Identification of Key Differentially Expressed Genes During Early Sex Determination in Chicken Embryos
Abstract
1. Introduction
2. Results
2.1. High-Throughput Sequencing and Read Mapping
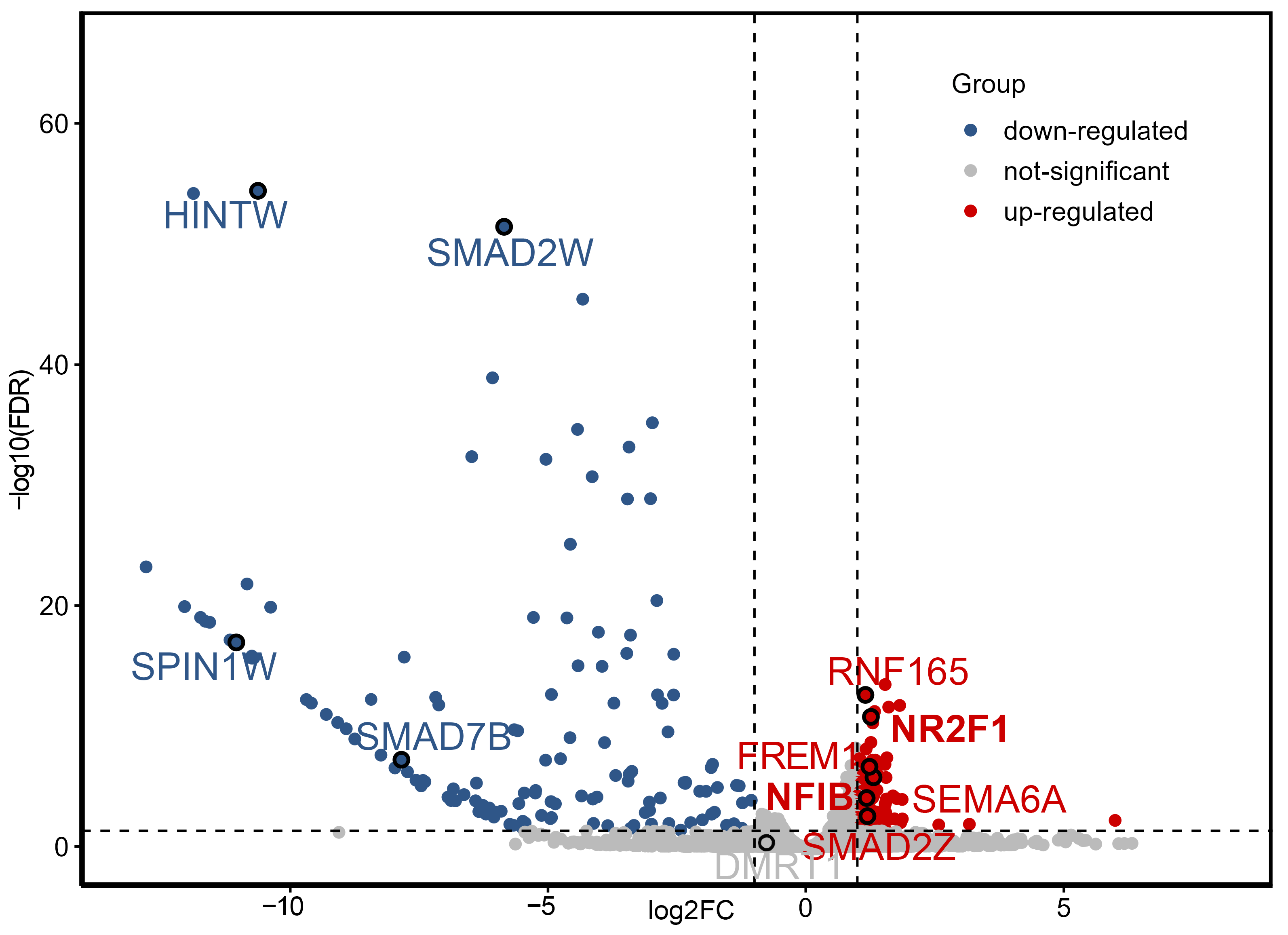
2.2. RNA-Seq and Data Analysis
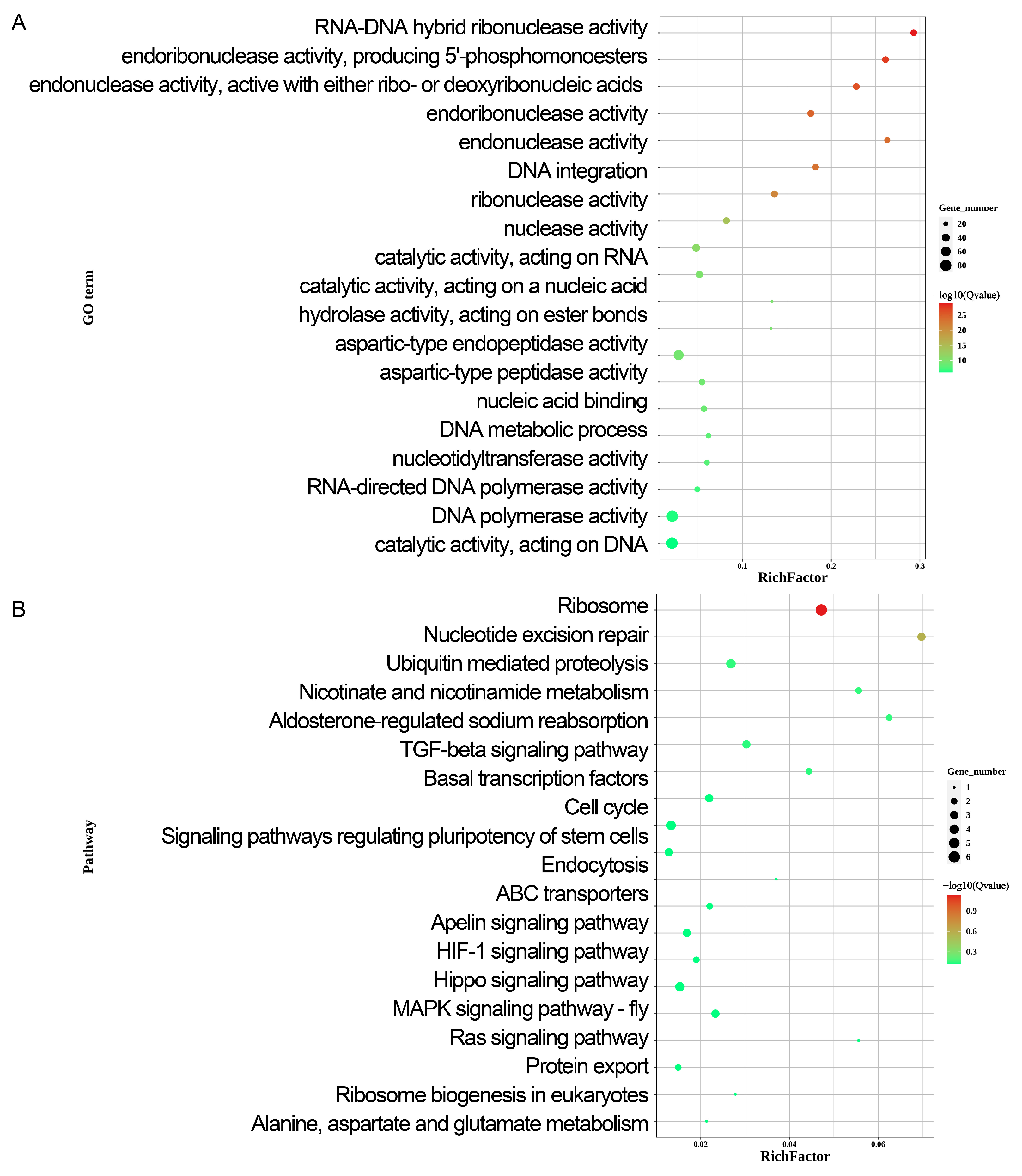
2.3. Gene Ontology, KEGG Pathway and GSEA Analysis
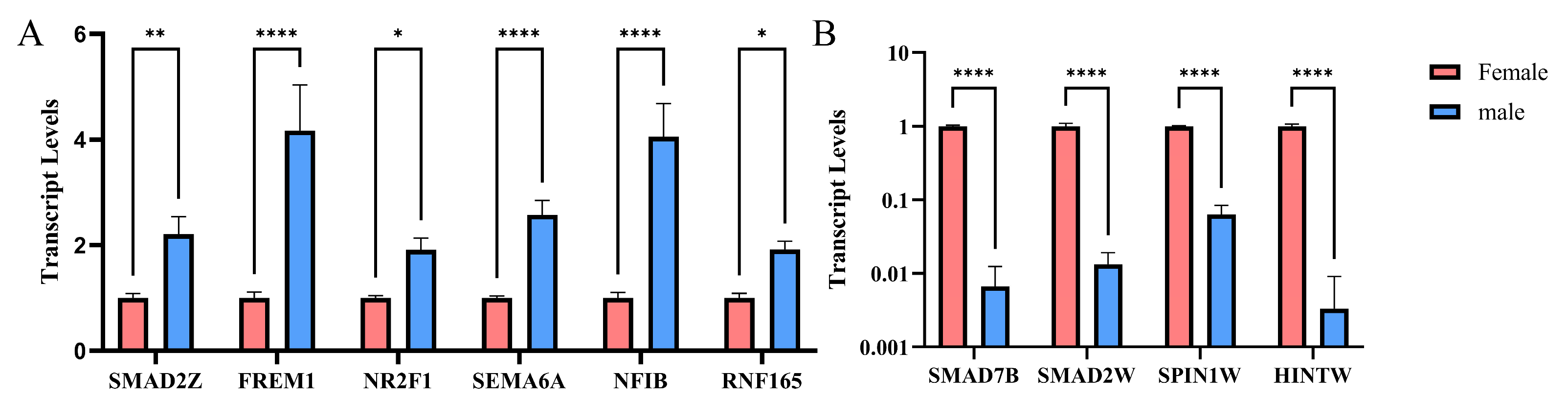
2.4. qPCR Results at DEGs
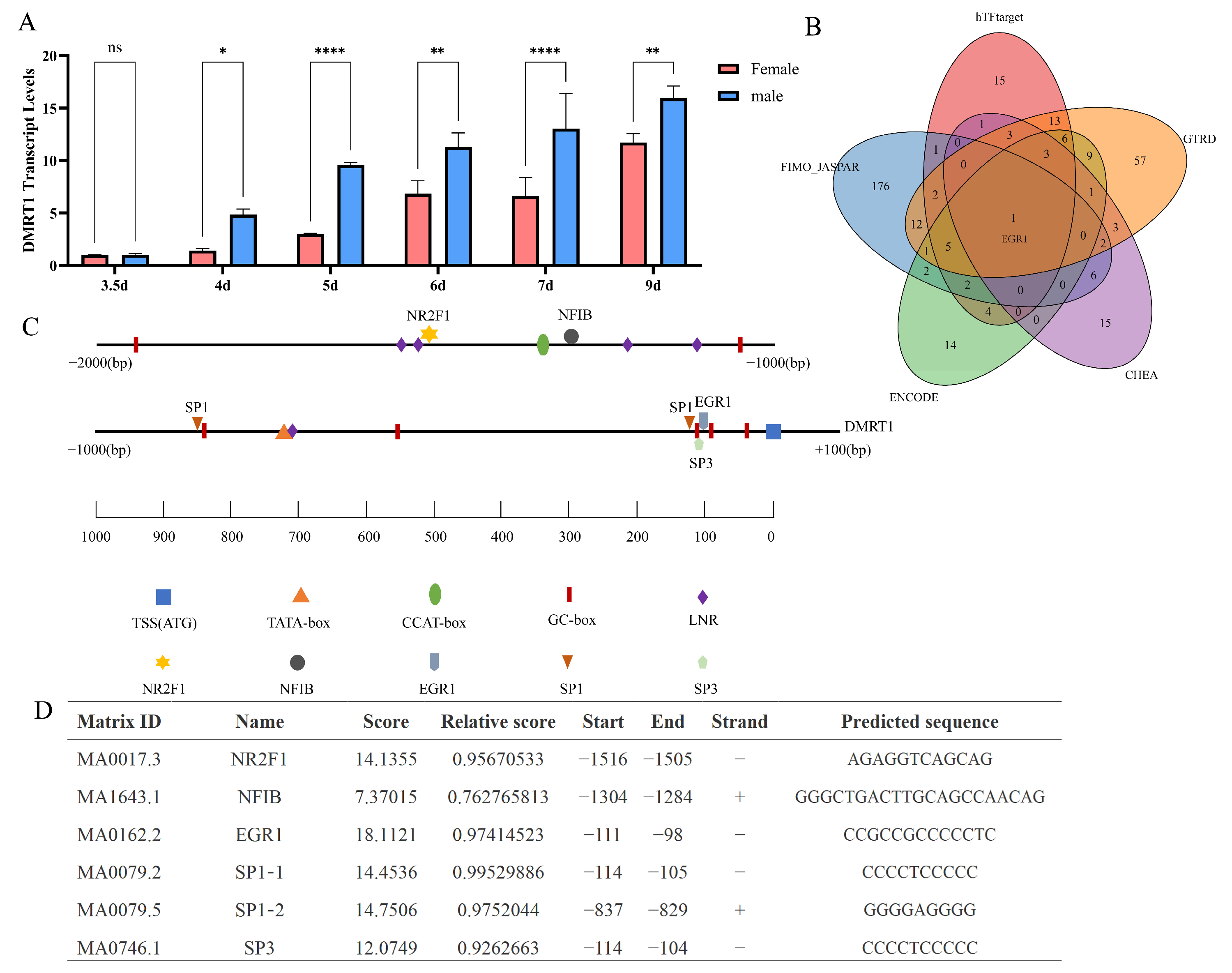
2.5. Prediction of Binding Between Transcription Factors NFIB and NR2F1 with DMRT1
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Sex Determination of Chicken Embryo
4.3. RNA Isolation and cDNA Preparation
4.4. RNA-Seq and Bioinformatics Analysis
4.5. Quantitative Real-Time PCR (qRT-PCR)
4.6. Prediction of Binding Sites for Transcription Factors NFIB and NR2F2
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fridolfsson, A.K.; Cheng, H.; Copeland, N.G.; Jenkins, N.A.; Liu, H.C.; Raudsepp, T.; Woodage, T.; Chowdhary, B.; Halverson, J.; Ellegren, H. Evolution of the avian sex chromosomes from an ancestral pair of autosomes. Proc. Natl. Acad. Sci. USA 1998, 95, 8147–8152. [Google Scholar] [CrossRef]
- Guioli, S.; Nandi, S.; Zhao, D.; Burgess-Shannon, J.; Lovell-Badge, R.; Clinton, M. Gonadal asymmetry and sex determination in birds. Sex. Dev. 2014, 8, 227–242. [Google Scholar] [CrossRef]
- Smith, C.A.; Sinclair, A.H. Sex determination: Insights from the chicken. Bioessays 2004, 26, 120–132. [Google Scholar] [CrossRef]
- Deviatiiarov, R.; Nagai, H.; Ismagulov, G.; Stupina, A.; Wada, K.; Ide, S.; Toji, N.; Zhang, H.; Sukparangsi, W.; Intarapat, S.; et al. Dosage compensation of Z sex chromosome genes in avian fibroblast cells. Genome Biol. 2023, 24, 213. [Google Scholar] [CrossRef]
- Nanda, I.; Schlegelmilch, K.; Haaf, T.; Schartl, M.; Schmid, M. Synteny conservation of the Z chromosome in 14 avian species (11 families) supports a role for Z dosage in avian sex determination. Cytogenet. Genome Res. 2008, 122, 150–156. [Google Scholar] [CrossRef]
- Huang, S.; Ye, L.; Chen, H. Sex determination and maintenance: The role of DMRT1 and FOXL2. Asian J. Androl. 2017, 19, 619–624. [Google Scholar] [CrossRef]
- Teranishi, M.; Shimada, Y.; Hori, T.; Nakabayashi, O.; Kikuchi, T.; Macleod, T.; Pym, R.; Sheldon, B.; Solovei, I.; Macgregor, H.; et al. Transcripts of the MHM region on the chicken Z chromosome accumulate as non-coding RNA in the nucleus of female cells adjacent to the locus. Chromosome Res. 2001, 9, 147–165. [Google Scholar] [CrossRef]
- Augstenová, B.; Ma, W.J. Decoding Dmrt1: Insights into vertebrate sex determination and gonadal sex differentiation. J. Evol. Biol. 2025, 38, 811–831. [Google Scholar] [CrossRef]
- Chue, J.; Smith, C.A. Sex determination and sexual differentiation in the avian model. FEBS J. 2011, 278, 1027–1034. [Google Scholar] [CrossRef]
- Xiang, X.; Yu, Z.; Liu, Y.; Huang, Y.; Wang, J.; Chen, L.; Ma, M. Differential proteomics between unhatched male and female egg yolks reveal the molecular mechanisms of sex-allocation and sex-determination in chicken. Poult. Sci. 2022, 101, 101906. [Google Scholar] [CrossRef]
- Smith, C.A.; Roeszler, K.N.; Ohnesorg, T.; Cummins, D.M.; Farlie, P.G.; Doran, T.J.; Sinclair, A.H. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 2009, 461, 267–271. [Google Scholar] [CrossRef]
- Li, J.; Zhang, X.; Wang, X.; Wang, Z.; Li, X.; Zheng, J.; Li, J.; Xu, G.; Sun, C.; Yi, G.; et al. Single-nucleus transcriptional and chromatin accessible profiles reveal critical cell types and molecular architecture underlying chicken sex determination. J. Adv. Res. 2025, 70, 29–43. [Google Scholar] [CrossRef]
- Luo, H.; Zhou, H.; Jiang, S.; He, C.; Xu, K.; Ding, J.; Liu, J.; Qin, C.; Chen, K.; Zhou, W.; et al. Gene Expression Profiling Reveals Potential Players of Sex Determination and Asymmetrical Development in Chicken Embryo Gonads. Int. J. Mol. Sci. 2023, 24, 14597. [Google Scholar] [CrossRef]
- Tajima, A.; Hayashi, H.; Kamizumi, A.; Ogura, J.; Kuwana, T.; Chikamune, T. Study on the concentration of circulating primordial germ cells (cPGCs) in early chick embryos. J. Exp. Zool. 1999, 284, 759–764. [Google Scholar] [CrossRef]
- Ichikawa, K.; Horiuchi, H. Fate Decisions of Chicken Primordial Germ Cells (PGCs): Development, Integrity, Sex Determination, and Self-Renewal Mechanisms. Genes 2023, 14, 612. [Google Scholar] [CrossRef]
- Lei, N.; Heckert, L.L. Sp1 and Egr1 regulate transcription of the gene in Sertoli cells. Biol. Reprod. 2002, 66, 675–684. [Google Scholar] [CrossRef]
- Lonn, P.; Moren, A.; Raja, E.; Dahl, M.; Moustakas, A. Regulating the stability of TGFbeta receptors and Smads. Cell Res. 2009, 19, 21–35. [Google Scholar] [CrossRef]
- Ceplitis, H.; Ellegren, H. Adaptive molecular evolution of HINTW, a female-specific gene in birds. Mol. Biol. Evol. 2004, 21, 249–254. [Google Scholar] [CrossRef]
- Monsivais, D.; Matzuk, M.M.; Pangas, S.A. The TGF-beta Family in the Reproductive Tract. Cold Spring Harb. Perspect. Biol. 2017, 9, a022251. [Google Scholar] [CrossRef]
- Mullen, A.C.; Wrana, J.L. TGF-beta Family Signaling in Embryonic and Somatic Stem-Cell Renewal and Differentiation. Cold Spring Harb. Perspect. Biol. 2017, 9, a022186. [Google Scholar] [CrossRef]
- Chang, H.; Brown, C.W.; Matzuk, M.M. Genetic analysis of the mammalian transforming growth factor-beta superfamily. Endocr. Rev. 2002, 23, 787–823. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 2018, 7, 1–6. [Google Scholar] [CrossRef]
- Kelly, C.E.; Thymiakou, E.; Dixon, J.E.; Tanaka, S.; Godwin, J.; Episkopou, V. Rnf165/Ark2C enhances BMP-Smad signaling to mediate motor axon extension. PLoS Biol. 2013, 11, e1001538. [Google Scholar] [CrossRef]
- Pallotta, M.M.; Fogliano, C.; Carotenuto, R. Temperature Incubation Influences Gonadal Gene Expression during Leopard Gecko Development. Animals 2022, 12, 3186. [Google Scholar] [CrossRef]
- Hori, T.; Asakawa, S.; Itoh, Y.; Shimizu, N.; Mizuno, S. Wpkci, encoding an altered form of PKCI, is conserved widely on the avian W chromosome and expressed in early female embryos: Implication of its role in female sex determination. Mol. Biol. Cell 2000, 11, 3645–3660. [Google Scholar] [CrossRef]
- O’Neill, M.; Binder, M.; Smith, C.; Andrews, J.; Reed, K.; Smith, M.; Millar, C.; Lambert, D.; Sinclair, A. ASW: A gene with conserved avian W-linkage and female specific expression in chick embryonic gonad. Dev. Genes. Evol. 2000, 210, 243–249. [Google Scholar] [CrossRef]
- Hrvatin, N.; Pereza, N.; Caljkusic-Mance, T.; Vuceric, T.M.; Ostojic, S.; Hodzic, A.; Maver, A.; Peterlin, B. Second Case of Gonadal Mosaicism and a Novel Nonsense NR2F1 Gene Variant as the Cause of Bosch-Boonstra-Schaaf Optic Atrophy Syndrome. Clin. Genet. 2024, 106, 786–787. [Google Scholar] [CrossRef]
- Mehanovic, S.; Mendoza-Villarroel, R.E.; de Mattos, K.; Talbot, P.; Viger, R.S.; Tremblay, J.J. Identification of novel genes and pathways regulated by the orphan nuclear receptor COUP-TFII in mouse MA-10 Leydig cellsdagger. Biol. Reprod. 2021, 105, 1283–1306. [Google Scholar] [CrossRef]
- Rada-Iglesias, A.; Bajpai, R.; Prescott, S.; Brugmann, S.A.; Swigut, T.; Wysocka, J. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell 2012, 11, 633–648. [Google Scholar] [CrossRef]
- Wu, S.P.; Lee, D.K.; Demayo, F.J.; Tsai, S.Y.; Tsai, M.J. Generation of ES cells for conditional expression of nuclear receptors and coregulators in vivo. Mol. Endocrinol. 2010, 24, 1297–1304. [Google Scholar] [CrossRef]
- Estermann, M.A.; Grimm, S.A.; Kitakule, A.S.; Rodriguez, K.F.; Brown, P.R.; McClelland, K.; Amato, C.M.; Yao, H.H. NR2F2 regulation of interstitial cell fate in the embryonic mouse testis and its impact on differences of sex development. Nat. Commun. 2025, 16, 3987. [Google Scholar] [CrossRef]
- Malaymar Pinar, D.; Goos, H.; Tan, Z.; Kumpula, E.P.; Chowdhury, I.; Wang, Z.; Zhang, Q.; Salokas, K.; Keskitalo, S.; Wei, G.H.; et al. Nuclear Factor I Family Members are Key Transcription Factors Regulating Gene Expression. Mol. Cell Proteom. 2025, 24, 100890. [Google Scholar] [CrossRef]
- Osada, S.; Matsubara, T.; Daimon, S.; Terazu, Y.; Xu, M.; Nishihara, T.; Imagawa, M. Expression, DNA-binding specificity and transcriptional regulation of nuclear factor 1 family proteins from rat. Biochem. J. 1999, 342 Pt 1, 189–198. [Google Scholar] [CrossRef]
- Chojnowski, J.L.; Braun, E.L. An unbiased approach to identify genes involved in development in a turtle with temperature-dependent sex determination. BMC Genom. 2012, 13, 308. [Google Scholar] [CrossRef]
- Scheider, J.; Afonso-Grunz, F.; Jessl, L.; Hoffmeier, K.; Winter, P.; Oehlmann, J. Morphological and transcriptomic effects of endocrine modulators on the gonadal differentiation of chicken embryos: The case of tributyltin (TBT). Toxicol. Lett. 2018, 284, 143–151. [Google Scholar] [CrossRef]
- Ayers, K.L.; Cutting, A.D.; Roeszler, K.N.; Sinclair, A.H.; Smith, C.A. DMRT1 is required for Mullerian duct formation in the chicken embryo. Dev. Biol. 2015, 400, 224–236. [Google Scholar] [CrossRef]
- Ioannidis, J.; Taylor, G.; Zhao, D.; Liu, L.; Idoko-Akoh, A.; Gong, D.; Lovell-Badge, R.; Guioli, S.; McGrew, M.J.; Clinton, M. Primary sex determination in birds depends on DMRT1 dosage, but gonadal sex does not determine adult secondary sex characteristics. Proc. Natl. Acad. Sci. USA 2021, 118, e2020909118. [Google Scholar] [CrossRef]
- Lambeth, L.S.; Raymond, C.S.; Roeszler, K.N.; Kuroiwa, A.; Nakata, T.; Zarkower, D.; Smith, C.A. Over-expression of DMRT1 induces the male pathway in embryonic chicken gonads. Dev. Biol. 2014, 389, 160–172. [Google Scholar] [CrossRef]
- Fridolfsson, A.K.; Ellegren, H. A simple and universal method for molecular sexing of non-ratite birds. J. Avian Biol. 1999, 30, 116–121. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Pan, H.; Huang, T.; Yu, L.; Wang, P.; Su, S.; Wu, T.; Bai, Y.; Teng, Y.; Wei, Y.; Zhou, L.; et al. Transcriptome Analysis of the Adipose Tissue of Luchuan and Duroc Pigs. Animals 2022, 12, 2258. [Google Scholar] [CrossRef] [PubMed]
- Dreos, R.; Ambrosini, G.; Groux, R.; Périer, R.C.; Bucher, P. The eukaryotic promoter database in its 30th year: Focus on non-vertebrate organisms. Nucleic Acids Res. 2017, 45, D51–D55. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. TFTF: An R-Based Integrative Tool for Decoding Human Transcription Factor-Target Interactions. Biomolecules 2024, 14, 749. [Google Scholar] [CrossRef]
| Sample | Clean Reads | Mapped Reads (%) | Uniq Mapped Reads (%) | Expressed_Gene | Q30 (%) | GC (%) |
|---|---|---|---|---|---|---|
| Female1 | 21,999,587 | 91.61 | 83.7 | 18,302 | 92.4 | 47.3 |
| Female2 | 20,677,927 | 91.34 | 81.7 | 18,160 | 91.3 | 48.4 |
| Female3 | 22,500,482 | 93.19 | 84.26 | 18,334 | 92.5 | 46.4 |
| Male1 | 20,478,251 | 90.95 | 82.29 | 18,564 | 91.5 | 47.2 |
| Male2 | 22,323,360 | 91.21 | 78.58 | 18,274 | 91.1 | 49.0 |
| Male3 | 21,026,280 | 91.47 | 82.69 | 18,210 | 91.7 | 48.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, R.; Miao, H.; Zhang, B.; Zhang, H. Identification of Key Differentially Expressed Genes During Early Sex Determination in Chicken Embryos. Int. J. Mol. Sci. 2025, 26, 9575. https://doi.org/10.3390/ijms26199575
Liu R, Miao H, Zhang B, Zhang H. Identification of Key Differentially Expressed Genes During Early Sex Determination in Chicken Embryos. International Journal of Molecular Sciences. 2025; 26(19):9575. https://doi.org/10.3390/ijms26199575
Chicago/Turabian StyleLiu, Ruijia, Huanhuan Miao, Bo Zhang, and Hao Zhang. 2025. "Identification of Key Differentially Expressed Genes During Early Sex Determination in Chicken Embryos" International Journal of Molecular Sciences 26, no. 19: 9575. https://doi.org/10.3390/ijms26199575
APA StyleLiu, R., Miao, H., Zhang, B., & Zhang, H. (2025). Identification of Key Differentially Expressed Genes During Early Sex Determination in Chicken Embryos. International Journal of Molecular Sciences, 26(19), 9575. https://doi.org/10.3390/ijms26199575







