OsPIP2;1 Positively Regulates Rice Tolerance to Water Stress Under Coupling of Partial Root-Zone Drying and Nitrogen Forms
Abstract
1. Introduction
2. Results
2.1. Expression Pattern of the OsPIP2;1 Gene
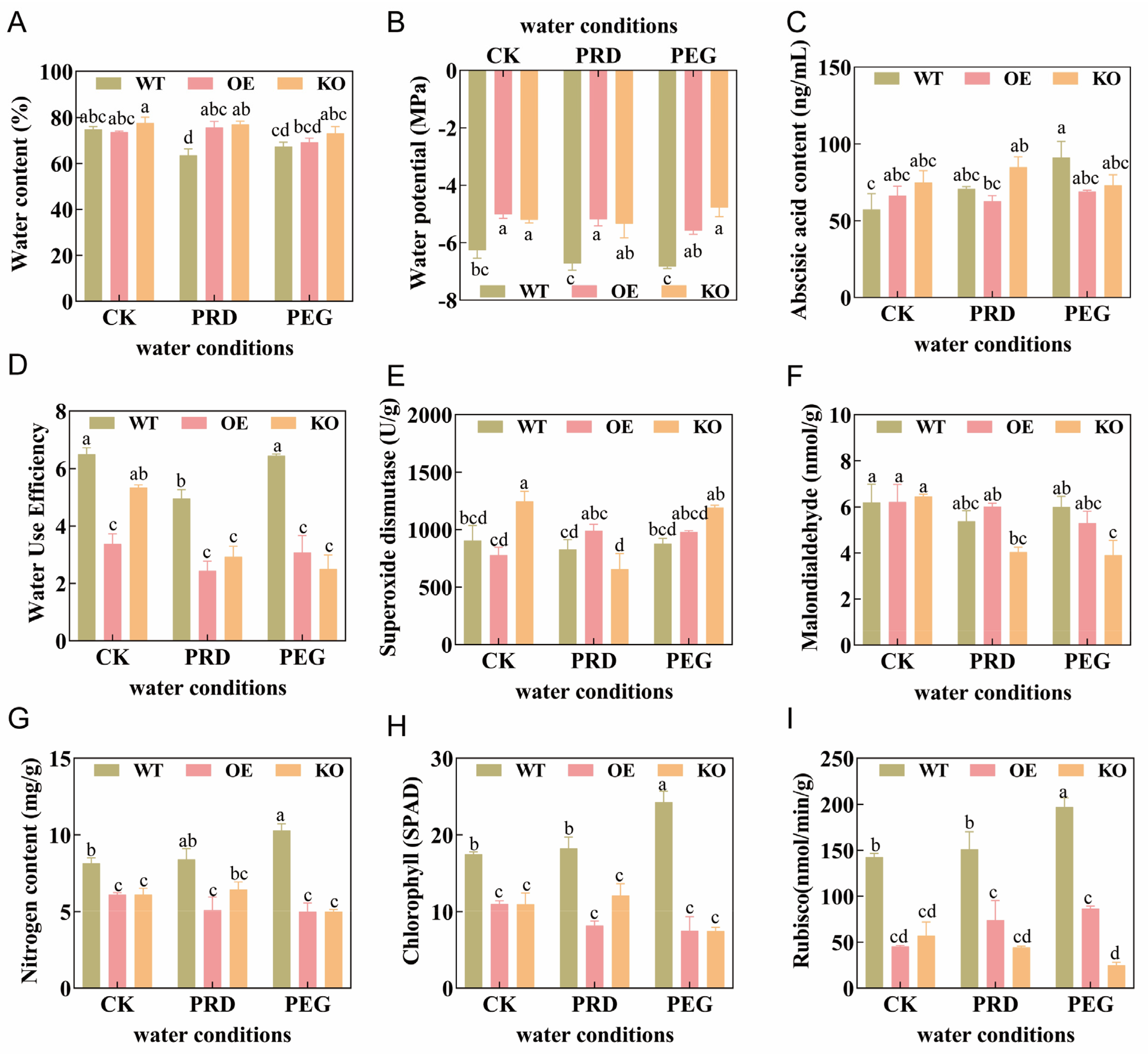
2.2. Effects of OsPIP2;1 on Water and Nitrogen Use of Rice Under Partial Root-Zone Drying and Nitrogen Form Coupling
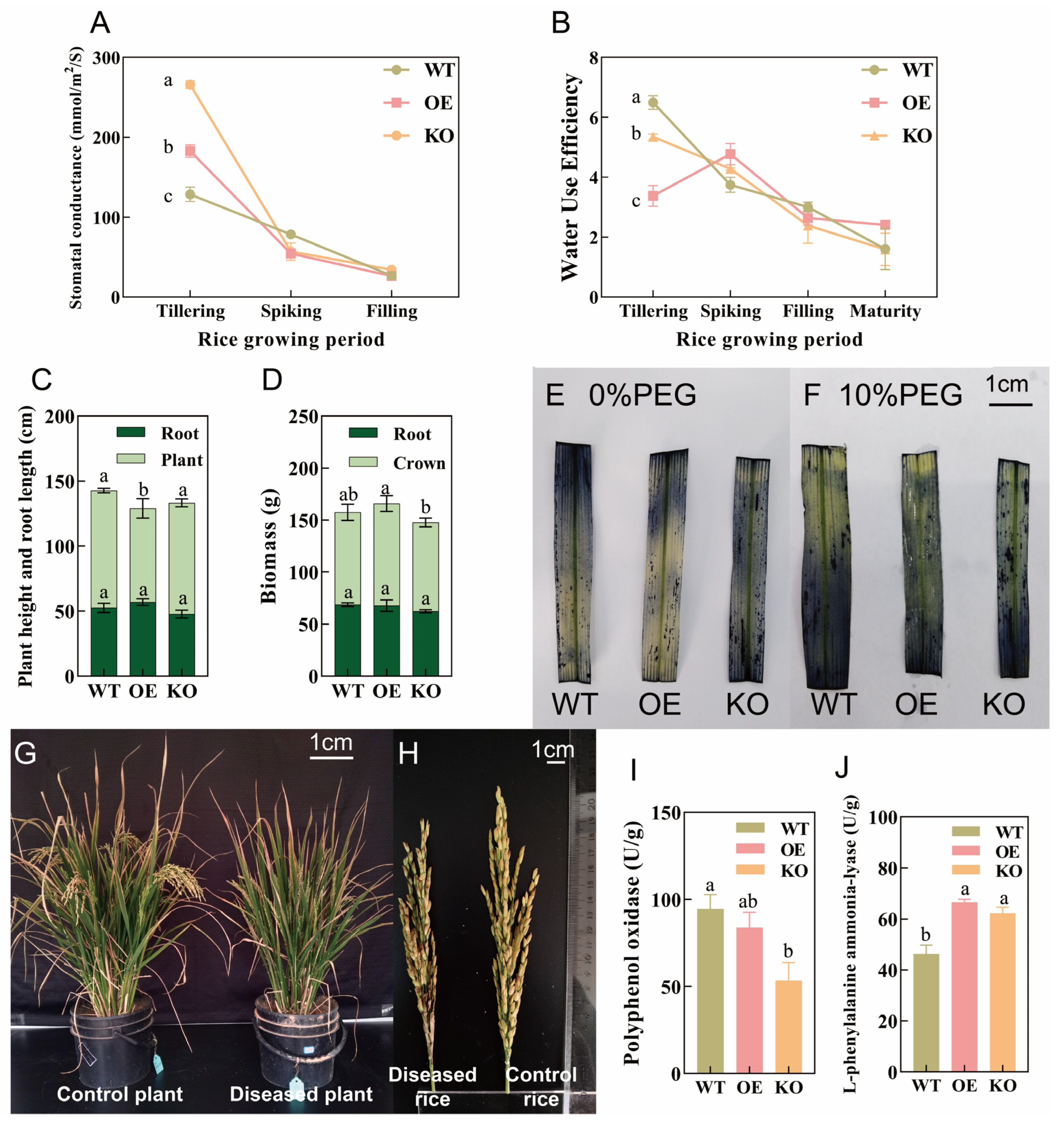
2.3. Effects of OsPIP2;1 on Rice Growth and Development and Stress Resistance
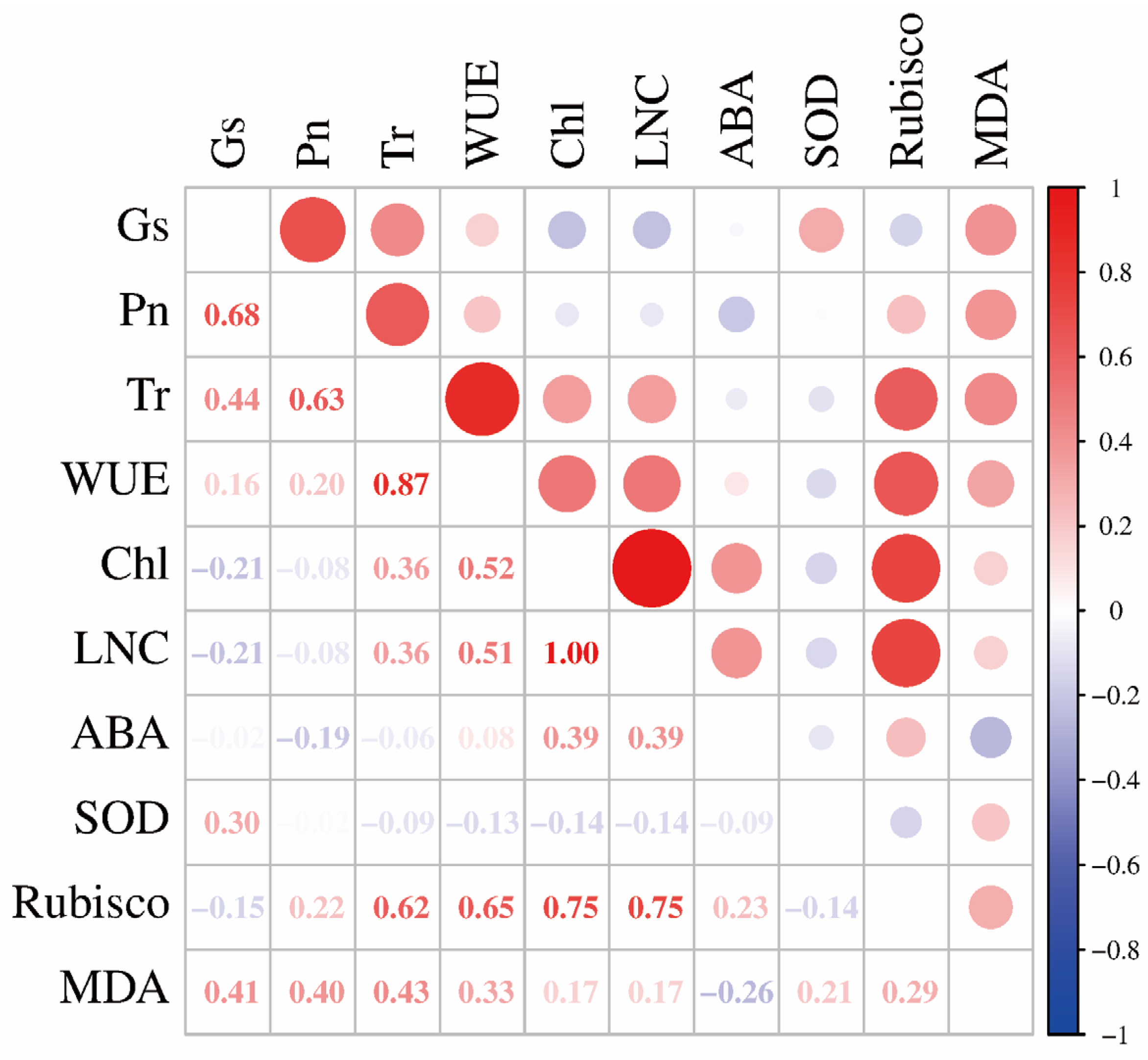
2.4. The Correlation of OsPIP2;1 Mediated Rice Physiological and Biochemical Processes
3. Discussion
3.1. PRD and Nitrogen Form Coupling Effects Influence the Expression Pattern of OsPIP2;1
3.2. OsPIP2;1 Affects Rice Nitrogen Utilization Under PRD and Nitrogen Form Coupling
3.3. OsPIP2;1 Affects Rice’s Response Ability to Water Stress Under PRD and Nitrogen Form Coupling
4. Materials and Methods
4.1. Experimental Materials
4.2. Reagents and Formulation
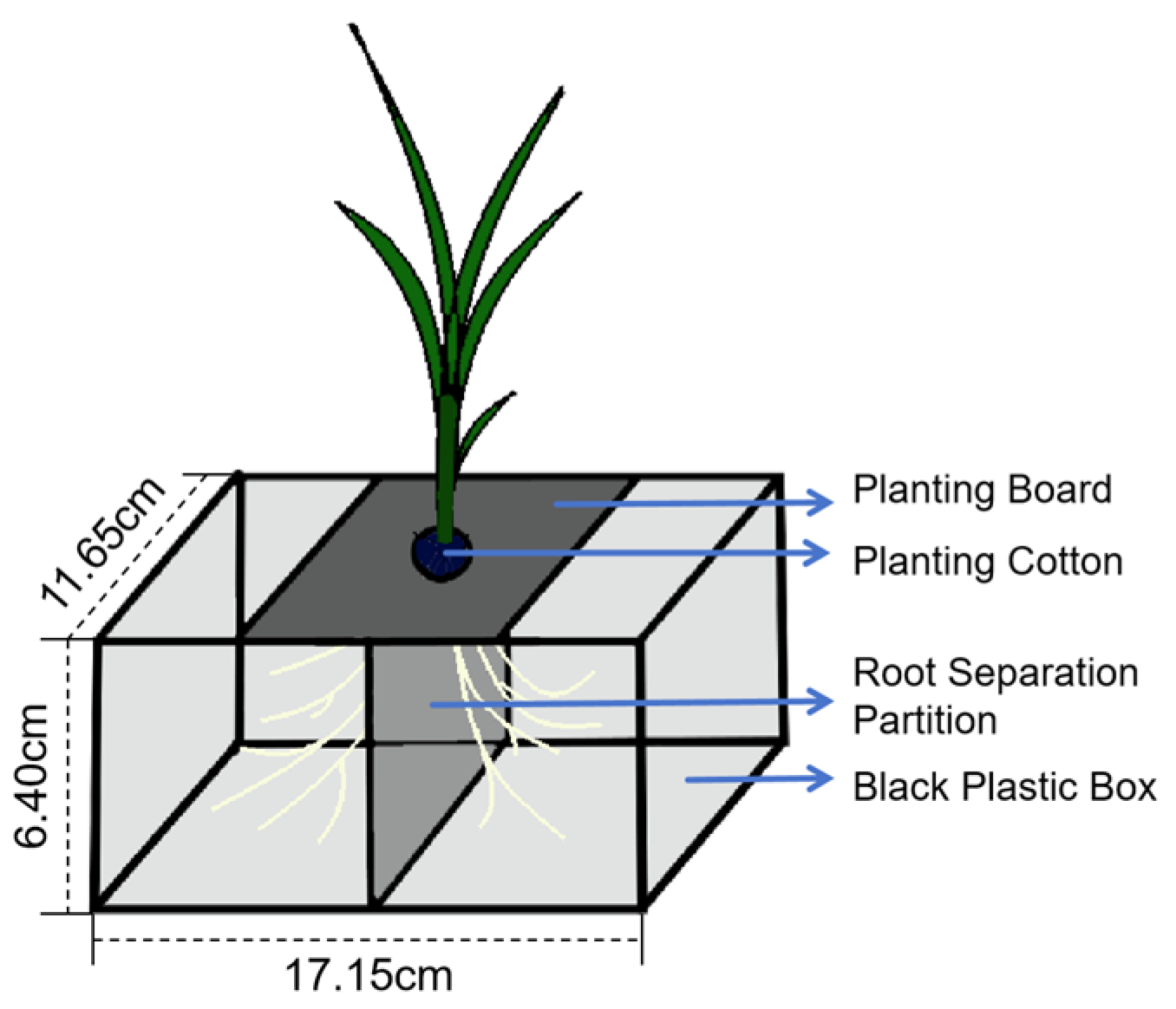
4.3. Experimental Design
4.4. Rice Seedling Raising Methods
4.5. Method for Determination of OsPIP2;1 Gene Expression Level
4.6. Method for Constructing an Overexpression Vector of the OsPIP2;1 Gene
4.7. Method for Constructing a Knockout Vector of the OsPIP2;1 Gene
4.8. Genetic Transformation Method of Rice for OsPIP2;1 Gene
4.9. Experimental Testing Indicators and Methods
4.10. Data Processing and Analysis Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PRD | Partial root-zone drying |
| AQP | Aquaporin |
| PEG | Polyethylene glycol |
| ABA | Abscisic acid |
| WUE | Water use efficiency |
| MDA | Malondialdehyde |
| ROS | Reactive oxygen species |
| SOD | Superoxide dismutase |
| PPO | Polyphenol oxidase |
| PAL | L-phenylalanine ammonia-lyase |
| PIPs | Plasma membrane intrinsic proteins |
| TIPs | Tonoplast intrinsic proteins |
| NIPs | Nodulin-26 intrinsic proteins |
| SIPs | Small and basic intrinsic proteins |
| NBT | Nitrotetrazolium blue chloride |
| FPKM | Fragments Per Kilobase Million |
| OE | Overexpression |
| KO | Knock out |
| WT | Wild type |
References
- Gobu, R.; Dash, G.K.; Lal, J.P.; Swain, P.; Mahender, A.; Anandan, A.; Ali, J. Unlocking the Nexus between Leaf-Level Water Use Efficiency and Root Traits Together with Gas Exchange Measurements in Rice (Oryza sativa L.). Plants 2022, 11, 1270. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, R.; Raza, M.A.S.; Toleikiene, M.; Ayaz, M.; Hashemi, F.; Habib-ur-Rahman, M.; Zaheer, M.S.; Ahmad, S.; Riaz, U.; Ali, M.; et al. Partial root-zone drying (PRD), its effects and agricultural significance: A review. Bull. Natl. Res. Cent. 2020, 44, 413. [Google Scholar] [CrossRef]
- Machekposhti, M.F.; Shahnazari, A.; Yousefian, M.; Ahmadi, M.Z.; Sarjaz, M.R.; Arabzadeh, B.; Akbarzadeh, A.; Leib, B.G. The effect of alternate partial root-zone drying and deficit irrigation on the yield, quality, and physiochemical parameters of milled rice. Agric. Water Manag. 2023, 289, 108546. [Google Scholar] [CrossRef]
- Yang, X.; Bornø, M.L.; Wei, Z.; Liu, F. Combined effect of partial root drying and elevated atmospheric CO2 on the physiology and fruit quality of two genotypes of tomato plants with contrasting endogenous ABA levels. Agric. Water Manag. 2021, 254, 106987. [Google Scholar] [CrossRef]
- Pérez-Pérez, J.G.; Navarro, J.M.; Robles, J.M.; Dodd, I.C. Prolonged drying cycles stimulate ABA accumulation in Citrus macrophylla seedlings exposed to partial rootzone drying. Agric. Water Manag. 2018, 210, 271–278. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, Y.; Wang, X.; Jiao, S.; Lu, Y.; Du, Y.; Zhang, W.; Kang, Y.; Liu, Y.; Qin, S. Differential responses of microstructure, antioxidant defense, and plant hormone signaling regulation in potato (Solanum tuberosum L.) under drought, alkaline salt, and combined stresses. Sci. Hortic. 2025, 341, 114014. [Google Scholar] [CrossRef]
- Li, W.; Jia, L.; Wang, L. Chemical signals and their regulations on the plant growth and water use efficiency of cotton seedlings under partial root-zone drying and different nitrogen applications. Saudi J. Biol. Sci. 2017, 24, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Dodd, I.C.; Egea, G.; Davies, W.J. Accounting for sap flow from different parts of the root system improves the prediction of xylem ABA concentration in plants grown with heterogeneous soil moisture. J. Exp. Bot. 2008, 59, 4083–4093. [Google Scholar] [CrossRef]
- Davies, W.J.; Wilkinson, S.; Loveys, B. Stomatal control by chemical signalling and the exploitation of this mechanism to increase water use efficiency in agriculture. New Phytol. 2002, 153, 449–460. [Google Scholar] [CrossRef]
- Zhang, Y.; Lv, H.; Wang, D.; Deng, J.; Song, W.; Makeen, K.; Shen, Q.; Xu, G. Partial nitrate nutrition amends photosynthetic characteristics in rice (Oryza sativa L. var. japonica) differing in nitrogen use efficiency. Plant Growth Regul. 2011, 63, 235–242. [Google Scholar]
- Zhang, J.; Lv, J.; Dawuda, M.M.; Xie, J.; Yu, J.; Li, J.; Zhang, X.; Tang, C.; Wang, C.; Gan, Y. Appropriate Ammonium-Nitrate Ratio Improves Nutrient Accumulation and Fruit Quality in Pepper (Capsicum annuum L.). Agronomy 2019, 9, 683. [Google Scholar] [CrossRef]
- Liu, G.; Du, Q.; Li, J. Interactive effects of nitrate-ammonium ratios and temperatures on growth, photosynthesis, and nitrogen metabolism of tomato seedlings. Sci. Hortic. 2017, 214, 41–50. [Google Scholar] [CrossRef]
- Singh, R.K.; Deshmukh, R.; Muthamilarasan, M.; Rani, R.; Prasad, M. Versatile roles of aquaporin in physiological processes and stress tolerance in plants. Plant Physiol. Biochem. 2020, 149, 178–189. [Google Scholar] [CrossRef]
- Nicolas-Espinosa, J.; Yepes-Molina, L.; Martinez-Bernal, F.; Fernandez-Pozurama, M.; Carvajal, M. Deciphering the effect of salinity and boron stress on broccoli plants reveals that membranes phytosterols and PIP aquaporins facilitate stress adaptation. Plant Sci. 2024, 338, 111923. [Google Scholar] [CrossRef]
- Sakurai, J.; Ishikawa, F.; Yamaguchi, T.; Uemura, M.; Maeshima, M. Identification of 33 Rice Aquaporin Genes and Analysis of Their Expression and Function. Plant Cell Physiol. 2005, 46, 1568–1577. [Google Scholar] [CrossRef]
- Pandey, B.; Sharma, P.; Pandey, D.M.; Sharma, I.; Chatrath, R. Identification of New Aquaporin Genes and Single Nucleotide Polymorphism in Bread Wheat. Evol. Bioinform. 2013, 9, EBO.S12568–EBO.S12576. [Google Scholar] [CrossRef] [PubMed]
- Ayadi, M.; Brini, F.; Masmoudi, K. Overexpression of a Wheat Aquaporin Gene, TdPIP2;1, Enhances Salt and Drought Tolerance in Transgenic Durum Wheat cv. Maali. Int. J. Mol. Sci. 2019, 20, 2389. [Google Scholar] [CrossRef] [PubMed]
- Kapilan, R.; Vaziri, M.; Zwiazek, J.J. Regulation of aquaporins in plants under stress. Biol. Res. 2018, 51, 152. [Google Scholar] [CrossRef]
- Zhang, G.; Yu, Z.; da Silva, J.A.T.; Wen, D. Identification of aquaporin members in Acacia auriculiformis and functional characterization of AaPIP1-2 involved in drought stress. Environ. Exp. Bot. 2021, 185, 104425. [Google Scholar] [CrossRef]
- Almadanim, M.C.; Alexandre, B.M.; Rosa, M.T.G.; Sapeta, H.; Leitão, A.E.; Ramalho, J.C.; Lam, T.T.; Negrão, S.; Abreu, I.A.; Oliveira, M.M. Rice calcium-dependent protein kinase OsCPK17 targets plasma membrane intrinsic protein and sucrose-phosphate synthase and is required for a proper cold stress response. Plant Cell Environ. 2017, 40, 1197–1213. [Google Scholar] [CrossRef]
- Hayashi, H.; Ishikawa-Sakurai, J.; Murai-Hatano, M.; Ahamed, A.; Uemura, M. Aquaporins in developing rice grains. Biosci. Biotechnol. Biochem. 2015, 79, 1422–1429. [Google Scholar] [CrossRef]
- Muto, Y.; Segami, S.; Hayashi, H.; Sakurai, J.; Murai-Hatano, M.; Hattori, Y.; Ashikari, M.; Maeshima, M. Vacuolar proton pumps and aquaporins involved in rapid internode elongation of deepwater rice. Biosci. Biotechnol. Biochem. 2011, 75, 114–122. [Google Scholar] [CrossRef]
- Liu, H.Y.; Yu, X.; Cui, D.Y.; Sun, M.H.; Sun, W.N.; Tang, Z.C.; Kwak, S.-S.; Su, W.-A. The role of water channel proteins and nitric oxide signaling in rice seed germination. Cell Res. 2007, 17, 638–649. [Google Scholar] [CrossRef]
- Chen, S.; Xu, K.; Kong, D.; Wu, L.; Chen, Q.; Ma, X.; Ma, S.; Li, T.; Xie, Q.; Liu, H.; et al. Ubiquitin Ligase OsRINGzf1 Regulates Drought Resistance by Controlling the Turnover of OsPIP2;1. Plant Biotechnol. J. 2022, 20, 1743–1755. [Google Scholar] [CrossRef]
- Kumari, A.; Bhatla, S.C. Nitric oxide modulates the expression of aquaporin isoforms (PIP2 and TIP1) on oil body membranes in sunflower (Helianthus annuus L.) seedling cotyledons in response to salt stress. J. Plant Biochem. Biotechnol. 2023, 32, 651–656. [Google Scholar] [CrossRef]
- Gao, Y.; Li, Y.; Yang, X.; Li, H.; Shen, Q.; Guo, S. Ammonium nutrition increases water absorption in rice seedlings (Oryza sativa L.) under water stress. Plant Soil 2010, 331, 193–201. [Google Scholar] [CrossRef]
- Ricardo, A.; Rosa, P.; Manuel, R.-L.J. Regulation of root water uptake under abiotic stress conditions. J. Exp. Bot. 2012, 63, 43–57. [Google Scholar]
- Luu, D.T.; Maurel, C. Aquaporins in a challenging environment: Molecular gears for adjusting plant water status. Plant Cell Environ. 2005, 28, 85–96. [Google Scholar] [CrossRef]
- Maurel, C.; Verdoucq, L.; Luu, D.-T.; Santoni, V. Plant Aquaporins: Membrane Channels with Multiple Integrated Functions. Annu. Rev. Plant Biol. 2008, 59, 595–624. [Google Scholar] [CrossRef] [PubMed]
- Shilpha, J.; Song, J.; Jeong, B.R. Ammonium Phytotoxicity and Tolerance: An Insight into Ammonium Nutrition to Improve Crop Productivity. Agronomy 2023, 13, 1544. [Google Scholar] [CrossRef]
- Anonymous. Changes in plasma membrane lipids, aquaporins, and proton pump of broccoli roots, as an adaptation mechanism to salinity. Int. News Fats Oils Relat. Mater. (INFORM) 2009, 20, 439. [Google Scholar]
- Ding, L.; Uehlein, N.; Kaldenhoff, R.; Guo, S.; Zhu, Y.; Kai, L. Aquaporin PIP2;1 affects water transport and root growth in rice (Oryza sativa L.). Plant Physiol. Biochem. 2019, 139, 152–160. [Google Scholar] [CrossRef]
- Ding, L.; Li, Y.; Wang, Y.; Gao, L.; Wang, M.; Chaumont, F.; Shen, Q.; Guo, S. Root ABA Accumulation Enhances Rice Seedling Drought Tolerance under Ammonium Supply: Interaction with Aquaporins. Front. Plant Sci. 2016, 7, 1206. [Google Scholar] [CrossRef]
- Schuurmans, J.A.; van Dongen, J.T.; Rutjens, B.P.; Boonman, A.; Pieterse, C.M.; Borstlap, A.C. Members of the aquaporin family in the developing pea seed coat include representatives of the PIP, TIP, and NIP subfamilies. Plant Mol. Biol. 2003, 53, 633–645. [Google Scholar] [CrossRef] [PubMed]
- Matiz, A.; Cambuí, C.A.; Richet, N.; Mioto, P.T.; Gomes, F.; Pikart, F.C.; Chaumont, F.; Gaspar, M.; Mercier, H. Involvement of aquaporins on nitrogen-acquisition strategies of juvenile and adult plants of an epiphytic tank-forming bromeliad. Planta 2019, 250, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Holm, L.M.; Jahn, T.P.; Møller, A.L.; Schjoerring, J.K.; Ferri, D.; Klaerke, D.A.; Zeuthen, T. NH3 and NH4+ permeability in aquaporin-expressing Xenopus oocytes. Pflugers Arch. 2005, 450, 415–428. [Google Scholar] [CrossRef]
- Liu, L.H.; Ludewig, U.; Gassert, B.; Frommer, W.B.; von Wirén, N. Urea transport by nitrogen-regulated tonoplast intrinsic proteins in Arabidopsis. Plant Physiol. 2003, 133, 1220–1228. [Google Scholar] [CrossRef] [PubMed]
- Kirscht, A.; Kaptan, S.S.; Bienert, G.P.; Chaumont, F.; Nissen, P.; de Groot, B.L.; Kjellbom, P.; Gourdon, P.; Johanson, U. Crystal Structure of an Ammonia-Permeable Aquaporin. PLoS Biol. 2016, 14, e1002411. [Google Scholar] [CrossRef]
- Groszmann, M.; Osborn, H.L.; Evans, J.R. Carbon dioxide and water transport through plant aquaporins. Plant Cell Environ. 2017, 40, 938–961. [Google Scholar] [CrossRef]
- Sade, N.; Moshelion, M. Plant Aquaporins and Abiotic Stress. In Plant Aquaporins: From Transport to Signaling; Chaumont, F., Tyerman, S.D., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 185–206. [Google Scholar]
- Xu, F.; Wang, K.; Yuan, W.; Xu, W.; Liu, S.; Kronzucker, H.J.; Chen, G.; Miao, R.; Zhang, M.; Ding, M.; et al. Overexpression of rice aquaporin OsPIP1;2 improves yield by enhancing mesophyll CO2 conductance and phloem sucrose transport. J. Exp. Bot. 2019, 70, 671–681, Erratum in J. Exp. Bot. 2019, 70, 4067. [Google Scholar] [CrossRef]
- Nada, R.M.; Abogadallah, G.M. Contrasting root traits and native regulation of aquaporin differentially determine the outcome of overexpressing a single aquaporin (OsPIP2;4) in two rice cultivars. Protoplasma 2020, 257, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.Y.; Lee, S.H.; Rhee, J.Y.; Chung, G.C.; Ahn, S.J.; Kang, H. Transgenic Arabidopsis and tobacco plants overexpressing an aquaporin respond differently to various abiotic stresses. Plant Mol. Biol. 2007, 64, 621–632. [Google Scholar] [CrossRef] [PubMed]
- Nada, R.M.; Abogadallah, G.M. Aquaporins are major determinants of water use efficiency of rice plants in the field. Plant Sci. 2014, 227, 165–180. [Google Scholar] [CrossRef]
- Alavilli, H.; Awasthi, J.P.; Rout, G.R.; Sahoo, L.; Lee, B.H.; Panda, S.K. Overexpression of a Barley Aquaporin Gene, HvPIP2;5 Confers Salt and Osmotic Stress Tolerance in Yeast and Plants. Front. Plant Sci. 2016, 7, 1566. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, W.; Liu, J.; Song, S.; Hou, X.; Jia, C.; Li, J.; Miao, H.; Wang, Z.; Tie, W.; et al. An aquaporin gene MaPIP2-7 is involved in tolerance to drought, cold and salt stresses in transgenic banana (Musa acuminata L.). Plant Physiol. Biochem. 2020, 147, 66–76. [Google Scholar] [CrossRef]
- Gechev, T.S.; Van Breusegem, F.; Stone, J.M.; Denev, I.; Laloi, C. Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. BioEssays 2006, 28, 1091–1101. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, J. Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up-regulates the activities of antioxidant enzymes in maize leaves. J. Exp. Bot. 2002, 53, 2401–2410. [Google Scholar] [CrossRef]
- Gad, M.; Nobuhiro, S.; Sultan, C.-Y.; Ron, M. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar]
- Yun, S.J.; Son, L.X.; Ullah, K.I.; Chen, W.X.; Min, Y.Z. OsPIP2;3 as an aquaporin contributes to rice resistance to water deficit but not to salt stress. Environ. Exp. Bot. 2021, 183, 104356. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Breusegem, F.V. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Junko, S.; Arifa, A.; Mari, M.; Masayoshi, M.; Matsuo, U. Tissue and cell-specific localization of rice aquaporins and their water transport activities. Plant Cell Physiol. 2008, 49, 30–39. [Google Scholar] [CrossRef]
- Sakurai-Ishikawa, J.; Murai-Hatano, M.; Hayashi, H.; Ahamed, A.; Fukushi, K.; Matsumoto, T.; Kitagawa, Y. Transpiration from shoots triggers diurnal changes in root aquaporin expression. Plant Cell Environ. 2011, 34, 1150–1163. [Google Scholar] [CrossRef] [PubMed]
- Rowe, J.H.; Topping, J.F.; Liu, J.; Lindsey, K. Abscisic acid regulates root growth under osmotic stress conditions via an interacting hormonal network with cytokinin, ethylene and auxin. New Phytol. 2016, 211, 225–239. [Google Scholar] [CrossRef]
- Schüssler, M.D.; Alexandersson, E.; Bienert, G.P.; Kichey, T.; Laursen, K.H.; Johanson, U.; Kjellbom, P.; Schjoerring, J.K.; Jahn, T.P. The effects of the loss of TIP1;1 and TIP1;2 aquaporins in Arabidopsis thaliana. Plant J. 2008, 56, 756–767. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, W.; Liu, J.; Zhang, J.; Jia, C.; Miao, H.; Xu, B.; Jin, Z. A banana aquaporin gene, MaPIP1;1, is involved in tolerance to drought and salt stresses. BMC Plant Biol. 2014, 14, 59. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Hu, Y.; Li, J.; Wu, Q.; Lin, Z. Sense and antisense expression of plasma membrane aquaporin BnPIP1 from Brassica napus in tobacco and its effects on plant drought resistance. Plant Sci. 2005, 169, 647–656. [Google Scholar] [CrossRef]
- Lian, H.L.; Yu, X.; Ye, Q.; Ding, X.S.; Kitagawa, Y.; Kwak, S.S.; Su, W.A.; Tang, Z.C. The role of aquaporin RWC3 in drought avoidance in rice. Plant Cell Physiol. 2004, 45, 481–491. [Google Scholar] [CrossRef]
- Li, R.; Wang, J.; Li, S.; Zhang, L.; Qi, C.; Weeda, S.; Zhao, B.; Ren, S.; Guo, Y.D. Plasma Membrane Intrinsic Proteins SlPIP2;1, SlPIP2;7 and SlPIP2;5 Conferring Enhanced Drought Stress Tolerance in Tomato. Sci. Rep. 2016, 6, 31814. [Google Scholar] [CrossRef]
- McLean, E.H.; Ludwig, M.; Grierson, P.F. Root hydraulic conductance and aquaporin abundance respond rapidly to partial root-zone drying events in a riparian Melaleuca species. New Phytol. 2011, 192, 664–675. [Google Scholar] [CrossRef]
- Javot, H.; Lauvergeat, V.; Santoni, V.; Martin-Laurent, F.; Güçlü, J.; Vinh, J.; Heyes, J.; Franck, K.I.; Schaffner, A.R.; Bouchez, D.; et al. Role of a single aquaporin isoform in root water uptake. Plant Cell 2003, 15, 509–522. [Google Scholar] [CrossRef]
- Ding, X.; Iwasaki, I.; Kitagawa, Y. Overexpression of a lily PIP1 gene in tobacco increased the osmotic water permeability of leaf cells. Plant Cell Environ. 2004, 27, 177–186. [Google Scholar] [CrossRef]
- Sade, N.; Vinocur, B.J.; Diber, A.; Shatil, A.; Ronen, G.; Nissan, H.; Wallach, R.; Karchi, H.; Moshelion, M. Improving plant stress tolerance and yield production: Is the tonoplast aquaporin SlTIP2;2 a key to isohydric to anisohydric conversion. New Phytol. 2009, 181, 651–661. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, L.; Ji, H.; Mo, X.; Li, P.; Wang, J.; Dong, H. Hpa1 is a type III translocator in Xanthomonas oryzae pv. oryzae. BMC Microbiol. 2018, 18, 105. [Google Scholar] [CrossRef]
- Chen, X.; Ma, J.; Wang, X.; Lu, K.; Liu, Y.; Zhang, L.; Peng, J.; Chen, L.; Yang, M.; Li, Y.; et al. Functional modulation of an aquaporin to intensify photosynthesis and abrogate bacterial virulence in rice. Plant J. 2021, 108, 330–346. [Google Scholar] [CrossRef] [PubMed]
- Boudet, A.M. Evolution and current status of research in phenolic compounds. Phytochemistry 2007, 68, 2722–2735. [Google Scholar] [CrossRef] [PubMed]
- Montillet, J.-L.; Leonhardt, N.; Mondy, S.; Tranchimand, S.; Rumeau, D.; Boudsocq, M.; Garcia, A.V.; Douki, T.; Bigeard, J.; Laurière, C.; et al. An abscisic acid-independent oxylipin pathway controls stomatal closure and immune defense in Arabidopsis. PLoS Biol. 2017, 11, e1001513. [Google Scholar] [CrossRef]
- Zhou, G.; Liao, S.; Zhang, J.; Meng, Z.; Wang, Z.; Long, L.; Li, W.; Qiu, Y. Establishment of Agrobacterium-mediated transformation system for elite indica rice. Plant Biotechnol. Rep. 2024, 18, 693–704. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuang, C.; Han, Z.; Zhang, X.; Chen, X.; Gao, Z.; Zhu, Y. OsPIP2;1 Positively Regulates Rice Tolerance to Water Stress Under Coupling of Partial Root-Zone Drying and Nitrogen Forms. Int. J. Mol. Sci. 2025, 26, 9782. https://doi.org/10.3390/ijms26199782
Kuang C, Han Z, Zhang X, Chen X, Gao Z, Zhu Y. OsPIP2;1 Positively Regulates Rice Tolerance to Water Stress Under Coupling of Partial Root-Zone Drying and Nitrogen Forms. International Journal of Molecular Sciences. 2025; 26(19):9782. https://doi.org/10.3390/ijms26199782
Chicago/Turabian StyleKuang, Chunyi, Ziying Han, Xiang Zhang, Xiaoyuan Chen, Zhihong Gao, and Yongyong Zhu. 2025. "OsPIP2;1 Positively Regulates Rice Tolerance to Water Stress Under Coupling of Partial Root-Zone Drying and Nitrogen Forms" International Journal of Molecular Sciences 26, no. 19: 9782. https://doi.org/10.3390/ijms26199782
APA StyleKuang, C., Han, Z., Zhang, X., Chen, X., Gao, Z., & Zhu, Y. (2025). OsPIP2;1 Positively Regulates Rice Tolerance to Water Stress Under Coupling of Partial Root-Zone Drying and Nitrogen Forms. International Journal of Molecular Sciences, 26(19), 9782. https://doi.org/10.3390/ijms26199782




