Transcriptomic-Driven Drug Repurposing Reveals SP600125 as a Promising Drug Candidate for the Treatment of Glial-Mesenchymal Transition in Glioblastoma
Abstract
1. Introduction
2. Results
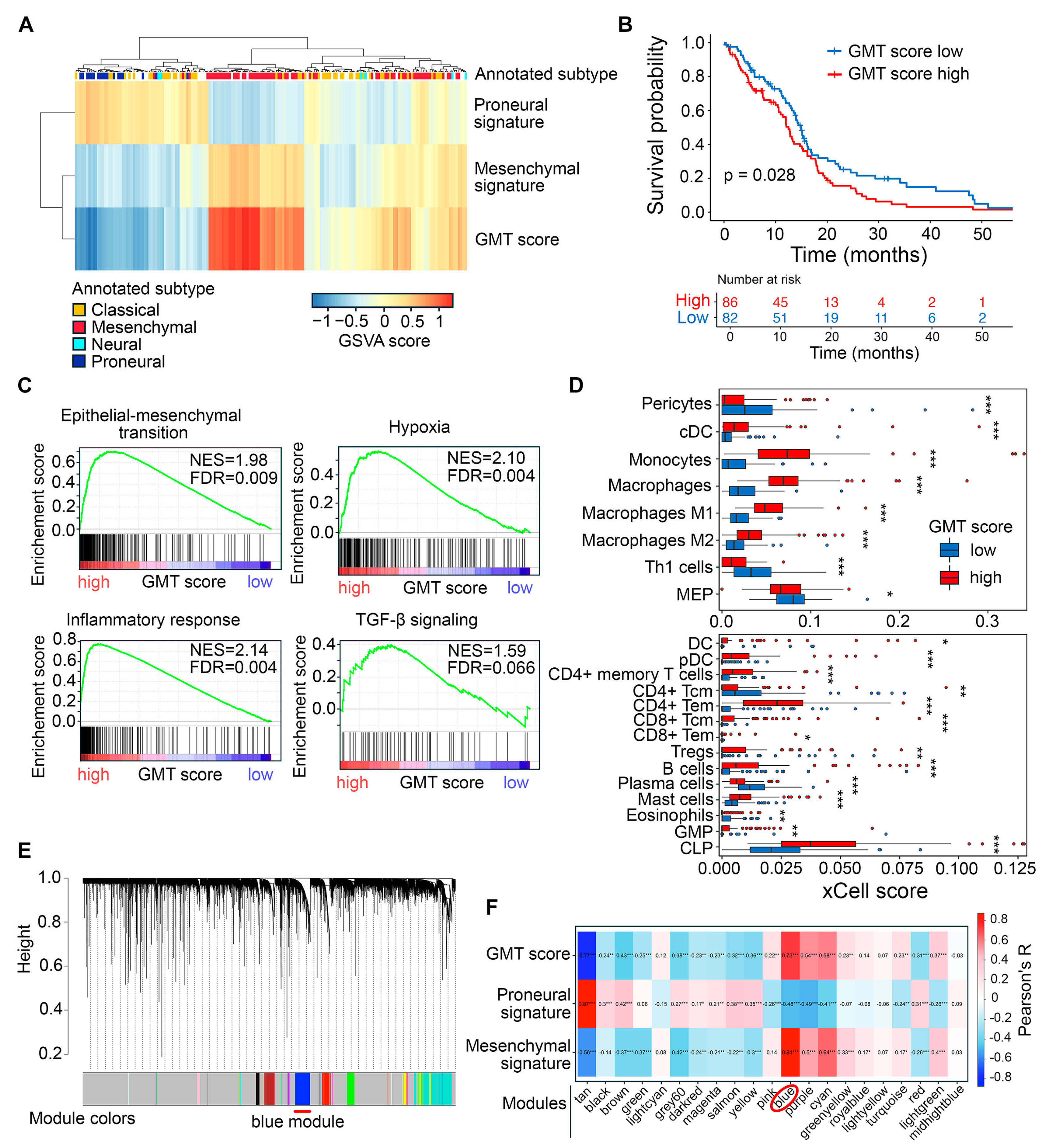
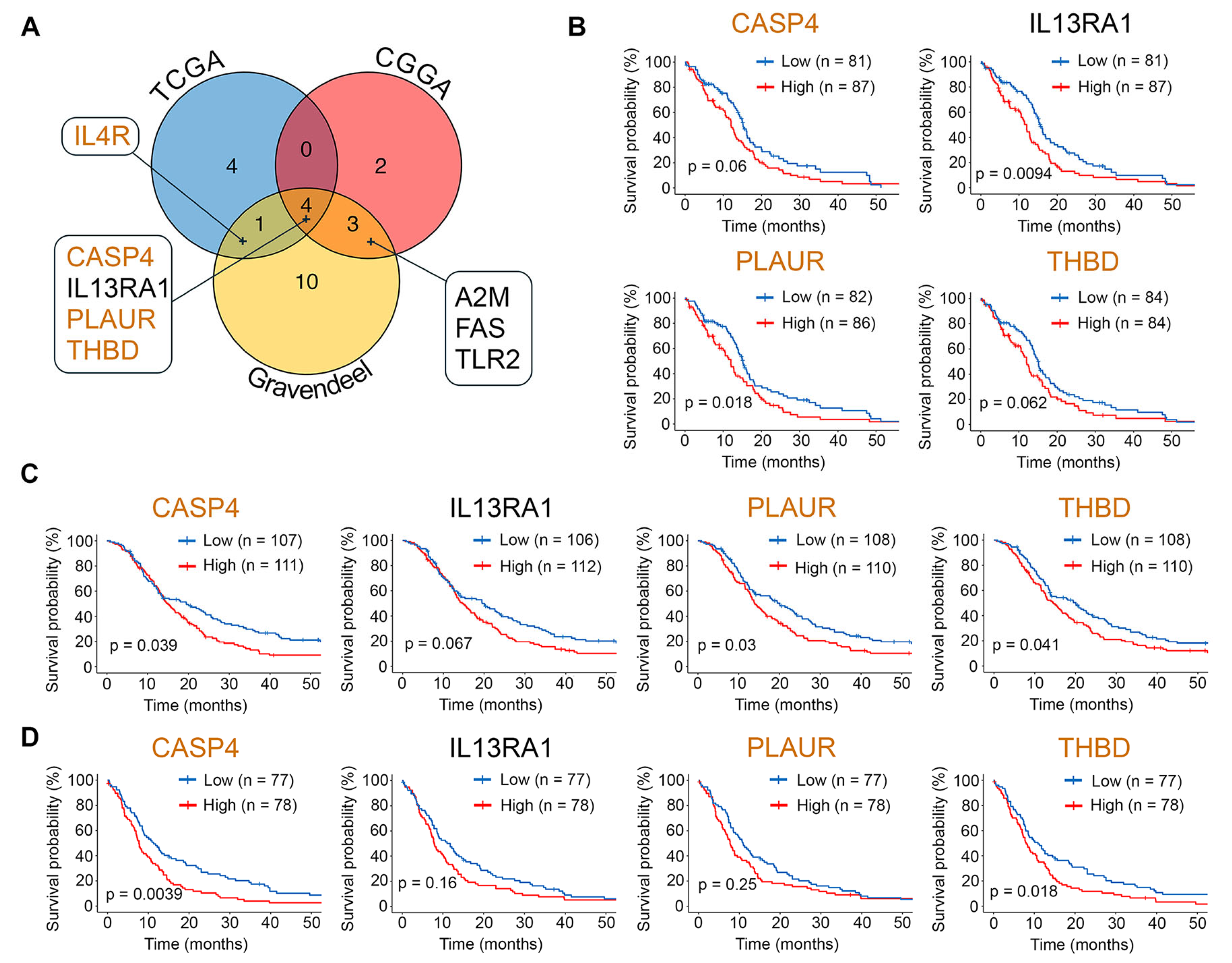
2.1. Exploring Glial-Mesenchymal Transition in TCGA-GBM Dataset
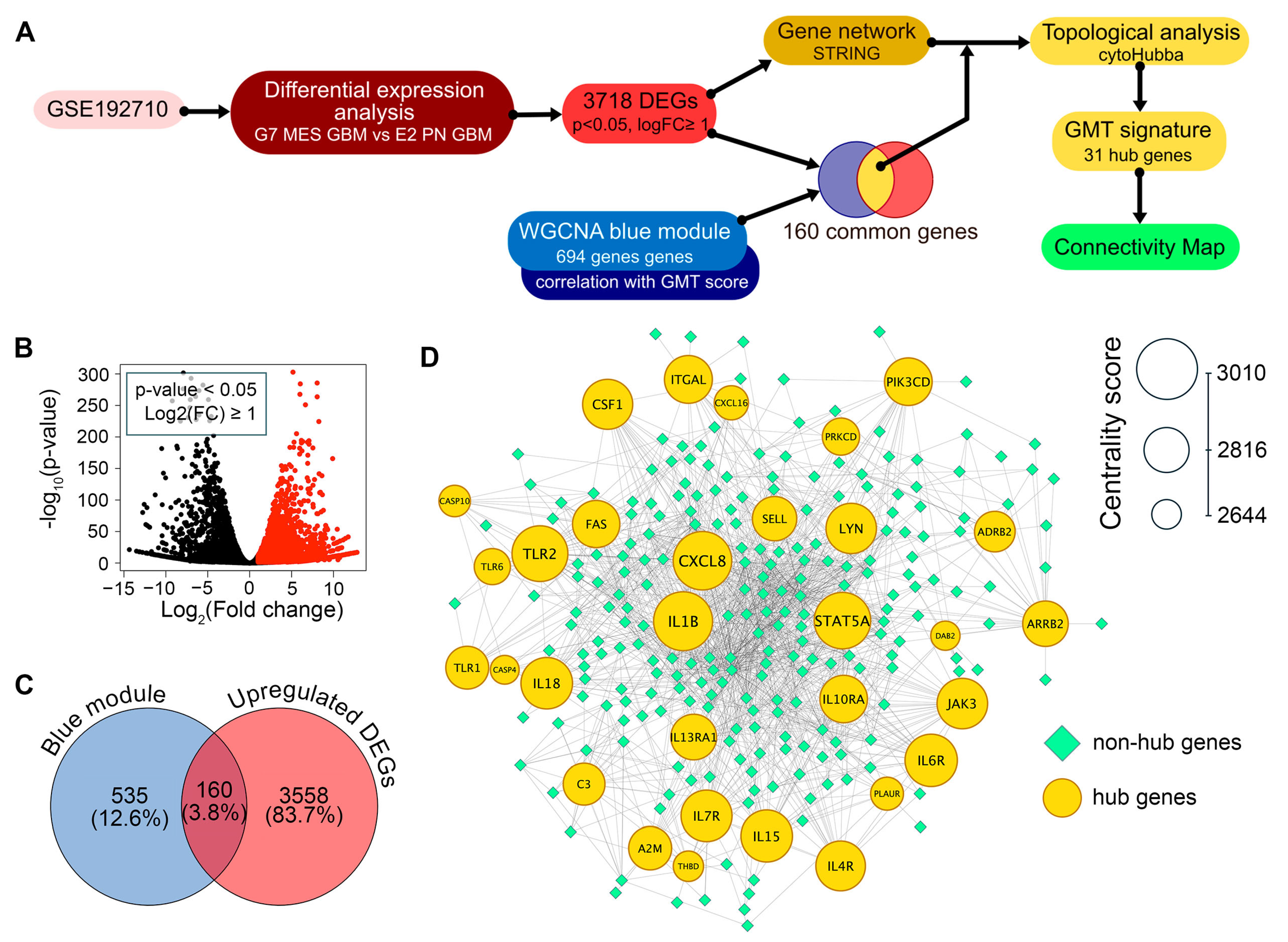
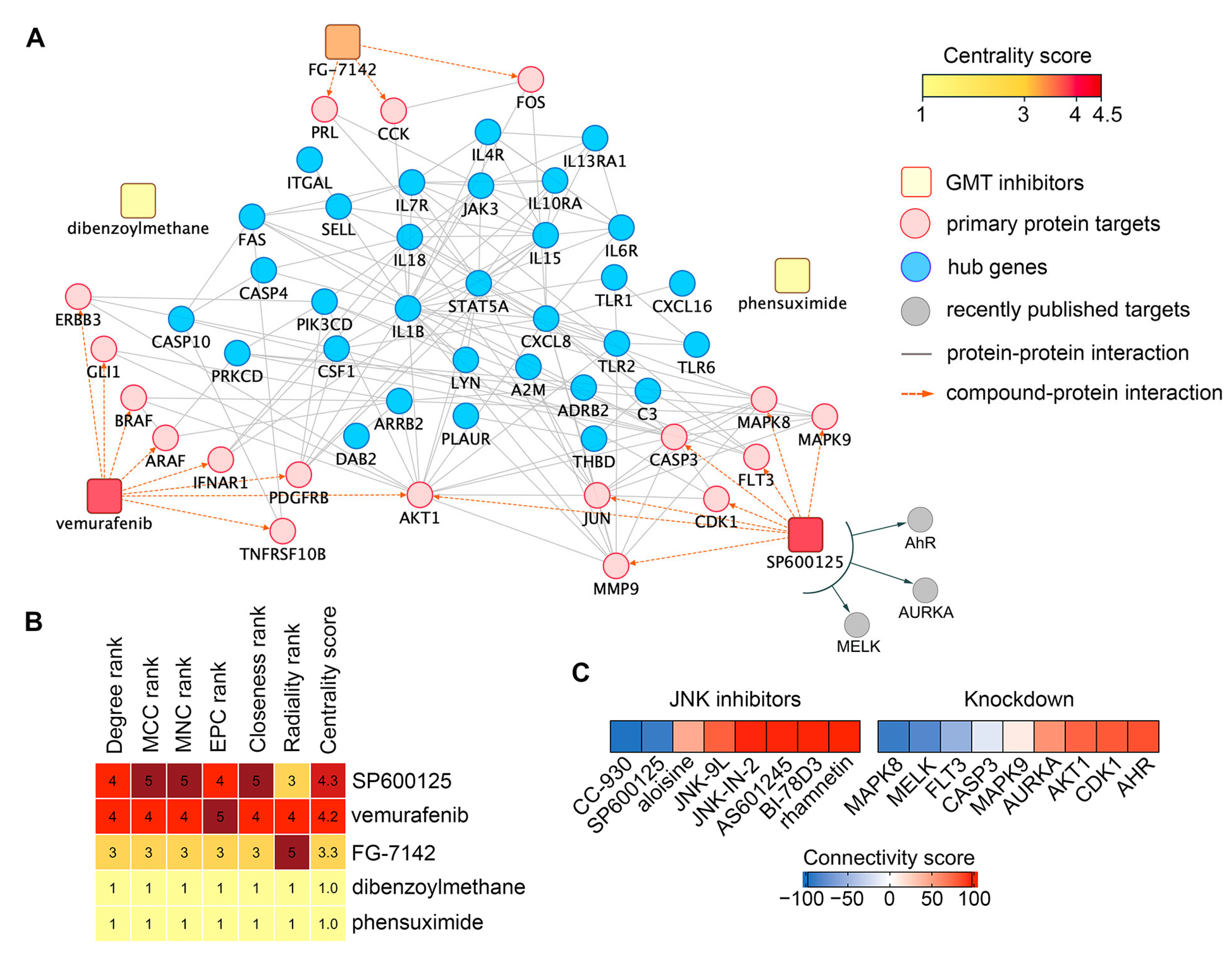
2.2. Identification of Hub Genes in the GMT Regulatory Network
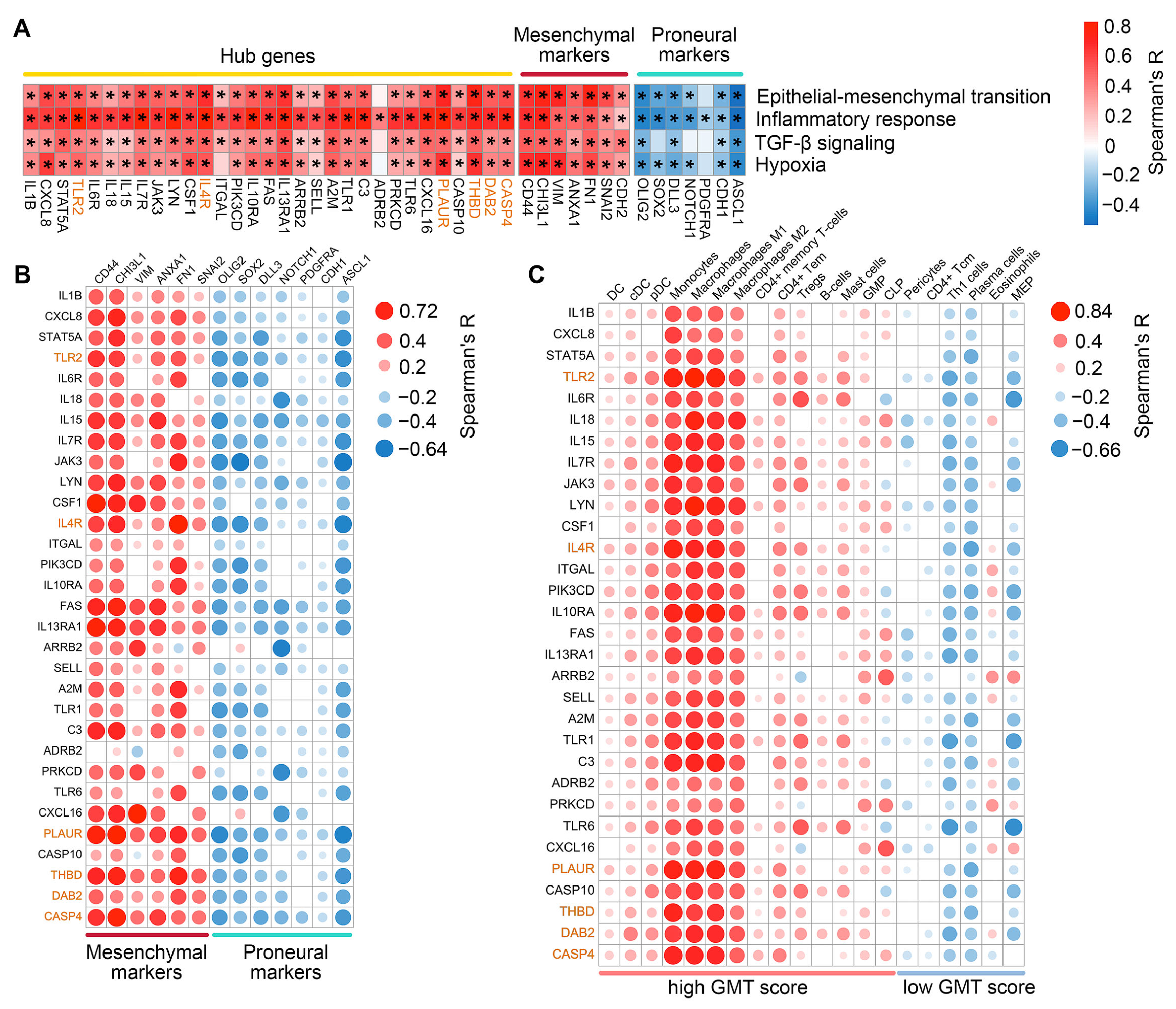
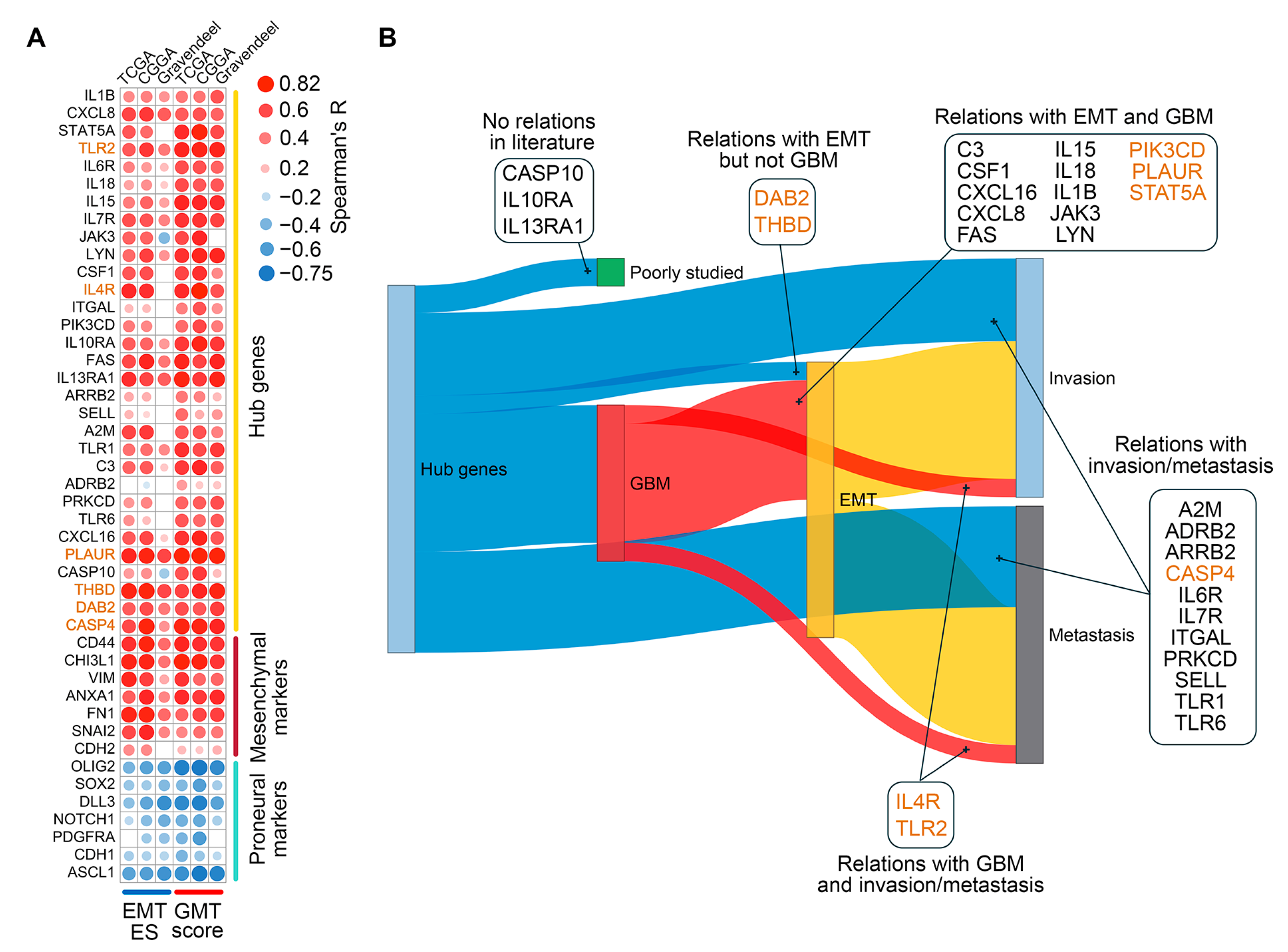
2.3. Assessment of Clinical Relevance for GMT-Associated Hub Genes
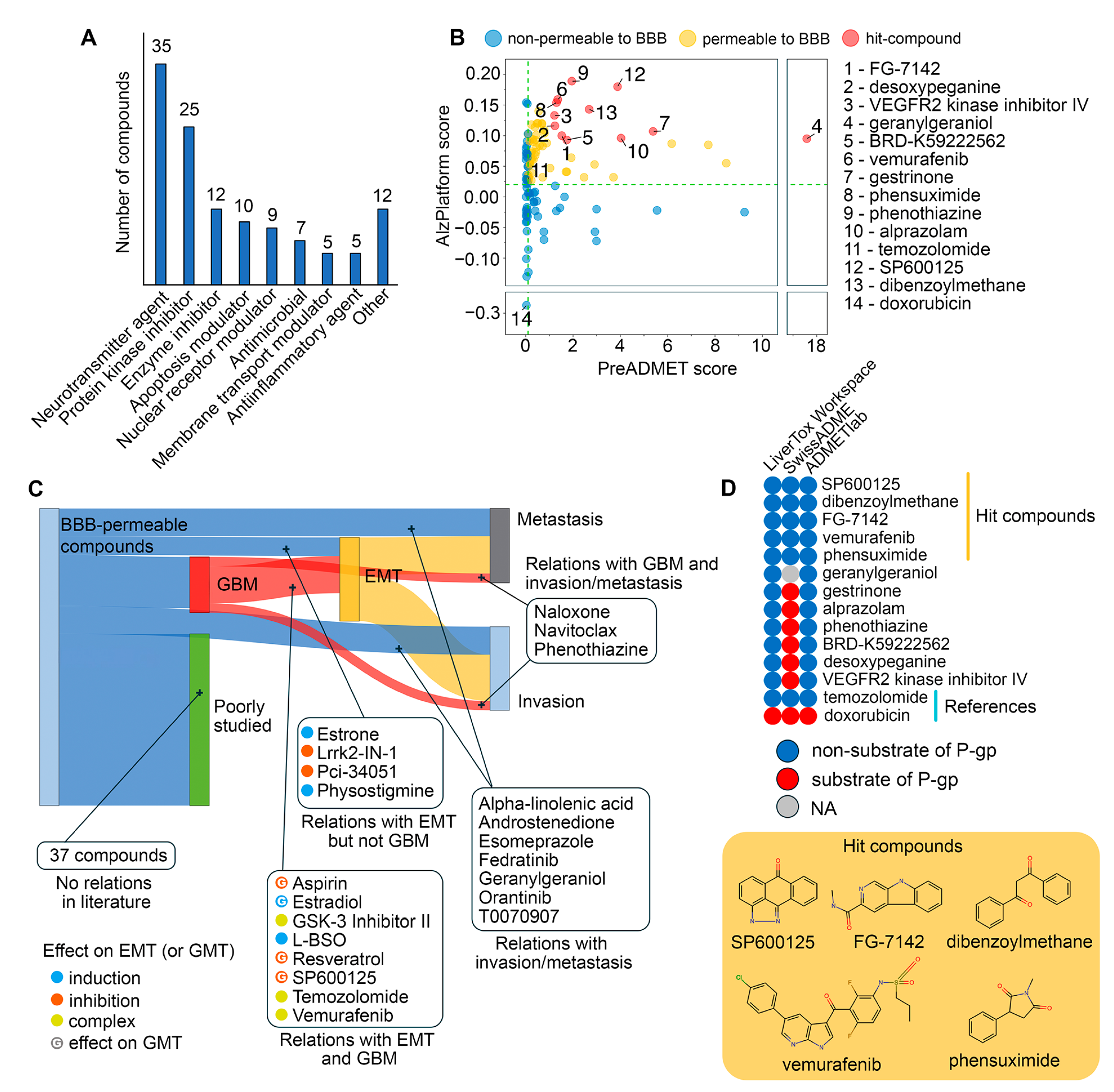
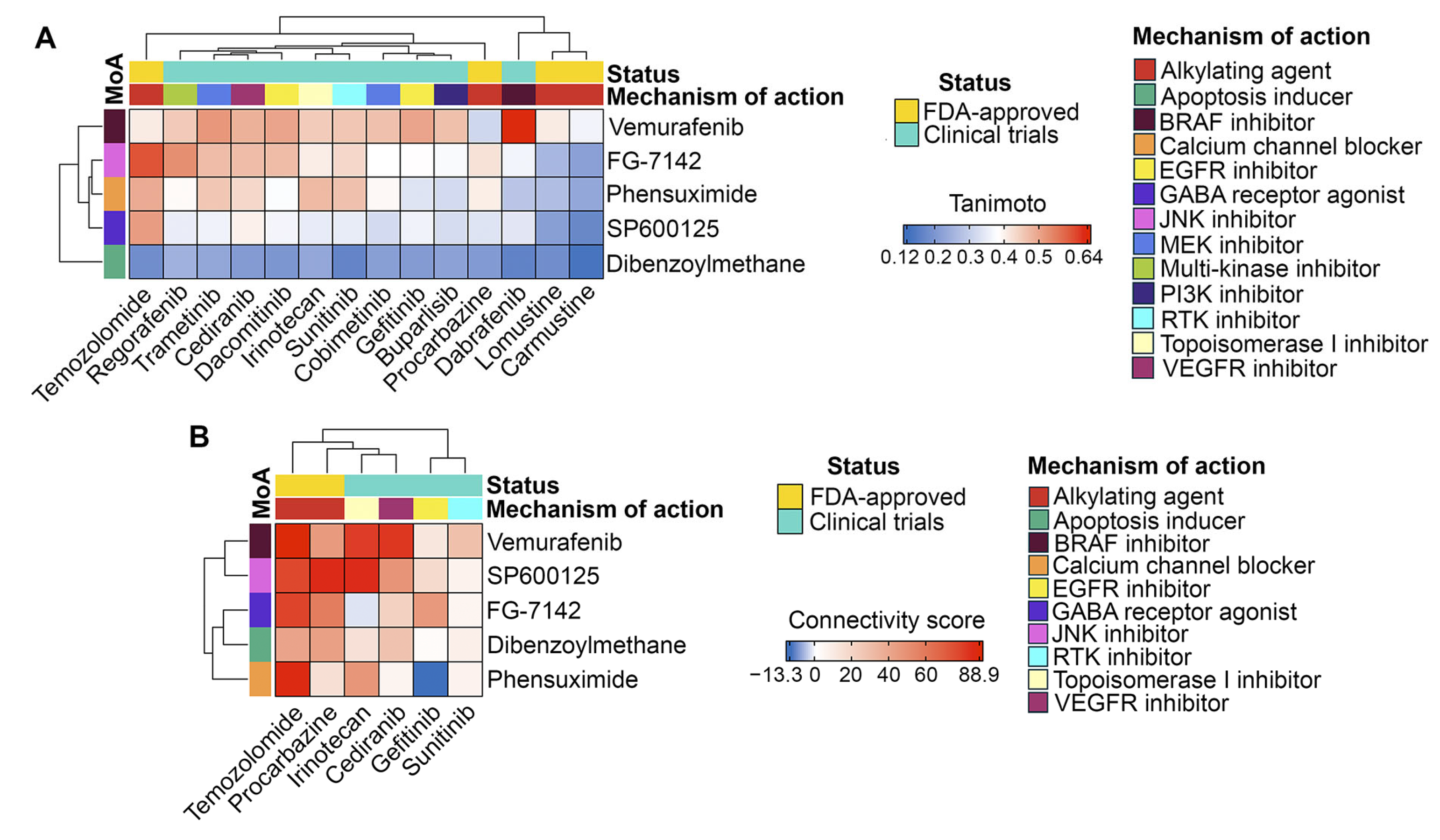
2.4. Drug Repurposing via Connectivity Mapping
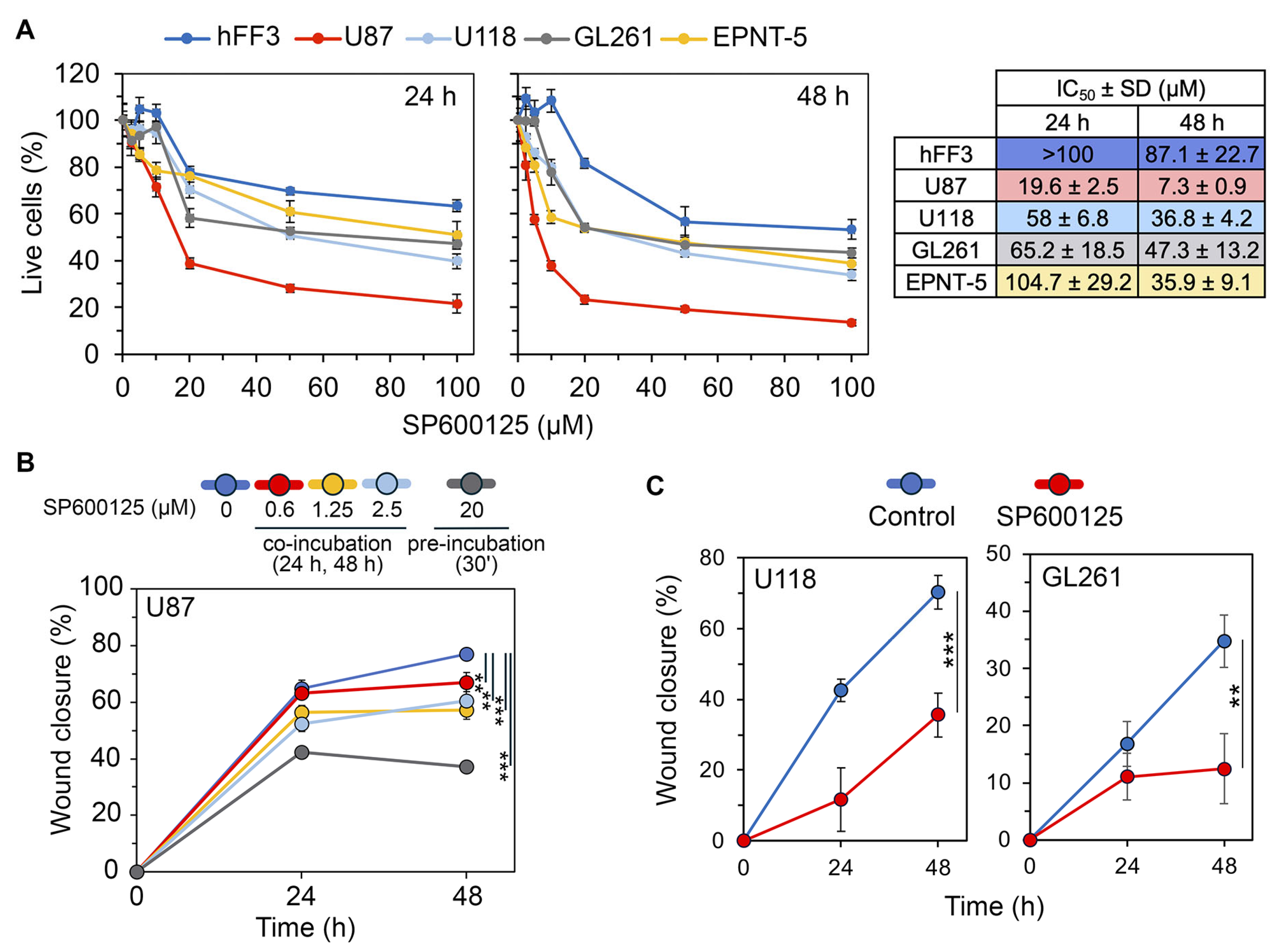
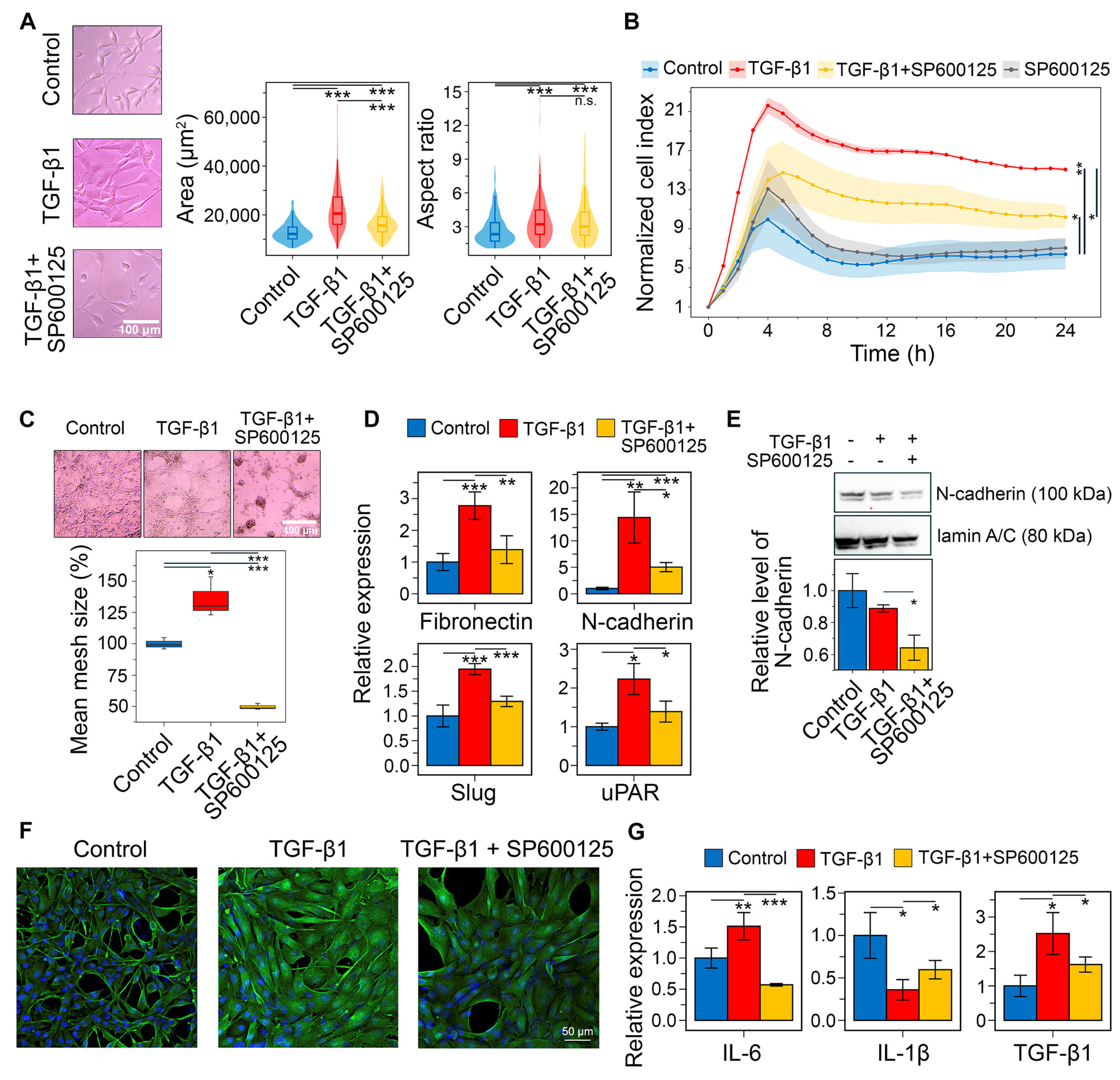
2.5. Effects of SP600125 on GMT Induced by TGF-β and Hypoxia
3. Discussion
- Vemurafenib: This BRAF inhibitor suppresses EMT in colorectal cancer [67] and melanoma [68] cells and demonstrates prolonged disease control in clinical trials for high-grade glioma [111]. However, since resistance to vemurafenib in melanoma led to a re-emergence of the mesenchymal phenotype [69], future evaluation of its anti-GMT potential should consider the chemoresistant status of GBM cells.
- FG-7142: Although there are no reports on the direct effect of this GABAA receptor agonist on GBM cells, its anti-GMT potency warrants investigation. Activation of the GABAA receptor suppresses brain cancer [115]. Furthermore, moxidectin, an analog of FG-7142, significantly inhibited cancer stem cell properties of medulloblastoma cells [116], which are closely associated with the mesenchymal phenotype of CNS tumors [117].
- Phensuximide: This anticonvulsant has not been previously investigated as an antitumor agent. However, network pharmacology and drug repurposing studies have identified it as a potential therapeutic agent for adrenal cortical [118] and breast [119] carcinomas, including a highly invasive form of the latter [120].
4. Materials and Methods
4.1. Bioinformatics
4.1.1. Data Acquisition
4.1.2. Gene Set Enrichment Analysis
4.1.3. Text Mining Analysis
4.1.4. Survival Analysis
4.1.5. Immune Cell Quantification
4.1.6. Weighted Gene Co-Expression Network Analysis (WGCNA)
4.1.7. Differential Expression Analysis
4.1.8. Gene Network Analysis
4.1.9. Connectivity Map Analysis
4.1.10. Chemoinformatics
4.2. Chemicals and Reagents
4.3. Cell Lines
4.4. Biological Evaluations
4.4.1. Cell Viability Analysis
4.4.2. Wound Healing Assay
4.4.3. Assessment of Cell Morphology
4.4.4. Transwell Migration Assay
4.4.5. Vasculogenic Mimicry Analysis
4.4.6. Western Blot
4.4.7. RT-qPCR
4.4.8. Immunofluorescence
4.4.9. Enzyme-Linked Immunosorbent Assay (ELISA)
4.5. Data Analysis and Visualization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BBB | Blood–brain barrier |
| CGGA | Chinese Glioma Genome Atlas |
| EMT | Epithelial–mesenchymal transition |
| GBM | Glioblastoma multiforme |
| GMT | Glial-mesenchymal transition |
| GSEA | Gene set enrichment analysis |
| GSVA | Gene set variation analysis |
| NCI | Normalized cell index |
| P-gp | P-glycoprotein |
| STITCH | Search Tool for Interactions of Chemicals |
| STRING | Search Tool for the Retrieval of Interacting Genes/Genomes |
| TCGA | The Cancer Genome Atlas |
| WGCNA | Weighted correlation network analysis |
References
- Obrador, E.; Moreno-Murciano, P.; Oriol-Caballo, M.; López-Blanch, R.; Pineda, B.; Gutiérrez-Arroyo, J.; Loras, A.; Gonzalez-Bonet, L.; Martinez-Cadenas, C.; Estrela, J.; et al. Glioblastoma Therapy: Past, Present and Future. Int. J. Mol. Sci. 2024, 25, 2529. [Google Scholar] [CrossRef]
- Poon, M.T.C.; Sudlow, C.L.M.; Figueroa, J.D.; Brennan, P.M. Longer-term (≥ 2 years) survival in patients with glioblastoma in population-based studies pre- and post-2005: A systematic review and meta-analysis. Sci. Rep. 2020, 10, 11622. [Google Scholar] [CrossRef]
- Vollmann-Zwerenz, A.; Leidgens, V.; Feliciello, G.; Klein, C.A.; Hau, P. Tumor Cell Invasion in Glioblastoma. Int. J. Mol. Sci. 2020, 21, 1932. [Google Scholar] [CrossRef]
- Verhaak, R.G.W.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Hu, B.; Hu, X.; Kim, H.; Squatrito, M.; Scarpace, L.; DeCarvalho, A.C.; Lyu, S.; Li, P.; Li, Y.; et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017, 32, 42–56. [Google Scholar] [CrossRef]
- Joseph, J.V.; Conroy, S.; Tomar, T.; Eggens-Meijer, E.; Bhat, K.; Copray, S.; Walenkamp, A.M.E.; Boddeke, E.; Balasubramanyian, V.; Wagemakers, M.; et al. TGF-β is an inducer of ZEB1-dependent mesenchymal transdifferentiation in glioblastoma that is associated with tumor invasion. Cell Death Dis. 2014, 5, e1443. [Google Scholar] [CrossRef]
- Azam, Z.; Shing-Shun Tony, T.O.; Tannous, B.A. Mesenchymal Transformation: The Rosetta Stone of Glioblastoma Pathogenesis and Therapy Resistance. Adv. Sci. 2020, 7, 2002015. [Google Scholar] [CrossRef]
- Odarenko, K.V.; Zenkova, M.A.; Markov, A.V. The Nexus of Inflammation-Induced Epithelial-Mesenchymal Transition and Lung Cancer Progression: A Roadmap to Pentacyclic Triterpenoid-Based Therapies. Int. J. Mol. Sci. 2023, 24, 17325. [Google Scholar] [CrossRef]
- Noronha, C.; Ribeiro, A.S.; Taipa, R.; Castro, D.S.; Reis, J.; Faria, C.; Paredes, J. Cadherin Expression and EMT: A Focus on Gliomas. Biomedicines 2021, 9, 1328. [Google Scholar] [CrossRef]
- Yin, W.; Zhu, H.; Tan, J.; Xin, Z.; Zhou, Q.; Cao, Y.; Wu, Z.; Wang, L.; Zhao, M.; Jiang, X.; et al. Identification of collagen genes related to immune infiltration and epithelial-mesenchymal transition in glioma. Cancer Cell Int. 2021, 21, 276. [Google Scholar] [CrossRef]
- Mao, J.; Liu, J.; Guo, G.; Mao, X.; Li, C. Glioblastoma vasculogenic mimicry: Signaling pathways progression and potential anti-angiogenesis targets. Biomark. Res. 2015, 3, 8. [Google Scholar] [CrossRef]
- Markov, A.V.; Moralev, A.D.; Odarenko, K.V. Sesquiterpene Lactones as Promising Anti-Glioblastoma Drug Candidates Exerting Complex Effects on Glioblastoma Cell Viability and Proneural–Mesenchymal Transition. Biomedicines 2025, 13, 133. [Google Scholar] [CrossRef]
- Kim, Y.; Varn, F.S.; Park, S.-H.H.; Yoon, B.W.; Park, H.R.; Lee, C.; Verhaak, R.G.W.W.; Paek, S.H. Perspective of mesenchymal transformation in glioblastoma. Acta Neuropathol. Commun. 2021, 9, 50. [Google Scholar] [CrossRef]
- Odarenko, K.V.; Sen’kova, A.V.; Salomatina, O.V.; Markov, O.V.; Salakhutdinov, N.F.; Zenkova, M.A.; Markov, A.V. Soloxolone para-methylanilide effectively suppresses aggressive phenotype of glioblastoma cells including TGF-β1-induced glial-mesenchymal transition in vitro and inhibits growth of U87 glioblastoma xenografts in mice. Front. Pharmacol. 2024, 15, 1428924. [Google Scholar] [CrossRef]
- Feng, Z.; Chen, G.; Huang, Y.; Zhang, K.; Wu, G.; Xing, W.; Wu, Y.; Zhou, Y.; Sun, C. TAK-242 inhibits glioblastoma invasion, migration, and proneural–mesenchymal transition by inhibiting TLR4 signaling. Exp. Cell Res. 2024, 439, 114091. [Google Scholar] [CrossRef]
- Haltom, A.R.; Hassen, W.E.; Hensel, J.; Kim, J.; Sugimoto, H.; Li, B.; McAndrews, K.M.; Conner, M.R.; Kirtley, M.L.; Luo, X.; et al. Engineered exosomes targeting MYC reverse the proneural-mesenchymal transition and extend survival of glioblastoma. Extracell. Vesicle 2022, 1, 100014. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Sun, M.; Huang, H.; Jin, W.-L. Drug repurposing for cancer therapy. Signal Transduct. Target. Ther. 2024, 9, 92. [Google Scholar] [CrossRef] [PubMed]
- Wen, P.Y.; Stein, A.; van den Bent, M.; De Greve, J.; Wick, A.; de Vos, F.Y.F.L.; von Bubnoff, N.; van Linde, M.E.; Lai, A.; Prager, G.W.; et al. Dabrafenib plus trametinib in patients with BRAFV600E-mutant low-grade and high-grade glioma (ROAR): A multicentre, open-label, single-arm, phase 2, basket trial. Lancet Oncol. 2022, 23, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, G.; De Salvo, G.L.; Brandes, A.A.; Eoli, M.; Rudà, R.; Faedi, M.; Lolli, I.; Pace, A.; Daniele, B.; Pasqualetti, F.; et al. Regorafenib compared with lomustine in patients with relapsed glioblastoma (REGOMA): A multicentre, open-label, randomised, controlled, phase 2 trial. Lancet Oncol. 2019, 20, 110–119. [Google Scholar] [CrossRef]
- Seok, H.J.; Choi, J.Y.; Lee, D.H.; Shin, I.; Bae, I.H. Atomoxetine suppresses radioresistance in glioblastoma via circATIC/miR-520d-5p/Notch2-Hey1 axis. Cell Commun. Signal. 2024, 22, 532. [Google Scholar] [CrossRef]
- Liang, M.-L.; Chen, C.-H.; Lin, Y.-C.; Lin, Y.-C.; Liu, Y.-R.; Ding, Y.-H.; Chu, C.-Y.; Hsieh, T.-H. Abemaciclib impairs glioblastoma sphere formation by targeting the GSK3β-mediated transcriptional regulation of CD44 and TCF7L2. Cancer Gene Ther. 2025, 32, 1120–1132. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e17. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.-W.; Liang, Y.-H.; Kuo, Y.-L.; Chuu, C.-P.; Lin, C.-Y.; Lee, M.-H.; Wu, A.T.H.; Yeh, C.-T.; Chen, E.I.-T.; Whang-Peng, J.; et al. Identification of thioridazine, an antipsychotic drug, as an antiglioblastoma and anticancer stem cell agent using public gene expression data. Cell Death Dis. 2015, 6, e1753. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.X.; Chen, R.Q.; Hu, D.X.; Xie, X.Q.; Yu, S.B.; Chen, X.Q. Identification of repaglinide as a therapeutic drug for glioblastoma multiforme. Biochem. Biophys. Res. Commun. 2017, 488, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Li, W.; Zheng, X.; Liu, J.; Yang, Y.; Li, S.; Zhang, S.; Fu, W.; Xiao, B.; Wang, J.; et al. Benzimidazoles induce concurrent apoptosis and pyroptosis of human glioblastoma cells via arresting cell cycle. Acta Pharmacol. Sin. 2022, 43, 194–208. [Google Scholar] [CrossRef]
- Chen, Y.; Pan, Y.; Gao, H.; Yi, Y.; Qin, S.; Ma, F.; Zhou, X.; Guan, M. Mechanistic insights into super-enhancer-driven genes as prognostic signatures in patients with glioblastoma. J. Cancer Res. Clin. Oncol. 2023, 149, 12315–12332. [Google Scholar] [CrossRef]
- Khan, R.B.; Tiwari, S.; Jarkharya, A.; Tiwari, A.; Chowdhary, R.; Shrivastava, A. Glioblastoma Multiforme miRNA based Comprehensive Study to Validate Phytochemicals for Effective Treatment against Deadly Tumour through In Silico Evaluation. MicroRNA 2024, 13, 240–250. [Google Scholar] [CrossRef]
- Dogra, N.; Singh, P.; Kumar, A. A Multistep In Silico Approach Identifies Potential Glioblastoma Drug Candidates via Inclusive Molecular Targeting of Glioblastoma Stem Cells. Mol. Neurobiol. 2024, 61, 9253–9271. [Google Scholar] [CrossRef]
- Johannessen, T.-C.; Hasan-Olive, M.M.; Zhu, H.; Denisova, O.; Grudic, A.; Latif, M.A.; Saed, H.; Varughese, J.K.; Røsland, G.V.; Yang, N.; et al. Thioridazine inhibits autophagy and sensitizes glioblastoma cells to temozolomide. Int. J. Cancer 2019, 144, 1735–1745. [Google Scholar] [CrossRef]
- Reka, A.K.; Kuick, R.; Kurapati, H.; Standiford, T.J.; Omenn, G.S.; Keshamouni, V.G. Identifying Inhibitors of Epithelial-Mesenchymal Transition by Connectivity Map–Based Systems Approach. J. Thorac. Oncol. 2011, 6, 1784–1792. [Google Scholar] [CrossRef]
- Zhang, B.; Zhao, R.; Wang, Q.; Zhang, Y.-J.; Yang, L.; Yuan, Z.-J.; Yang, J.; Wang, Q.-J.; Yao, L. An EMT-Related Gene Signature to Predict the Prognosis of Triple-Negative Breast Cancer. Adv. Ther. 2023, 40, 4339–4357. [Google Scholar] [CrossRef] [PubMed]
- Meng, K.; Zhao, Z.; Gao, Y.; Wu, K.; Liu, W.; Wang, X.; Zheng, Y.; Zhao, W.; Wang, B. The synergistic effects of anoikis-related genes and EMT-related genes in the prognostic prediction of Wilms tumor. Front. Mol. Biosci. 2024, 11, 1469775. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Guan, G.; Li, X.; Wei, C.; Wu, J.; Cheng, P.; Wu, A.; Cheng, W. Profiling pro-neural to mesenchymal transition identifies a lncRNA signature in glioma. J. Transl. Med. 2020, 18, 378. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.W.; Verhaak, R.G.W.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The Somatic Genomic Landscape of Glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, N.; Regev, A.; He, S.; Hemann, M.T. Integrated regulatory models for inference of subtype-specific susceptibilities in glioblastoma. Mol. Syst. Biol. 2020, 16, e9506. [Google Scholar] [CrossRef]
- Mukerjee, N.; Nag, S.; Bhattacharya, B.; Alexiou, A.; Mirgh, D.; Mukherjee, D.; Adhikari, M.D.; Anand, K.; Muthusamy, R.; Gorai, S.; et al. Clinical impact of epithelial–mesenchymal transition for cancer therapy. Clin. Transl. Discov. 2024, 4, e260. [Google Scholar] [CrossRef]
- Xu, C.; Hou, P.; Li, X.; Xiao, M.; Zhang, Z.; Li, Z.; Xu, J.; Liu, G.; Tan, Y.; Fang, C. Comprehensive understanding of glioblastoma molecular phenotypes: Classification, characteristics, and transition. Cancer Biol. Med. 2024, 21, 363–381. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Johansson, J.; Tabor, V.; Wikell, A.; Jalkanen, S.; Fuxe, J. TGF-β1-Induced Epithelial–Mesenchymal Transition Promotes Monocyte/Macrophage Properties in Breast Cancer Cells. Front. Oncol. 2015, 5, 3. [Google Scholar] [CrossRef]
- Chen, H.; Wang, Q.; Li, J.; Li, Y.; Chen, A.; Zhou, J.; Zhao, J.; Mao, Z.; Zhou, Z.; Zhang, J.; et al. IFNγ Transcribed by IRF1 in CD4+ Effector Memory T Cells Promotes Senescence-Associated Pulmonary Fibrosis. Aging Dis. 2023, 14, 2215. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, K.-N.; Wang, Q.; Li, G.; Zeng, F.; Zhang, Y.; Wu, F.; Chai, R.; Wang, Z.; Zhang, C.; et al. Chinese Glioma Genome Atlas (CGGA): A Comprehensive Resource with Functional Genomic Data from Chinese Glioma Patients. Genom. Proteom. Bioinform. 2021, 19, 1–12. [Google Scholar] [CrossRef]
- Gravendeel, L.A.M.; Kouwenhoven, M.C.M.; Gevaert, O.; de Rooi, J.J.; Stubbs, A.P.; Duijm, J.E.; Daemen, A.; Bleeker, F.E.; Bralten, L.B.C.; Kloosterhof, N.K.; et al. Intrinsic Gene Expression Profiles of Gliomas Are a Better Predictor of Survival than Histology. Cancer Res. 2009, 69, 9065–9072. [Google Scholar] [CrossRef] [PubMed]
- Sethi, B.; Kumar, V.; Mahato, K.; Coulter, D.W.; Mahato, R.I. Recent advances in drug delivery and targeting to the brain. J. Control. Release 2022, 350, 668–687. [Google Scholar] [CrossRef] [PubMed]
- Dréan, A.; Goldwirt, L.; Verreault, M.; Canney, M.; Schmitt, C.; Guehennec, J.; Delattre, J.-Y.; Carpentier, A.; Idbaih, A. Blood–brain barrier, cytotoxic chemotherapies and glioblastoma. Expert Rev. Neurother. 2016, 16, 1285–1300. [Google Scholar] [CrossRef] [PubMed]
- Ying, J.; Zhou, H.; Liu, P.; You, Q.; Kuang, F.; Shen, Y.; Hu, Z. Aspirin inhibited the metastasis of colon cancer cells by inhibiting the expression of toll-like receptor 4. Cell Biosci. 2018, 8, 1. [Google Scholar] [CrossRef]
- Nam, M.-H.; Smith, A.J.O.; Pantcheva, M.B.; Park, K.U.; Brzezinski, J.A.; Galligan, J.J.; Fritz, K.; Wormstone, I.M.; Nagaraj, R.H. Aspirin inhibits TGFβ2-induced epithelial to mesenchymal transition of lens epithelial cells: Selective acetylation of K56 and K122 in histone H3. Biochem. J. 2020, 477, 75–97. [Google Scholar] [CrossRef]
- Baribeau, S.; Chaudhry, P.; Parent, S.; Asselin, É. Resveratrol Inhibits Cisplatin-Induced Epithelial-to-Mesenchymal Transition in Ovarian Cancer Cell Lines. PLoS ONE 2014, 9, e86987. [Google Scholar] [CrossRef]
- Ferraresi, A.; Esposito, A.; Girone, C.; Vallino, L.; Salwa, A.; Ghezzi, I.; Thongchot, S.; Vidoni, C.; Dhanasekaran, D.N.; Isidoro, C. Resveratrol Contrasts LPA-Induced Ovarian Cancer Cell Migration and Platinum Resistance by Rescuing Hedgehog-Mediated Autophagy. Cells 2021, 10, 3213. [Google Scholar] [CrossRef]
- Cai, J.; Du, S.; Wang, H.; Xin, B.; Wang, J.; Shen, W.; Wei, W.; Guo, Z.; Shen, X. Tenascin-C induces migration and invasion through JNK/c-Jun signalling in pancreatic cancer. Oncotarget 2017, 8, 74406–74422. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, K.; Chen, Y.; Qiu, Y.; Zhu, J.; Li, B.; Wang, Z.; Chen, J. NKCC1 promotes proliferation, invasion and migration in human gastric cancer cells via activation of the MAPK-JNK/EMT signaling pathway. J. Cancer 2021, 12, 253–263. [Google Scholar] [CrossRef]
- Weygant, N.; Qu, D.; Berry, W.L.; May, R.; Chandrakesan, P.; Owen, D.B.; Sureban, S.M.; Ali, N.; Janknecht, R.; Houchen, C.W. Small molecule kinase inhibitor LRRK2-IN-1 demonstrates potent activity against colorectal and pancreatic cancer through inhibition of doublecortin-like kinase 1. Mol. Cancer 2014, 13, 103. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, H.; Shi, Y.; Li, J.; Ma, X.; Du, L.; Hu, Y.; Tao, M.; Zhong, Q.; Yan, D.; et al. Histone deacetylase 8 inhibition prevents the progression of peritoneal fibrosis by counteracting the epithelial-mesenchymal transition and blockade of M2 macrophage polarization. Front. Immunol. 2023, 14, 1137332. [Google Scholar] [CrossRef]
- Qi, S.; Yan, L.; Liu, Z.; Mu, Y.; Li, M.; Zhao, X.; Chen, Z.-J.; Zhang, H. Melatonin inhibits 17β-estradiol-induced migration, invasion and epithelial-mesenchymal transition in normal and endometriotic endometrial epithelial cells. Reprod. Biol. Endocrinol. 2018, 16, 62. [Google Scholar] [CrossRef]
- Pacheco-Velázquez, S.C.; Ortega-Mejía, I.I.; Vargas-Navarro, J.L.; Padilla-Flores, J.A.; Robledo-Cadena, D.X.; Tapia-Martínez, G.; Peñalosa-Castro, I.; Aguilar-Ponce, J.L.; Granados-Rivas, J.C.; Moreno-Sánchez, R.; et al. 17-β Estradiol up-regulates energy metabolic pathways, cellular proliferation and tumor invasiveness in ER+ breast cancer spheroids. Front. Oncol. 2022, 12, 1018137. [Google Scholar] [CrossRef]
- Qureshi, R.; Picon-Ruiz, M.; Sho, M.; Van Booven, D.; Nunes de Paiva, V.; Diaz-Ruano, A.B.; Ince, T.A.; Slingerland, J. Estrone, the major postmenopausal estrogen, binds ERa to induce SNAI2, epithelial-to-mesenchymal transition, and ER+ breast cancer metastasis. Cell Rep. 2022, 41, 111672. [Google Scholar] [CrossRef] [PubMed]
- Whitson, J.A.; Zhang, X.; Medvedovic, M.; Chen, J.; Wei, Z.; Monnier, V.M.; Fan, X. Transcriptome of the GSH-Depleted Lens Reveals Changes in Detoxification and EMT Signaling Genes, Transport Systems, and Lipid Homeostasis. Investig. Opthalmology Vis. Sci. 2017, 58, 2666. [Google Scholar] [CrossRef] [PubMed]
- Whitson, J.A.; Wilmarth, P.A.; Klimek, J.; Monnier, V.M.; David, L.; Fan, X. Proteomic analysis of the glutathione-deficient LEGSKO mouse lens reveals activation of EMT signaling, loss of lens specific markers, and changes in stress response proteins. Free Radic. Biol. Med. 2017, 113, 84–96. [Google Scholar] [CrossRef]
- Yang, K.; Song, Y.; Tang, Y.-B.; Xu, Z.-P.; Zhou, W.; Hou, L.-N.; Zhu, L.; Yu, Z.-H.; Chen, H.-Z.; Cui, Y.-Y. mAChRs activation induces epithelial-mesenchymal transition on lung epithelial cells. BMC Pulm. Med. 2014, 14, 53. [Google Scholar] [CrossRef]
- Thar Min, A.K.; Okayama, H.; Saito, M.; Ashizawa, M.; Aoto, K.; Nakajima, T.; Saito, K.; Hayase, S.; Sakamoto, W.; Tada, T.; et al. Epithelial-mesenchymal transition-converted tumor cells can induce T-cell apoptosis through upregulation of programmed death ligand 1 expression in esophageal squamous cell carcinoma. Cancer Med. 2018, 7, 3321–3330. [Google Scholar] [CrossRef]
- Fukuda, K.; Takeuchi, S.; Arai, S.; Kita, K.; Tanimoto, A.; Nishiyama, A.; Yano, S. Glycogen synthase kinase-3 inhibition overcomes epithelial-mesenchymal transition-associated resistance to osimertinib in EGFR-mutant lung cancer. Cancer Sci. 2020, 111, 2374–2384. [Google Scholar] [CrossRef]
- Zheng, S.-X.; Chen, J.; Zhuang, B.-B.; Zhang, Q.; Shi, S.-S.; Zhang, G.-L. Cordycepin improves sensitivity to temozolomide in glioblastoma cells by down-regulating MYC. J. Cancer Res. Clin. Oncol. 2023, 149, 16055–16067. [Google Scholar] [CrossRef]
- Lan, B.; Zhuang, Z.; Zhang, J.; He, Y.; Wang, N.; Deng, Z.; Mei, L.; Li, Y.; Gao, Y. Triggering of endoplasmic reticulum stress via ATF4-SPHK1 signaling promotes glioblastoma invasion and chemoresistance. Cell Death Dis. 2024, 15, 552. [Google Scholar] [CrossRef]
- Yi, G.; Liu, Y.; Xiang, W.; Wang, H.; Chen, Z.; Xie, S.; Qi, S. Akt and β-catenin contribute to TMZ resistance and EMT of MGMT negative malignant glioma cell line. J. Neurol. Sci. 2016, 367, 101–106. [Google Scholar] [CrossRef]
- Gao, Z.; Xu, J.; Fan, Y.; Qi, Y.; Wang, S.; Zhao, S.; Guo, X.; Xue, H.; Deng, L.; Zhao, R.; et al. PDIA3P1 promotes Temozolomide resistance in glioblastoma by inhibiting C/EBPβ degradation to facilitate proneural-to-mesenchymal transition. J. Exp. Clin. Cancer Res. 2022, 41, 223. [Google Scholar] [CrossRef]
- Shi, Z.-F.; Li, G.-Z.; Zhai, Y.; Pan, C.-Q.; Wang, D.; Yu, M.-C.; Liu, C.; Zhang, W.; Yu, X.-G. EGFRvIII Promotes the Proneural–Mesenchymal Transition of Glioblastoma Multiforme and Reduces Its Sensitivity to Temozolomide by Regulating the NF-κB/ALDH1A3 Axis. Genes 2023, 14, 651. [Google Scholar] [CrossRef]
- Radić, M.; Vlašić, I.; Jazvinšćak Jembrek, M.; Horvat, A.; Tadijan, A.; Sabol, M.; Dužević, M.; Herak Bosnar, M.; Slade, N. Characterization of Vemurafenib-Resistant Melanoma Cell Lines Reveals Novel Hallmarks of Targeted Therapy Resistance. Int. J. Mol. Sci. 2022, 23, 9910. [Google Scholar] [CrossRef]
- Padder, R.A.; Bhat, Z.I.; Ahmad, Z.; Singh, N.; Husain, M. DRP1 Promotes BRAFV600E-Driven Tumor Progression and Metabolic Reprogramming in Colorectal Cancer. Front. Oncol. 2021, 10, 592130. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Fan, W.; Li, Y.; Tang, R.; Tian, C.; Yang, Q.; Zhu, T.; Diao, C.; Hu, S.; Chen, M.M.; et al. Melatonin synergizes BRAF-targeting agent vemurafenib in melanoma treatment by inhibiting iNOS/hTERT signaling and cancer-stem cell traits. J. Exp. Clin. Cancer Res. 2019, 38, 48. [Google Scholar] [CrossRef] [PubMed]
- Jandova, J.; Wondrak, G.T. Vemurafenib Drives Epithelial-to-Mesenchymal Transition Gene Expression in BRAF Inhibitor—Resistant BRAFV600E/NRASQ61K Melanoma Enhancing Tumor Growth and Metastasis in a Bioluminescent Murine Model. J. Investig. Dermatol. 2022, 142, 1456–1465.e1. [Google Scholar] [CrossRef] [PubMed]
- Ming, J.; Sun, B.; Li, Z.; Lin, L.; Meng, X.; Han, B.; Wang, R.; Wu, P.; Li, J.; Cai, J.; et al. Aspirin inhibits the SHH/GLI1 signaling pathway and sensitizes malignant glioma cells to temozolomide therapy. Aging 2017, 9, 1233–1247. [Google Scholar] [CrossRef]
- Song, Y.; Chen, Y.; Li, Y.; Lyu, X.; Cui, J.; Cheng, Y.; Zheng, T.; Zhao, L.; Zhao, G. Resveratrol Suppresses Epithelial-Mesenchymal Transition in GBM by Regulating Smad-Dependent Signaling. Biomed Res. Int. 2019, 2019, 1321973. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Li, Y.; Liu, M.; Li, Q.; Liu, Q.; Li, R. The Interleukin-33/ST2 axis promotes glioma mesenchymal transition, stemness and TMZ resistance via JNK activation. Aging 2020, 12, 1685–1703. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Vega, A.M.; Del Moral-Morales, A.; Zamora-Sánchez, C.J.; Piña-Medina, A.G.; González-Arenas, A.; Camacho-Arroyo, I. Estradiol Induces Epithelial to Mesenchymal Transition of Human Glioblastoma Cells. Cells 2020, 9, 1930. [Google Scholar] [CrossRef] [PubMed]
- Pilotto Heming, C.; Muriithi, W.; Wanjiku Macharia, L.; Niemeyer Filho, P.; Moura-Neto, V.; Aran, V. P-glycoprotein and cancer: What do we currently know? Heliyon 2022, 8, e11171. [Google Scholar] [CrossRef]
- Joiakim, A.; Mathieu, P.A.; Palermo, C.; Gasiewicz, T.A.; Reiners, J.J. The Jun N-terminal kinase inhibitor SP600125 is a ligand and antagonist of the aryl hydrocarbon receptor. Drug Metab. Dispos. 2003, 31, 1279–1282. [Google Scholar] [CrossRef]
- Kim, J.A.; Lee, J.; Margolis, R.L.; Fotedar, R. SP600125 suppresses Cdk1 and induces endoreplication directly from G2 phase, independent of JNK inhibition. Oncogene 2010, 29, 1702–1716. [Google Scholar] [CrossRef]
- Colombo, R.; Caldarelli, M.; Mennecozzi, M.; Giorgini, M.L.; Sola, F.; Cappella, P.; Perrera, C.; Depaolini, S.R.; Rusconi, L.; Cucchi, U.; et al. Targeting the Mitotic Checkpoint for Cancer Therapy with NMS-P715, an Inhibitor of MPS1 Kinase. Cancer Res. 2010, 70, 10255–10264. [Google Scholar] [CrossRef]
- Tam, S.Y.; Wu, V.W.C.; Law, H.K.W. JNK Pathway Mediates Low Oxygen Level Induced Epithelial–Mesenchymal Transition and Stemness Maintenance in Colorectal Cancer Cells. Cancers 2020, 12, 224. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Gao, Y.; Kang, J.; Wang, W.; Yong, Y.-L.; Qu, X.; Dang, X.; Shang, D.; Shao, Y.; et al. Fine particulate matter exposure disturbs autophagy, redox balance and mitochondrial homeostasis via JNK activation to inhibit proliferation and promote EMT in human alveolar epithelial A549 cells. Ecotoxicol. Environ. Saf. 2023, 262, 115134. [Google Scholar] [CrossRef]
- Al-Mutairi, M.S.; Habashy, H.O. DUSP4 Silencing Enhances the Sensitivity of Breast Cancer Cells to Doxorubicin through the Activation of the JNK/c-Jun Signalling Pathway. Molecules 2022, 27, 6146. [Google Scholar] [CrossRef]
- Ling, G.; Ji, Q.; Ye, W.; Ma, D.; Wang, Y. Epithelial-mesenchymal transition regulated by p38/MAPK signaling pathways participates in vasculogenic mimicry formation in SHG44 cells transfected with TGF-β cDNA loaded lentivirus in vitro and in vivo. Int. J. Oncol. 2016, 49, 2387–2398. [Google Scholar] [CrossRef]
- Zhang, X.; Song, Q.; Wei, C.; Qu, J. LRIG1 inhibits hypoxia-induced vasculogenic mimicry formation via suppression of the EGFR/PI3K/AKT pathway and epithelial-to-mesenchymal transition in human glioma SHG-44 cells. Cell Stress Chaperones 2015, 20, 631–641. [Google Scholar] [CrossRef]
- Yao, Y.; Ye, H.; Qi, Z.; Mo, L.; Yue, Q.; Baral, A.; Hoon, D.S.B.; Vera, J.C.; Heiss, J.D.; Chen, C.C.; et al. B7-H4(B7x)–Mediated Cross-talk between Glioma-Initiating Cells and Macrophages via the IL6/JAK/STAT3 Pathway Lead to Poor Prognosis in Glioma Patients. Clin. Cancer Res. 2016, 22, 2778–2790. [Google Scholar] [CrossRef]
- Tan, S.K.; Jermakowicz, A.; Mookhtiar, A.K.; Nemeroff, C.B.; Schürer, S.C.; Ayad, N.G. Drug Repositioning in Glioblastoma: A Pathway Perspective. Front. Pharmacol. 2018, 9, 218. [Google Scholar] [CrossRef]
- Lai, Y.; Lu, X.; Liao, Y.; Ouyang, P.; Wang, H.; Zhang, X.; Huang, G.; Qi, S.; Li, Y. Crosstalk between glioblastoma and tumor microenvironment drives proneural–mesenchymal transition through ligand-receptor interactions. Genes Dis. 2024, 11, 874–889. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Sun, Y.; Zhang, Q.; Jin, W.; Gordon, R.E.; Zhang, Y.; Wang, J.; Sun, C.; Wang, Z.J.; Qi, X.; et al. Pro-inflammatory and proliferative microglia drive progression of glioblastoma. Cell Rep. 2021, 36, 109718. [Google Scholar] [CrossRef] [PubMed]
- Basheer, A.S.; Abas, F.; Othman, I.; Naidu, R. Role of Inflammatory Mediators, Macrophages, and Neutrophils in Glioma Maintenance and Progression: Mechanistic Understanding and Potential Therapeutic Applications. Cancers 2021, 13, 4226. [Google Scholar] [CrossRef] [PubMed]
- Zanotto-Filho, A.; Gonçalves, R.M.; Klafke, K.; de Souza, P.O.; Dillenburg, F.C.; Carro, L.; Gelain, D.P.; Moreira, J.C.F. Inflammatory landscape of human brain tumors reveals an NFκB dependent cytokine pathway associated with mesenchymal glioblastoma. Cancer Lett. 2017, 390, 176–187. [Google Scholar] [CrossRef]
- Ji, H.; Ba, Y.; Ma, S.; Hou, K.; Mi, S.; Gao, X.; Jin, J.; Gong, Q.; Liu, T.; Wang, F.; et al. Construction of Interferon-Gamma-Related Gene Signature to Characterize the Immune-Inflamed Phenotype of Glioblastoma and Predict Prognosis, Efficacy of Immunotherapy and Radiotherapy. Front. Immunol. 2021, 12, 729359. [Google Scholar] [CrossRef]
- Li, X.-Y.; Zhang, L.-Y.; Li, X.-Y.; Yang, X.-T.; Su, L.-X. A Pyroptosis-Related Gene Signature for Predicting Survival in Glioblastoma. Front. Oncol. 2021, 11, 697198. [Google Scholar] [CrossRef]
- Pilchova, I.; Klacanova, K.; Dibdiakova, K.; Saksonova, S.; Stefanikova, A.; Vidomanova, E.; Lichardusova, L.; Hatok, J.; Racay, P. Proteasome Stress Triggers Death of SH-SY5Y and T98G Cells via Different Cellular Mechanisms. Neurochem. Res. 2017, 42, 3170–3185. [Google Scholar] [CrossRef] [PubMed]
- Kardosh, A.; Golden, E.B.; Pyrko, P.; Uddin, J.; Hofman, F.M.; Chen, T.C.; Louie, S.G.; Petasis, N.A.; Schönthal, A.H. Aggravated Endoplasmic Reticulum Stress as a Basis for Enhanced Glioblastoma Cell Killing by Bortezomib in Combination with Celecoxib or Its Non-Coxib Analogue, 2,5-Dimethyl-Celecoxib. Cancer Res. 2008, 68, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Banik, N.L.; Ray, S.K. Flavonoids activated caspases for apoptosis in human glioblastoma T98G and U87MG cells but not in human normal astrocytes. Cancer 2010, 116, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Banik, N.L.; Ray, S.K. Garlic compounds generate reactive oxygen species leading to activation of stress kinases and cysteine proteases for apoptosis in human glioblastoma T98G and U87MG cells. Cancer 2007, 110, 1083–1095. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, M.; Ji, F.; Peng, Y.; Wang, B.; Zhao, J.; Wu, J.; Zhao, H. A Novel Glucose Metabolism-Related Gene Signature for Overall Survival Prediction in Patients with Glioblastoma. Biomed Res. Int. 2021, 2021, 8872977. [Google Scholar] [CrossRef]
- Han, J.; Puri, R.K. Analysis of the cancer genome atlas (TCGA) database identifies an inverse relationship between interleukin-13 receptor α1 and α2 gene expression and poor prognosis and drug resistance in subjects with glioblastoma multiforme. J. Neurooncol. 2018, 136, 463–474. [Google Scholar] [CrossRef]
- Dong, J.; Peng, Y.; Zhong, M.; Xie, Z.; Jiang, Z.; Wang, K.; Wu, Y. Implication of lncRNA ZBED3-AS1 downregulation in acquired resistance to Temozolomide and glycolysis in glioblastoma. Eur. J. Pharmacol. 2023, 938, 175444. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Y.; Wang, M. Identification of a Pyroptosis-Related Gene Signature and Effect of Silencing the CHMP4C and CASP4 in Pancreatic Adenocarcinoma. Int. J. Gen. Med. 2022, 15, 3199–3213. [Google Scholar] [CrossRef]
- Papoff, G.; Presutti, D.; Lalli, C.; Bolasco, G.; Santini, S.; Manelfi, C.; Fustaino, V.; Alemà, S.; Ruberti, G. CASP4 gene silencing in epithelial cancer cells leads to impairment of cell migration, cell-matrix adhesion and tissue invasion. Sci. Rep. 2018, 8, 17705. [Google Scholar] [CrossRef]
- Chiba, Y.; Doi, T.; Obayashi, K.; Sumida, K.; Nagasaka, S.; Wang, K.-Y.; Yamasaki, K.; Masago, K.; Matsushita, H.; Kuroda, H.; et al. Caspase-4 promotes metastasis and interferon-γ-induced pyroptosis in lung adenocarcinoma. Commun. Biol. 2024, 7, 699. [Google Scholar] [CrossRef]
- Cao, H.; Zhang, J.; Liu, H.; Wan, L.; Zhang, H.; Huang, Q.; Xu, E.; Lai, M. IL-13/STAT6 signaling plays a critical role in the epithelial-mesenchymal transition of colorectal cancer cells. Oncotarget 2016, 7, 61183–61198. [Google Scholar] [CrossRef]
- Zhang, Y.; Lin, W.; Yang, Y.; Zhu, S.; Chen, Y.; Wang, H.; Teng, L. MEF2D facilitates liver metastasis of gastric cancer cells through directly inducing H1X under IL-13 stimulation. Cancer Lett. 2024, 591, 216878. [Google Scholar] [CrossRef]
- Zheng, N.; Huo, Z.; Zhang, B.; Meng, M.; Cao, Z.; Wang, Z.; Zhou, Q. Thrombomodulin reduces tumorigenic and metastatic potential of lung cancer cells by up-regulation of E-cadherin and down-regulation of N-cadherin expression. Biochem. Biophys. Res. Commun. 2016, 476, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-T.; Chang, Y.-H.; Lin, P.-Y.; Chen, W.-C.; Chen, M.-F. Thrombomodulin expression regulates tumorigenesis in bladder cancer. BMC Cancer 2014, 14, 375. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-J.; Cheng, Y.-W.; Lin, R.-K.; Huang, C.-C.; Chen, W.T.-L.; Ke, T.-W.; Wei, P.-L. Thrombomodulin Influences the Survival of Patients with Non-Metastatic Colorectal Cancer through Epithelial-To-Mesenchymal Transition (EMT). PLoS ONE 2016, 11, e0160550. [Google Scholar] [CrossRef] [PubMed]
- Dadi, K.; Varoquaux, G.; Houenou, J.; Bzdok, D.; Thirion, B.; Engemann, D. Population modeling with machine learning can enhance measures of mental health. Gigascience 2021, 10, giab071. [Google Scholar] [CrossRef]
- Kwon, H.; Ali, Z.A.; Wong, B.M. Harnessing Semi-Supervised Machine Learning to Automatically Predict Bioactivities of Per- and Polyfluoroalkyl Substances (PFASs). Environ. Sci. Technol. Lett. 2023, 10, 1017–1022. [Google Scholar] [CrossRef]
- Tang, M.; Jiang, S.; Huang, X.; Ji, C.; Gu, Y.; Qi, Y.; Xiang, Y.; Yao, E.; Zhang, N.; Berman, E.; et al. Integration of 3D bioprinting and multi-algorithm machine learning identified glioma susceptibilities and microenvironment characteristics. Cell Discov. 2024, 10, 39. [Google Scholar] [CrossRef]
- Lee, S.; Weiss, T.; Bühler, M.; Mena, J.; Lottenbach, Z.; Wegmann, R.; Sun, M.; Bihl, M.; Augustynek, B.; Baumann, S.P.; et al. High-throughput identification of repurposable neuroactive drugs with potent anti-glioblastoma activity. Nat. Med. 2024, 30, 3196–3208. [Google Scholar] [CrossRef]
- Ghafoor, N.A.; Yildiz, A. Targeting MDM2–p53 Axis through Drug Repurposing for Cancer Therapy: A Multidisciplinary Approach. ACS Omega 2023, 8, 34583–34596. [Google Scholar] [CrossRef]
- Di Nunno, V.; Gatto, L.; Tosoni, A.; Bartolini, S.; Franceschi, E. Implications of BRAF V600E mutation in gliomas: Molecular considerations, prognostic value and treatment evolution. Front. Oncol. 2023, 12, 1067252. [Google Scholar] [CrossRef] [PubMed]
- Dhandapani, K.M.; Mahesh, V.B.; Brann, D.W. Curcumin suppresses growth and chemoresistance of human glioblastoma cells via AP-1 and NFκB transcription factors. J. Neurochem. 2007, 102, 522–538. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.-F.; Tzeng, Y.-M.; Hung, H.-C.; Liu, G.-Y. Dibenzoylmethane, hydroxydibenzoylmethane and hydroxymethyldibenzoylmethane inhibit phorbol-12-myristate 13-acetate-induced breast carcinoma cell invasion. Mol. Med. Rep. 2015, 11, 4597–4604. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Davidson, A.J.; Wu, D.; Marshall, F.F.; Chung, L.W.K.; Zhau, H.E.; He, D.; Wang, R. Phorbol ester phorbol-12-myristate-13-acetate induces epithelial to mesenchymal transition in human prostate cancer ARCaP E cells. Prostate 2010, 70, 1119–1126. [Google Scholar] [CrossRef]
- Huang, D.; Alexander, P.B.; Li, Q.-J.; Wang, X.-F. GABAergic signaling beyond synapses: An emerging target for cancer therapy. Trends Cell Biol. 2023, 33, 403–412. [Google Scholar] [CrossRef]
- Kaushik, I.; Srivastava, S.K. GABAA receptor agonist suppresses pediatric medulloblastoma progression by inhibiting PKA-Gli1 signaling axis. Mol. Ther. 2022, 30, 2584–2602. [Google Scholar] [CrossRef]
- Han, Y.-P.; Lin, H.-W.; Li, H. Cancer Stem Cells in Tumours of the Central Nervous System in Children: A Comprehensive Review. Cancers 2023, 15, 3154. [Google Scholar] [CrossRef]
- Ma, C.; Xiong, J.; Su, H.; Li, H. The underlying molecular mechanism and drugs for treatment in adrenal cortical carcinoma. Int. J. Med. Sci. 2021, 18, 3026–3038. [Google Scholar] [CrossRef]
- Firoozbakht, F.; Rezaeian, I.; Rueda, L.; Ngom, A. Computationally repurposing drugs for breast cancer subtypes using a network-based approach. BMC Bioinform. 2022, 23, 143. [Google Scholar] [CrossRef]
- Wang, F.; Tang, C.; Gao, X.; Xu, J. Identification of a six-gene signature associated with tumor mutation burden for predicting prognosis in patients with invasive breast carcinoma. Ann. Transl. Med. 2020, 8, 453. [Google Scholar] [CrossRef]
- Xue, H.; Yuan, G.; Guo, X.; Liu, Q.; Zhang, J.; Gao, X.; Guo, X.; Xu, S.; Li, T.; Shao, Q.; et al. A novel tumor-promoting mechanism of IL6 and the therapeutic efficacy of tocilizumab: Hypoxia-induced IL6 is a potent autophagy initiator in glioblastoma via the p-STAT3- MIR155-3p -CREBRF pathway. Autophagy 2016, 12, 1129–1152. [Google Scholar] [CrossRef]
- Herwartz, C.; Castillo-Juárez, P.; Schröder, L.; Barron, B.L.; Steger, G. The Transcription Factor ZNF395 Is Required for the Maximal Hypoxic Induction of Proinflammatory Cytokines in U87-MG Cells. Mediat. Inflamm. 2015, 2015, 804264. [Google Scholar] [CrossRef]
- Shen, C.-K.; Huang, B.-R.; Yeh, W.-L.; Chen, C.-W.; Liu, Y.-S.; Lai, S.-W.; Tseng, W.-P.; Lu, D.-Y.; Tsai, C.-F. Regulatory effects of IL-1β in the interaction of GBM and tumor-associated monocyte through VCAM-1 and ICAM-1. Eur. J. Pharmacol. 2021, 905, 174216. [Google Scholar] [CrossRef]
- Yan, H.; He, L.; Lv, D.; Yang, J.; Yuan, Z. The Role of the Dysregulated JNK Signaling Pathway in the Pathogenesis of Human Diseases and Its Potential Therapeutic Strategies: A Comprehensive Review. Biomolecules 2024, 14, 243. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, J.; Wu, Q.; Wang, L.; Lu, W.; Feng, Y. ANKRD22 promotes glioma proliferation, migration, invasion, and epithelial-mesenchymal transition by upregulating E2F1-mediated MELK expression. J. Neuropathol. Exp. Neurol. 2023, 82, 631–640. [Google Scholar] [CrossRef]
- Shee, K.; Chambers, M.; Hughes, E.G.; Almiron, D.A.; Deharvengt, S.J.; Green, D.; Lefferts, J.A.; Andrew, A.S.; Hickey, W.F.; Tsongalis, G.J. Molecular genetic profiling reveals novel association between FLT3 mutation and survival in glioma. J. Neurooncol. 2020, 148, 473–480. [Google Scholar] [CrossRef]
- Andrae, N.; Kirches, E.; Hartig, R.; Haase, D.; Keilhoff, G.; Kalinski, T.; Mawrin, C. Sunitinib targets PDGF-receptor and Flt3 and reduces survival and migration of human meningioma cells. Eur. J. Cancer 2012, 48, 1831–1841. [Google Scholar] [CrossRef] [PubMed]
- Ennis, B.W.; Fultz, K.E.; Smith, K.A.; Westwick, J.K.; Zhu, D.; Boluro-Ajayi, M.; Bilter, G.K.; Stein, B. Inhibition of Tumor Growth, Angiogenesis, and Tumor Cell Proliferation by a Small Molecule Inhibitor of c-Jun N-terminal Kinase. J. Pharmacol. Exp. Ther. 2005, 313, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Wu, C.-L.; Wang, X.; Ban, Q.; Quan, C.; Liu, M.; Dong, H.; Li, J.; Kim, G.-Y.; Choi, Y.H.; et al. SP600125 enhances C-2-induced cell death by the switch from autophagy to apoptosis in bladder cancer cells. J. Exp. Clin. Cancer Res. 2019, 38, 448. [Google Scholar] [CrossRef]
- Wang, W.; Shi, L.; Xie, Y.; Ma, C.; Li, W.; Su, X.; Huang, S.; Chen, R.; Zhu, Z.; Mao, Z.; et al. SP600125, a new JNK inhibitor, protects dopaminergic neurons in the MPTP model of Parkinson’s disease. Neurosci. Res. 2004, 48, 195–202. [Google Scholar] [CrossRef]
- Gagnani, R.; Singh, H.; Suri, M.; Bali, A. JNK inhibition mitigates sepsis-associated encephalopathy via attenuation of neuroinflammation, oxidative stress and apoptosis. Metab. Brain Dis. 2025, 40, 148. [Google Scholar] [CrossRef]
- Zhong, B.; Zhang, Y.; Zheng, H.; Chen, Q.; Lu, H.; Chen, X. SP600125, a selective JNK inhibitor, is a potent inhibitor of NAD(P)H: Quinone oxidoreductase 1 (NQO1). Acta Pharmacol. Sin. 2025, 46, 1137–1144. [Google Scholar] [CrossRef]
- Yao, C.; Shen, Z.; Shen, L.; Kadier, K.; Zhao, J.; Guo, Y.; Xu, L.; Cao, J.; Dong, X.; Yang, B. Identification of Potential JNK3 Inhibitors: A Combined Approach Using Molecular Docking and Deep Learning-Based Virtual Screening. Pharmaceuticals 2023, 16, 1459. [Google Scholar] [CrossRef]
- Prasad, K.D.; Trinath, J.; Biswas, A.; Sekar, K.; Balaji, K.N.; Guru Row, T.N. Anthrapyrazolone analogues intercept inflammatory JNK signals to moderate endotoxin induced septic shock. Sci. Rep. 2014, 4, 7214. [Google Scholar] [CrossRef] [PubMed]
- Ganduri, R.; Singh, V.; Biswas, A.; Karothu, D.P.; Sekar, K.; Balaji, K.N.; Guru Row, T.N. Structural and biological evaluation of halogen derivatives of 1,9-pyrazoloanthrones towards the design of a specific potent inhibitor of c-Jun-N-terminal kinase (JNK). New J. Chem. 2018, 42, 10651–10660. [Google Scholar] [CrossRef]
- Kusakabe, K.; Ide, N.; Daigo, Y.; Tachibana, Y.; Itoh, T.; Yamamoto, T.; Hashizume, H.; Hato, Y.; Higashino, K.; Okano, Y.; et al. Indazole-Based Potent and Cell-Active Mps1 Kinase Inhibitors: Rational Design from Pan-Kinase Inhibitor Anthrapyrazolone (SP600125). J. Med. Chem. 2013, 56, 4343–4356. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Zhang, D.; Du, F.; An, W.; Ge, J.; Yu, C.; Yang, N.; Zhang, C.; Lim, K.; Li, L. Design and Synthesis of a Mitochondrial—Targeted JNK Inhibitor and Its Protective Effect on Parkinson’s Disease Phenotypes. ChemBioChem 2023, 24, e202200748. [Google Scholar] [CrossRef]
- Wu, Q.; Wu, W.; Jacevic, V.; Franca, T.C.C.; Wang, X.; Kuca, K. Selective inhibitors for JNK signalling: A potential targeted therapy in cancer. J. Enzym. Inhib. Med. Chem. 2020, 35, 574–583. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Fertig, E.J.; Jaffe, A.E.; Zhang, Y.; Storey, J.D.T.L. Sva: Surrogate Variable Analysis. Available online: https://bioconductor.org/packages/sva (accessed on 6 November 2024).
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-Seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Wang, J.-H.; Zhao, L.-F.; Wang, H.-F.; Wen, Y.-T.; Jiang, K.-K.; Mao, X.-M.; Zhou, Z.-Y.; Yao, K.-T.; Geng, Q.-S.; Guo, D.; et al. GenCLiP 3: Mining human genes’ functions and regulatory networks from PubMed based on co-occurrences and natural language processing. Bioinformatics 2020, 36, 1973–1975. [Google Scholar] [CrossRef]
- Allot, A.; Chen, Q.; Kim, S.; Vera Alvarez, R.; Comeau, D.C.; Wilbur, W.J.; Lu, Z. LitSense: Making sense of biomedical literature at sentence level. Nucleic Acids Res. 2019, 47, W594–W599. [Google Scholar] [CrossRef] [PubMed]
- Kassambara, A.; Kosinski, M.; Biecek, P. Survminer: Drawing Survival Curves Using Ggplot2. Available online: https://cran.r-project.org/web/packages/survminer/index.html (accessed on 6 November 2024).
- Therneau, T.M. Survival: Survival Analysis. Available online: https://cran.r-project.org/web/packages/survival/index.html (accessed on 6 November 2024).
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef]
- Chin, C.-H.H.; Chen, S.-H.H.; Wu, H.-H.H.; Ho, C.-W.W.; Ko, M.-T.T.; Lin, C.-Y.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Santos, A.; Von Mering, C.; Jensen, L.J.; Bork, P.; Kuhn, M. STITCH 5: Augmenting protein–chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016, 44, D380–D384. [Google Scholar] [CrossRef]
- Liu, H.; Wang, L.; Lv, M.; Pei, R.; Li, P.; Pei, Z.; Wang, Y.; Su, W.; Xie, X.-Q.Q. AlzPlatform: An Alzheimer’s Disease Domain-Specific Chemogenomics Knowledgebase for Polypharmacology and Target Identification Research. J. Chem. Inf. Model. 2014, 54, 1050–1060. [Google Scholar] [CrossRef]
- Montanari, F.; Knasmüller, B.; Kohlbacher, S.; Hillisch, C.; Baierová, C.; Grandits, M.; Ecker, G.F. Vienna LiverTox Workspace—A Set of Machine Learning Models for Prediction of Interactions Profiles of Small Molecules With Transporters Relevant for Regulatory Agencies. Front. Chem. 2020, 7, 899. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Dong, J.; Wang, N.N.; Yao, Z.J.; Zhang, L.; Cheng, Y.; Ouyang, D.; Lu, A.P.; Cao, D.S. Admetlab: A platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J. Cheminform. 2018, 10, 29. [Google Scholar] [CrossRef]
- Markov, A.V.; Ilyina, A.A.; Salomatina, O.V.; Sen’kova, A.V.; Okhina, A.A.; Rogachev, A.D.; Salakhutdinov, N.F.; Zenkova, M.A. Novel Soloxolone Amides as Potent Anti-Glioblastoma Candidates: Design, Synthesis, In Silico Analysis and Biological Activities In Vitro and In Vivo. Pharmaceuticals 2022, 15, 603. [Google Scholar] [CrossRef]
- Karatzas, E.; Zamora, J.E.; Athanasiadis, E.; Dellis, D.; Cournia, Z.; Spyrou, G.M. ChemBioServer 2.0: An advanced web server for filtering, clustering and networking of chemical compounds facilitating both drug discovery and repurposing. Bioinformatics 2020, 36, 2602–2604. [Google Scholar] [CrossRef]
- Moralev, A.D.; Salomatina, O.V.; Salakhutdinov, N.F.; Zenkova, M.A.; Markov, A.V. Soloxolone N-3-(Dimethylamino)propylamide Restores Drug Sensitivity of Tumor Cells with Multidrug-Resistant Phenotype via Inhibition of P-Glycoprotein Efflux Function. Molecules 2024, 29, 4939. [Google Scholar] [CrossRef]
- Carpentier, G.; Berndt, S.; Ferratge, S.; Rasband, W.; Cuendet, M.; Uzan, G.; Albanese, P. Angiogenesis Analyzer for ImageJ—A comparative morphometric analysis of “Endothelial Tube Formation Assay” and “Fibrin Bead Assay”. Sci. Rep. 2020, 10, 11568. [Google Scholar] [CrossRef]
- Wickham, H. Data Analysis. In Ggplot2. Use R! Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Yan, L. Ggvenn: Draw Venn Diagram by Ggplot2. Available online: https://cran.r-project.org/web/packages/ggvenn/index.html (accessed on 6 November 2024).
- Allaire, J.J.; Gandrud, C.; Russell, K.; Yetman, C.J. NetworkD3: D3 JavaScript Network Graphs from R. Available online: https://cran.r-project.org/package=networkD3 (accessed on 6 November 2024).
- Sievert, C. Interactive Web-Based Data Visualization with R, Plotly, and Shiny; Chapman and Hall/CRC: Boca Raton, FL, USA, 2020; ISBN 9780429447273. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Odarenko, K.V.; Zenkova, M.A.; Markov, A.V. Transcriptomic-Driven Drug Repurposing Reveals SP600125 as a Promising Drug Candidate for the Treatment of Glial-Mesenchymal Transition in Glioblastoma. Int. J. Mol. Sci. 2025, 26, 9772. https://doi.org/10.3390/ijms26199772
Odarenko KV, Zenkova MA, Markov AV. Transcriptomic-Driven Drug Repurposing Reveals SP600125 as a Promising Drug Candidate for the Treatment of Glial-Mesenchymal Transition in Glioblastoma. International Journal of Molecular Sciences. 2025; 26(19):9772. https://doi.org/10.3390/ijms26199772
Chicago/Turabian StyleOdarenko, Kirill V., Marina A. Zenkova, and Andrey V. Markov. 2025. "Transcriptomic-Driven Drug Repurposing Reveals SP600125 as a Promising Drug Candidate for the Treatment of Glial-Mesenchymal Transition in Glioblastoma" International Journal of Molecular Sciences 26, no. 19: 9772. https://doi.org/10.3390/ijms26199772
APA StyleOdarenko, K. V., Zenkova, M. A., & Markov, A. V. (2025). Transcriptomic-Driven Drug Repurposing Reveals SP600125 as a Promising Drug Candidate for the Treatment of Glial-Mesenchymal Transition in Glioblastoma. International Journal of Molecular Sciences, 26(19), 9772. https://doi.org/10.3390/ijms26199772






