Genome-Wide Analysis of Terpene Synthase Genes in Crocus sativus Reveals Their Regulatory Roles in Terpenoid Biosynthesis and Abiotic Stress Tolerance
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of CsTPS Genes
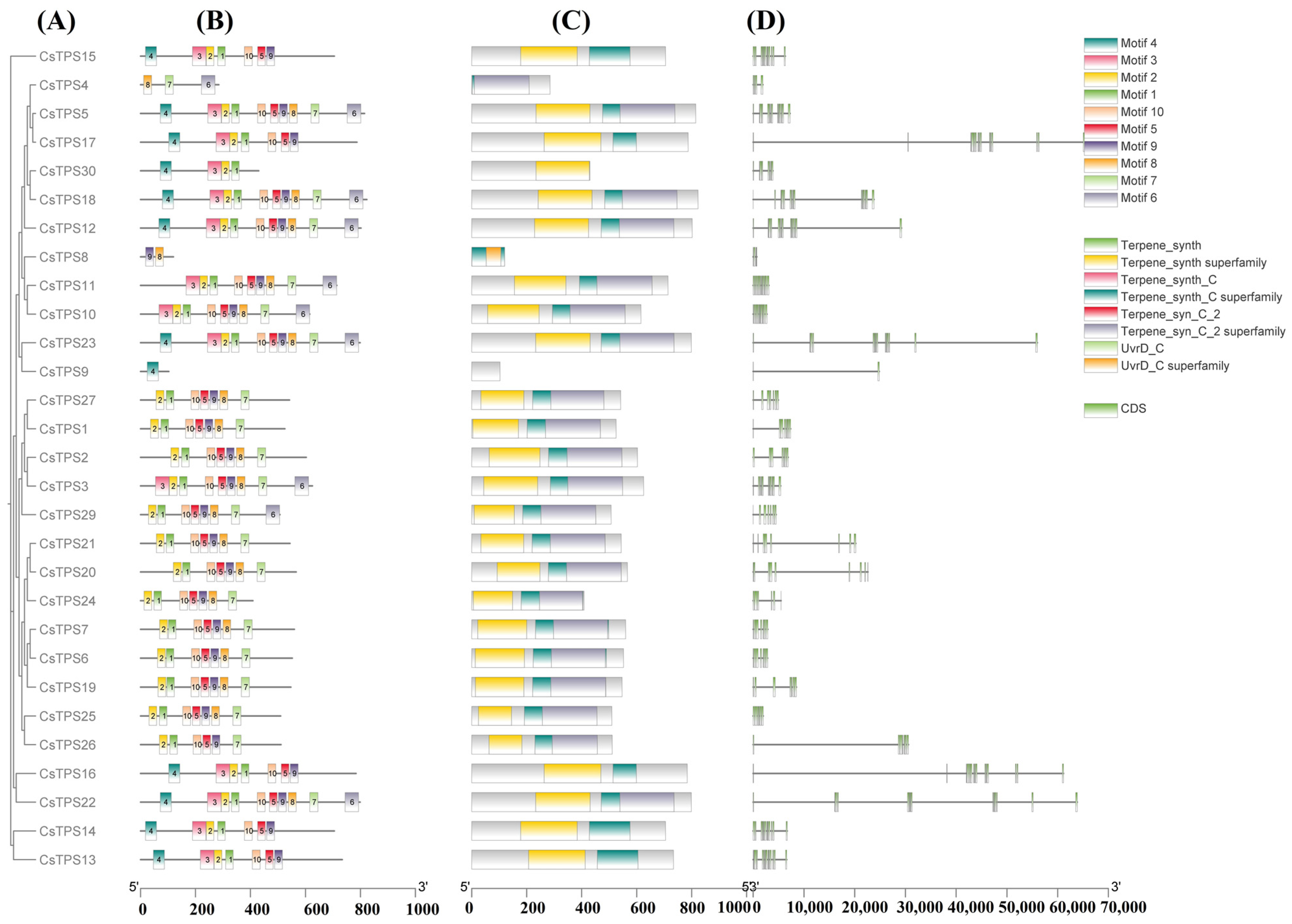
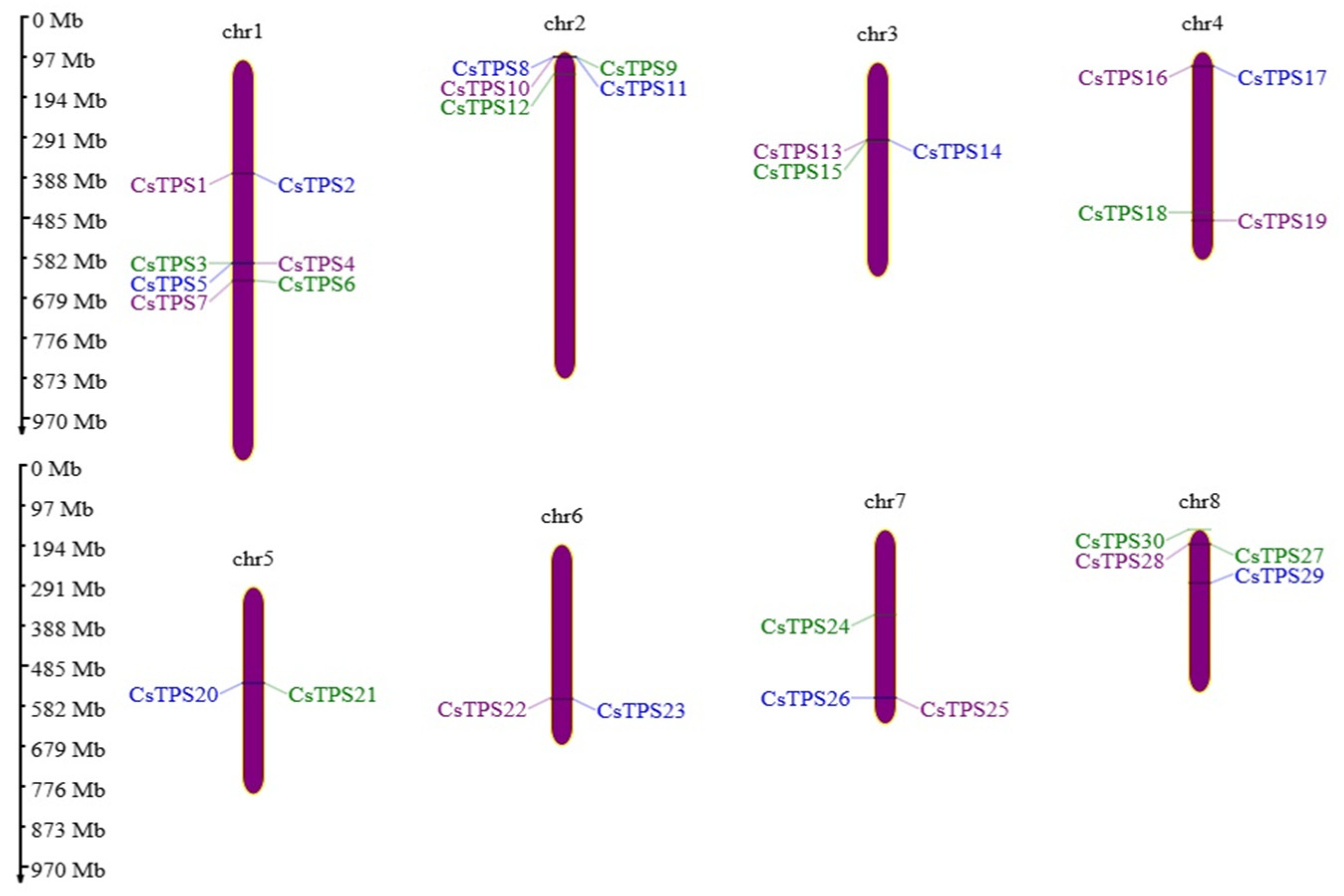
2.2. Gene Structure, Motifs, and Chromosomal Distribution
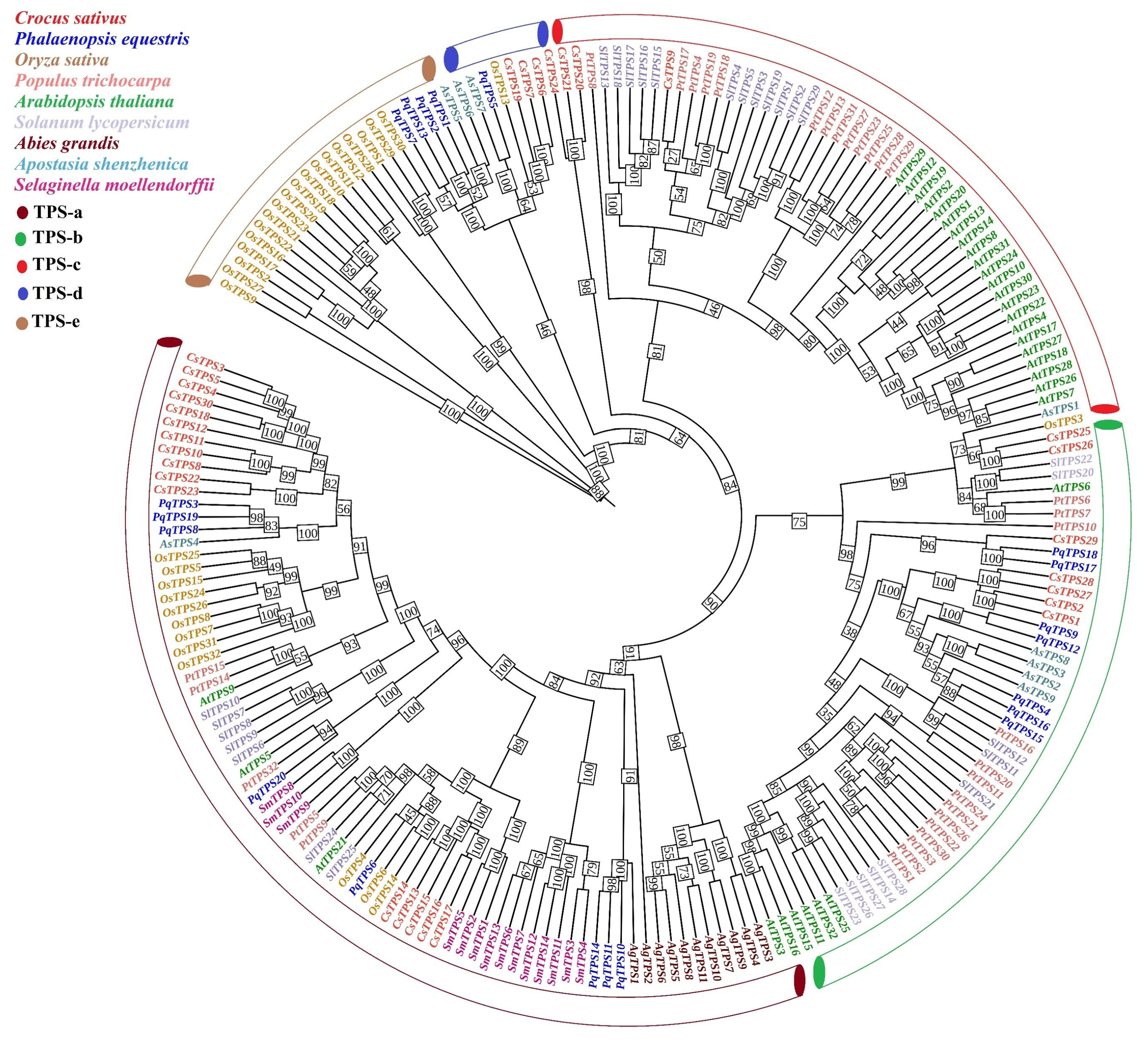
2.3. Phylogenetic and Evolutionary Dynamics
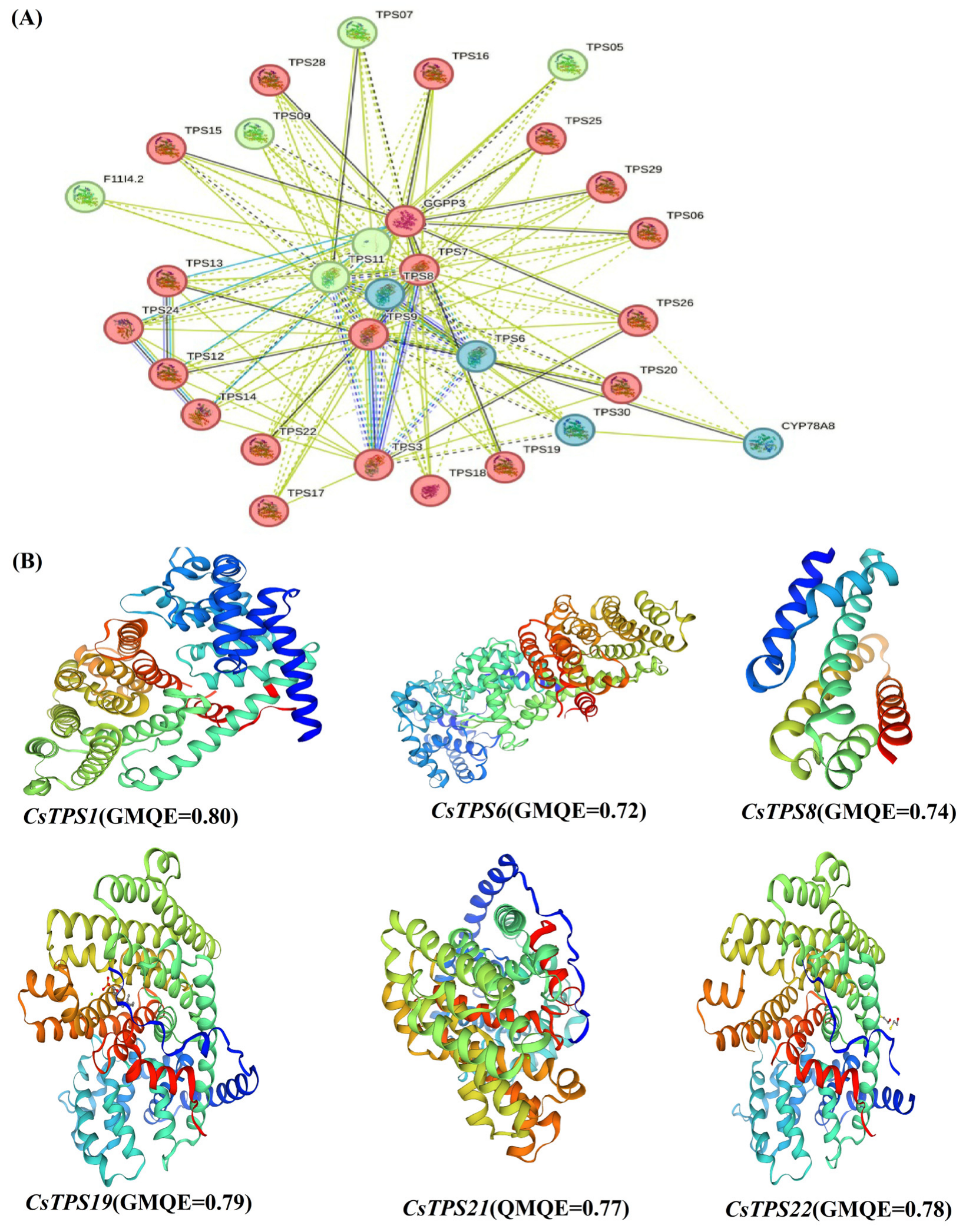
2.4. Protein Structure and Interaction Networks
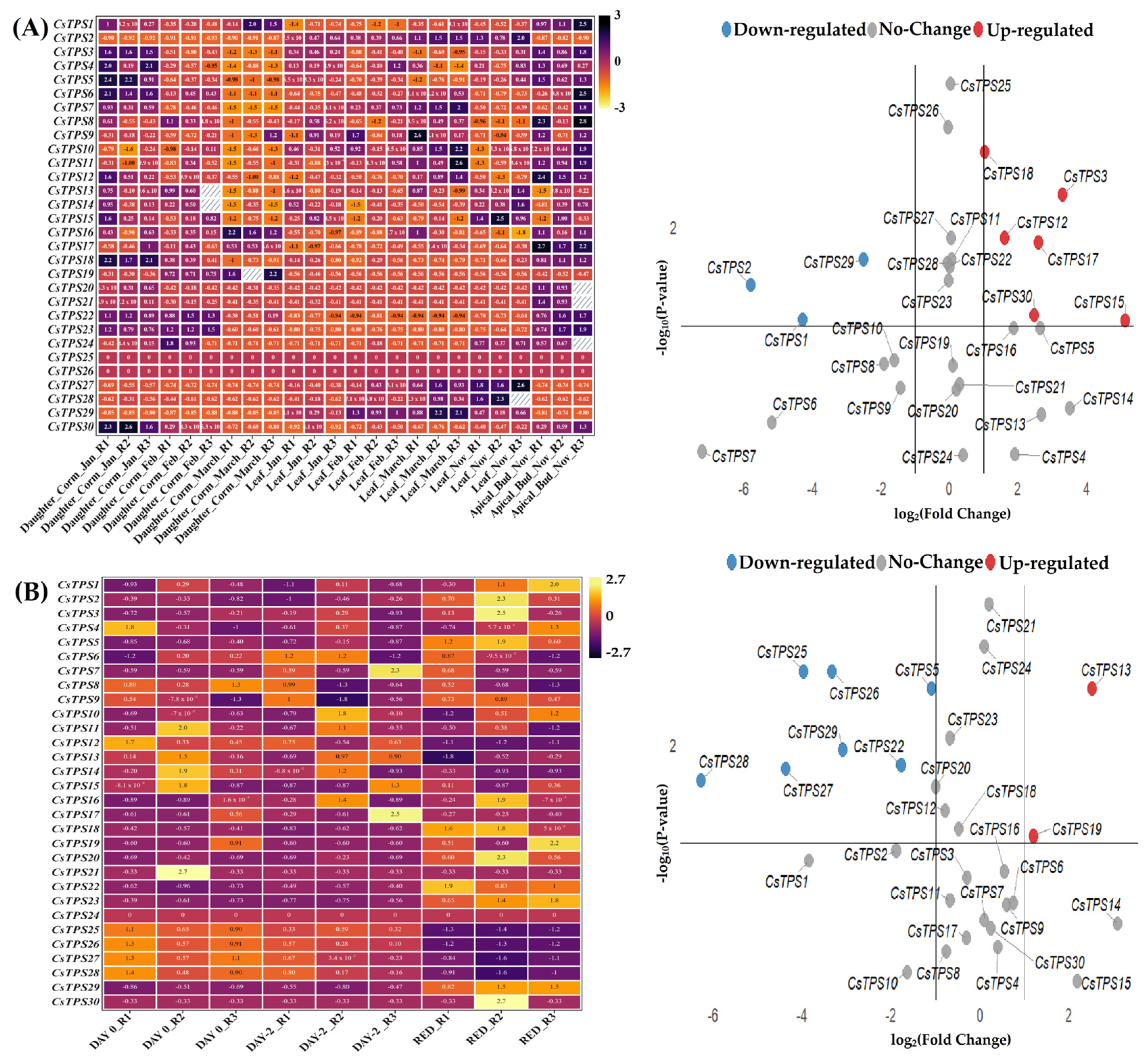
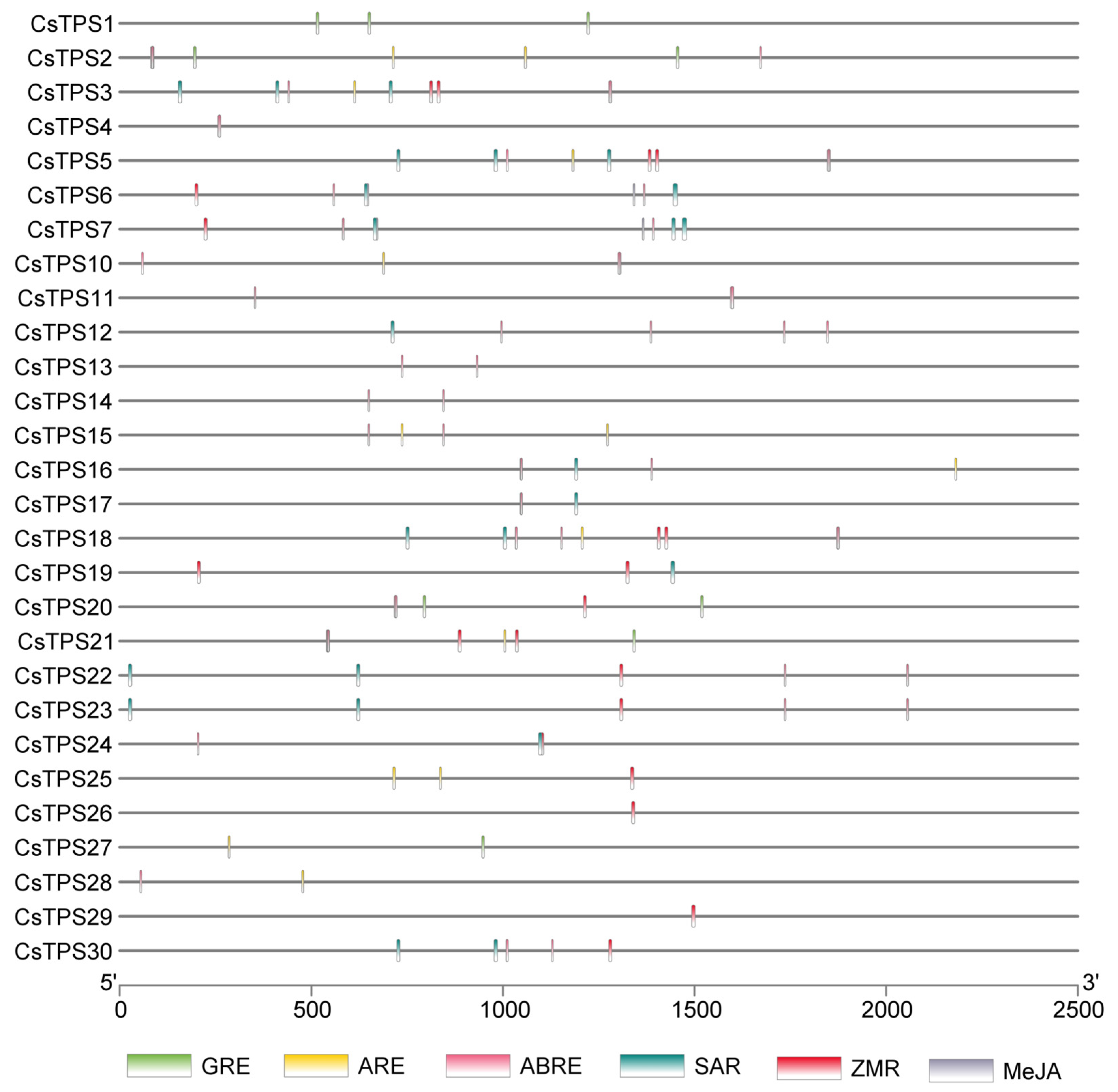
2.5. Expression Profiles and Cis-Regulatory Elements
2.6. In Silico Validation and Vector Optimization for CsTPS1
3. Discussion
4. Materials and Methods
4.1. Identification and Annotation of Terpene Synthase Genes
4.2. Gene Structure, Motif, and Chromosomal Analysis
4.3. Phylogenetic and Evolutionary Analysis
4.4. Protein Structure and Interaction Analysis
4.5. Transcriptomic Expression Analysis
4.6. Computational Validation and Vector Design for Expression of CsTPS1
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, J.-K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Loreto, F.; Schnitzler, J.-P. Abiotic stresses and induced BVOCs. Trends Plant Sci. 2010, 15, 154–166. [Google Scholar] [CrossRef] [PubMed]
- Nagegowda, D.A. Plant volatile terpenoid metabolism: Biosynthetic genes, transcriptional regulation and subcellular compartmentation. FEBS Lett. 2010, 584, 2965–2973. [Google Scholar] [CrossRef] [PubMed]
- Gershenzon, J.; Dudareva, N. The function of terpene natural products in the natural world. Nat. Chem. Biol. 2007, 3, 408–414. [Google Scholar] [CrossRef]
- Husaini, A.M. Challenges of climate change: Omics-based biology of saffron plants and organic agricultural biotechnology for sustainable saffron production. GM Crops Food 2014, 5, 97–105. [Google Scholar] [CrossRef]
- Melnyk, J.P.; Wang, S.; Marcone, M.F. Chemical and biological properties of the world’s most expensive spice: Saffron. Food Res. Int. 2010, 43, 1981–1989. [Google Scholar] [CrossRef]
- Gu, J.N.; Zhu, J.; Yu, Y.; Teng, X.D.; Lou, Y.; Xu, X.F.; Liu, J.L.; Yang, Z.N. DYT 1 directly regulates the expression of TDF 1 for tapetum development and pollen wall formation in A rabidopsis. Plant J. 2014, 80, 1005–1013. [Google Scholar] [CrossRef]
- Ahrazem, O.; Trapero, A.; Gómez, M.D.; Rubio-Moraga, A.; Gómez-Gómez, L. Genomic analysis and gene structure of the plant carotenoid dioxygenase 4 family: A deeper study in Crocus sativus and its allies. Genomics 2010, 96, 239–250. [Google Scholar] [CrossRef]
- Moghaddasi, M.S. Saffron chemicals and medicine usage. J. Med. Plants Res. 2010, 4, 427–430. [Google Scholar]
- Bathaie, S.Z.; Mousavi, S.Z. New applications and mechanisms of action of saffron and its important ingredients. Crit. Rev. Food Sci. Nutr. 2010, 50, 761–786. [Google Scholar] [CrossRef]
- Borowska-Zuchowska, N.; Kwasniewski, M.; Hasterok, R. Cytomolecular analysis of ribosomal DNA evolution in a natural allotetraploid Brachypodium hybridum and its putative ancestors—Dissecting complex repetitive structure of intergenic spacers. Front. Plant Sci. 2016, 7, 1499. [Google Scholar] [CrossRef]
- Sun, Y.; Zou, X.; Guo, X.; Yang, C.; Hung, T.; Lu, Z. CELO Fiber1 knob is a promising candidate to modify the tropism of adenoviral vectors. Genes 2022, 13, 2316. [Google Scholar] [CrossRef]
- Bohlmann, J.; Meyer-Gauen, G.; Croteau, R. Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. USA 1998, 95, 4126–4133. [Google Scholar] [CrossRef]
- Aubourg, S.; Lecharny, A.; Bohlmann, J. Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol. Genet. Genom. 2002, 267, 730–745. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Pichersky, E. The complete functional characterisation of the terpene synthase family in tomato. New Phytol. 2020, 226, 1341–1360. [Google Scholar] [CrossRef] [PubMed]
- Tholl, D.; Lee, S. Terpene specialized metabolism in Arabidopsis thaliana. In The Arabidopsis Book; American Society of Plant Biologists: Rockville, MD, USA, 2011; Volume 9, p. e0143. [Google Scholar]
- Baba, S.A.; Mohiuddin, T.; Basu, S.; Swarnkar, M.K.; Malik, A.H.; Wani, Z.A.; Abbas, N.; Singh, A.K.; Ashraf, N. Comprehensive transcriptome analysis of Crocus sativus for discovery and expression of genes involved in apocarotenoid biosynthesis. BMC Genom. 2015, 16, 698. [Google Scholar] [CrossRef]
- Rubio-Moraga, A.; Rambla, J.L.; Fernández-de-Carmen, A.; Trapero-Mozos, A.; Ahrazem, O.; Orzáez, D.; Granell, A.; Gómez-Gómez, L. New target carotenoids for CCD4 enzymes are revealed with the characterization of a novel stress-induced carotenoid cleavage dioxygenase gene from Crocus sativus. Plant Mol. Biol. 2014, 86, 555–569. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37 (Suppl. 2), W202–W208. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39 (Suppl. S2), W29–W37. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega, Accurate Alignment of Very Large Numbers of Sequences. In Multiple Sequence Alignment Methods; Springer: Berlin/Heidelberg, Germany, 2013; pp. 105–116. [Google Scholar]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Hammond, R.W.; Zhao, Y. Modification of tobacco plant development by sense and antisense expression of the tomato viroid-induced AGC VIIIa protein kinase PKV suggests involvement in gibberellin signaling. BMC Plant Biol. 2009, 9, 108. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.M.; Aubourg, S.; Schouwey, M.B.; Daviet, L.; Schalk, M.; Toub, O.; Lund, S.T.; Bohlmann, J. Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol. 2010, 10, 226. [Google Scholar] [CrossRef] [PubMed]
- Dudareva, N.; Klempien, A.; Muhlemann, J.K.; Kaplan, I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013, 198, 16–32. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011, 66, 212–229. [Google Scholar] [CrossRef]
- Jain, M.; Srivastava, P.L.; Verma, M.; Ghangal, R.; Garg, R. De novo transcriptome assembly and comprehensive expression profiling in Crocus sativus to gain insights into apocarotenoid biosynthesis. Sci. Rep. 2016, 6, 22456. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, S.; Wang, Y.; Tian, Y.; Wang, X.; Xin, T.; Li, Z.; Hua, X.; Tan, S.; Sun, W.; et al. Crocus genome reveals the evolutionary origin of crocin biosynthesis. Acta Pharm. Sin. B 2024, 14, 1878–1891. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35 (Suppl. 2), W585–W587. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2012, 41, D36–D42. [Google Scholar] [CrossRef]
- Goodstein, D.; Batra, S.; Carlson, J.; Hayes, R.; Phillips, J.; Shu, S.; Schmutz, J.; Rokhsar, D. Phytozome Comparative Plant Genomics Portal; LBNL Report#: LBNL-7048E; Lawrence Berkeley National Laboratory: Berkeley, CA, USA, 2014. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Kiefer, F.; Arnold, K.; Künzli, M.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009, 37 (Suppl. 1), D387–D392. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef]
- Jose-Santhi, J.; Sheikh, F.R.; Kalia, D.; Sood, R.; Kumar, R.; Acharya, V.; Singh, R.K. Transcriptional dynamics in source-sink tissues identifies molecular factors regulating the corm development process in saffron (Crocus sativus L.). Physiol. Plant. 2024, 176, e14285. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Chen, X.; Liang, N.; Chen, R.; Chen, J.; Hu, C.; Li, Q.; Li, Q.; Pei, W.; Xiao, W.; et al. Transcriptome analysis reveals novel enzymes for apo-carotenoid biosynthesis in saffron and allows construction of a pathway for crocetin synthesis in yeast. J. Exp. Bot. 2019, 70, 4819–4834. [Google Scholar] [CrossRef] [PubMed]
- Pearson, W.R. Using the FASTA program to search protein and DNA sequence databases. In Computer Analysis of Sequence Data: Part I; Springer: Berlin/Heidelberg, Germany, 1994; pp. 307–331. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Dwight, Z.; Palais, R.; Wittwer, C.T. uMELT: Prediction of high-resolution melting curves and dynamic melting profiles of PCR products in a rich web application. Bioinformatics 2011, 27, 1019–1020. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, A.; Javed, M.R.; Ali, S.W.; Tusleem, K.; ul Qamar, M.T. PCR Primer Design for In-Silico Rapid Detection of Ocular Infection Caused by Candida Species in Humans. Open J. Bioinform. Biostat. 2017, 1, 4–9. [Google Scholar]
- Hassan, A.; Gul, I.; Chikan, N.A.; Nazki, S.; Fazili, K.M.; Ganai, N.A.; Ahmad, S.M.; Shah, R.A.; Careem, M.F.A.; Shabir, N. Utilizing Vet-Informatics for Developing Reverse Genetics-Based Vaccine Platforms Against RNA Viruses Affecting Poultry and Livestock. In Bioinformatics in Veterinary Science: Vetinformatics; Springer: Berlin/Heidelberg, Germany, 2025; pp. 415–438. [Google Scholar]
- Grote, A.; Hiller, K.; Scheer, M.; Münch, R.; Nörtemann, B.; Hempel, D.C.; Jahn, D. JCat: A novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic. Acids Acids Res. 2005, 33 (Suppl. S2), W526–W531. [Google Scholar] [CrossRef] [PubMed]
| Gene Name | No. of AA 1 | PMW 2 | PI 3 | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|
| CsTPS1 | 525 | 60,627.36 | 5.34 | −0.28 | Cyto 4 |
| CsTPS2 | 603 | 69,739.58 | 6.17 | −0.406 | Nu 5 |
| CsTPS3 | 625 | 72,119.86 | 5.54 | −0.341 | Cyto |
| CsTPS4 | 285 | 32,743.56 | 5.83 | −0.28 | Cyto |
| CsTPS5 | 815 | 93,156.69 | 5.53 | −0.301 | Cyto, PM 6, Nu |
| CsTPS6 | 552 | 65,077.44 | 5.3 | −0.404 | Cyto |
| CsTPS7 | 560 | 65,816.22 | 5.21 | −0.381 | Cyto |
| CsTPS8 | 120 | 13,888.93 | 4.99 | −0.239 | EC 7, Cyto |
| CsTPS9 | 103 | 11,548.16 | 6.49 | −0.288 | Cyto |
| CsTPS10 | 616 | 70,628.38 | 5.72 | −0.291 | Cyto, PM |
| CsTPS11 | 714 | 81,662.06 | 5.72 | −0.238 | Cyto, PM |
| CsTPS12 | 802 | 91,554.43 | 6.02 | −0.267 | Cyto, PM, Nu |
| CsTPS13 | 734 | 83,579.96 | 5.37 | −0.336 | Cyto |
| CsTPS14 | 705 | 80,237.33 | 5.77 | −0.322 | Cyto |
| CsTPS15 | 705 | 80,132.29 | 5.73 | −0.299 | Cyto |
| CsTPS16 | 784 | 89,194.93 | 5.57 | −0.3 | Cyto |
| CsTPS17 | 787 | 89,620.48 | 5.73 | −0.3 | Cyto |
| CsTPS18 | 823 | 94,009.96 | 5.73 | −0.275 | Cyto, PM, Nu |
| CsTPS19 | 547 | 64,287.29 | 5.34 | −0.403 | Cyto, Nu |
| CsTPS20 | 566 | 66,011.99 | 5.37 | −0.449 | Cyto |
| CsTPS21 | 544 | 63,205.22 | 5.32 | −0.299 | Cyto, Nu |
| CsTPS22 | 799 | 91,777.64 | 5.49 | −0.276 | Cyto, PM |
| CsTPS23 | 799 | 91,775.78 | 5.59 | −0.259 | Cyto, PM |
| CsTPS24 | 409 | 47,964.55 | 5.16 | −0.382 | Cyto |
| CsTPS25 | 510 | 59,631.9 | 5.05 | −0.344 | Nu |
| CsTPS26 | 511 | 59,518.71 | 5.36 | −0.405 | Nu |
| CsTPS27 | 542 | 62,857.37 | 5.19 | −0.387 | Cyto, Nu |
| CsTPS28 | 606 | 70,150.71 | 5.12 | −0.336 | Cyto, PM, Nu |
| CsTPS29 | 507 | 58,383.77 | 5.29 | −0.22 | Cyto |
| CsTPS30 | 430 | 48,502.76 | 5.67 | −0.29 | Nu |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bano, M.; Li, X.; Ali, A.; Khan, M.; Chen, L.; Zhang, X. Genome-Wide Analysis of Terpene Synthase Genes in Crocus sativus Reveals Their Regulatory Roles in Terpenoid Biosynthesis and Abiotic Stress Tolerance. Int. J. Mol. Sci. 2025, 26, 9548. https://doi.org/10.3390/ijms26199548
Bano M, Li X, Ali A, Khan M, Chen L, Zhang X. Genome-Wide Analysis of Terpene Synthase Genes in Crocus sativus Reveals Their Regulatory Roles in Terpenoid Biosynthesis and Abiotic Stress Tolerance. International Journal of Molecular Sciences. 2025; 26(19):9548. https://doi.org/10.3390/ijms26199548
Chicago/Turabian StyleBano, Muqaddas, Xingnuo Li, Ahmad Ali, Mohsin Khan, Liang Chen, and Xiujun Zhang. 2025. "Genome-Wide Analysis of Terpene Synthase Genes in Crocus sativus Reveals Their Regulatory Roles in Terpenoid Biosynthesis and Abiotic Stress Tolerance" International Journal of Molecular Sciences 26, no. 19: 9548. https://doi.org/10.3390/ijms26199548
APA StyleBano, M., Li, X., Ali, A., Khan, M., Chen, L., & Zhang, X. (2025). Genome-Wide Analysis of Terpene Synthase Genes in Crocus sativus Reveals Their Regulatory Roles in Terpenoid Biosynthesis and Abiotic Stress Tolerance. International Journal of Molecular Sciences, 26(19), 9548. https://doi.org/10.3390/ijms26199548








