Custom Gene Panel Analysis Identifies Novel Polymorphisms Associated with Clopidogrel Response in Patients Undergoing Percutaneous Coronary Intervention with Stent
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics
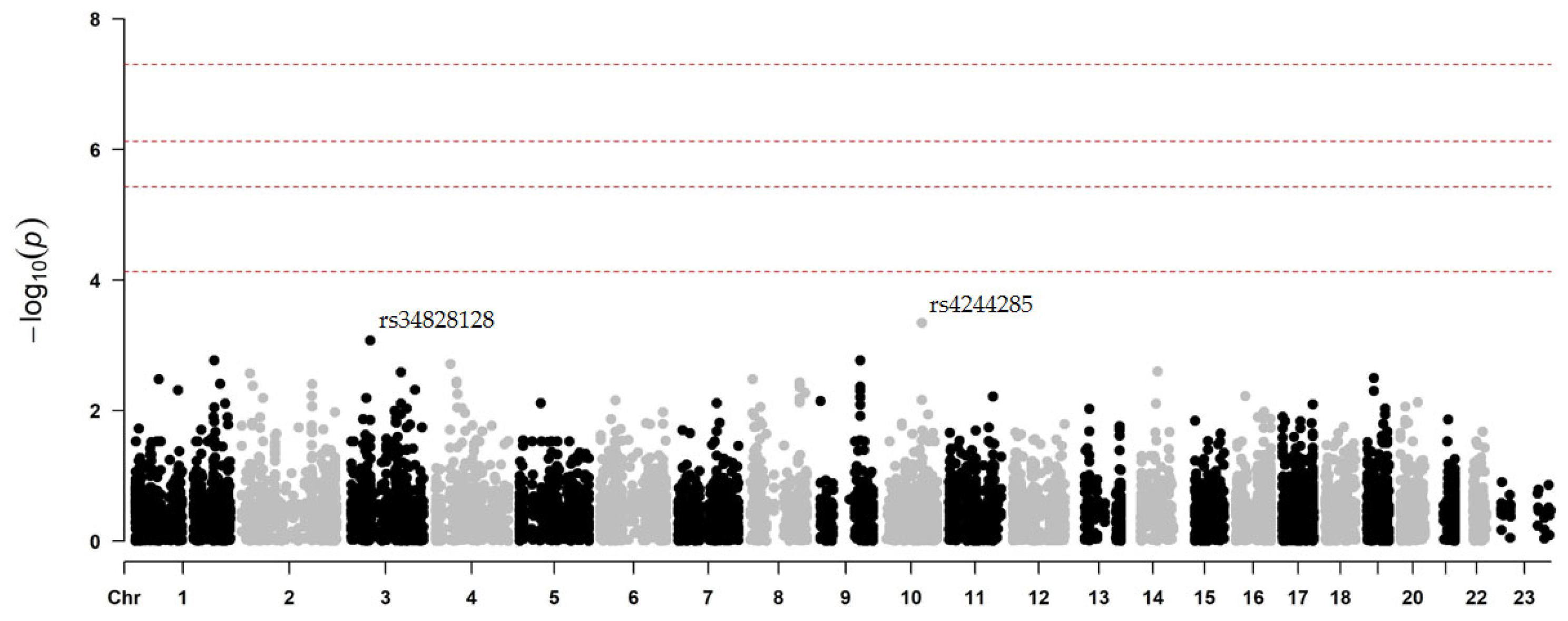
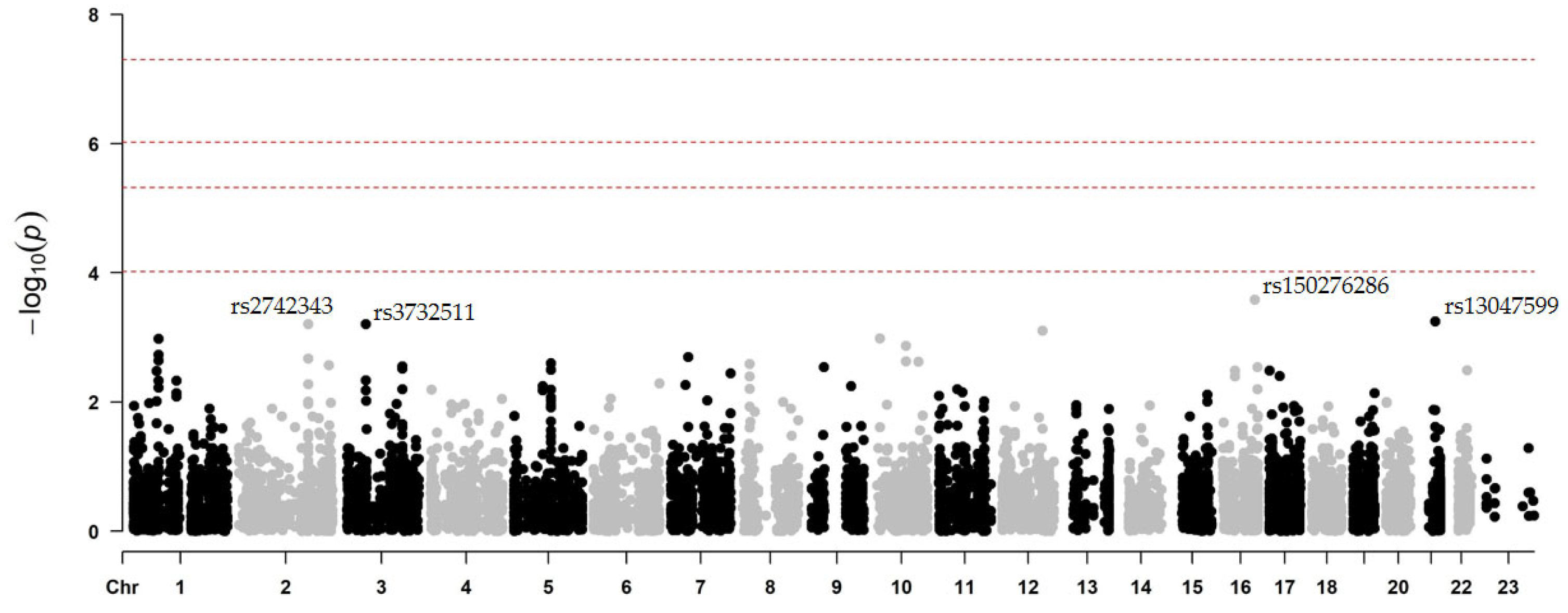
2.2. Two-Stage Association Study
- ○
- “Intervention vs. non-intervention” (see Supplementary Data, Supplementary Results, p. 7)
- ○
- “Event vs. non-event” (see Supplementary Data, Supplementary Results, p. 9 or in more detail in the publication by Antúnez-Rodríguez et al. [2])
- ○
- “Case vs. control” (see Supplementary Data, Supplementary Results, p. 11)
2.2.1. Secondary CV Events After Clopidogrel Treatment
2.2.2. Secondary CV Events After Prasugrel Treatment
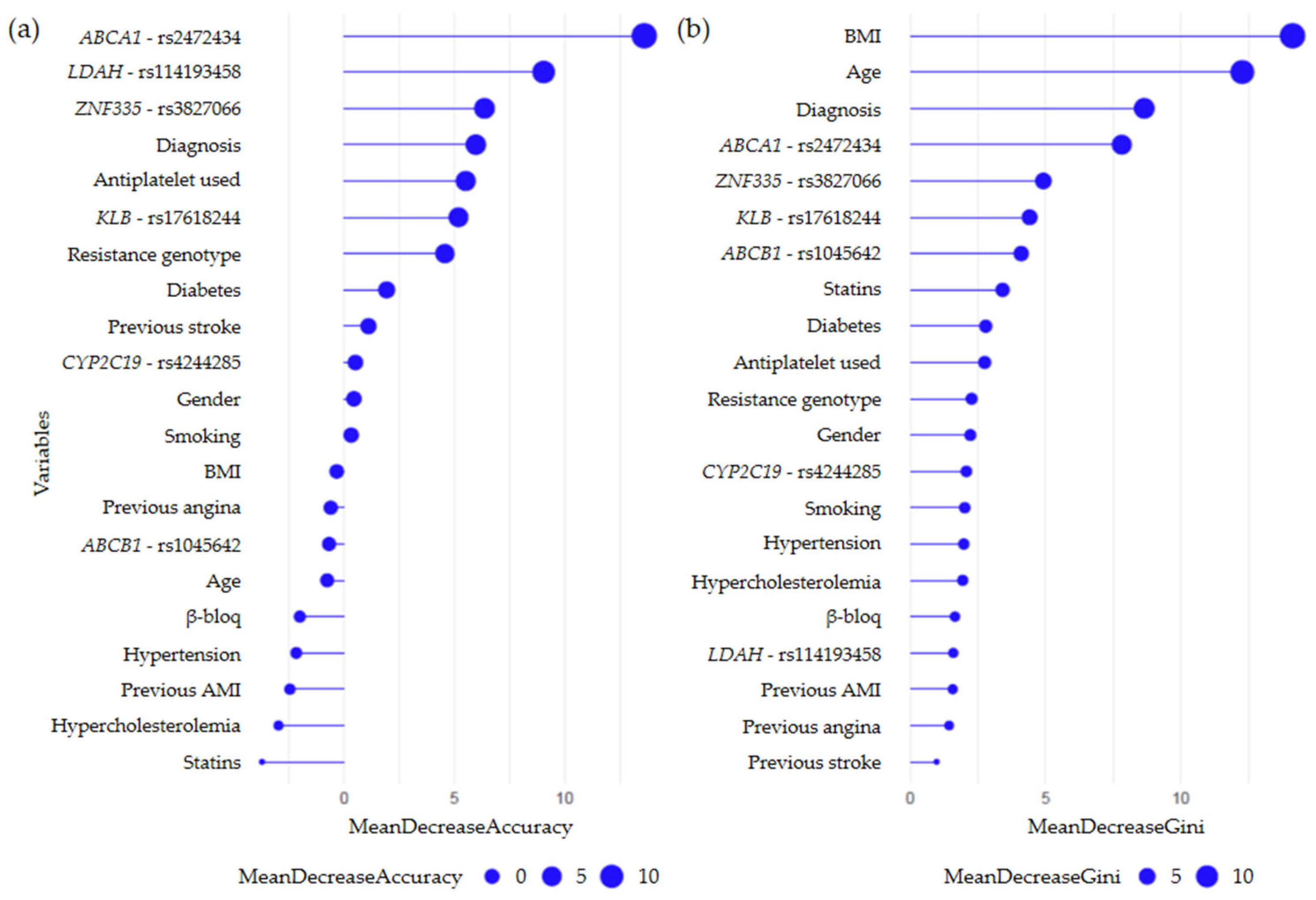
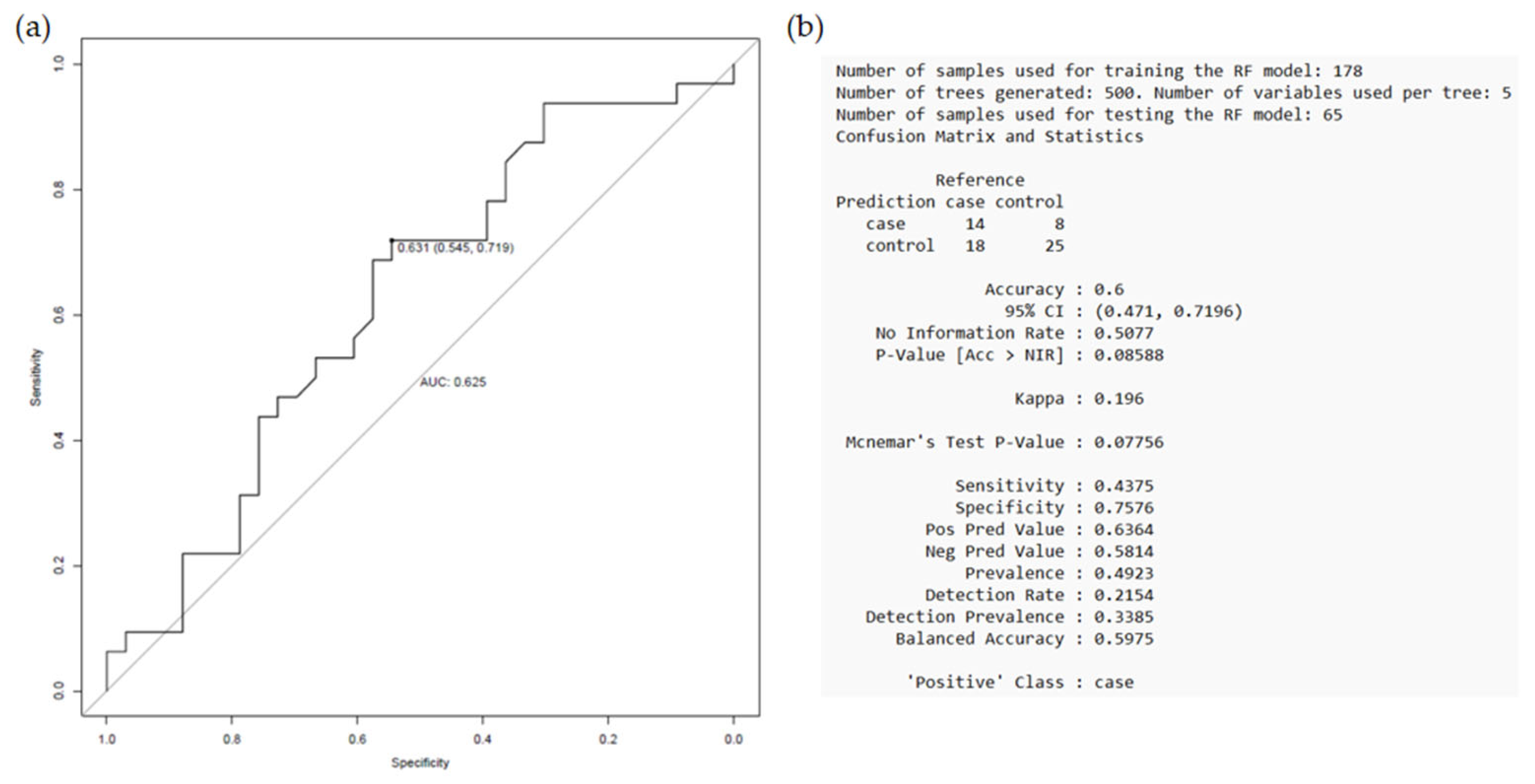
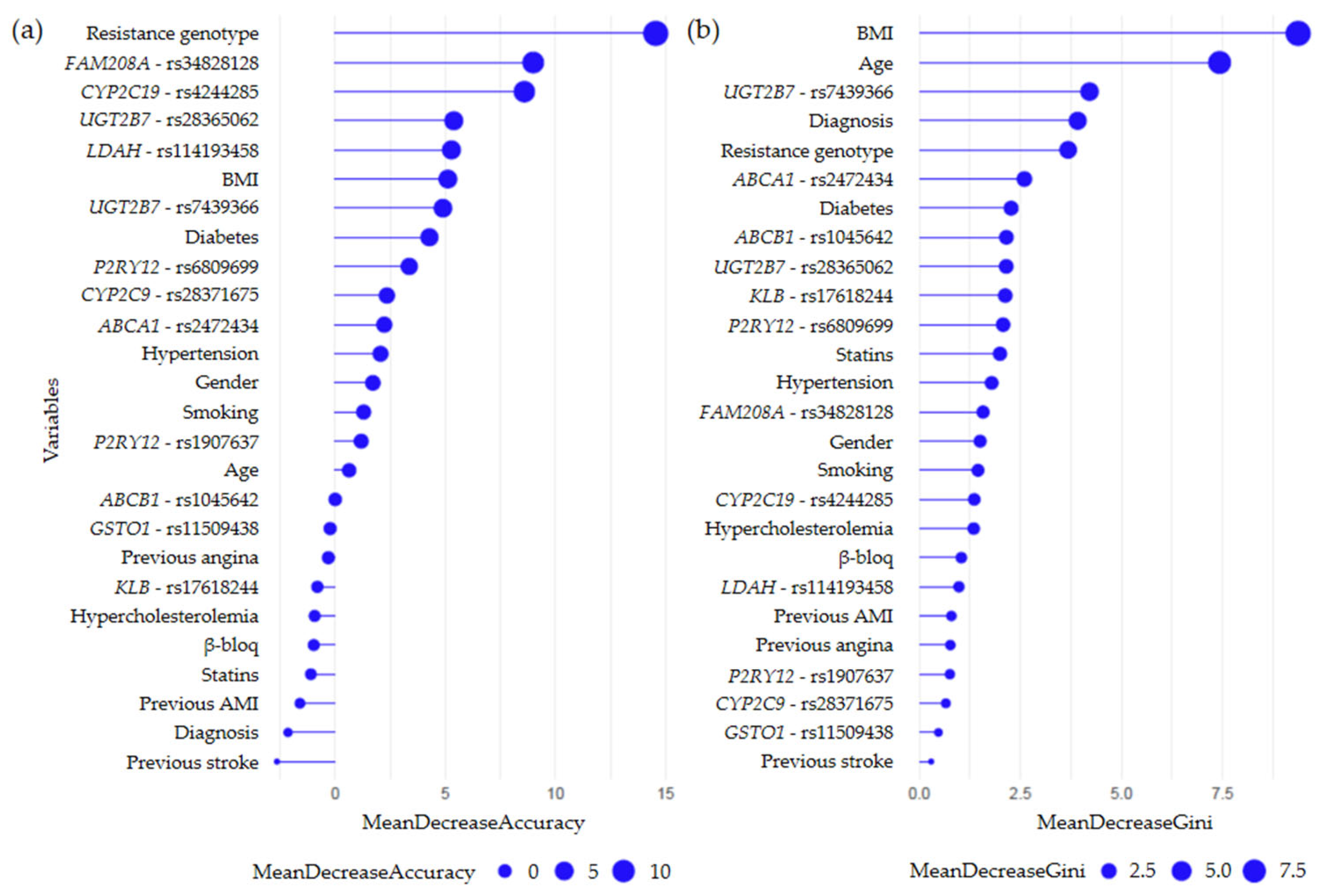
2.3. Random Forest Models
2.3.1. Secondary CV Events Regardless of Prescribed Therapy
2.3.2. Secondary CV Events Following Clopidogrel Treatment
2.3.3. Secondary CV Events Following Prasugrel Treatment
3. Discussion
4. Methods
4.1. Study Population
- ○
- G1 group: a cohort of ACS-PCI-stent patients on antiplatelet therapy (guided or not by genetic testing) who experienced MACEs and/or bleeding during one-year follow-up (n = 109). This group was divided into two subgroups:
- “intervention group” (G1.1), in which carriers of CYP2C19 LoF alleles (*2 or *3) and/or the TT risk genotype for ABCB1 C3435T received prasugrel or ticagrelor, while patients with normal CYP2C19 and ABCB1 gene function received clopidogrel (n = 50).
- “non-intervention group” (G1.2), in which all patients were primarily treated with clopidogrel, regardless of their genetic profile (n = 59).
- ○
- G2 group: a cohort of ACS-PCI-stent patients on PGx-guided antiplatelet therapy who did not experience MACEs and/or bleeding (n = 135).
- ○
- G3 group: a cohort of patients without structural CV disease (n = 99).
4.2. Genetic Analysis
4.3. Statistical Analysis
4.3.1. Association Study
- ○
- “intervention” vs. “non-intervention” (G1.1 vs. G1.2);
- ○
- “event” vs. “non-event” (G1 vs. G2);
- ○
- “case” vs. “control” (G1&G2 vs. G3).
4.3.2. Random Forest Analysis
- ○
- Event vs. non-event” comparison, regardless of the antiplatelet drug received;
- ○
- “Event vs. non-event” comparison in patients taking clopidogrel;
- ○
- “Event vs. non-event” comparison in patients taking prasugrel.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACS | acute coronary syndrome |
| BMI | body mass index |
| CAD | coronary artery disease |
| CV | cardiovascular |
| CYP | cytochrome P450 |
| DAPT | dual antiplatelet therapy |
| GTEx | Genotype-Tissue Expression Project |
| HDL | high-density lipoprotein |
| HTPR | high on-treatment platelet reactivity |
| LD | linkage disequilibrium |
| LDL | low-density lipoprotein |
| LoF | loss-of-function |
| MACE | major adverse cardiovascular event |
| MAF | minor allele frequency |
| PCI | percutaneous coronary intervention |
| PGx | pharmacogenomics |
| RCT | reverse cholesterol transport |
| SNP | single nucleotide polymorphisms |
| UGT | uridine diphosphate (UDP)-glucuronosyltransferase |
References
- Zhang, Y.J.; Li, M.P.; Tang, J.; Chen, X.P. Pharmacokinetic and pharmacodynamic responses to clopidogrel: Evidences and perspectives. Int. J. Environ. Res. Public Health 2017, 14, 301. [Google Scholar] [CrossRef]
- Antúnez-Rodríguez, A.; García-Rodríguez, S.; Pozo-Agundo, A.; Sánchez-Ramos, J.G.; Moreno-Escobar, E.; Triviño-Juárez, J.M.; Martínez-González, L.J.; Dávila-Fajardo, C.L. Targeted next-generation sequencing panel to investigate antiplatelet adverse reactions in acute coronary syndrome patients undergoing percutaneous coronary intervention with stenting. Thromb. Res. 2024, 240, 109060. [Google Scholar] [CrossRef] [PubMed]
- Strisciuglio, T.; Franco, D.; Di Gioia, G.; De Biase, C.; Morisco, C.; Trimarco, B.; Barbato, E. Impact of genetic polymorphisms on platelet function and response to anti platelet drugs. Cardiovasc. Diagn. Ther. 2018, 8, 610–620. [Google Scholar] [CrossRef] [PubMed]
- Echeverría, O.; Angulo-Aguado, M.; Vela, R.; Calderón-Ospina, C.; Parra, K.; Contreras, N.; Morel, A.; Cabrera, R.; Restrepo, C.; Ramírez-Santana, C.; et al. The polygenic implication of clopidogrel responsiveness: Insights from platelet reactivity analysis and next-generation sequencing. PLoS ONE 2024, 19, e0306445. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.S.; Bergmeijer, T.O.; Gong, L.; Reny, J.L.; Lewis, J.P.; Mitchell, B.D.; Alexopoulos, D.; Aradi, D.; Altman, R.B.; Bliden, K.; et al. Genome-wide Association Study of Platelet Reactivity and Cardiovascular Response in Patients Treated With Clopidogrel: A Study by the International Clopidogrel Pharmacogenomics Consortium. Clin. Pharmacol. Ther. 2020, 108, 1067–1077. [Google Scholar] [CrossRef]
- Tunehag, K.R.; Thomas, C.D.; Franchi, F.; Rossi, J.S.; Keeley, E.C.; Anderson, R.D.; Beitelshees, A.L.; Duarte, J.D.; Gong, Y.; Kerensky, R.A.; et al. CYP2C19 Genotype Is Associated With Adverse Cardiovascular Outcomes in Black Patients Treated With Clopidogrel Undergoing Percutaneous Coronary Intervention. J. Am. Heart Assoc. 2024, 13, e033791. [Google Scholar] [CrossRef]
- Lee, C.; Sriramoju, V.; Cervantes, A.; Howell, L.; Varunok, N.; Shivanshu, M.; Hamrick, K.; Polasek, M.; Lee, J.; Clarke, M.; et al. Clinical Outcomes and Sustainability of Using CYP2C19 Genotype–Guided Antiplatelet Therapy After Percutaneous Coronary Intervention. Circ. Genomic Precis. Med. 2018, 11, e002069. [Google Scholar] [CrossRef]
- Gower, M.N.; Ratner, L.R.; Williams, A.K.; Rossi, J.S.; Stouffer, G.A.; Lee, C.R. Clinical Utility of CYP2C19 Genotype-Guided Antiplatelet Therapy in Patients at Risk of Adverse Cardiovascular and Cerebrovascular Events: A Review of Emerging Evidence. Pharmgenomics. Pers. Med. 2020, 13, 239–252. [Google Scholar] [CrossRef]
- Sánchez-Ramos, J.; Dávila-Fajardo, C.L.; Toledo-Frías, P.; Díaz-Villamarín, X.; Martínez-González, L.J.; Martínez-Huertas, S.; Burillo-Gómez, F.; Caballero-Borrego, J.; Bautista-Pavés, A.; Marín-Guzmán, M.C.; et al. Results of genotype-guided antiplatelet therapy in patients who undergone percutaneous coronary intervention with stent. Int. J. Cardiol. 2016, 225, 289–295. [Google Scholar] [CrossRef]
- Duarte, J.D.; Cavallari, L.H. Pharmacogenetics to guide cardiovascular drug therapy. Nat. Rev. Cardiol. 2021, 18, 649–665. [Google Scholar] [CrossRef]
- Lewis, J.P.; Backman, J.D.; Reny, J.L.; Bergmeijer, T.O.; Mitchell, B.D.; Ritchie, M.D.; Déry, J.P.; Pakyz, R.E.; Gong, L.; Ryan, K.; et al. Pharmacogenomic polygenic response score predicts ischaemic events and cardiovascular mortality in clopidogrel-treated patients. Eur. Heart J.-Cardiovasc. Pharmacother. 2020, 6, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Lauschke, V.M.; Ingelman-Sundberg, M. Prediction of drug response and adverse drug reactions: From twin studies to Next Generation Sequencing. Eur. J. Pharm. Sci. 2019, 130, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Pirmohamed, M. Pharmacogenomics: Current status and future perspectives. Nat. Rev. Genet. 2023, 24, 350–362. [Google Scholar] [CrossRef] [PubMed]
- Antúnez-Rodríguez, A. Aplicación de la tecnología NGS para conocer la respuesta a los antiagregantes plaquetarios en pacientes con síndrome coronario agudo (SCA) sometidos a intervención coronaria percutánea (ICP) con implantación de stent. Ph.D. Dissertation, Universidad de Granada, Granada, Spain, 2025. [Google Scholar]
- Schumacher, T.; Benndorf, R.A. ABC transport proteins in cardiovascular disease—A brief summary. Molecules 2017, 22, 589. [Google Scholar] [CrossRef]
- Zwarts, K.Y.; Clee, S.M.; Zwinderman, A.H.; Engert, J.C.; Singaraja, R.; Loubser, O.; James, E.; Roomp, K.; Hudson, T.J.; Jukema, J.W.; et al. ABCA1 regulatory variants influence coronary artery disease independent of effects on plasma lipid levels. Clin. Genet. 2002, 61, 115–125. [Google Scholar] [CrossRef]
- Suresh, R.; Li, X.; Chiriac, A.; Goel, K.; Terzic, A.; Perez-Terzic, C.; Nelson, T.J. Transcriptome from circulating cells suggests dysregulated pathways associated with long-term recurrent events following first-time myocardial infarction. J. Mol. Cell. Cardiol. 2014, 74, 13–21. [Google Scholar] [CrossRef]
- Ðukanović, N.; Obradović, S.; Zdravković, M.; Ðurašević, S.; Stojković, M.; Tosti, T.; Jasnić, N.; Ðorđević, J.; Todorović, Z. Lipids and antiplatelet therapy: Important considerations and future perspectives. Int. J. Mol. Sci. 2021, 22, 3180. [Google Scholar] [CrossRef]
- Mega, J.; Close, S.; Wiviott, S.; Shen, L.; Walker, J.; Simon, T.; Antman, E.; Braunwald, E.; Sabatine, M. Genetic variants in ABCB1 and CYP2C19 and cardiovascular outcomes after treatment with clopidogrel and prasugrel in the TRITON-TIMI 38 trial: A pharmacogenetic analysis. Lancet 2010, 376, 1312–1319. [Google Scholar] [CrossRef]
- Castrichini, M.; Luzum, J.A.; Pereira, N. Pharmacogenetics of Antiplatelet Therapy. Annu. Rev. Pharmacol. Toxicol. 2023, 63, 211–229. [Google Scholar] [CrossRef]
- Lee, C.R.; Luzum, J.A.; Sangkuhl, K.; Gammal, R.S.; Sabatine, M.S.; Stein, C.M.; Kisor, D.F.; Limdi, N.A.; Lee, Y.M.; Scott, S.A.; et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2C19 Genotype and Clopidogrel Therapy: 2022 Update. Clin. Pharmacol. Ther. 2022, 112, 959–967. [Google Scholar] [CrossRef]
- Galeazzi, R.; Olivieri, F.; Spazzafumo, L.; Rose, G.; Montesanto, A.; Giovagnetti, S.; Cecchini, S.; Malatesta, G.; Di Pillo, R.; Antonicelli, R. Clustering of ABCB1 and CYP2C19 Genetic Variants Predicts Risk of Major Bleeding and Thrombotic Events in Elderly Patients with Acute Coronary Syndrome Receiving Dual Antiplatelet Therapy with Aspirin and Clopidogrel. Drugs Aging 2018, 35, 649–656. [Google Scholar] [CrossRef]
- So, D.; Wells, G.; McPherson, R.; Labinaz, M.; Le May, M.; Glover, C.; Dick, A.; Froeschl, M.; Marquis, J.; Gollob, M.; et al. A prospective randomized evaluation of a pharmacogenomic approach to antiplatelet therapy among patients with ST-elevation myocardial infarction: The RAPID STEMI study. Pharmacogenomics J. 2016, 16, 71–78. [Google Scholar] [CrossRef]
- Park, M.W.; Her, S.H.; Kim, C.J.; Suncho, J.; Park, G.M.; Kim, T.S.; Choi, Y.S.; Park, C.S.; Koh, Y.S.; Park, H.J.; et al. Evaluation of the incremental prognostic value of the combination of CYP2C19 poor metabolizer status and ABCB1 3435 TT polymorphism over conventional risk factors for cardiovascular events after drug-eluting stent implantation in East Asians. Genet. Med. 2016, 18, 833–841. [Google Scholar] [CrossRef] [PubMed]
- Jarrar, Y.; Lee, S.-J. The Functionality of UDP-Glucuronosyltransferase Genetic Variants and their Association with Drug Responses and Human Diseases. J. Pers. Med. 2021, 11, 554. [Google Scholar] [CrossRef] [PubMed]
- Kahma, H.; Filppula, A.M.; Neuvonen, M.; Tarkiainen, E.K.; Tornio, A.; Holmberg, M.T.; Itkonen, M.K.; Finel, M.; Neuvonen, P.J.; Niemi, M.; et al. Clopidogrel carboxylic acid glucuronidation is mediated mainly by UGT2B7, UGT2B4, and UGT2B17: Implications for pharmacogenetics and drug-drug interactions. Drug Metab. Dispos. 2018, 46, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, Y.; Takechi, S.; Ishii, Y. Functional Interaction between Cytochrome P450 and UDP-Glucuronosyltransferase on the Endoplasmic Reticulum Membrane: One of Post-translational Factors Which Possibly Contributes to Their Inter-Individual Differences. Biol. Pharm. Bull. 2021, 44, 1635–1644. [Google Scholar] [CrossRef]
- Liu, X.; Xu, H.; Xu, H.; Geng, Q.; Mak, W.H.; Ling, F.; Su, Z.; Yang, F.; Zhang, T.; Chen, J.; et al. New genetic variants associated with major adverse cardiovascular events in patients with acute coronary syndromes and treated with clopidogrel and aspirin. Pharmacogenomics J. 2021, 21, 664–672. [Google Scholar] [CrossRef]
- Yang, G.; Alarcon, C.; Chanfreau, C.; Lee, N.H.; Friedman, P.; Nutescu, E.; Tuck, M.; O’Brien, T.; Gong, L.; Klein, T.E.; et al. Investigation of Genomic and Transcriptomic Risk Factors of Clopidogrel Response in African Americans. Clin. Pharmacol. Ther. 2025, 117, 1313–1324. [Google Scholar] [CrossRef]
- Dávila-Fajardo, C.L.; Sánchez-Ramos, J.; Díaz-Villamarín, X.; Martínez-González, L.J.; Toledo-Frías, P.; Martínez-Huertas, S.; Burillo-Gómez, F.; Caballero-Borrego, J.; Bautista-Pavés, A.; Marín-Guzmán, M.C.; et al. The study protocol for a non-randomized controlled clinical trial using a genotype-guided strategy in a dataset of patients who undergone percutaneous coronary intervention with stent. Data Brief 2016, 10, 518–524. [Google Scholar] [CrossRef]
- Freeman, B.; Smith, N.; Curtis, C.; Huckett, L.; Mill, J.; Craig, I.W. DNA from buccal swabs recruited by mail: Evaluation of storage effects on long-term stability and suitability for multiplex polymerase chain reaction genotyping. Behav. Genet. 2003, 33, 67–72. [Google Scholar] [CrossRef]
- Gómez-Martín, A.; Hernández, A.F.; Martínez-González, L.J.; González-Alzaga, B.; Rodríguez-Barranco, M.; López-Flores, I.; Aguilar-Garduno, C.; Lacasana, M. Polymorphisms of pesticide-metabolizing genes in children living in intensive farming communities. Chemosphere 2015, 139, 534–540. [Google Scholar] [CrossRef]
- Su, R.; Li, Y.; Zink, D.; Loo, L.H. Supervised prediction of drug-induced nephrotoxicity based on interleukin-6 and -8 expression levels. BMC Bioinform. 2014, 15, S16. [Google Scholar] [CrossRef] [PubMed]
- Sangkuhl, K.; Klein, T.E.; Altman, R.B. Clopidogrel pathway. Pharmacogenet Genom. 2010, 20, 463–465. [Google Scholar] [CrossRef] [PubMed]
- Bindesbøll, C.; Aas, A.; Ogmundsdottir, M.H.; Pankiv, S.; Reine, T.; Zoncu, R.; Simonsen, A. NBEAL1 controls SREBP2 processing and cholesterol metabolism and is a susceptibility locus for coronary artery disease. Sci. Rep. 2020, 10, 4528. [Google Scholar] [CrossRef] [PubMed]
- Zabalza, M.; Subirana, I.; Lluis-Ganella, C.; Sayols-Baixeras, S.; de Groot, E.; Arnold, R.; Cenarro, A.; Ramos, R.; Marrugat, J.; Elosua, R. Association Between Coronary Artery Disease Genetic Variants and Subclinical Atherosclerosis: An Association Study and Meta-analysis. Rev. Espanola Cardiol. 2015, 68, 869–877. [Google Scholar] [CrossRef]
- Wang, L.; Hauser, E.R.; Shah, S.H.; Pericak-Vance, M.A.; Haynes, C.; Crosslin, D.; Harris, M.; Nelson, S.; Hale, A.B.; Granger, C.B.; et al. Peakwide Mapping on Chromosome 3q13 Identifies the Kalirin Gene as a Novel Candidate Gene for Coronary Artery Disease. Am. J. Hum. Genet. 2007, 80, 650–663. [Google Scholar] [CrossRef]
- Wang, J.; Xiao, Q.; Wang, L.; Wang, Y.; Wang, D.; Ding, H. Role of ABCA1 in Cardiovascular Disease. J. Pers. Med. 2022, 12, 1010. [Google Scholar] [CrossRef]
- Jacobo-Albavera, L.; Domínguez-Pérez, M.; Medina-Leyte, D.J.; González-Garrido, A.; Villarreal-Molina, T. The Role of the ATP-Binding Cassette A1 (ABCA1) in Human Disease. Int. J. Mol. Sci. 2021, 22, 1593. [Google Scholar] [CrossRef]
- Aaldijk, A.S.; Verzijl, C.R.C.; Jonker, J.W.; Struik, D. Biological and pharmacological functions of the FGF19- and FGF21-coreceptor beta klotho. Front. Endocrinol. 2023, 14, 1150222. [Google Scholar] [CrossRef]
- Panera, N.; Meroni, M.; Longo, M.; Crudele, A.; Valenti, L.; Bellacchio, E.; Miele, L.; D’ORia, V.; Paolini, E.; Maggioni, M.; et al. The KLB rs17618244 gene variant is associated with fibrosing MAFLD by promoting hepatic stellate cell activation. EBioMedicine 2021, 65, 103249. [Google Scholar] [CrossRef]
- Kathiresan, S.; Willer, C.J.; Peloso, G.M.; Demissie, S.; Musunuru, K.; Schadt, E.E.; Kaplan, L.; Bennett, D.; Li, Y.; Tanaka, T.; et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 2009, 41, 56–65. [Google Scholar] [CrossRef]
- Lazarenko, V.; Churilin, M.; Azarova, I.; Klyosova, E.; Bykanova, M.; Ob’Edkova, N.; Churnosov, M.; Bushueva, O.; Mal, G.; Povetkin, S.; et al. Comprehensive Statistical and Bioinformatics Analysis in the Deciphering of Putative Mechanisms by Which Lipid-Associated GWAS Loci Contribute to Coronary Artery Disease. Biomedicines 2022, 10, 259. [Google Scholar] [CrossRef]
- Rautureau, Y.; Deschambault, V.; Higgins, M.; Rivas, D.; Mecteau, M.; Geoffroy, P.; Miquel, G.; Uy, K.; Sanchez, R.; Lavoie, V.; et al. ADCY9 (Adenylate Cyclase Type 9) Inactivation Protects From Atherosclerosis Only in the Absence of CETP (Cholesteryl Ester Transfer Protein). Circulation 2018, 138, 1677–1692. [Google Scholar] [CrossRef]
- Ebrahimi, N.; Asadikaram, G.; Mohammadi, A.; Jahani, Y.; Moridi, M.; Masoumi, M. The association of endothelin-1 gene polymorphism and its plasma levels with hypertension and coronary atherosclerosis. Arch. Med Sci. 2021, 17, 613–620. [Google Scholar] [CrossRef]
- Ponasenko, A.; Sinitskaya, A.; Sinitsky, M.; Khutornaya, M.; Barbarash, O. The Role of Polymorphism in the Endothelial Homeostasis and Vitamin D Metabolism Genes in the Severity of Coronary Artery Disease. Biomedicines 2023, 11, 2382. [Google Scholar] [CrossRef]
- Lee, W.-S.; Ham, W.; Kim, J. Roles of NAD(P)H:quinone Oxidoreductase 1 in Diverse Diseases. Life 2021, 11, 1301. [Google Scholar] [CrossRef] [PubMed]

| ACS Patients | Controls | |||||
|---|---|---|---|---|---|---|
| All (N = 343) | G1 (N = 109) | G2 (N = 135) | p-Value * | G3 (N = 99) | p-Value ** | |
| Age | 65.9 (11.1) | 66.8 (11.4) | 64.8 (12.3) | 0.202 | 66.4 (8.8) | 0.339 |
| Gender | ||||||
| Male | 232 (67.6) | 78 (71.6) | 101 (74.8) | 0.670 | 53 (53.5) | 0.002 |
| Female | 111 (32.4) | 31(28.4) | 34 (25.2) | 46 (46.5) | ||
| BMI | 28.6 (4.6) | 28.9 (4.6) | 28.4 (4.5) | 0.369 | NA | NA |
| Ethnic origin | ||||||
| Caucasian | 335 (97.7) | 105 (96.3) | 132 (97.8) | 0.517 | 98 (99.0) | 0.423 |
| Gypsy | 6 (1.7) | 3 (2.8) | 3 (2.2) | 0 (0.0) | ||
| Moroccan | 2 (0.6) | 1 (0.9) | 0 (0.0) | 1 (1.0) | ||
| Cv history | 93 (27.1) | 52 (47.7) | 41 (30.4) | 0.008 | 0 | <0.001 |
| All (N = 244) | G1 (N = 109) | G2 (N = 135) | p-Value | |
|---|---|---|---|---|
| Acetylsalicylic acid | 244 (100) | 109 (100) | 135 (100) | 1.000 |
| Antiplatelet therapy | ||||
| Clopidogrel | 169 (69.3) | 82 (75.2) | 87 (64.4) | 0.035 |
| Prasugrel | 73 (29.9) | 25 (22.9) | 48 (35.6) | |
| Ticagrelor | 2 (0.8) | 2 (1.8) | 0 (0.0) | |
| DAPT duration | ||||
| 1 month | 14 (5.7) | 11 (10.1) | 3 (2.2) | 0.006 |
| 3 months | 2 (0.8) | 2 (1.8) | 0 (0.0) | |
| 6 months | 2 (0.8) | 2 (1.8) | 0 (0.0) | |
| 12 months or more | 226 (92.6) | 94 (86.2) | 132 (97.8) | |
| Comparison | SNP | Alleles | Gene | Functional Consequence | Biological Process Involved |
|---|---|---|---|---|---|
| Intervention vs. non-intervention | rs72934556 | T>G | NBEAL1 | synonymous | Lipid metabolism |
| rs2289843 | A>T | KALRN | splice region | Endothelial dysfunction, atherosclerosis | |
| Event vs. non-event | rs2472434 | A>C | ABCA1 | intronic | Lipid metabolism, inflammation |
| rs17618244 | G>A | KLB | missense | Lipid metabolism | |
| rs3827066 | C>T | ZNF335 | intronic | Lipid metabolism | |
| Case vs. control | rs11076799 | G>A | ADCY9 | intronic | Endothelial dysfunction |
| rs5370 | G>T | EDN1 | missense | Endothelial dysfunction, inflammation, lipid metabolism | |
| rs1800566 | G>A | NQO1 | missense | Inflammation, atherogenesis |
| SNP | Chr | Position | Ref | Alt | Gene | Gene Role | Func. | MAF | p-Value | Beta | SD |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs4244285 | 10 | 96541616 | G | A | CYP2C19 | drug metabolism | synon. | 0.15 | 0.00046 | −1.9297 | 0.5785 |
| rs4986894 | 10 | 96522365 | T | C | CYP2C19 | drug metabolism | upstream | 0.15 | 0.00046 | −1.9297 | 0.5785 |
| rs34828128 | 3 | 56667213 | T | C | FAM208A | unknown | synon. | 0.02 | 0.00085 | −2.2830 | 0.7476 |
| rs2472434 | 9 | 107623249 | A | C | ABCA1 | disease predisposition | intronic | 0.28 | 0.00171 | −0.8835 | 0.2836 |
| rs17618244 | 4 | 39448529 | G | A | KLB | disease predisposition | missense | 0.19 | 0.00194 | 0.8797 | 0.2859 |
| rs4986938 | 14 | 64699816 | C | T | ESR2 | disease predisposition | UTR3 | 0.38 | 0.00251 | −0.6907 | 0.2300 |
| SNP | Chr | Position | Ref | Alt | Gene | Func. | MAF | p-Value | Beta | SD |
|---|---|---|---|---|---|---|---|---|---|---|
| rs2235048 | 7 | 87138511 | G | A | ABCB1 | intronic | 0.47 | 0.03322 | 0.6291 | 0.2961 |
| rs28365062 | 4 | 69964271 | A | G | UGT2B7 | synon. | 0.14 | 0.00917 | 0.8886 | 0.3429 |
| rs7439366 | 4 | 69964338 | T | C | UGT2B7 | missense | 0.48 | 0.03090 | 0.5158 | 0.2394 |
| rs4244285 | 10 | 96541616 | G | A | CYP2C19 | synon. | 0.15 | 0.00046 | −1.9297 | 0.5785 |
| rs4986894 | 10 | 96522365 | T | C | upstream | |||||
| rs11509438 | 10 | 106027059 | G | A | GSTO1 | missense | 0.03 | 0.02745 | −1.4006 | 0.6430 |
| rs1907637 | 3 | 151104838 | A | G | P2RY12 | upstream | 0.87 | 0.01814 | 0.9126 | 0.3876 |
| rs6809699 | 3 | 151056598 | A | C | P2RY12 | synon. | 0.83 | 0.03925 | 0.6609 | 0.3215 |
| rs2046934 | 3 | 151057642 | G | A | intronic | |||||
| rs10935838 | 3 | 151058247 | A | G | intronic |
| SNP | Chr | Position | Ref | Alt | Gene | Gene Role | Func. | MAF | p-Value | Beta | SD |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs150276286 | 16 | 81902879 | C | G | PLCG2 | disease predisposition | synon. | 0.01 | 0.00026 | −3.6496 | 1.0675 |
| rs13047599 | 21 | 34926260 | C | T | SON | unknown | missense | 0.67 | 0.00057 | −1.3065 | 0.3844 |
| rs2742343 | 2 | 179569436 | A | G | TTN | disease predisposition | synon. | 0.03 | 0.00062 | −3.1466 | 1.0184 |
| rs3732511 | 3 | 56766435 | C | G | ARHGEF3 | disease predisposition | synon. | 0.12 | 0.00062 | −1.8055 | 0.5398 |
| rs2888805 | 12 | 104380734 | G | A | TDG | unknown | missense | 0.10 | 0.00079 | −2.5707 | 0.8042 |
| rs6686 | 10 | 12209752 | T | C | NUDT5 | unknown | synon. | 0.50 | 0.00104 | −1.2772 | 0.3958 |
| rs61781311 | 1 | 66087620 | G | T | LEPR | disease predisposition | intronic | 0.17 | 0.00105 | −1.3725 | 0.4255 |
| rs2472434 | 9 | 107623249 | A | C | ABCA1 | disease predisposition | intronic | 0.28 | 0.00572 | −1.2671 | 0.4631 |
| rs3827066 | 20 | 44586023 | C | T | ZNF335 | disease predisposition | intronic | 0.17 | 0.02880 | 1.0502 | 0.4836 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antúnez-Rodríguez, A.; García-Rodríguez, S.; Pozo-Agundo, A.; Sánchez-Ramos, J.G.; Moreno-Escobar, E.; Triviño-Juárez, J.M.; Álvarez-Cubero, M.J.; Martínez-González, L.J.; Dávila-Fajardo, C.L. Custom Gene Panel Analysis Identifies Novel Polymorphisms Associated with Clopidogrel Response in Patients Undergoing Percutaneous Coronary Intervention with Stent. Int. J. Mol. Sci. 2025, 26, 9766. https://doi.org/10.3390/ijms26199766
Antúnez-Rodríguez A, García-Rodríguez S, Pozo-Agundo A, Sánchez-Ramos JG, Moreno-Escobar E, Triviño-Juárez JM, Álvarez-Cubero MJ, Martínez-González LJ, Dávila-Fajardo CL. Custom Gene Panel Analysis Identifies Novel Polymorphisms Associated with Clopidogrel Response in Patients Undergoing Percutaneous Coronary Intervention with Stent. International Journal of Molecular Sciences. 2025; 26(19):9766. https://doi.org/10.3390/ijms26199766
Chicago/Turabian StyleAntúnez-Rodríguez, Alba, Sonia García-Rodríguez, Ana Pozo-Agundo, Jesús Gabriel Sánchez-Ramos, Eduardo Moreno-Escobar, José Matías Triviño-Juárez, María Jesús Álvarez-Cubero, Luis Javier Martínez-González, and Cristina Lucía Dávila-Fajardo. 2025. "Custom Gene Panel Analysis Identifies Novel Polymorphisms Associated with Clopidogrel Response in Patients Undergoing Percutaneous Coronary Intervention with Stent" International Journal of Molecular Sciences 26, no. 19: 9766. https://doi.org/10.3390/ijms26199766
APA StyleAntúnez-Rodríguez, A., García-Rodríguez, S., Pozo-Agundo, A., Sánchez-Ramos, J. G., Moreno-Escobar, E., Triviño-Juárez, J. M., Álvarez-Cubero, M. J., Martínez-González, L. J., & Dávila-Fajardo, C. L. (2025). Custom Gene Panel Analysis Identifies Novel Polymorphisms Associated with Clopidogrel Response in Patients Undergoing Percutaneous Coronary Intervention with Stent. International Journal of Molecular Sciences, 26(19), 9766. https://doi.org/10.3390/ijms26199766









