Transcriptome and Metabolome Analyses Reveal Molecular Mechanisms Regulating Growth Traits in Large Yellow Croaker (Larimichthys crocea)
Abstract
1. Introduction
2. Results
2.1. Transcriptome Analysis
2.1.1. Transcriptome Data Processing and Quality Control
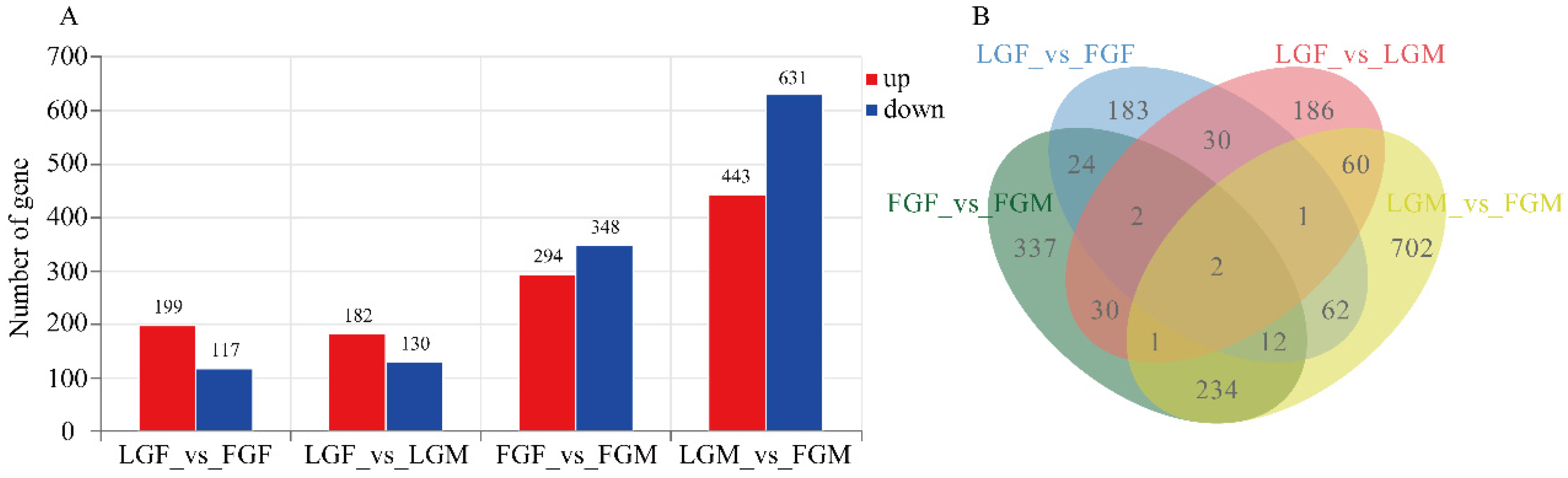
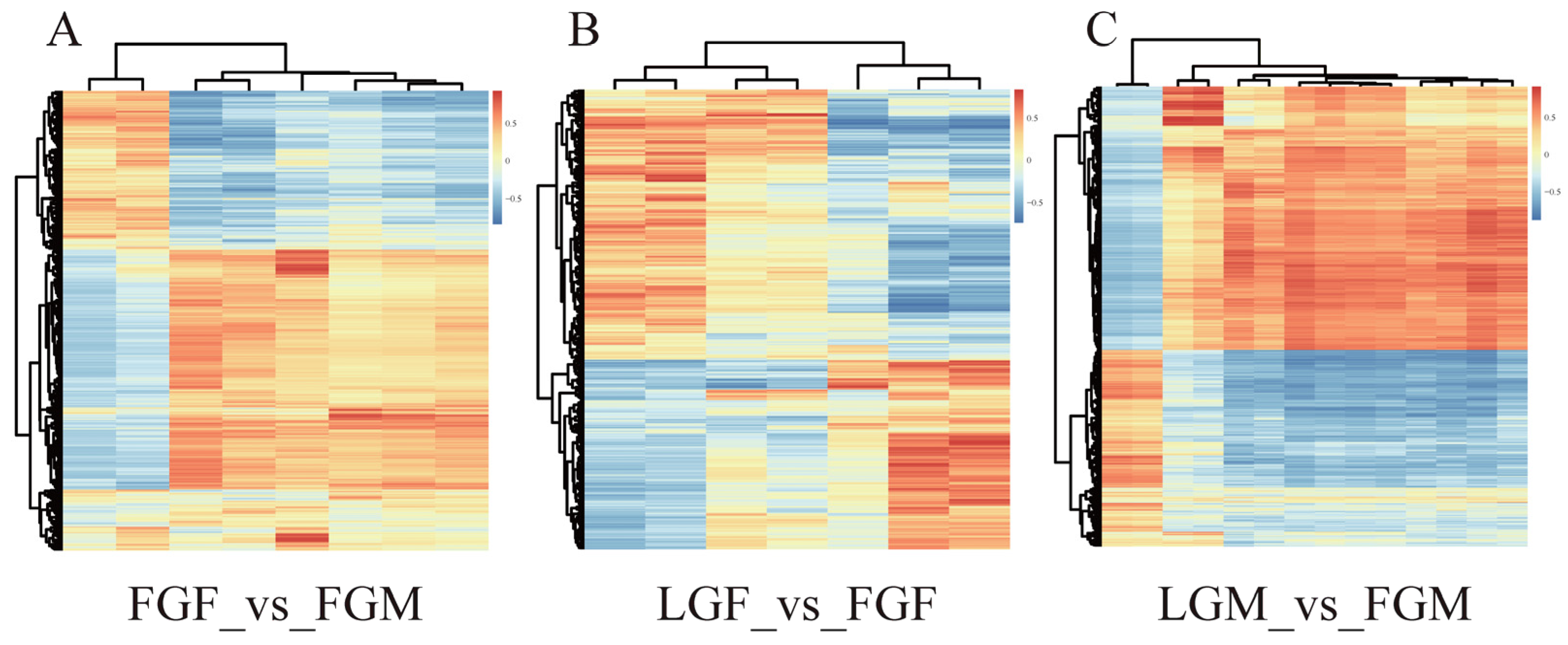
2.1.2. DEGs Analysis
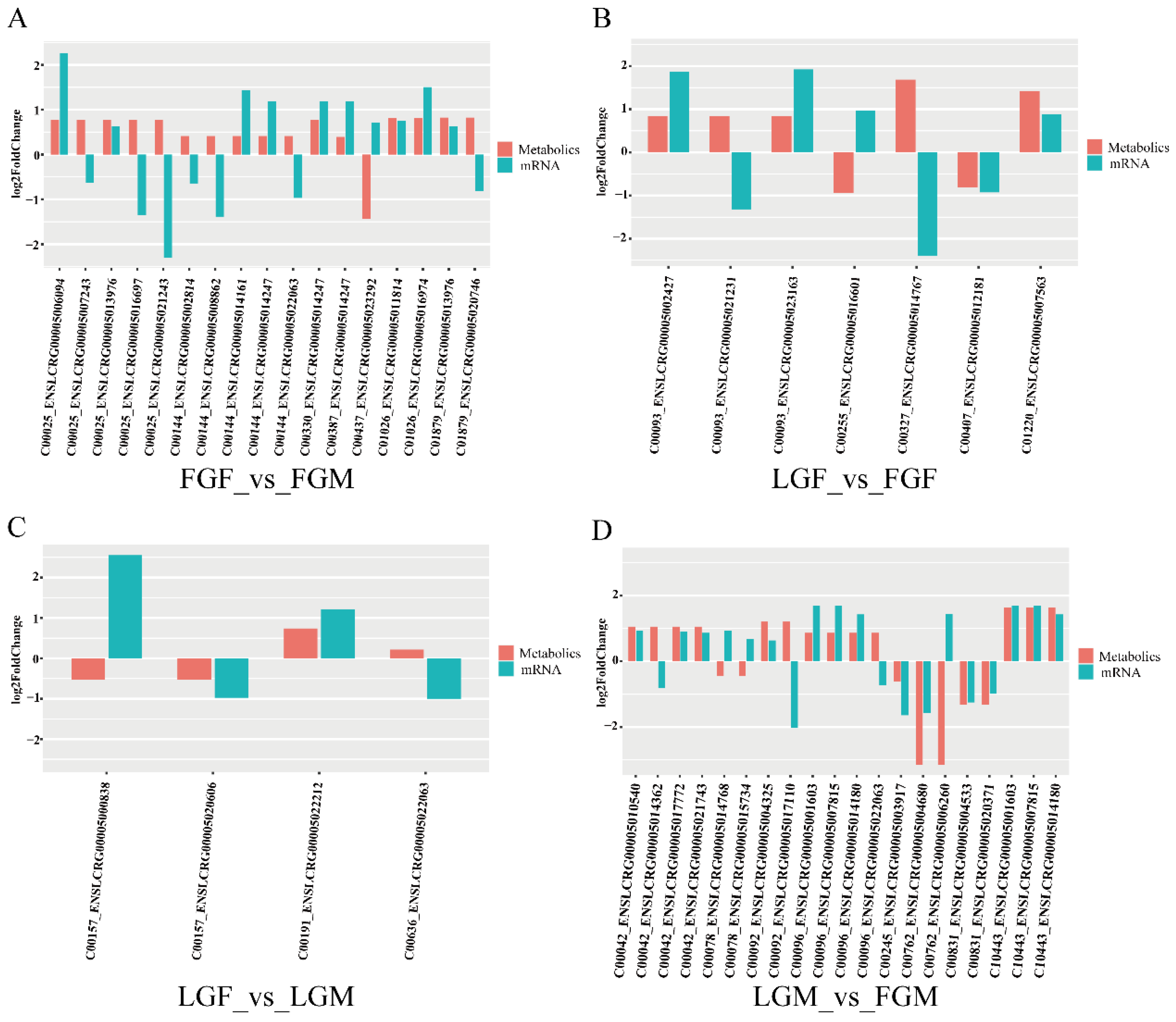
2.1.3. Validation by qRT-PCR
2.2. Metabolome Analysis
2.2.1. Multivariate Statistical Analysis
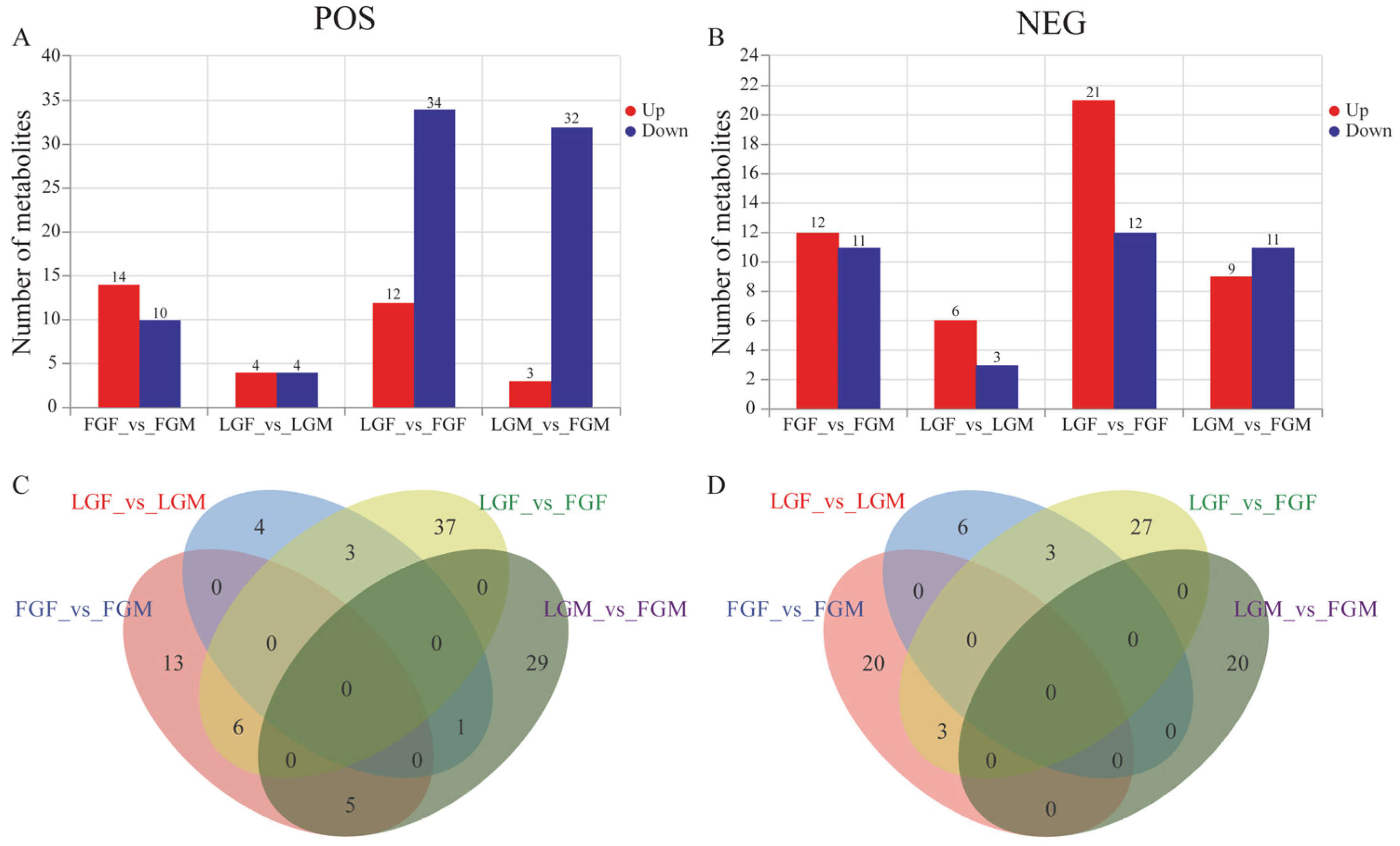
2.2.2. DEMs Analysis
2.3. Metabolome and Transcriptome Association Analysis
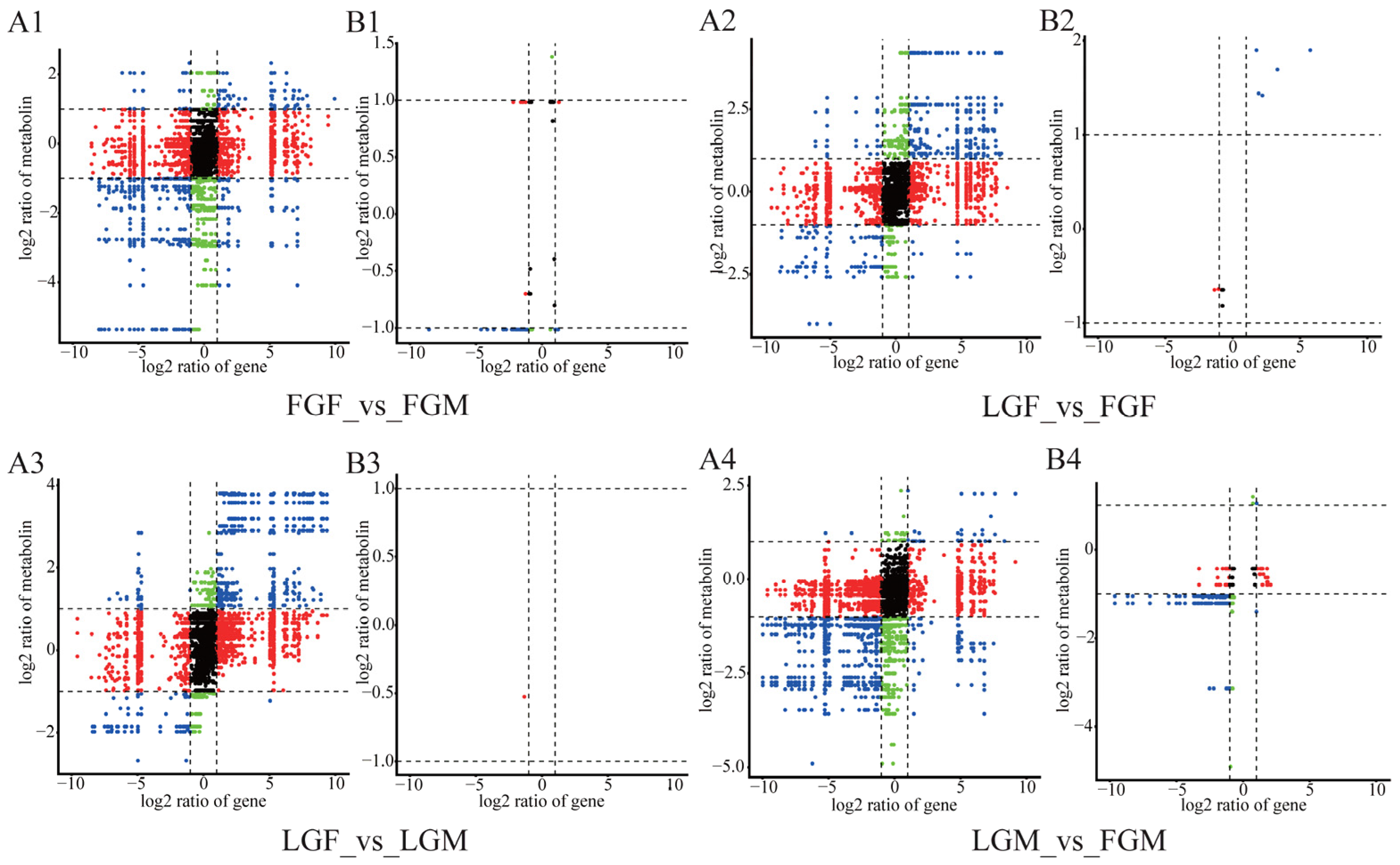
2.3.1. Correlation Analysis of Metabolites and Corresponding Transcripts
2.3.2. Differential Expression Results of Metabolites and Related Transcripts
2.3.3. Transcriptional and Metabolomic Co-Enrichment Pathways
3. Discussion
4. Materials and Methods
4.1. Experimental Fish
4.2. Experimental Design
4.3. Transcriptome Analysis
4.4. qRT-PCR Verification
4.5. Metabolomic Analysis
4.6. Metabolome and Transcriptome Association Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, X.; Lai, J.; Chen, Y.; Liu, Y.; Song, M.; Li, F.; Li, P.; Li, Q.; Gong, Q. Combination of Metabolome and Proteome Analyses Provides Insights into the Mechanism Underlying Growth Differences in Acipenser dabryanus. iScience 2023, 26, 107413. [Google Scholar] [CrossRef]
- Wang, Z.-R.; Li, S.-Y.; Zhang, Y.-Z.; Li, Y.-A.; Huo, H.-H.; Yu, C.-Q.; Zhou, Q.-B. Metabolomic and Transcriptomic Profiling Reveals the Effect of Dietary Protein and Lipid Levels on Growth Performance in Loach (Paramisgurnus dabryanus). Front. Immunol. 2023, 14, 1236812. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; An, X.; Bao, M.; He, J.; Wu, C.; Liu, H. Cloning of Catalase Gene in Large Yellow Croaker (Larimichthys crocea) and Its Response to Vibrio anguillarum Infection. J. Fish. China 2017, 41, 641–648. [Google Scholar]
- Yu, Y.; Huang, W.; Yin, F.; Liu, H.; Cui, M. Aquaculture in an Offshore Ship: An On-Site Test of Large Yellow Croaker (Larimichthys crocea). J. Mar. Sci. Eng. 2023, 11, 101. [Google Scholar] [CrossRef]
- Gao, Y.; Huang, X.; Liu, Y.; Lv, H.; Yin, X.; Li, W.; Chu, Z. Transcriptome Analysis of Large Yellow Croaker (Larimichthys crocea) at Different Growth Rates. Fish Physiol. Biochem. 2024, 50, 1745–1757. [Google Scholar] [CrossRef]
- Xiao, S.; Wang, P.; Zhang, Y.; Fang, L.; Liu, Y.; Li, J.-T.; Wang, Z.-Y. Gene Map of Large Yellow Croaker (Larimichthys crocea) Provides Insights into Teleost Genome Evolution and Conserved Regions Associated with Growth. Sci. Rep. 2015, 5, 18661. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Dong, L.; Xiao, S.; Han, Z.; Wang, X.; Wang, Z. Genomewide Association Study for Economic Traits in the Large Yellow Croaker with Different Numbers of Extreme Phenotypes. J. Genet. 2018, 97, 887–895. [Google Scholar] [CrossRef]
- Ye, H.; Liu, Y.; Liu, X.; Wang, X.; Wang, Z. Genetic Mapping and QTL Analysis of Growth Traits in the Large Yellow Croaker (Larimichthys crocea). Mar. Biotechnol. 2014, 16, 729–738. [Google Scholar] [CrossRef]
- Lin, H.; Zhou, Z.; Zhao, J.; Zhou, T.; Bai, H.; Ke, Q.; Pu, F.; Zheng, W.; Xu, P. Genome-Wide Association Study Identifies Genomic Loci of Sex Determination and Gonadosomatic Index Traits in Large Yellow Croaker (Larimichthys crocea). Mar. Biotechnol. 2021, 23, 127–139. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Yebra, L. Aminoacyl-tRNA Synthetases Activity as a Growth Index in Zooplankton. J. Plankton Res. 2004, 26, 351–356. [Google Scholar] [CrossRef]
- Wang, P.; Ng, Q.X.; Zhang, H.; Zhang, B.; Ong, C.N.; He, Y. Metabolite Changes behind Faster Growth and Less Reproduction of Daphnia similis Exposed to Low-Dose Silver Nanoparticles. Ecotoxicol. Environ. Saf. 2018, 163, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Scanlan, L.D.; Reed, R.B.; Loguinov, A.V.; Antczak, P.; Tagmount, A.; Aloni, S.; Nowinski, D.T.; Luong, P.; Tran, C.; Karunaratne, N.; et al. Silver Nanowire Exposure Results in Internalization and Toxicity to Daphnia Magna. ACS Nano 2013, 7, 10681–10694. [Google Scholar] [CrossRef]
- Saavedra, M.; Conceição, L.E.C.; Barr, Y.; Helland, S.; Pousão-Ferreira, P.; Yúfera, M.; Dinis, M.T. Tyrosine and Phenylalanine Supplementation on Diplodus Sargus Larvae: Effect on Growth and Quality. Aquac. Res. 2010, 41, 1523–1532. [Google Scholar] [CrossRef]
- Papoutsoglou, S.E.; Karakatsouli, N.; Koustas, P. Effects of Dietary L-Tryptophan and Lighting Conditions on Growth Performance of European Sea Bass (Dicentrarchus labrax) Juveniles Reared in a Recirculating Water System. J. Appl. Ichthyol. 2005, 21, 520–524. [Google Scholar] [CrossRef]
- Biswas, P.; Rawat, P.; Patel, A.B.; Jena, A.K. Dietary Supplementation of L-Tryptophan: Effect on Growth and Survival of Pabda, Ompok bimaculatus (Bloch) Fry. J. Appl. Aquac. 2019, 31, 322–336. [Google Scholar] [CrossRef]
- Shang, X.D.; Luo, L.; Wen, H.; Gao, W.; Wang, Q.S.; Xu, H. Dietary isoleucine requirement of juvenile grass carp (Ctenopharyngodon idella). J. Fish. China 2009, 33, 813–822. [Google Scholar]
- Rahimnejad, S.; Lee, K.-J. Dietary Isoleucine Influences Non-Specific Immune Response in Juvenile Olive Flounder (Paralichthys Olivaceus). Turk. J. Fish. Aquat. Sci. 2014, 14, 853–862. [Google Scholar] [CrossRef]
- Ahmed, I.; Khan, M.A. Dietary Branched-Chain Amino Acid Valine, Isoleucine and Leucine Requirements of Fingerling Indian Major Carp, Cirrhinus Mrigala (Hamilton). Br. J. Nutr. 2006, 96, 450–460. [Google Scholar] [CrossRef]
- Wang, Z.; Mai, K.; Xu, W.; Zhang, Y.; Liu, Y.; Ai, Q. Dietary Methionine Level Influences Growth and Lipid Me-tabolism via GCN2 Pathway in Cobia (Rachycentron canadum). Aquaculture 2016, 454, 148–156. [Google Scholar] [CrossRef]
- Zhou, Y.; He, J.; Su, N.; Masagounder, K.; Xu, M.; Chen, L.; Liu, Q.; Ye, H.; Sun, Z.; Ye, C. Effects of DL-Methionine and a Methionine Hydroxy Analogue (MHA-Ca) on Growth, Amino Acid Profiles and the Ex-pression of Genes Related to Taurine and Protein Synthesis in Common Carp (Cyprinus carpio). Aquaculture 2021, 532, 735962. [Google Scholar] [CrossRef]
- Zou, X.; Liu, Q.; Guan, Q.; Zhao, M.; Zhu, X.; Pan, Y.; Liu, L.; Gao, Z. Muscle Fiber Characteristics and Transcrip-tome Analysis in Slow- and Fast-Growing Megalobrama amblycephala. Genes 2024, 15, 179. [Google Scholar] [CrossRef]
- Valente, L.M.P.; Moutou, K.A.; Conceição, L.E.C.; Engrola, S.; Fernandes, J.M.O.; Johnston, I.A. What Determines Growth Potential and Juvenile Quality of Farmed Fish Species? Rev. Aquac. 2013, 5, S168–S193. [Google Scholar] [CrossRef]
- Ohno-Shosaku, T.; Tanimura, A.; Hashimotodani, Y.; Kano, M. Endocannabinoids and Retrograde Modulation of Synaptic Transmission. Neuroscientist 2012, 18, 119–132. [Google Scholar] [CrossRef]
- Harkany, T.; Horvath, T.L. (S)Pot on Mitochondria: Cannabinoids Disrupt Cellular Respiration to Limit Neuronal Activity. Cell Metab. 2017, 25, 8–10. [Google Scholar] [CrossRef]
- Brandt, U. Energy Converting NADH: Quinone Oxidoreductase (Complex I). Annu. Rev. Biochem. 2006, 75, 69–92. [Google Scholar] [CrossRef]
- Yagi, T.; Matsuno-Yagi, A. The Proton-Translocating NADH−Quinone Oxidoreductase in the Respiratory Chain: The Secret Unlocked. Biochemistry 2003, 42, 2266–2274. [Google Scholar] [CrossRef]
- Nakamaru-Ogiso, E.; Han, H.; Matsuno-Yagi, A.; Keinan, E.; Sinha, S.C.; Yagi, T.; Ohnishi, T. The ND2 Subunit Is Labeled by a Photoaffinity Analogue of Asimicin, a Potent Complex I Inhibitor. FEBS Lett. 2010, 584, 883–888. [Google Scholar] [CrossRef] [PubMed]
- Eya, J.C.; Ashame, M.F.; Pomeroy, C.F.; Manning, B.B.; Peterson, B.C. Genetic Variation in Feed Consumption, Growth, Nutrient Utilization Efficiency and Mitochondrial Function within a Farmed Population of Channel Catfish (Ictalurus punctatus). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2012, 163, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Albarracin, S.L.; Baldeon, M.E.; Sangronis, E.; Petruschina, A.C.; Reyes, F.G.R. L-Glutamate: A Key Amino Acid for Senory and Metabolic Functions. Arch. Latinoam. Nutr. 2016, 66, 101–112. [Google Scholar]
- Li, X.; Zheng, S.; Wu, G. Nutrition and Metabolism of Glutamate and Glutamine in Fish. Amino Acids 2020, 52, 671–691. [Google Scholar] [CrossRef]
- Zhao, Y.; Hu, Y.; Zhou, X.-Q.; Zeng, X.-Y.; Feng, L.; Liu, Y.; Jiang, W.-D.; Li, S.-H.; Li, D.-B.; Wu, X.-Q.; et al. Effects of Dietary Glutamate Supplementation on Growth Performance, Digestive Enzyme Activities and Antioxidant Capacity in Intestine of Grass Carp (Ctenopharyngodon idella). Aquac. Nutr. 2015, 21, 935–941. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Wang, J.; Chen, X.; Chu, S.; You, Y.; Chi, Y.; Wang, R.; Yang, X.; Hayat, K.; Zhang, D.; Zhou, P. Comparative Cytology Combined with Transcriptomic and Metabolomic Analyses of Solanum nigrum L. in Response to Cd Toxicity. J. Hazard. Mater. 2022, 423, 127168. [Google Scholar] [CrossRef] [PubMed]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A Cross-Platform Toolkit for Mass Spectrometry and Proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Le, X.C.; Zhu, L. Metabolomics and Transcriptomics Reveal Defense Mechanism of Rice (Oryza Sativa) Grains under Stress of 2,2′,4,4′-Tetrabromodiphenyl Ether. Environ. Int. 2019, 133, 105154. [Google Scholar] [CrossRef]



| Pathway | DEGs | DEMs | ||
|---|---|---|---|---|
| LGF_vs_FGF | LGM_vs_FGM | LGF_vs_FGF | LGM_vs_FGM | |
| Aminoacyl-tRNA biosynthesis | (−) arginyl-tRNA synthetase 1 | (−) L-Isoleucine | (−) L-Tyrosine | |
| (−) asparaginyl-tRNA synthetase 1 | (−) L-Tryptophan | |||
| Alanine, aspartate, and glutamate metabolism | (−) asparagine synthetase | (−) N-Acetyl-L-aspartate | (+) Succinate | |
| (−) argininosuccinate synthase 1 | (+) D-Glucosamine 6-phosphate | |||
| Inositol phosphate metabolism | (+) phospholipase C, epsilon 1 | (+) 1D-myo-Inositol 1,4-bisphosphate | (+) D-Glucose 6-phosphate | |
| (+) triosephosphate isomerase 1b | ||||
| (−) phospholipase C, epsilon 1 | (−) D-Glucuronate | |||
| Central carbon metabolism in cancer | (+) phosphofructokinase, muscle b | (−) L-Leucine | (−) L-Leucine | |
| (+) lactate dehydrogenase A4 | (+) D-Glucose 6-phosphate | |||
| (+) phosphoglycerate mutase 2 | (+) Succinate | |||
| Mineral absorption | (−) hephaestin like 1 | (−) L-Isoleucine | (−) L-Tryptophan | |
| Protein digestion and absorption | (−) solute carrier family 16 member 10 | (−) L-Isoleucine | (−) L-Tyrosine | |
| (−) L-Tryptophan | ||||
| Amino sugar and nucleotide sugar metabolism | (−) UDP-N-acetylglucosamine pyrophosphorylase 1, like 1 | (+) D-Glucosamine 6-phosphate | (+) alpha-D-Galactose 1-phosphate | |
| (+) glucose-6-phosphate isomerase b | ||||
| (+) phosphoglucomutase-1 | (+) GDP-mannose | |||
| Thyroid hormone synthesis | (+) ATPase Na+/K+ transporting subunit beta 1b | (−) ATPase Na+/K+ transporting subunit beta 1b | (+) Glutathione disulfide | (+) D-Glucose 6-phosphate |
| Insulin resistance | (+) protein kinase, AMP-activated, gamma 3a non-catalytic subunit | (+) D-Glucosamine 6-phosphate | (+) D-Glucose 6-phosphate | |
| Starch and sucrose metabolism | (+) glucose-6-phosphate isomerase b | (+) alpha-D-Glucose 1,6-bisphosphate | (+) D-Glucose 6-phosphate | |
| (+) phosphoglucomutase-1 | ||||
| (+) glycogen debranching enzyme | ||||
| Pathway | DEGs | DEMs | ||
|---|---|---|---|---|
| FGF_vs_FGM | LGF_vs_LGM | FGF_vs_FGM | LGF_vs_LGM | |
| Retrograde endocannabinoid signaling | (+) mitogen-activated protein kinase 8a | (+) NADH:ubiquinone oxidoreductase subunit B1 | (+) L-Glutamate | (−) Phosphatidylcholine |
| (+) NADH dehydrogenase subunit 1 | ||||
| (−) NADH:ubiquinone oxidoreductase subunit S5 | ||||
| (+) NADH dehydrogenase subunit 3 | ||||
| (+) fatty acid amide hydrolase | ||||
| (+) NADH dehydrogenase subunit 4L | ||||
| (+) calcium voltage-gated channel subunit alpha1 S | ||||
| (+) NADH dehydrogenase subunit 4 | ||||
| (−) NADH:ubiquinone oxidoreductase core subunit S1 | ||||
| (+) NADH dehydrogenase subunit 5 | ||||
| (+) N-acyl phosphatidylethanolamine phospholipase D | ||||
| (+) NADH dehydrogenase subunit 6 | ||||
| beta-Alanine metabolism | (−) carnosine dipeptidase 2 | (+) glutamate decarboxylase 1 | (−) Pantothenate | (−) Carnosine |
| Ascorbate and aldarate metabolism | (+) UDP-glucuronosyltransferase | (+) UDP-glucuronosyltransferase | (+) D-Galactarate | (+) D-Glucuronate |
| (−) UDP-glucose 6-dehydrogenase | ||||
| (+) regucalcin | ||||
| Arachidonic acid metabolism | (+) gamma-glutamyltransferase 5a | - | (−) 5(S)-HETE | (−) Phosphatidylcholine |
| (−) prostaglandin E synthase 3b (cytosolic) | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, J.; Wang, Y.; Qin, J.; Qiao, G.; Ke, Q.; Li, B.; Wang, X.; Liu, S.; Peng, S. Transcriptome and Metabolome Analyses Reveal Molecular Mechanisms Regulating Growth Traits in Large Yellow Croaker (Larimichthys crocea). Int. J. Mol. Sci. 2025, 26, 9473. https://doi.org/10.3390/ijms26199473
Fang J, Wang Y, Qin J, Qiao G, Ke Q, Li B, Wang X, Liu S, Peng S. Transcriptome and Metabolome Analyses Reveal Molecular Mechanisms Regulating Growth Traits in Large Yellow Croaker (Larimichthys crocea). International Journal of Molecular Sciences. 2025; 26(19):9473. https://doi.org/10.3390/ijms26199473
Chicago/Turabian StyleFang, Jiayi, Yabing Wang, Jianguang Qin, Guangde Qiao, Qiaozhen Ke, Bingfei Li, Xiaoshan Wang, Shengyu Liu, and Shiming Peng. 2025. "Transcriptome and Metabolome Analyses Reveal Molecular Mechanisms Regulating Growth Traits in Large Yellow Croaker (Larimichthys crocea)" International Journal of Molecular Sciences 26, no. 19: 9473. https://doi.org/10.3390/ijms26199473
APA StyleFang, J., Wang, Y., Qin, J., Qiao, G., Ke, Q., Li, B., Wang, X., Liu, S., & Peng, S. (2025). Transcriptome and Metabolome Analyses Reveal Molecular Mechanisms Regulating Growth Traits in Large Yellow Croaker (Larimichthys crocea). International Journal of Molecular Sciences, 26(19), 9473. https://doi.org/10.3390/ijms26199473






