Unsupervised Machine Learning Reveals Temporal Components of Gene Expression in HeLa Cells Following Release from Cell Cycle Arrest
Abstract
1. Introduction
2. Results
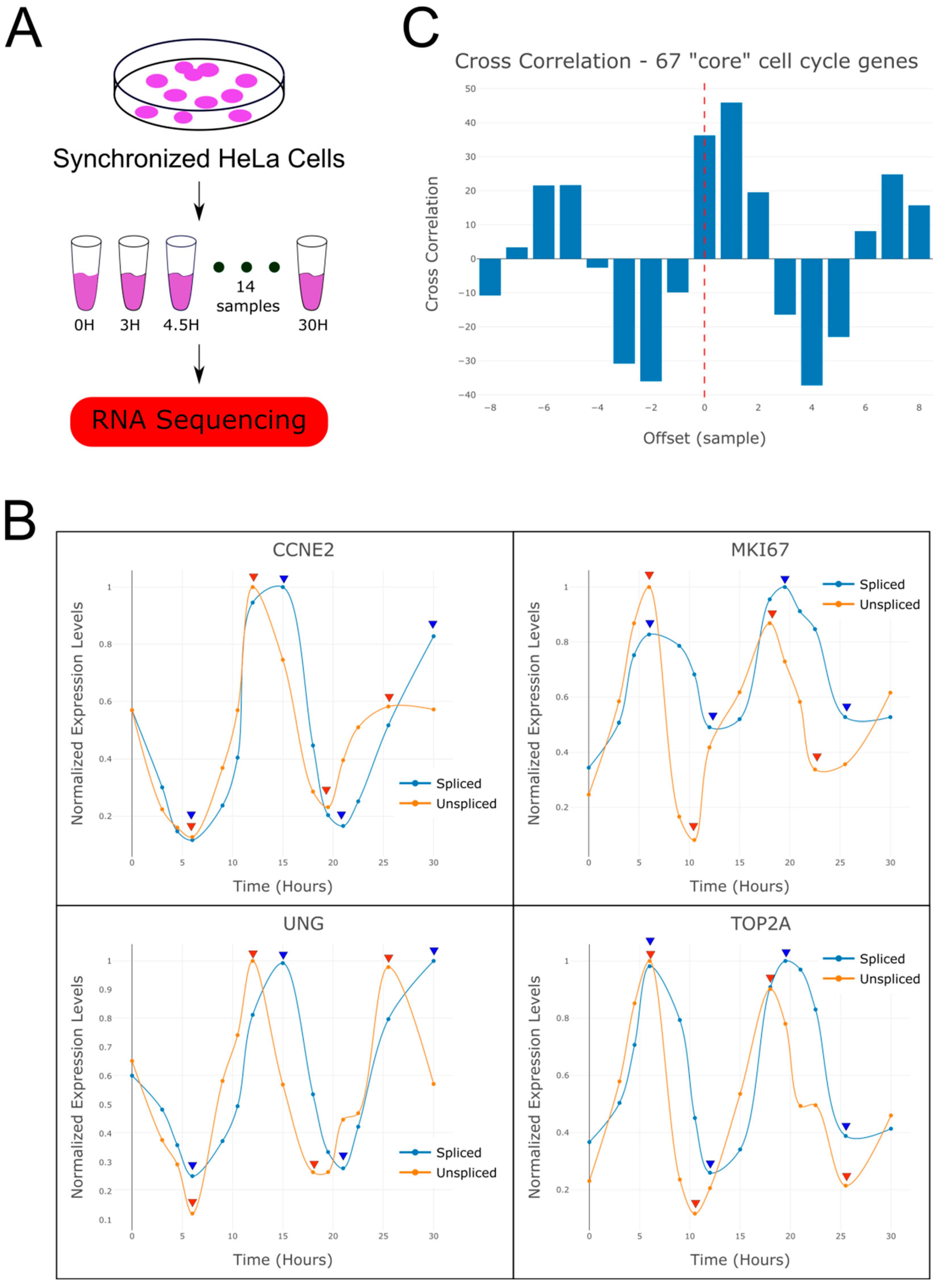
2.1. RNA Velocity Analysis of Periodically Expressed Genes Reveals a Time Lag Between Spliced and Un-Spliced mRNA
2.2. Fourier Analysis Identifies Sets of Genes with Potentially Transient and Oscillatory Behaviors over Time
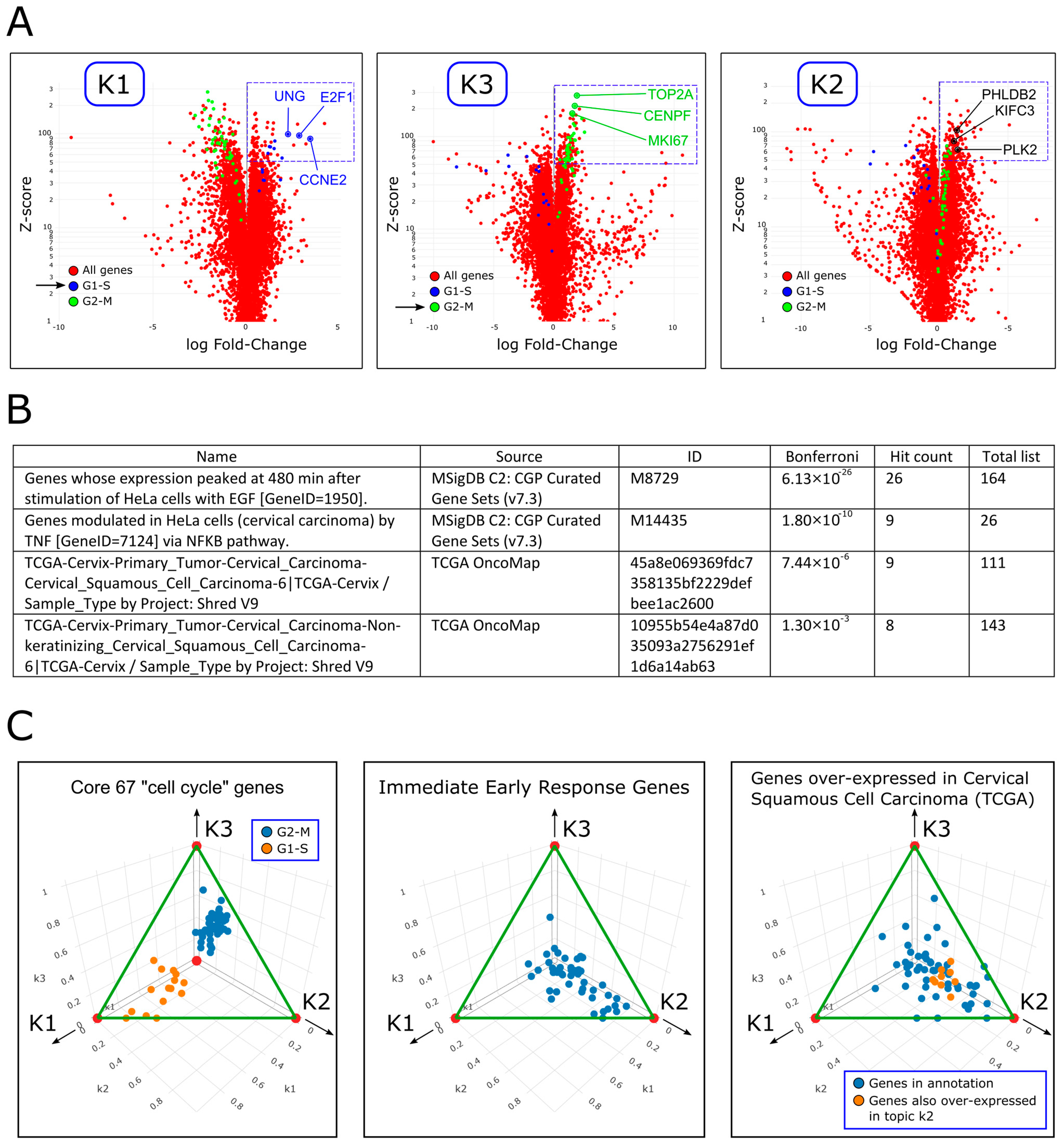
2.3. Topic Modeling Reveals Three Temporal Components, Two of Which Are Periodic, Corresponding to the G1-S and G2-M Phases of the Cell Cycle, and a Third, Transient Component, Related to Immediate Early Response, Regulation of Cell Proliferation, and Cervical Cancer
3. Discussion
4. Materials and Methods
4.1. Datasets and Preprocessing
4.2. RNA Velocity
4.3. Fourier Transform
4.4. Topic Modeling
4.5. Gene Ontology (GO) and Gene Set Enrichment Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dominguez, D.; Tsai, Y.H.; Weatheritt, R.; Wang, Y.; Blencowe, B.J.; Wang, Z. An Extensive Program of Periodic Alternative Splicing Linked to Cell Cycle Progression. eLife 2016, 5, e10288. [Google Scholar] [CrossRef]
- Dominguez, D.; Tsai, Y.H.; Gomez, N.; Jha, D.K.; Davis, I.; Wang, Z. A High-Resolution Transcriptome Map of Cell Cycle Reveals Novel Connections between Periodic Genes and Cancer. Cell Res. 2016, 26, 946–962. [Google Scholar] [CrossRef]
- La Manno, G.; Soldatov, R.; Zeisel, A.; Braun, E.; Hochgerner, H.; Petukhov, V.; Lidschreiber, K.; Kastriti, M.E.; Lönnerberg, P.; Furlan, A.; et al. RNA Velocity of Single Cells. Nature 2018, 560, 494–498. [Google Scholar] [CrossRef] [PubMed]
- Dey, K.K.; Hsiao, C.J.; Stephens, M. Visualizing the Structure of RNA-Seq Expression Data Using Grade of Membership Models. PLoS Genet. 2017, 13, e1006599, Correction in PLoS Genet. 2017,13, e1006759. [Google Scholar] [CrossRef] [PubMed]
- Blei, D.M.; Ng, A.Y.; Jordan, M.I. Latent Dirichlet Allocation. J. Mach. Learn. Res. 2003, 3, 993–1022. [Google Scholar]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for Gene List Enrichment Analysis and Candidate Gene Prioritization. Nucleic Acids Res. 2009, 37 (Suppl. S2), W305–W311. [Google Scholar] [CrossRef]
- Herschman, H.R. Primary Response Genes Induced by Growth Factors and Tumor Promoters. Annu. Rev. Biochem. 1991, 60, 281–319. [Google Scholar] [CrossRef]
- O’Donnell, A.; Odrowaz, Z.; Sharrocks, A.D. Immediate early Gene Activation by the MAPK Pathways: What Do and Don’t We Know? Biochem. Soc. Trans. 2012, 40, 58–66. [Google Scholar] [CrossRef]
- Tullai, J.W.; Schaffer, M.E.; Mullenbrock, S.; Sholder, G.; Kasif, S.; Cooper, G.M. Immediate early and Delayed Primary Response Genes Are Distinct in Function and Genomic Architecture. J. Biol. Chem. 2007, 282, 23981–23995. [Google Scholar] [CrossRef]
- Winkles, J.A. Serum- and Polypeptide Growth Factor-Inducible Gene Expression in Mouse Fibroblasts. In Progress in Nucleic Acid Research and Molecular Biology; Moldave, K., Ed.; Academic Press: Cambridge, MA, USA, 1997; pp. 41–78. [Google Scholar] [CrossRef]
- de Cárcer, G.; Manning, G.; Malumbres, M. From Plk1 to Plk5: Functional evolution of polo-like kinases. Cell Cycle 2011, 10, 2255–2262. [Google Scholar] [CrossRef]
- Casalino, L.; Talotta, F.; Cimmino, A.; Verde, P. The Fra-1/AP-1 Oncoprotein: From the ‘Undruggable’ Transcription Factor to Therapeutic Targeting. Cancers 2022, 14, 1480. [Google Scholar] [CrossRef]
- Milde-Langosch, K. The Fos Family of Transcription Factors and Their Role in Tumourigenesis. Eur. J. Cancer 2005, 41, 2449–2461. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, R.M.G.; Bastos, M.M.S.M.; Medeiros, R.; Oliveira, P.A. The NFκB Signaling Pathway in Papillomavirus-Induced Lesions: Friend or Foe? Anticancer Res. 2016, 36, 2073–2083. [Google Scholar]
- Tilborghs, S.; Corthouts, J.; Verhoeven, Y.; Arias, D.; Rolfo, C.; Trinh, X.B.; van Dam, P.A. The Role of Nuclear Factor-Kappa B Signaling in Human Cervical Cancer. Crit. Rev. Oncol. Hematol. 2017, 120, 141–150. [Google Scholar] [CrossRef]
- Silke, J.; Vaux, D.L. Two Kinds of BIR-Containing Protein—Inhibitors of Apoptosis, or Required for Mitosis. J. Cell Sci. 2001, 114, 1821–1827. [Google Scholar] [CrossRef]
- Theilgaard-Mönch, K.; Pundhir, S.; Reckzeh, K.; Su, J.; Tapia, M.; Furtwängler, B.; Jendholm, J.; Jakobsen, J.S.; Hasemann, M.S.; Knudsen, K.J.; et al. Transcription Factor-Driven Coordination of Cell Cycle Exit and Lineage-Specification in Vivo during Granulocytic Differentiation. Nat. Commun. 2022, 13, 3595. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Carbonetto, P.; Luo, K.; Dey, K.; Hsiao, J.; Stephens, M. fastTopics: Fast Algorithms for Fitting Topic Models and Non-Negative Matrix Factorizations to Count Data, R Package Version 0.4–11; R Foundation: Vienna, Austria, 2021. [Google Scholar]
- Chen, J.; Xu, H.; Aronow, B.J.; Jegga, A.G. Improved Human Disease Candidate Gene Prioritization Using Mouse Phenotype. BMC Bioinform. 2007, 8, 392. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Bi, L.; Ma, F.; Tian, R.; Zhou, Y.; Lan, W.; Song, Q.; Cheng, X. AJUBA increases the cisplatin resistance through hippo pathway in cervical cancer. Gene 2018, 644, 148–154. [Google Scholar] [CrossRef]
- Kalan, S.; Matveyenko, A.; Loayza, D. LIM Protein Ajuba Participates in the Repression of the ATR-Mediated DNA Damage Response. Front. Genet. 2013, 4, 95. [Google Scholar] [CrossRef]
- Mehdi, H.K.; Raju, K.; Sheela, S.R. Association of P16, Ki-67, and CD44 expression in high-grade squamous intraepithelial neoplasia and squamous cell carcinoma of the cervix. J. Cancer Res. Ther. 2023, 19, S260–S267. [Google Scholar] [CrossRef] [PubMed]
- Meyerson, M.; Harlow, E. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol. Cell. Biol. 1994, 14, 2077–2086. [Google Scholar] [CrossRef]
- Huang, C.; Chen, Z.; He, Y.; He, Z.; Ban, Z.; Zhu, Y.; Ding, L.; Yang, C.; Jeong, J.; Yuan, W.; et al. EphA2 promotes tumorigenicity of cervical cancer by up-regulating CDK6. J. Cell. Mol. Med. 2021, 25, 2967–2975. [Google Scholar] [CrossRef]
- Xie, H.; Zhao, Y.; Caramuta, S.; Larsson, C.; Lui, W.-O. miR-205 Expression Promotes Cell Proliferation and Migration of Human Cervical Cancer Cells. PLoS ONE 2012, 7, e46990. [Google Scholar] [CrossRef]
- Wong, Y.; Cheung, T.; Tsao, G.S.; Lo, K.W.; Yim, S.; Wang, V.W.; Heung, M.M.; Chan, S.C.; Chan, L.K.; Ho, T.W.; et al. Genome-wide gene expression profiling of cervical cancer in Hong Kong women by oligonucleotide microarray. Int. J. Cancer 2006, 118, 2461–2469. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Avila, L.; Castro-Amaya, A.M.; Molina-Pineda, A.; Hernández-Gutiérrez, R.; Jave-Suarez, L.F.; Aguilar-Lemarroy, A. The Value of CXCL1, CXCL2, CXCL3, and CXCL8 as Potential Prognosis Markers in Cervical Cancer: Evidence of E6/E7 from HPV16 and 18 in Chemokines Regulation. Biomedicines 2023, 11, 2655. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Kim, Y.T.; Kim, D.K.; Song, C.H.; Lee, J.W. Expression of Epidermal Growth Factor Receptor in Carcinoma of the Cervix. Gynecol. Oncol. 1996, 60, 283–287. [Google Scholar] [CrossRef]
- Shen, L.; Shui, Y.; Wang, X.; Sheng, L.; Yang, Z.; Xue, D.; Wei, Q. EGFR and HER2 expression in primary cervical cancers and corresponding lymph node metastases: Implications for targeted radiotherapy. BMC Cancer 2008, 8, 232. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, Q.; Hu, J.; Chen, Y.; Liu, C.; Xu, C. Expression of HIF-2αand VEGF in Cervical Squamous Cell Carcinoma and Its Clinical Significance. BioMed Res. Int. 2016, 2016, 5631935. [Google Scholar] [CrossRef]
- Fujimoto, J.; Ichigo, S.; Hori, M.; Hirose, R.; Sakaguchi, H.; Tamaya, T. Expression of basic fibroblast growth factor and its mRNA in advanced uterine cervical cancers. Cancer Lett. 1997, 111, 21–26. [Google Scholar] [CrossRef]
- Yee, G.P.C.; De Souza, P.L.; Khachigian, L.M. Reducing invasion potential of cervical cancer cells via targeted knockdown of c-Jun. J. Clin. Oncol. 2013, 31, e22005. [Google Scholar] [CrossRef]
- Prusty, B.K.; Das, B.C. Constitutive activation of transcription factor AP-1 in cervical cancer and suppression of human papillomavirus (HPV) transcription and AP-1 activity in HeLa cells by curcumin. Int. J. Cancer 2004, 113, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Zhang, L.; Lu, S.; Li, W.; Dong, W. KIFC3 Promotes Proliferation, Migration, and Invasion in Colorectal Cancer via PI3K/AKT/mTOR Signaling Pathway. Front. Genet. 2022, 13, 848926. [Google Scholar] [CrossRef] [PubMed]
- Halle, M.K.; Sødal, M.; Forsse, D.; Engerud, H.; Woie, K.; Lura, N.G.; Wagner-Larsen, K.S.; Trovik, J.; Bertelsen, B.I.; Haldorsen, I.S.; et al. A 10-gene prognostic signature points to LIMCH1 and HLA-DQB1 as important players in aggressive cervical cancer disease. Br. J. Cancer 2021, 124, 1690–1698. [Google Scholar] [CrossRef]
- Wang, L.; Zhong, Y.; Yang, B.; Zhu, Y.; Zhu, X.; Xia, Z.; Xu, J.; Xu, L. LINC00958 facilitates cervical cancer cell proliferation and metastasis by sponging miR-625-5p to upregulate LRRC8E expression. J. Cell. Biochem. 2019, 121, 2500–2509. [Google Scholar] [CrossRef]
- van de Weerdt, B.C.; Medema, R.H. Polo-Like Kinases: A Team in Control of the Division. Cell Cycle 2006, 5, 853–864. [Google Scholar] [CrossRef]
- Li, J.; Jia, H.; Xie, L.; Wang, X.; Wang, X.; He, H.; Lin, Y.; Hu, L. Association of Constitutive Nuclear Factor-JB Activation with Aggressive Aspects and Poor Prognosis in Cervical Cancer. Int. J. Gynecol. Cancer 2009, 19, 1421–1426. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, R.; Zhou, S.; Ji, Y. Overexpression of Notch1 is associated with the progression of cervical cancer. Oncol. Lett. 2015, 9, 2750–2756, Erratum in Oncol. Lett. 2021, 21, 134. [Google Scholar] [CrossRef]
- Zagouras, P.; Stifani, S.; Blaumueller, C.M.; Carcangiu, M.L.; Artavanis-Tsakonas, S. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proc. Natl. Acad. Sci. USA 1995, 92, 6414–6418. [Google Scholar] [CrossRef]
- Kim, H.-S.; Yoon, G.; Ryu, J.-Y.; Cho, Y.-J.; Choi, J.-J.; Lee, Y.-Y.; Kim, T.-J.; Choi, C.-H.; Song, S.Y.; Kim, B.-G.; et al. Sphingosine kinase 1 is a reliable prognostic factor and a novel therapeutic target for uterine cervical cancer. Oncotarget 2015, 6, 26746–26756. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, Y.; Cheng, H.; Yang, G.; Tan, W. Stanniocalcin 2 promotes cell proliferation and cisplatin resistance in cervical cancer. Biochem. Biophys. Res. Commun. 2015, 466, 362–368. [Google Scholar] [CrossRef]
- Caffarel, M.M.; Chattopadhyay, A.; Araujo, A.M.; Bauer, J.; Scarpini, C.G.; Coleman, N. Tissue transglutaminase mediates the pro-malignant effects of oncostatin M receptor over-expression in cervical squamous cell carcinoma. J. Pathol. 2013, 231, 168–179. [Google Scholar] [CrossRef]
- Liu, S.; Meng, F.; Ding, J.; Ji, H.; Lin, M.; Zhu, J.; Ma, R. High TRIM44 expression as a valuable biomarker for diagnosis and prognosis in cervical cancer. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Zhang, X.; Qin, G.; Chen, G.; Li, T.; Gao, L.; Huang, L.; Zhang, Y.; Ouyang, K.; Wang, Y.; Pang, Y.; et al. Variants inTRIM44Cause Aniridia by ImpairingPAX6Expression. Hum. Mutat. 2015, 36, 1164–1167. [Google Scholar] [CrossRef]
- van Rijssel, J.; van Buul, J.D. The many faces of the guanine-nucleotide exchange factor trio. Cell Adhes. Migr. 2012, 6, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.; Zhuang, Z.; Deng, X.; Xu, Y.; Zhang, P.; Zhu, L. Knockdown of Trio by CRISPR/Cas9 suppresses migration and invasion of cervical cancer cells. Oncol. Rep. 2017, 39, 795–801. [Google Scholar] [CrossRef]
- Lin, L.; Liu, Y.; Zhao, W.; Sun, B.; Chen, Q. Wnt5A expression is associated with the tumor metastasis and clinical survival in cervical cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 6072–6078. [Google Scholar]
- Song, Z.; Lin, Y.; Ye, X.; Feng, C.; Lu, Y.; Yang, G.; Dong, C. Expression of IL-1α and IL-6 is Associated with Progression and Prognosis of Human Cervical Cancer. Med. Sci. Monit. 2016, 22, 4475–4481. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maimon, T.; Trink, Y.; Goldberger, J.; Kalisky, T. Unsupervised Machine Learning Reveals Temporal Components of Gene Expression in HeLa Cells Following Release from Cell Cycle Arrest. Int. J. Mol. Sci. 2025, 26, 9491. https://doi.org/10.3390/ijms26199491
Maimon T, Trink Y, Goldberger J, Kalisky T. Unsupervised Machine Learning Reveals Temporal Components of Gene Expression in HeLa Cells Following Release from Cell Cycle Arrest. International Journal of Molecular Sciences. 2025; 26(19):9491. https://doi.org/10.3390/ijms26199491
Chicago/Turabian StyleMaimon, Tom, Yaron Trink, Jacob Goldberger, and Tomer Kalisky. 2025. "Unsupervised Machine Learning Reveals Temporal Components of Gene Expression in HeLa Cells Following Release from Cell Cycle Arrest" International Journal of Molecular Sciences 26, no. 19: 9491. https://doi.org/10.3390/ijms26199491
APA StyleMaimon, T., Trink, Y., Goldberger, J., & Kalisky, T. (2025). Unsupervised Machine Learning Reveals Temporal Components of Gene Expression in HeLa Cells Following Release from Cell Cycle Arrest. International Journal of Molecular Sciences, 26(19), 9491. https://doi.org/10.3390/ijms26199491





