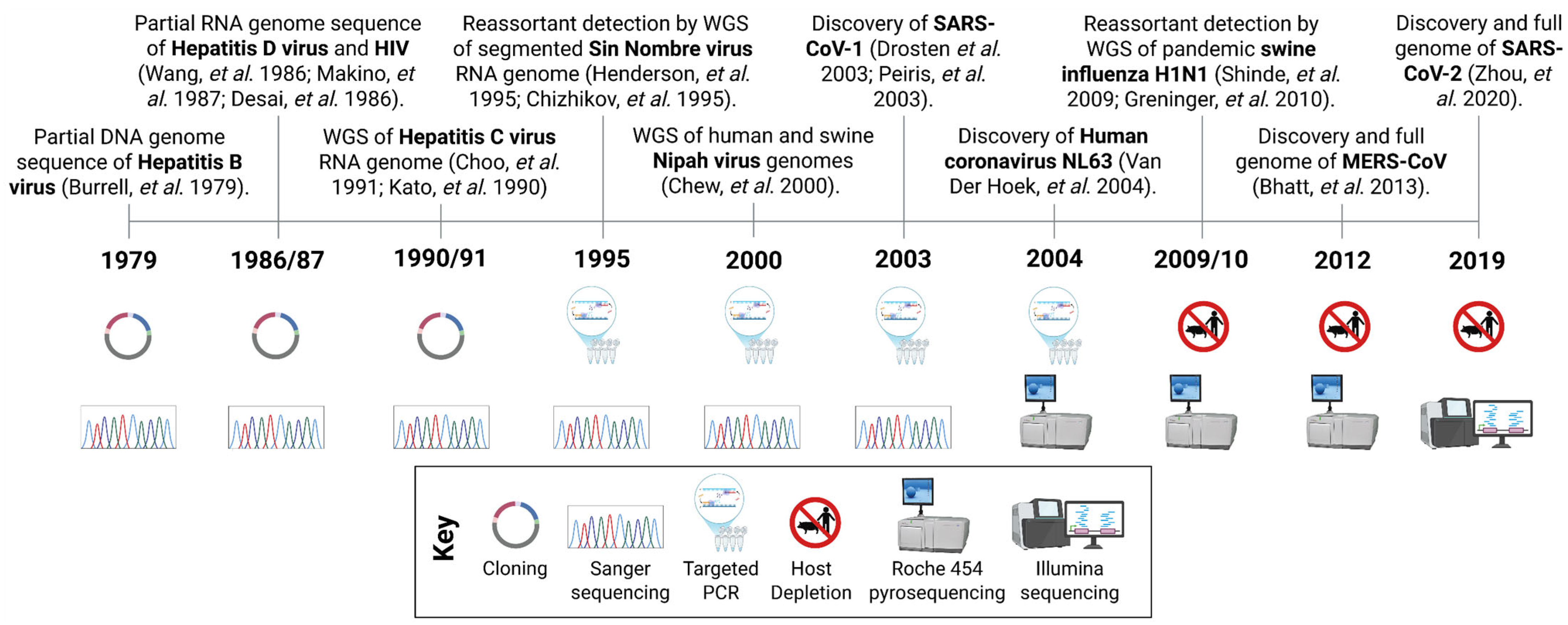

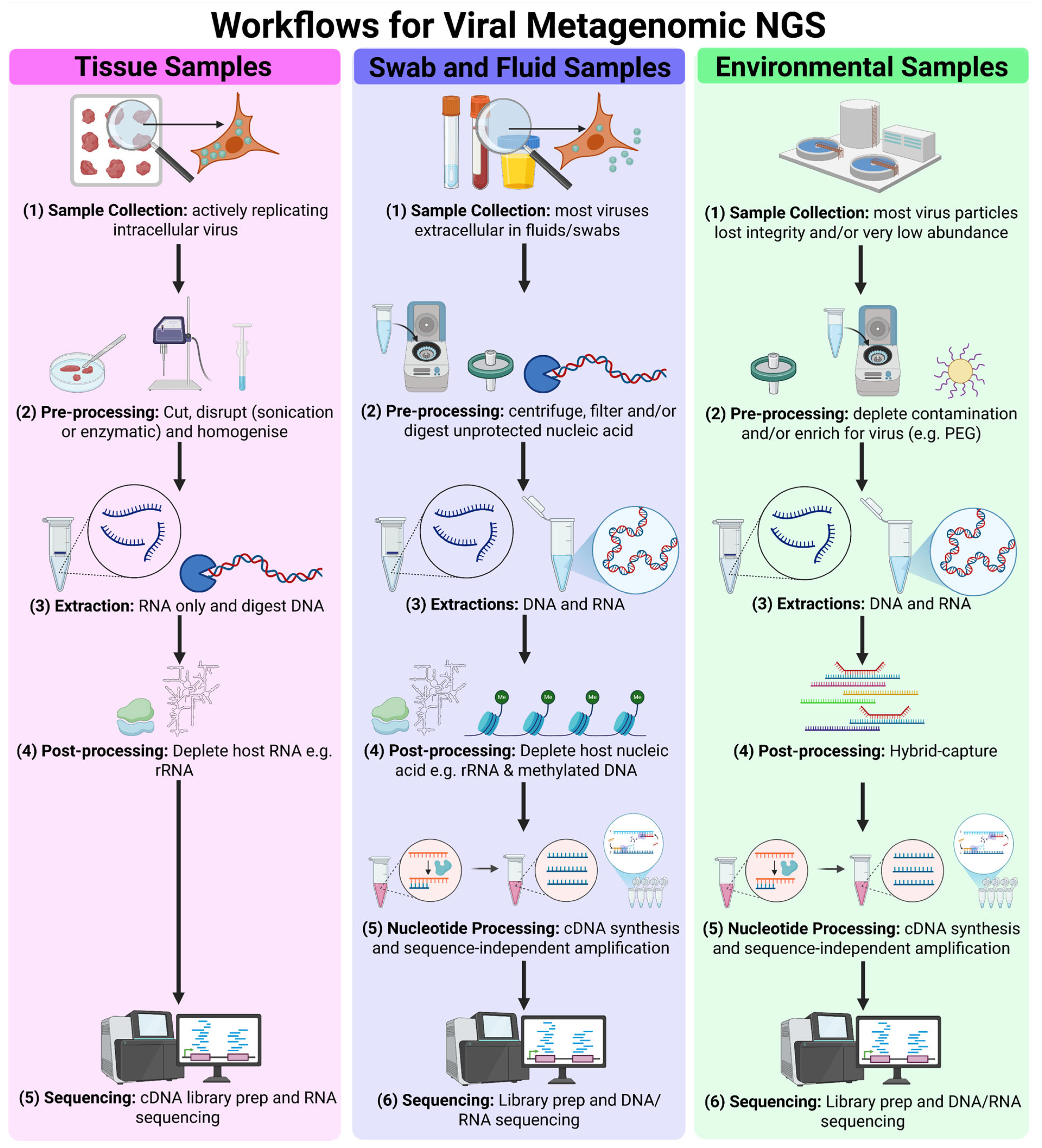
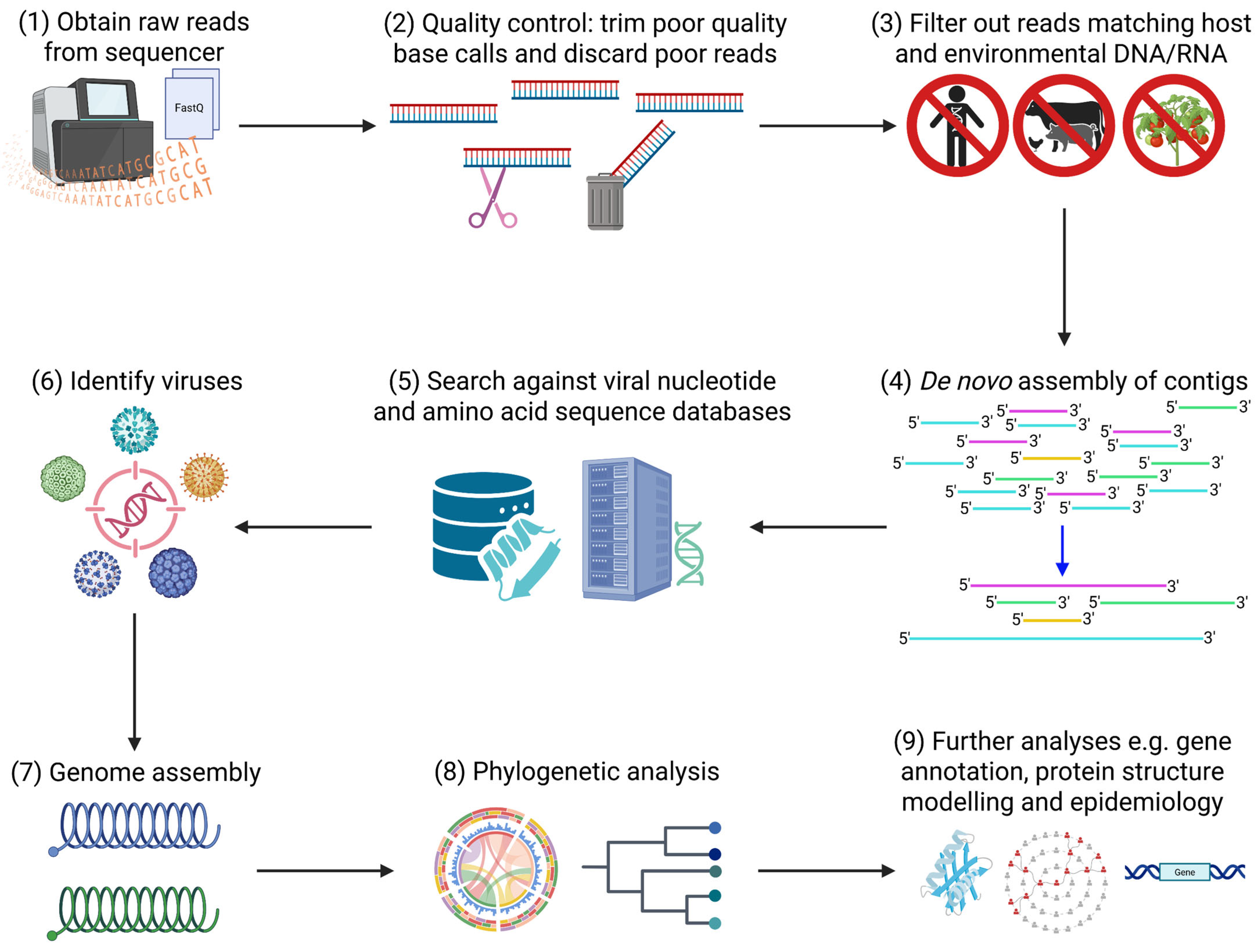
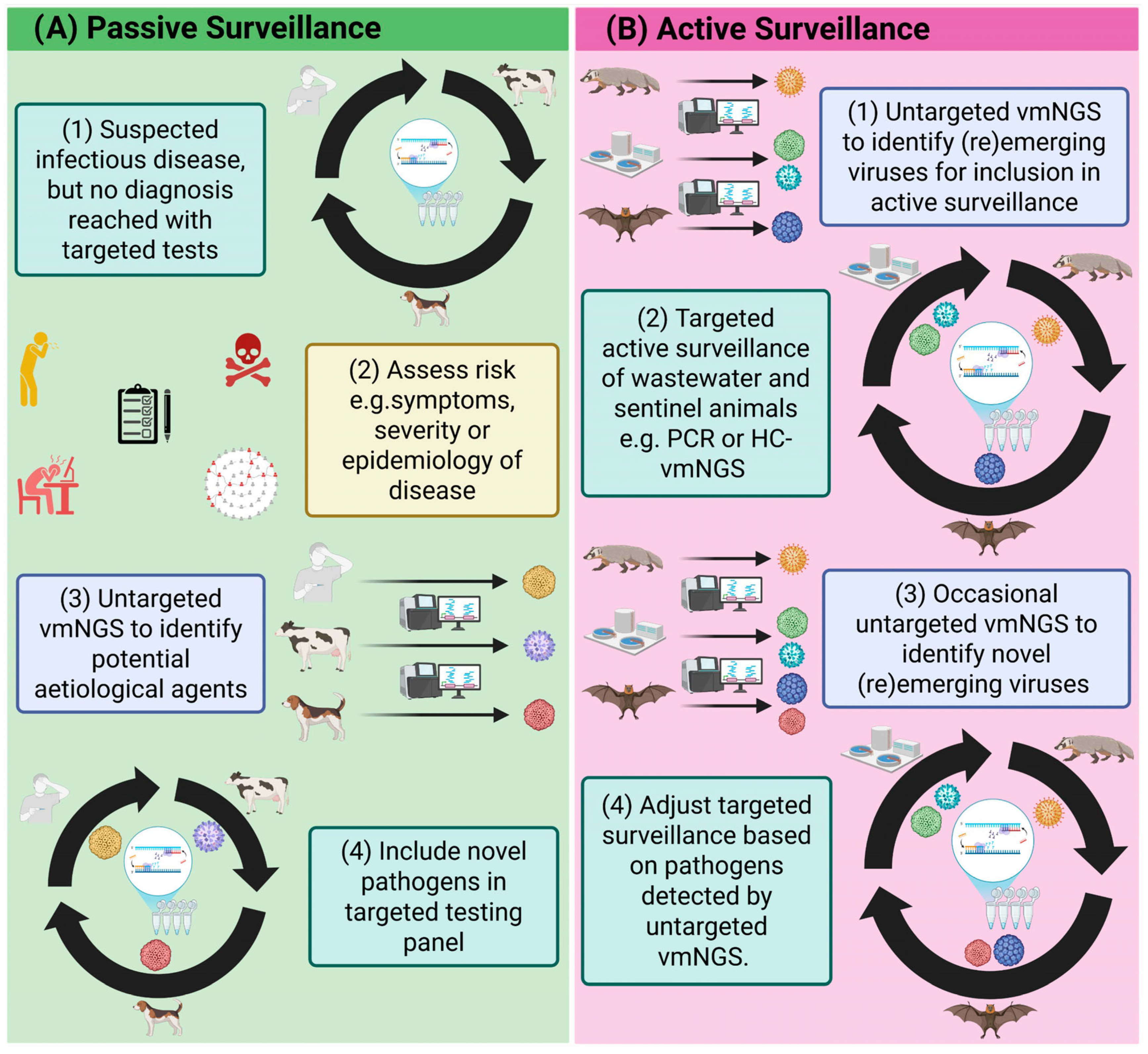
| Generation | First (Sanger) | Second (Illumina) | Third (ONT) |
|---|---|---|---|
| Other Platforms | Maxam-Gilbert | Roche 454, Ion Torrent, SOLiD | PacBio |
| Cost per Kb ($) 1 | 500–1000 | 0.01–0.10 | 0.10–10.00 |
| Error Rate (%) | 0.001 | 0.1–1.0 | 1–15 |
| Output (bases per run) | 1000 bp | 60 Gb–6 Tb | 10 Gb–10 Tb |
| Read Length | 1000 bp | 50–300 bp | Up to 1+ Mb |
| Requirement for PCR | Yes (sequence-dependent to enrich target) | Yes (sequence-independent for cluster generation on flow cell, can also be used in library preparation and to enrich targets) | No (can be used to enrich targets) |
| Preparation Methods | PCR or clonal amplification of target | Generate library: fragmentation, ds-cDNA synthesis, adapter/barcode ligation and purification | Generate library: adapter/barcode ligation and purification |
| Data Storage Requirements | Low | High | High |
| Portable | No | No | Yes |
| Suitable for Metagenomics | No | Yes (including degraded samples) | Yes (unsuitable for degraded samples) |
| Other Applications | Genotyping and Targeted Sequencing | WGS, SNP Variant Calling and Transcriptomics | WGS, Splice Variant Detection, Real-time Sequencing, Direct RNA Sequencing, Epigenetic Modification Detection and Adaptive Sequencing 2 |
| Main Strengths | High sensitivity (for target only), high specificity and high accuracy | High accuracy, high sensitivity, high depth, high throughput and suitability for degraded samples | Long read length, high throughput, high sensitivity and high coverage |
| Main Weakness | Low throughput | Short read-length | Poor accuracy |
| Virus | Original Location | New Location | Year | Cause | Refs. |
|---|---|---|---|---|---|
| Mpox IIb | Nigeria | Worldwide | 2022 | See main text | [124] |
| Mpox Ib | DRC | Worldwide | 2023 | See main text | [125] |
| Zika virus | Africa, Asia and French Polynesia | Brazil | 2015 | See main text | [126] |
| Oropouche virus | Brazil, Caribbean, Peru and Panama | Ecuador | 2016 | Unknown, PCR recognised mutated region | [122,123] |
| St Louis Encephalitis virus | Argentina | USA | 2015 | Migratory birds | [127] |
| Ebola virus | DRC | Guinea, Liberia, Sierra Leone | 2014 | Zoonotic spillover, probably bats | [128] |
| Surveillance Objective | Sample Type | NGS Methods |
|---|---|---|
| Active or passive surveillance for (re)emerging respiratory disease agents | Nasopharyngeal swabs, lung biopsy (postmortem only) | Untargeted 1 or semi-targeted vmNGS |
| Active or passive surveillance for (re)emerging gastrointestinal disease agents. | Cloacal swabs, faecal swabs/samples, gut biopsy | Untargeted 1 or semi-targeted vmNGS |
| Active or passive surveillance for neurological disease agents | Cerebral spinal fluid (CSF) or brain biopsy (postmortem) | Untargeted 1 or semi-targeted vmNGS |
| Surveillance of undifferentiated illnesses or sentinel animals | Spleen biopsy or blood | Untargeted 1 or semi-targeted vmNGS |
| Environmental surveillance, early outbreak detection | Wastewater, farm effluents, soils | Semi-targeted vmNGS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Russell, T.; Formiconi, E.; Casey, M.; McElroy, M.; Mallon, P.W.G.; Gautier, V.W. Viral Metagenomic Next-Generation Sequencing for One Health Discovery and Surveillance of (Re)Emerging Viruses: A Deep Review. Int. J. Mol. Sci. 2025, 26, 9831. https://doi.org/10.3390/ijms26199831
Russell T, Formiconi E, Casey M, McElroy M, Mallon PWG, Gautier VW. Viral Metagenomic Next-Generation Sequencing for One Health Discovery and Surveillance of (Re)Emerging Viruses: A Deep Review. International Journal of Molecular Sciences. 2025; 26(19):9831. https://doi.org/10.3390/ijms26199831
Chicago/Turabian StyleRussell, Tristan, Elisa Formiconi, Mícheál Casey, Maíre McElroy, Patrick W. G. Mallon, and Virginie W. Gautier. 2025. "Viral Metagenomic Next-Generation Sequencing for One Health Discovery and Surveillance of (Re)Emerging Viruses: A Deep Review" International Journal of Molecular Sciences 26, no. 19: 9831. https://doi.org/10.3390/ijms26199831
APA StyleRussell, T., Formiconi, E., Casey, M., McElroy, M., Mallon, P. W. G., & Gautier, V. W. (2025). Viral Metagenomic Next-Generation Sequencing for One Health Discovery and Surveillance of (Re)Emerging Viruses: A Deep Review. International Journal of Molecular Sciences, 26(19), 9831. https://doi.org/10.3390/ijms26199831






