The NME7 Gene Is Involved in the Kinetics of Glucose Processing
Abstract
1. Introduction
1.1. Factors Disrupting Glucose Homeostasis
1.2. Oral Glucose Tolerance Test Course
1.3. The NME7 Gene
1.4. Aims
1.5. Study Subjects
2. Results
2.1. Metabolic Profile of the Cohort
2.2. Representation of Individual Types of Glycemic Curves
2.3. Genetic Determination of Glycemic Curve Shapes
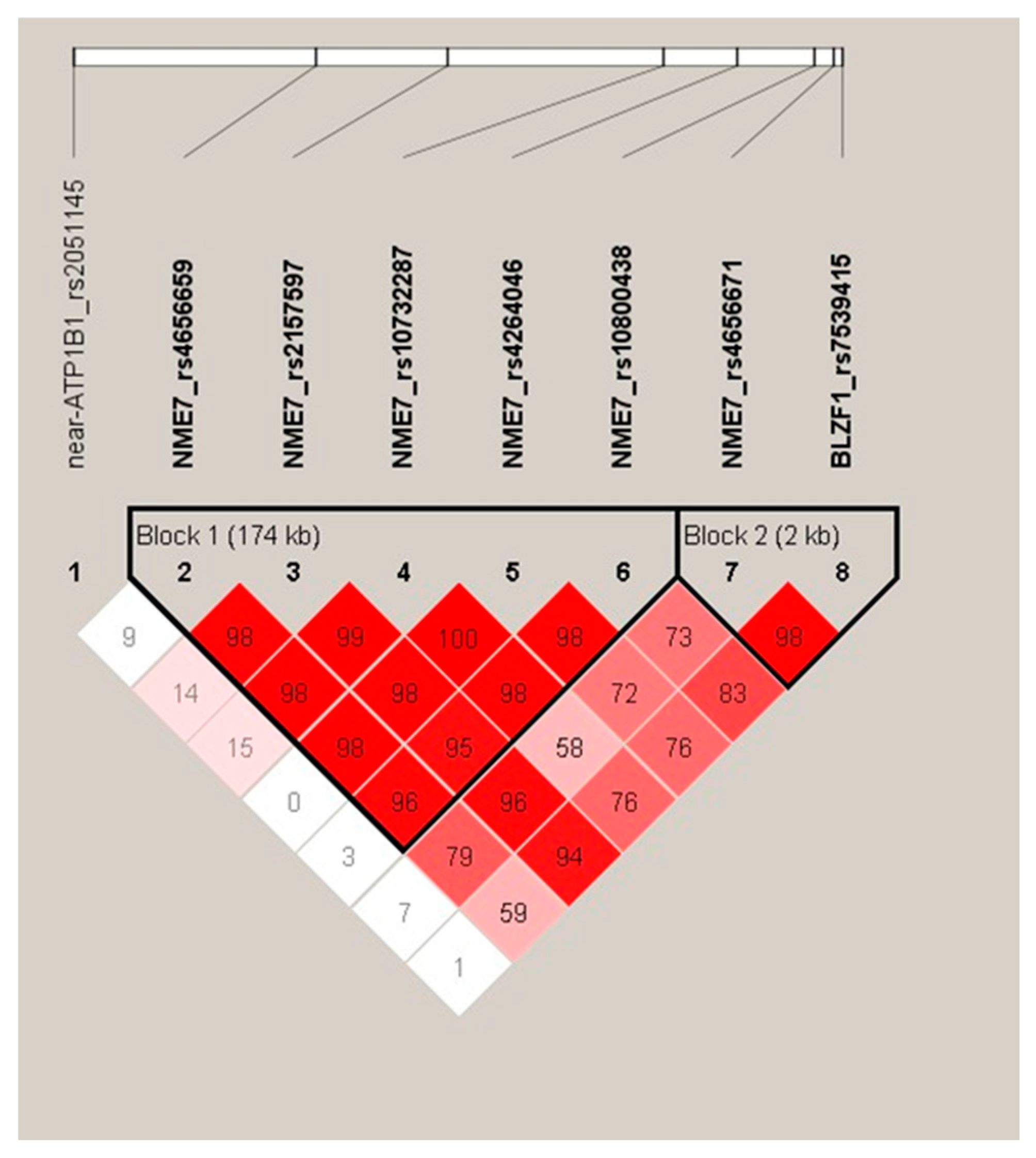
2.4. Haplotype Analysis
2.5. Haplotypes and Biochemical Parameters
3. Discussion
4. Materials and Methods
4.1. Anthropometric and Metabolic Characterization of the Subjects
4.2. Classification of the OGTT Curves
4.3. Molecular Genetic Analysis
4.4. Calculations and Statistical Evaluation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALT | alanine aminotransferase |
| AST | aspartate aminotransferase |
| ATP1B1 | sodium/potassium-transporting ATPase subunit beta-1 |
| ATP | adenosine triphosphate |
| AUCC-peptide | area under the C-peptide curve |
| AUCGlycemia | area under the glycemic curve |
| AUCInsulin | area under the insulinemic curve |
| BLZF1 | basic leucine zipper nuclear factor 1 |
| BMI | body mass index |
| df | degrees of freedom |
| DNA | deoxyribonucleic acid |
| fT3 | free triiodothyronine |
| fT4 | free thyroxine |
| GDM | gestational diabetes mellitus |
| GGT | γ-glutamyl transferase |
| HDL | high-density lipoprotein |
| HOMA B | homeostatic model assessment of beta cell function |
| HOMA IR | homeostatic model assessment of insulin resistance |
| IFG | impaired fasting glucose |
| IGI | insulinogenic index |
| IGT | impaired glucose tolerance |
| IR | insulin resistance |
| ISICOMP | insulin sensitivity composite index |
| LCL | lower confidence limit |
| LD | linkage disequilibrium |
| LDL | low-density lipoprotein |
| NME7 | non-metastatic cells 7 |
| OGIS | oral glucose insulin sensitivity index |
| OGTT | oral glucose tolerance test |
| PCOS | polycystic ovary syndrome |
| PREDIM | peripheral insulin sensitivity index |
| SNP | single nucleotide polymorphism |
| T2DM | type 2 diabetes mellitus |
| TAG | triacylglycerols |
| TSH | thyrotropin |
| UCL | upper confidence limit |
| WHR | waist-hip ratio |
References
- The World Obesity Federation Website. Available online: https://www.worldobesity.org/ (accessed on 16 May 2024).
- Institute of Health Information and Statistics of the Czech Republic. Available online: https://www.uzis.cz/ (accessed on 25 August 2025).
- Kajantie, E.; Strang-Karlsson, S.; Hovi, P.; Wehkalampi, K.; Lahti, J.; Kaseva, N.; Järvenpää, A.-L.; Räikkönen, K.; Eriksson, J.G.; Andersson, S. Insulin sensitivity and secretory response in adults born preterm: The Helsinki Study of Very Low Birth Weight Adults. J. Clin. Endocrinol. Metab. 2015, 100, 244–250. [Google Scholar] [CrossRef]
- Palatianou, M.E.; Simos, Y.V.; Andronikou, S.K.; Kiortsis, D.N. Long-term metabolic effects of high birth weight: A critical review of the literature. Horm. Metab. Res. 2014, 46, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Harder, T.; Rodekamp, E.; Schellong, K.; Dudenhausen, J.W.; Plageman, A. Birth weight and subsequent risk of type 2 diabetes: A meta-analysis. Am. J. Epidemiol. 2007, 165, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Vaag, A.; Brøns, C.; Gillberg, L.; Hansen, N.S.; Hjort, L.; Arora, G.P.; Thomas, N.; Broholm, C.; Ribel-Madsen, R.; Grunnet, L.G. Genetic, nongenetic and epigenetic risk determinants in developmental programming of type 2 diabetes. Acta Obstet. Gynecol. Scand. 2014, 93, 1099–1108. [Google Scholar] [CrossRef] [PubMed]
- Shojima, N.; Yamauchi, T. Progress in genetics of type 2 diabetes and diabetic complications. J. Diabetes Investig. 2023, 14, 503–515. [Google Scholar] [CrossRef]
- Hu, C.; Jia, W. Multi-omics profiling: The way towards precision medicine in metabolic diseases. J. Mol. Cell Biol. 2021, 13, 576–593. [Google Scholar] [CrossRef]
- Stumvoll, M.; Mitrakou, A.; Pimenta, W.; Jenssen, T.; Yki-Järvinen, H.; Van Haeften, T.; Renn, W.; Gerich, J. Use of the oral glucose tolerance test to assess insulin release and insulin sensitivity. Diabetes Care 2000, 23, 295–301. [Google Scholar] [CrossRef]
- Chung, S.T.; Ha, J.; Onuzuruike, A.U.; Kasturi, K.; Galvan-De La Cruz, M.; Bingham, B.A.; Baker, R.L.; Utumatwishima, J.N.; Mabundo, L.S.; Ricks, M.; et al. Time to glucose peak during an oral glucose tolerance test identifies prediabetes risk. Clin. Endocrinol. 2017, 87, 484–491. [Google Scholar] [CrossRef]
- Kim, J.Y.; Michaliszyn, S.F.; Nasr, A.; Lee, S.; Tfayli, H.; Hannon, T.; Hughan, K.S.; Bacha, F.; Arslanian, S. The Shape of the Glucose Response Curve During an Oral Glucose Tolerance Test Heralds Biomarkers of Type 2 Diabetes Risk in Obese Youth. Diabetes Care 2016, 39, 1431–1439. [Google Scholar] [CrossRef]
- Bervoets, L.; Mewis, A.; Massa, G. The shape of the plasma glucose curve during an oral glucose tolerance test as an indicator of Beta cell function and insulin sensitivity in end-pubertal obese girls. Horm. Metab. Res. 2015, 47, 445–451. [Google Scholar] [CrossRef]
- Tura, A.; Morbiducci, U.; Sbrignadello, S.; Winhofer, Y.; Pacini, G.; Kautzky-Willer, A. Shape of glucose, insulin, C-peptide curves during a 3-h oral glucose tolerance test: Any relationship with the degree of glucose tolerance? Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 300, R941–R948. [Google Scholar] [CrossRef]
- Vejrazkova, D.; Vankova, M.; Lukasova, P.; Hill, M.; Vcelak, J.; Tura, A.; Chocholova, D.; Bendlova, B. The Glycemic Curve during the Oral Glucose Tolerance Test: Is It Only Indicative of Glycoregulation? Biomedicines 2023, 11, 1278. [Google Scholar] [CrossRef] [PubMed]
- Engle, S.E.; Bansal, R.; Antonellis, P.J.; Berbari, N.F. Cilia signaling and obesity. Semin. Cell Dev. Biol. 2021, 110, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Mujahid, S.; Hunt, K.F.; Cheah, Y.S.; Forsythe, E.; Hazlehurst, J.M.; Sparks, K.; Mohammed, S.; Tomlinson, J.W.; Amiel, S.A.; Carroll, P.V.; et al. The Endocrine and Metabolic Characteristics of a Large Bardet-Biedl Syndrome Clinic Population. J. Clin. Endocrinol. Metab. 2018, 103, 1834–1841. [Google Scholar] [CrossRef] [PubMed]
- Collin, G.B.; Marshall, J.D.; Ikeda, A.; So, W.V.; Russell-Eggitt, I.; Maffei, P.; Beck, S.; Boerkoel, C.F.; Sicolo, N.; Martin, M.; et al. Mutations in ALMS1 cause obesity, type 2 diabetes and neurosensory degeneration in Alström syndrome. Nat. Genet. 2002, 31, 74–78. [Google Scholar] [CrossRef]
- Siljee, J.E.; Wang, Y.; Bernard, A.A.; Ersoy, B.A.; Zhang, S.; Marley, A.; Von Zastrow, M.; Reiter, J.F.; Vaisse, C. Subcellular localization of MC4R with ADCY3 at neuronal primary cilia underlies a common pathway for genetic predisposition to obesity. Nat. Genet. 2018, 50, 180–185. [Google Scholar] [CrossRef]
- Lee, E.Y.; Hughes, J.W. Rediscovering Primary Cilia in Pancreatic Islets. Diabetes Metab. J. 2023, 47, 454–469. [Google Scholar] [CrossRef]
- Idevall-Hagren, O.; Nilsson, C.I.; Sanchez, G. Keeping pace: The primary cilium as the conducting baton of the islet. Diabetologia 2024, 67, 773–782. [Google Scholar] [CrossRef]
- Starks, R.D.; Beyer, A.M.; Guo, D.F.; Boland, L.; Zhang, Q.; Sheffield, V.C.; Rahmouni, K. Regulation of Insulin Receptor Trafficking by Bardet Biedl Syndrome Proteins. PLoS Genet. 2015, 11, e1005311. [Google Scholar] [CrossRef]
- Cho, J.H.; Li, Z.A.; Zhu, L.; Muegge, B.D.; Roseman, H.F.; Lee, E.Y.; Utterback, T.; Woodhams, L.G.; Bayly, P.V.; Hughes, J.W. Islet primary cilia motility controls insulin secretion. Sci. Adv. 2022, 8, eabq8486. [Google Scholar] [CrossRef]
- Walker, J.T.; Saunders, D.C.; Rai, V.; Chen, H.H.; Orchard, P.; Dai, C.; Pettway, Y.D.; Hopkirk, A.L.; Reihsmann, C.V.; Tao, Y.; et al. Genetic risk converges on regulatory networks mediating early type 2 diabetes. Nature 2023, 624, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Šedová, L.; Buková, I.; Bažantová, P.; Petrezsélyová, S.; Prochazka, J.; Školníková, E.; Zudová, D.; Včelák, J.; Makovický, P.; Bendlová, B.; et al. Semi-Lethal Primary Ciliary Dyskinesia in Rats Lacking the Nme7 Gene. Int. J. Mol. Sci. 2021, 22, 3810. [Google Scholar] [CrossRef] [PubMed]
- Šedová, L.; Prochazka, J.; Zudová, D.; Bendlová, B.; Včelák, J.; Sedlacek, R.; Šeda, O. Heterozygous Nme7 Mutation Affects Glucose Tolerance in Male Rats. Genes 2021, 12, 1087. [Google Scholar] [CrossRef] [PubMed]
- Ferrannini, E.; Škrha, J.; Eizirik, D.L.; Gale, E.; Jörgens, V. PS4 Genome-wide association scans and bioinformatic tools for identifying candidate pathways. Abstracts of the 43rd European Association for the Study of Diabetes (EASD) annual meeting, 18–21 September 2007, Amsterdam, The Netherlands. Diabetologia 2007, 50, S128. [Google Scholar] [CrossRef]
- Piccinini, F.; Dalla Man, C.; Vella, A.; Cobelli, C. A Model for the Estimation of Hepatic Insulin Extraction After a Meal. IEEE Trans. Biomed. Eng. 2016, 63, 1925–1932. [Google Scholar] [CrossRef]
- Tura, A.; Chemello, G.; Szendroedi, J.; Göbl, C.; Færch, K.; Vrbíková, J.; Pacini, G.; Ferrannini, E.; Roden, M. Prediction of clamp-derived insulin sensitivity from the oral glucose insulin sensitivity index. Diabetologia 2018, 61, 1135–1141. [Google Scholar] [CrossRef]
- Retnakaran, R.; Qi, Y.; Goran, M.I.; Hamilton, J.K. Evaluation of proposed oral disposition index measures in relation to the actual disposition index. Diabet. Med. 2009, 26, 1198–1203. [Google Scholar] [CrossRef]
- Tura, A.; Kautzky-Willer, A.; Pacini, G. Insulinogenic indices from insulin and C-peptide: Comparison of beta-cell function from OGTT and IVGTT. Diabetes Res. Clin. Pract. 2006, 72, 298–301. [Google Scholar] [CrossRef]
- Pacini, G.; Mari, A. Methods for clinical assessment of insulin sensitivity and beta-cell function. Best Pract. Res. Clin. Endocrinol. Metab. 2003, 17, 305–322. [Google Scholar] [CrossRef]
- Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults. Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III). JAMA 2001, 285, 2486–2497. [Google Scholar] [CrossRef]
- Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil. Steril. 2004, 81, 19–25. [Google Scholar] [CrossRef]
- Tura, A.; Ludvik, B.; Nolan, J.J.; Pacini, G.; Thomaseth, K. Insulin and C-peptide secretion and kinetics in humans: Direct and model-based measurements during OGTT. Am. J. Physiol. Endocrinol. Metab. 2001, 281, E966–E974. [Google Scholar] [CrossRef]
- Stephens, M.; Scheet, P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am. J. Hum. Genet. 2005, 76, 449–462. [Google Scholar] [CrossRef]
- Stephens, M.; Donnelly, P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 2003, 73, 1162–1169. [Google Scholar] [CrossRef]
- Stephens, M.; Smith, N.J.; Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001, 68, 978–989. [Google Scholar] [CrossRef]

| Parameter (Units) | Median [95% LCL; 95% UCL] | Reference Limits |
|---|---|---|
| number | 1262 | N/A |
| age (years) | 33.3 [32.6; 34.0] | N/A |
| systolic blood pressure (mmHg) | 115.0 [114.0; 115.0] | 100–140 |
| diastolic blood pressure (mmHg) | 72.0 [71.0; 73.0] | 65–90 |
| BMI (kg/m2) | 23.9 [23.5; 24.3] | 18.5–24.9 |
| WHR women | 0.764 [0.759; 0.768] | <0.85 |
| WHR men | 0.864 [0.849; 0.877] | <0.95 |
| glucose metabolism | ||
| basal glycemia (mmol/L) | 4.8 [4.7; 4.8] | 3.9–5.5 |
| AUCGlycemia(mmol × min/L) | 1046 [1031; 1059] | n.s. * |
| basal insulinemia (mIU/L) | 6.16 [5.90; 6.30] | n.s. * |
| AUCInsulin (pmol × min/L) | 33,885 [32,598; 35,091] | n.s. * |
| basal C-peptide (nmol/L) | 0.59 [0.58; 0.60] | n.s. * |
| AUCC-peptide (pmol × min/L) | 3.6 × 105 [3.5 × 105; 3.7 × 105] | n.s. * |
| hepatic insulin extraction (%) | 68.1 [67.5; 68.8] | n.s. * |
| insulin sensitivity/resistance | ||
| HOMA IR | 1.30 [1.26; 1.36] | n.s. * |
| OGIS 120 min (mL/min/m2) | 457.7 [453.5; 462.1] | n.s. * |
| ISICOMP ([(mg/dl)2(μU/)mL2]−1/2) | 8.48 [8.18; 8.81] | n.s. * |
| MCRest (mL/min/kg) | 9.87 [9.75; 9.99] | n.s. * |
| Si(oral) ((mL/min/kg)/(μU/)mL) | 0.15 [0.15; 0.16] | n.s. * |
| PREDIM (mg/min/kg) | 6.97 [6.83; 7.17] | n.s. * |
| beta cell function | ||
| HOMA B (mIU/mmol) | 103.5 [98.8; 106.2] | n.s. * |
| IGI (pmol/mmol) | 88.5 [84.2; 95.1] | n.s. * |
| Ins0/Glc0 (pmol/mmol) | 7.68 [7.40; 8.00] | n.s. * |
| AUCInsulin/AUCGlc (pmol/mmol) | 32.7 [31.9; 33.9] | n.s. * |
| IGI × ISICOMP | 264.9 [257.9; 270.2] | n.s. * |
| lipid spectrum | ||
| total cholesterol (mmol/L) | 4.59 [4.54; 4.65] | 2.9–5.0 |
| HDL cholesterol in women (mmol/L) | 1.57 [1.54; 1.60] | 1.2–2.7 |
| HDL cholesterol in men (mmol/L) | 1.26 [1.21; 1.32] | 1.0–2.1 |
| LDL cholesterol (mmol/L) | 2.56 [2.52; 2.63] | 1.2–3.0 |
| TAG (mmol/L) | 0.86 [0.82; 0.89] | 0.45–1.70 |
| liver enzymes | ||
| ALT (ukat/L) | 0.30 [0.30; 0.31] | 0.17–0.58 |
| AST (ukat/L) | 0.35 [0.34; 0.36] | 0.17–0.60 |
| GGT (ukat/L) | 0.23 [0.22; 0.24] | 0.10–0.70 |
| thyroid hormones | ||
| TSH (mIU/L) | 2.25 [2.17; 2.33] | 0.27–4.20 |
| fT3 (pmol/L) | 4.90 [4.85; 4.97] | 3.10–6.80 |
| fT4 (pmol/L) | 15.2 [15.0; 15.3] | 12.0–22.0 |
| n = 1262 | Monophasic n = 633 | Biphasic n = 221 | Triphasic n = 351 | Multiphasic n = 57 | p-Level |
|---|---|---|---|---|---|
| age (years) | 34.1 [33.2; 35.1] | 31.4 [28.3; 32.9] | 34.0 [32.4; 34.7] | 28.7 [26.6; 30.6] | <0.01 |
| BMI (kg/m2) | 24.6 [24.1; 25.1] | 23.3 [22.6; 24.1] | 23.2 [22.7; 23.8] | 23.5 [22.2; 24.2] | <0.01 |
| NME7_rs4656659 | GENOTYPES (%) | ALLELES (%) | ||||
| TT | CT | CC | T | C | STATGenotype distribution: | |
| monophasic (n = 632) | 45 | 43 | 12 | 67 | 33 | Chi2 = 16.43 Power = 0.88 p-level = 0.012 |
| biphasic (n = 219) | 34 | 54 | 12 | 61 | 39 | |
| triphasic (n = 351) | 39 | 53 | 8 | 66 | 34 | |
| multiphasic (n = 56) | 48 | 40 | 12 | 68 | 32 | |
| NME7_rs2157597 | GENOTYPES (%) | ALLELES (%) | ||||
| CC | CT | TT | C | T | STATGenotype distribution: | |
| monophasic (n = 631) | 52 | 40 | 8 | 71 | 29 | Chi2 = 12.64 Power = 0.76 p-level = 0.049 |
| biphasic (n = 219) | 42 | 48 | 10 | 66 | 34 | |
| triphasic (n = 350) | 48 | 47 | 5 | 72 | 28 | |
| multiphasic (n = 57) | 52 | 41 | 7 | 73 | 27 | |
| NME7_rs10732287 | GENOTYPES (%) | ALLELES (%) | ||||
| CC | CT | TT | C | T | STATGenotype distribution: | |
| monophasic (n = 632) | 46 | 43 | 11 | 67 | 33 | Chi2 = 19.39 Power = 0.93 p-level = 0.003 |
| biphasic (n = 219) | 59 | 33 | 8 | 76 | 24 | |
| triphasic (n = 351) | 43 | 48 | 9 | 67 | 33 | |
| multiphasic (n = 57) | 37 | 48 | 15 | 61 | 39 | |
| NME7_rs4264046 | GENOTYPES (%) | ALLELES (%) | ||||
| CC | CT | TT | C | T | STATGenotype distribution: | |
| monophasic (n = 628) | 29 | 49 | 22 | 53 | 47 | Chi2 = 16.42 Power = 0.88 p-level = 0.012 |
| biphasic (n = 217) | 39 | 44 | 17 | 61 | 39 | |
| triphasic (n = 348) | 28 | 56 | 16 | 56 | 44 | |
| multiphasic (n = 57) | 25 | 53 | 22 | 52 | 48 | |
| NME7_rs10800438 | GENOTYPES (%) | ALLELES (%) | ||||
| GG | GT | TT | G | T | STATGenotype distribution: | |
| monophasic (n = 631) | 31 | 49 | 20 | 56 | 44 | Chi2 = 14.21 Power = 0.82 p-level = 0.027 |
| biphasic (n = 219) | 42 | 43 | 15 | 64 | 36 | |
| triphasic (n = 351) | 32 | 53 | 15 | 59 | 41 | |
| multiphasic (n = 56) | 24 | 57 | 19 | 53 | 47 | |
| Haplotype | Number of Carriers | ||
|---|---|---|---|
| Women (n = 1033) | Men (n = 226) | All (n = 1259) | |
| 1. TCTTT | 550 (53.2%) | 107 (47.3%) | 657 (52.2%) |
| 2. CTCCG | 509 (49.3%) | 117 (51.8%) | 626 (49.7%) |
| 3. TCCCG | 377 (36.5%) | 91 (40.3%) | 468 (37.2%) |
| 4. TCCTT | 209 (20.2%) | 47 (20.8%) | 256 (20.3%) |
| 5. CCCCG | 110 (10.6%) | 29 (12.8%) | 139 (11.0%) |
| 6. TCCTG | 54 (5.2%) | 11 (4.9%) | 65 (5.2%) |
| Curve Type | Number of Carriers | |||||
|---|---|---|---|---|---|---|
| 1. TCTTT | 2. CTCCG | 3. TCCCG | 4. TCCTT | 5. CCCCG | 6.TCCTG | |
| monophasic | 339 (54%) | 298 (47%) | 224 (35%) | 137 (22%) | 63 (10%) | 33 (5%) |
| biphasic | 88 (40%) | 124 (57%) | 87 (40%) | 50 (23%) | 23 (11%) | 14 (6%) |
| triphasic | 197 (56%) | 179 (51%) | 134 (38%) | 59 (17%) | 47 (13%) | 18 (5%) |
| multiphasic | 33 (58%) | 25 (44%) | 23 (40%) | 10 (18%) | 6 (11%) | 0 (0%) |
| Parameter * (Units) | CTCCG Haplotype + | CTCCG Haplotype − | p-Level |
|---|---|---|---|
| number | 626 | 633 | n/a |
| age (years) | 33.2 [32.3; 34.3] | 33.5 [32.4; 34.1] | 0.58 |
| systolic blood pressure (mmHg) | 114 [113; 115] | 115 [114; 116] | 0.65 |
| diastolic blood pressure (mmHg) | 72 [71; 73] | 72 [71; 74] | 0.78 |
| BMI (kg/m2) | 23.7 [23.2; 24.1] | 24.2 [23.6; 24.5] | 0.10 |
| WHR women | 0.760 [0.751; 0.765] | 0.769 [0.760; 0.777] | 0.11 |
| WHR men | 0.862 [0.845; 0.877] | 0.867 [0.848; 0.888] | 0.49 |
| glucose metabolism | |||
| basal glycemia (mmol/L) | 4.7 [4.7; 4.8] | 4.8 [4.8; 4.8] | 0.01 |
| AUCGlycemia(mmol × min/L) | 1029 [1008; 1052] | 1059 [1041; 1079] | <0.01 |
| basal insulinemia (mIU/L) | 5.9 [5.5; 6.2] | 6.3 [6.0; 6.7] | 0.04 |
| AUCInsulin (pmol × min/L) | 31,757 [30,510; 33,309] | 35,793 [34,209; 38,421] | <0.01 |
| basal C-peptide (nmol/L) | 0.57 [0.56; 0.59] | 0.61 [0.59; 0.63] | <0.01 |
| AUCC-peptide (pmol × min/L) | 3.5 × 105 [3.4 × 105; 3.6 × 105] | 3.7 × 105 [3.6 × 105; 3.8 × 105] | <0.01 |
| hepatic insulin extraction (%) | 68.5 [67.7; 69.4] | 67.7 [66.8; 68.7] | 0.14 |
| insulin sensitivity/resistance | |||
| HOMA IR | 1.24 [1.17; 1.32] | 1.37 [1.29; 1.46] | 0.02 |
| OGIS 120 min (ml/min/m2) | 462.5 [455.8; 468.6] | 453.0 [448.0; 459.3] | <0.01 |
| ISICOMP ([(mg/dl)2(μU/)mL2]−1/2) | 8.91 [8.56; 9.24] | 7.98 [7.52; 8.38] | <0.01 |
| MCRest (mL/min/kg) | 9.99 [9.81; 10.17] | 9.76 [9.54; 9.93] | <0.01 |
| Si(oral) ((mL/min/kg)/(μU/)mL) | 0.16 [0.15; 0.17] | 0.15 [0.14; 0.16] | <0.01 |
| PREDIM (mg/min/kg) | 7.18 [6.90; 7.38] | 6.83 [6.64; 7.00] | 0.03 |
| beta cell function | |||
| HOMA B (mIU/mmol) | 103.7 [95.7; 108.3] | 103.2 [97.8; 107.3] | 0.78 |
| IGI (pmol/mmol) | 89.1 [83.1; 95.8] | 88.3 [83.2; 98.3] | 0.59 |
| Ins0/Glc0 (pmol/mmol) | 7.40 [7.00; 7.85] | 7.92 [7.56; 8.37] | 0.11 |
| AUCInsulin/AUCGlc (pmol/mmol) | 31.3 [29.9; 32.7] | 34.5 [32.7; 35.8] | <0.01 |
| IGI × ISICOMP | 267.9 [257.7; 275.5] | 260.6 [253.3; 269.9] | 0.21 |
| lipid spectrum | |||
| total cholesterol (mmol/L) | 4.61 [4.55; 4.70] | 4.58 [4.47; 4.65] | 0.54 |
| HDL chol. in women (mmol/L) | 1.58 [1.56; 1.64] | 1.54 [1.50; 1.59] | 0.03 |
| HDL chol. in men (mmol/L) | 1.30 [1.23; 1.35] | 1.21 [1.15; 1.31] | 0.28 |
| LDL cholesterol (mmol/L) | 2.57 [2.47; 2.64] | 2.56 [2.52; 2.64] | 0.35 |
| TAG (mmol/L) | 0.84 [0.79; 0.88] | 0.89 [0.83; 0.93] | 0.09 |
| liver enzymes | |||
| ALT (ukat/L) | 0.30 [0.29; 0.31] | 0.30 [0.29; 0.31] | 0.59 |
| AST (ukat/L) | 0.36 [0.35; 0.37] | 0.34 [0.34; 0.35] | 0.07 |
| GGT (ukat/L) | 0.23 [0.22; 0.25] | 0.23 [0.22; 0.24] | 0.65 |
| thyroid hormones | |||
| TSH (mIU/L) | 2.19 [2.11; 2.33] | 2.26 [2.19; 2.38] | 0.99 |
| fT3 (pmol/L) | 4.89 [4.80; 4.99] | 4.91 [4.84; 5.00] | 0.71 |
| fT4 (pmol/L) | 15.0 [14.8; 15.2] | 15.4 [15.1; 15.6] | 0.13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vejražková, D.; Včelák, J.; Vaňková, M.; Lukášová, P.; Svojtková, M.; Grimmichová, T.; Kvasničková, H.; Tura, A.; Šedová, L.; Šeda, O.; et al. The NME7 Gene Is Involved in the Kinetics of Glucose Processing. Int. J. Mol. Sci. 2025, 26, 9821. https://doi.org/10.3390/ijms26199821
Vejražková D, Včelák J, Vaňková M, Lukášová P, Svojtková M, Grimmichová T, Kvasničková H, Tura A, Šedová L, Šeda O, et al. The NME7 Gene Is Involved in the Kinetics of Glucose Processing. International Journal of Molecular Sciences. 2025; 26(19):9821. https://doi.org/10.3390/ijms26199821
Chicago/Turabian StyleVejražková, Daniela, Josef Včelák, Markéta Vaňková, Petra Lukášová, Michaela Svojtková, Tereza Grimmichová, Hana Kvasničková, Andrea Tura, Lucie Šedová, Ondřej Šeda, and et al. 2025. "The NME7 Gene Is Involved in the Kinetics of Glucose Processing" International Journal of Molecular Sciences 26, no. 19: 9821. https://doi.org/10.3390/ijms26199821
APA StyleVejražková, D., Včelák, J., Vaňková, M., Lukášová, P., Svojtková, M., Grimmichová, T., Kvasničková, H., Tura, A., Šedová, L., Šeda, O., Škultéty, K., & Bendlová, B. (2025). The NME7 Gene Is Involved in the Kinetics of Glucose Processing. International Journal of Molecular Sciences, 26(19), 9821. https://doi.org/10.3390/ijms26199821





_Svojtková.JPG)


