Detection of Targetable Genetic Abnormalities in Neuroblastoma Circulating Tumour DNA
Abstract
1. Introduction
2. Results
2.1. Pathogenic Variants Identified Following ctDNA Variant Filtering
2.2. A High Proportion of Variants Were Identified in NB ctDNA but Not in NB Tumours
2.3. Spatial and Temporal Heterogeneity of Heterozygous ALK Mutations
2.4. Pathogenic, Homozygous Somatic TP53 Mutations
2.5. Heterozygous, Pathogenic PTPN11 Variants
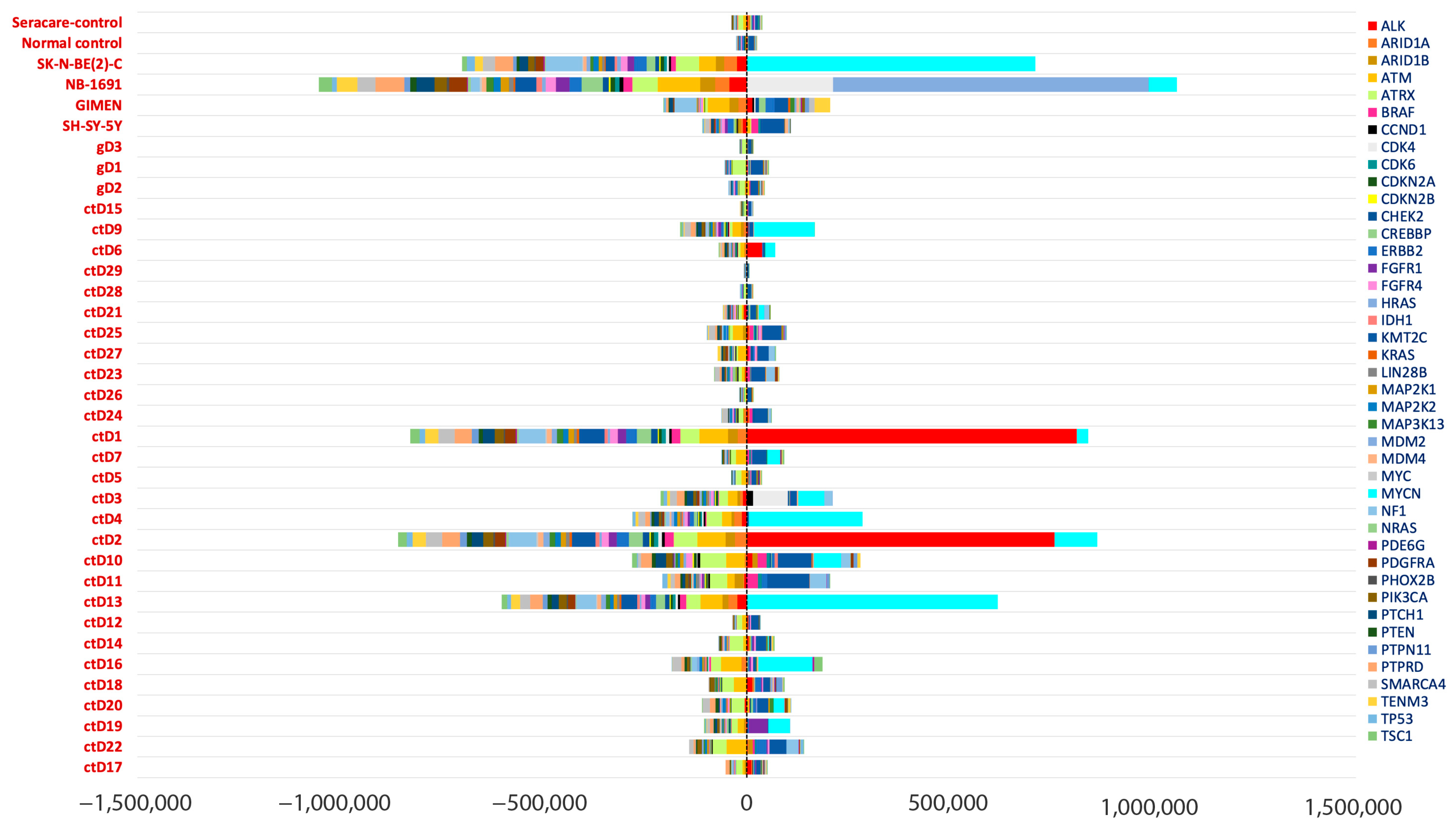
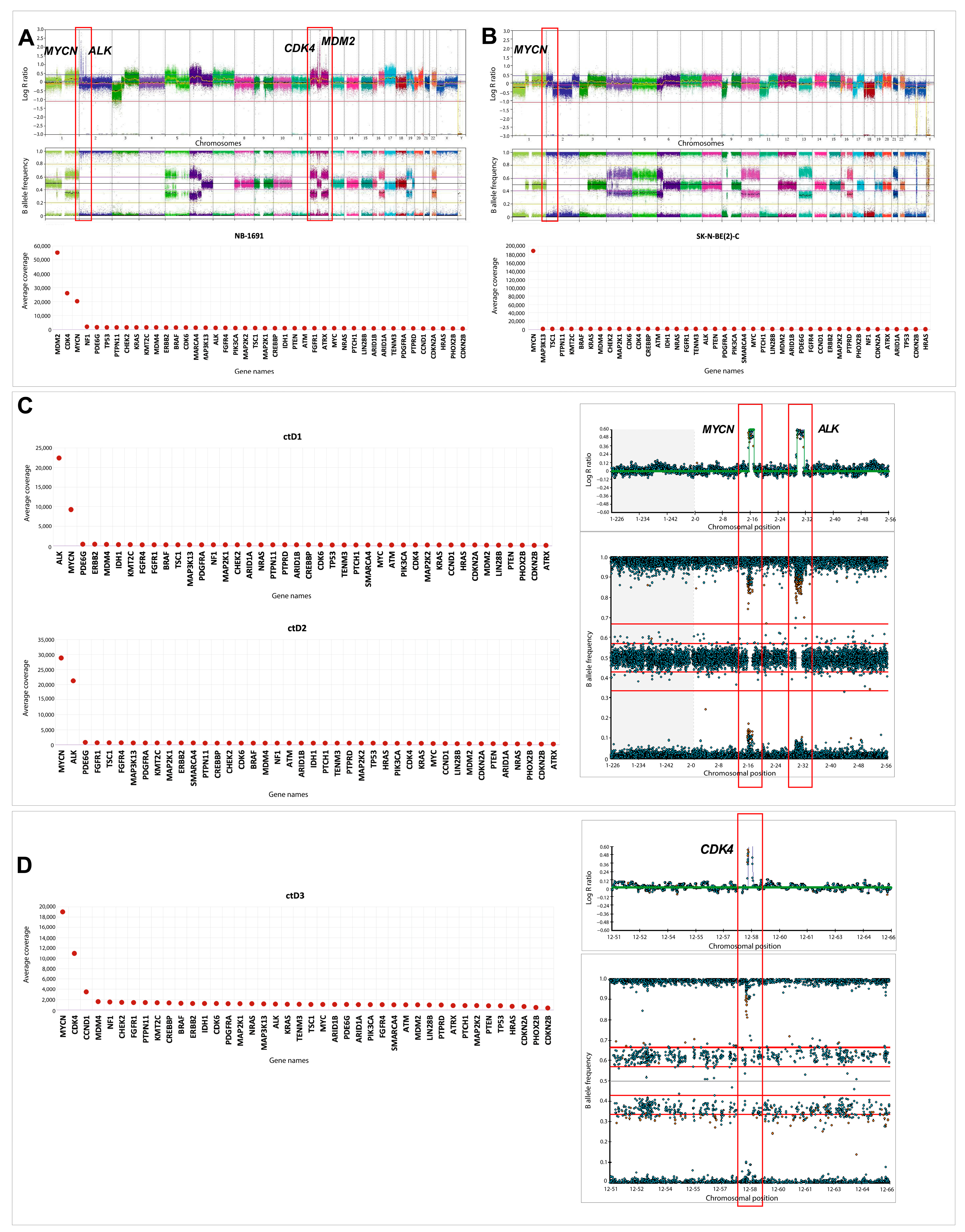
2.6. Oncogene Amplifications Detected in ctDNA
3. Discussion
4. Materials and Methods
4.1. Plasma and Cell-Line Samples
4.2. DNA Extraction
4.3. Library Preparation and Next-Generation Sequencing
4.4. Variant Filtering
4.5. Coverage Analysis
4.6. Classification of Somatic Variants
4.7. ddPCR
4.8. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1
| Sample Name | Sample Type | NB Risk Group and MYCN Status | Time Point | SNP Array Data | |
|---|---|---|---|---|---|
| Gain | Loss | ||||
| a ctD1 | Plasma (PB) | High MNA | R | * 1q, 17q, 18q. ALK amplification | * 1p, 5p, 18p. |
| a ctD2 | Plasma (PB) | High MNA | D | ALK amplification 11q, 17q. | 1p, 10q. |
| ctD3 | Plasma (PB) | High MNA | D | 1q13.1q32.1, 2p16.1p25.3, CDK4 amplification, CCND1 amplification 6p21.3q27, 12p13.3q24.3, 14q11.2q32.12. | X, 11, 9, 10, 4, 5p. |
| ctD4 | Plasma (PB) | High MNA | D | 17q. | 1p, 3p, 10, 15q. |
| ctD5 | Plasma (PB) | High Non-MNA | D | 11pq, 17q, 18. | 11q. |
| ctD6 | Plasma (PB) | High MNA | D | 2p23.1(ALK), 17q. | 1p, 9, 11. |
| ctD7 | Plasma (PB) | High MNA | D | 17q22q25.3. | 1p31.2p36.33. |
| ctD9 | Plasma (PB) | High MNA | D | 16q, 17q, 21. | 1p, 3, 4q, 5pq, 6, 8, 14, 15q, 16q, 17p, |
| CN-LOH: 9, 10, 11, 19, X. | |||||
| ctD10 | Plasma (PB) | High MNA | D | 17p12p11.2, 17q12q22. | 3, 5, 4q, 9, 10, 11, 13q, 21q. |
| d ctD11 | Plasma (PB) | High Non-MNA | D | 6p, 7p, 13q, 21 & 12. | 6p, 6q, 8p, 8q, 11, 17p. |
| ctD12 | Plasma (PB) | High Non-MNA | D | 2p, 6pq, 7p, 11p, 12q, 16q, 17q, 22q, 13 & 18. | 9p, 11q. |
| ctD13 | Plasma (PB) | High MNA | D | 17q. | 1p, 3p. |
| ctD14 | Plasma (PB) | Low Non-MNA | D | Whole-chromosome abnormalities only | |
| e ctD15 | Plasma (PB) | Low Non-MNA | D | Whole-chromosome abnormalities only | |
| ctD16 | Plasma (PB) | High MNA | D | 1q, 2p, 9q, 11q, 17q, 18p. | 1p, 3p, 11q, X. |
| f ctD17 | Plasma (PB) | Int ** Non-MNA | D | 1q, 2p, 5p, 11q, 16q, 17q, 20q. | 3p, 5q, 6p, 6q, 9p (PTPRD), 9q, Y. |
| ctD18 | Plasma (PB) | High Non-MNA | D | 1q, 2p (MYCN), 12q, 17q, | 3p, 3q, 11q, 19q, |
| 20q. | 22q, Xq21.1 (ATRX). | ||||
| ctD19 | Plasma (PB) | High MNA | D | 2, 4, 7, 8, 8p11.22p11.21, 12, 15, 17p11.2q25.3, 18, 20, 21, 22, FGFR1 amplification. | 10 |
| b ctD20 | Plasma (PB) | High MNA | D | 3, 11, 14, 20q13.2q13.33, 21, 22. | 1, 2, 8p23.3q21.3, 9, 10p15.3q21.1, 13, 16q11.2q24.3, 18, 17p. |
| b ctD21 | Plasma (PB) | High MNA | R | 3, 11, 14, 20q13 2q13.33, 21, 22. | 1, 2, 8p23, 3q21.3, 9, 10p15, 3q21.1, 13, 16q11, 2q24.3, 18, 17p. |
| ctD22 | Plasma (PB) | High Non-MNA | D | 1q21.1q42.2,6, 7, 8p23.3p22,10p15.3q11.21, 13, 17, 18, 20, 21. | 1q42.2q44, 3, 9p24.1 (PTPRD), 11, 11q14.1q22.3, 15q13.2q26.3, 19. |
| ctD23 | Plasma (PB) | High Non-MNA | D | 4p16, 3q21.23, 7, 13q11q31.3, 13q32, 1q34, 17, 18. | 1q44, 2q44, 2q21, 3q37.3, 3, 5q15q35.3, 9p24, 3p22.3, 9p21, 3q34.3, 11, 14, 16, 19, 22, X, LOH: 2q21.3q37.3, 5q15q35.3, 11, 16, 22. |
| ctD24 | Plasma (PB) | High Non-MNA | D | 6p, 7pq, 13q, 17pq, 18, 21. | 6q, 7p, 17p. |
| ctD25 | Plasma (PB) | High Non-MNA | D | 1q, 3q, 6p, 3q, 7, 11q, 1q, 12, 16q,17q, 18. | 3p26, 3p12.2, 3p12, 2q29, 4, 6q22, 33q27, 8, 11q13, 3q25, 14, 17p13, 3q21.31, |
| 19p13, 3p13.11. | |||||
| ctD26 | Plasma (PB) | Low Non-MNA | D | 1, 2, 7, 12, 17. | No loss |
| ctD27 | Plasma (PB) | Int Non-MNA | D | 2, 5, 7, 13, 17, 22, X. | 3, 4, 11, 14, 21 LOH: 3, 14. |
| ctD28 | Plasma (PB) | Int ** Non-MNA | D | Tetraploid. | 3, 4, 14, 15, 16. |
| ctD29 | Plasma (PB) | Low Non-MNA | D | No gains. | 2, 3, 4, 11, 12, 14, 15, 16, 19, 21, X |
| CN-LOH: 11. | |||||
| c ctD30 | Plasma (BM) | High Non-MNA | D | * 2p, 6p, 7, 17q, 18. | 3q, 5, 6q, |
| 9p, 10, 11q, 17p, 19. | |||||
| c ctD31 | Plasma (PB) | High Non-MNA | D | * 2p, 6p, 7, 17q, 18. | 3q, 5, 6q, |
| 9p, 10, 11q, 17p, 19. | |||||
| ctD32 | Plasma (PB) | High MNA | R | * 1q, 2p, 11q, 17q,19q, 3, 5, 6, 7, 8, 12, 13, 14, 16, 20, 21. | * 1p, 11q, 19p, 22q. |
| d gD1 | DNA | High | D | - | - |
| e gD2 | DNA | Low | D | - | - |
| f gD3 | DNA | Int ** | D | - | - |
| SH-SY-5Y | DNA | High Non-MNA | R | 1q, 7, 17q. | 14p, 22q. |
| GIMEN | DNA | High Non-MNA | R | 2p. | 1p, 6q, 11q. |
| NB-1691 | DNA | High MNA | R | ALK, CDK4, MDM2 amp 3q, 5p, 6p, 12, 17. | 3p. |
| SK-N-BE-(2)C | DNA | High MNA | R | 2p. | 1p, 17, 18, X, 11p, 9p, 3p. |
| Control1 | Synthetic plasma | - | - | - | - |
| Control2 | Normal control DNA | - | - | - | - |
Appendix A.2
| Variant Gene | Chromosomal Location | Transcript | Function |
|---|---|---|---|
| ALK | 2p23.1 | NM_004304.4 | Tyrosine kinase |
| ARID1A | 1p36.11 | NM_006015.4 | Chromatin remodelling factor |
| ARID1B | 6q25.3 | NM_020732.3 | Chromatin remodelling factor |
| ATM | 11q22.3 | NM_000051.3 | Serine/threonine kinase |
| ATRX | Xq21.1 | NM_000489.3 | Chromatin remodelling factor |
| BRAF | 7q34 | NM_004333.4 | Protein kinase |
| CCND1 | 11q13.3 | NM_053056.2 | Cyclin dependent protein kinase |
| CDK4 | 12q14.1 | NM_000075.3 | Cyclin dependent protein kinase |
| CDK6 | 7q21.2 | NM_001259.6 | Cyclin-dependent protein kinase |
| CDKN2A | 9p21.3 | NM_001195132.1 | Cyclin-dependent protein kinase |
| CDKN2B | 9p21.3 | NM_004936.3 | Cyclin-dependent protein kinase |
| CHEK2 | 22q22.1 | NM_007194.4 | Protein kinase |
| CREBBP | 16p13.3 | NM_004380.3 | Acetyltransferase |
| ERBB2 | 17q12 | NM_004448.2 | Tyrosine kinase |
| FGFR1 | 8p11.23 | NM_001174067.1 | Tyrosine kinase growth factor |
| FGFR4 | 5q35.2 | NM_002011.3 | Growth factor receptor tyrosine kinase |
| HRAS | 11p15.5 | NM_005343.2 | GTPase |
| IDH1 | 2q34 | NM_005896.2 | Catalytic isozyme |
| KMT2C | 7q36.1 | NM_170606.3 | Histone methyltransferase |
| KRAS | 12p12.1 | NM_033360.2 | GTPase |
| LIN28B | 6q16-6q21 | NM_001004317.3 | RNA-binding protein |
| MAP2K1 | 15q22.31 | NM_002755.3 | MAP kinase |
| MAP2K2 | 19p13.3 | NM_030662.3 | Tyrosine kinase |
| MAP3K13 | 3q27.3 | NM_001242314.1 | Serine/threonine kinase |
| MDM2 | 12q15 | NM_002392.4 | TP53 regulator (negative) |
| MDM4 | 1q32.1 | NM_002393.4 | TP53 regulator (negative) |
| MYC | 8q24.21 | NM_002467.4 | Transcription factor |
| MYCN | 2p24.3 | NM_005378.4 | Transcription factor |
| NF1 | 17q11.2 | NM_001042492.2 | Tumour suppressor |
| NRAS | 1p13.2 | NM_002524.4 | GTPase |
| PDE6G | 17q25.3 | NM_002602.3 | Phosphodiesterase |
| PDGFRA | 4q12 | NM_006206.4 | Tyrosine kinase |
| PHOX2B | 4p13 | NM_003924.3 | Transcription factor |
| PIK3CA | 3q26.32 | NM_006218.2 | Lipid kinase |
| PTCH1 | 9q22.32 | NM_000264.3 | Receptor for Hedgehog genes |
| PTEN | 10q23.31 | NM_000314.4 | Phosphatase |
| PTPN11 | 12q24.13 | NM_002834.3 | Protein tyrosine phosphatase |
| PTPRD | 9p23 | NM_002839.3 | Tyrosine phosphatase |
| SMARCA4 | 19p13.2 | NM_003072.5 | Chromatin regulator |
| TENM3 | 4q34.3 | NM_001080477.1 | Neuronal development protein |
| TP53 | 17p13.1 | NM_000546.6 | Tumour suppressor |
| TSC1 | 5q32-q33 | NM_000368.5 | Tumour suppressor |
References
- Tolbert, V.P.; Matthay, K.K. Neuroblastoma: Clinical and biological approach to risk stratification and treatment. Cell Tissue Res. 2018, 372, 195–209. [Google Scholar] [CrossRef]
- Matthay, K.K.; Maris, J.M.; Schleiermacher, G.; Nakagawara, A.; Mackall, C.L.; Diller, L.; Weiss, W.A. Neuroblastoma. Nat. Rev. Dis. Primers 2016, 2, 16078. [Google Scholar] [CrossRef] [PubMed]
- Lone, S.N.; Nisar, S.; Masoodi, T.; Singh, M.; Rizwan, A.; Hashem, S.; El-Rifai, W.; Bedognetti, D.; Batra, S.K.; Haris, M.; et al. Liquid biopsy: A step closer to transform diagnosis, prognosis and future of cancer treatments. Mol. Cancer 2022, 21, 79. [Google Scholar] [CrossRef] [PubMed]
- Russano, M.; Napolitano, A.; Ribelli, G.; Iuliani, M.; Simonetti, S.; Citarella, F.; Pantano, F.; Dell’Aquila, E.; Anesi, C.; Silvestris, N.; et al. Liquid biopsy and tumor heterogeneity in metastatic solid tumors: The potentiality of blood samples. J. Exp. Clin. Cancer Res. 2020, 39, 95, Erratum in J. Exp. Clin. Cancer Res. 2020, 39, 120. [Google Scholar] [CrossRef]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Lodrini, M.; Graef, J.; Thole-Kliesch, T.M.; Astrahantseff, K.; Sprüssel, A.; Grimaldi, M.; Peitz, C.; Linke, R.B.; Hollander, J.F.; Lankes, E.; et al. Targeted Analysis of Cell-free Circulating Tumor DNA is Suitable for Early Relapse and Actionable Target Detection in Patients with Neuroblastoma. Clin. Cancer Res. 2022, 28, 1809–1820. [Google Scholar] [CrossRef]
- Kahana-Edwin, S.; Cain, L.E.; McCowage, G.; Darmanian, A.; Wright, D.; Mullins, A.; Saletta, F.; Karpelowsky, J. Neuroblastoma Molecular Risk-Stratification of DNA Copy Number and ALK Genotyping via Cell-Free Circulating Tumor DNA Profiling. Cancers 2021, 13, 3365. [Google Scholar] [CrossRef]
- Lodrini, M.; Wünschel, J.; Thole-Kliesch, T.M.; Grimaldi, M.; Sprüssel, A.; Linke, R.B.; Hollander, J.F.; Tiburtius, D.; Künkele, A.; Schulte, J.H.; et al. Circulating Cell-Free DNA Assessment in Biofluids from Children with Neuroblastoma Demonstrates Feasibility and Potential for Minimally Invasive Molecular Diagnostics. Cancers 2022, 14, 2080. [Google Scholar] [CrossRef]
- Chan, K.C.; Jiang, P.; Zheng, Y.W.; Liao, G.J.; Sun, H.; Wong, J.; Siu, S.S.; Chan, W.C.; Chan, S.L.; Chan, A.T.; et al. Cancer genome scanning in plasma: Detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin. Chem. 2013, 59, 211–224. [Google Scholar] [CrossRef]
- Leary, R.J.; Sausen, M.; Kinde, I.; Papadopoulos, N.; Carpten, J.D.; Craig, D.; O’Shaughnessy, J.; Kinzler, K.W.; Parmigiani, G.; Vogelstein, B.; et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci. Transl. Med. 2012, 4, 162ra54. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- Ioannidis, N.M.; Rothstein, J.H.; Pejaver, V.; Middha, S.; McDonnell, S.K.; Baheti, S.; Musolf, A.; Li, Q.; Holzinger, E.; Karyadi, D.; et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet. 2016, 99, 877–885. [Google Scholar] [CrossRef]
- Allinson, L.M.; Potts, A.; Goodman, A.; Bown, N.; Bashton, M.; Thompson, D.; Basta, N.O.; Gabriel, A.S.; McCorkindale, M.; Ng, A.; et al. Loss of ALK hotspot mutations in relapsed neuroblastoma. Genes Chromosomes Cancer 2022, 61, 747–753. [Google Scholar] [CrossRef]
- George, S.L.; Izquierdo, E.; Campbell, J.; Koutroumanidou, E.; Proszek, P.; Jamal, S.; Hughes, D.; Yuan, L.; Marshall, L.V.; Carceller, F.; et al. A tailored molecular profiling programme for children with cancer to identify clinically actionable genetic alterations. Eur. J. Cancer 2019, 121, 224–235. [Google Scholar] [CrossRef] [PubMed]
- George, S.L.; Lynn, C.; Stankunaite, R.; Hughes, D.; Sauer, C.M.; Chalker, J.; Waqar Ahmed, S.; Oostveen, M.; Proszek, P.Z.; Yuan, L.; et al. Stratified Medicine Pediatrics: Cell-Free DNA and Serial Tumor Sequencing Identifies Subtype-Specific Cancer Evolution and Epigenetic States. Cancer Discov. 2025, 15, 717–732. [Google Scholar] [CrossRef] [PubMed]
- Jongmans, M.C.J.; van der Burgt, I.; Hoogerbrugge, P.M.; Noordam, K.; Yntema, H.G.; Nillesen, W.M.; Kuiper, R.P.; Ligtenberg, M.J.; van Kessel, A.G.; van Krieken, J.H.; et al. Cancer risk in patients with Noonan syndrome carrying a PTPN11 mutation. Eur. J. Hum. Genet. 2011, 19, 870–874. [Google Scholar] [CrossRef]
- Mutesa, L.; Pierquin, G.; Janin, N.; Segers, K.; Thomée, C.; Provenzi, M.; Bours, V. Germline PTPN11 missense mutation in a case of Noonan syndrome associated with mediastinal and retroperitoneal neuroblastic tumors. Cancer Genet. Cytogenet. 2008, 182, 40–42. [Google Scholar] [CrossRef]
- Ijiri, R.; Tanaka, Y.; Keisuke, K.; Masuno, M.; Imaizumi, K. A case of Noonan’s syndrome with possible associated neuroblastoma. Pediatr. Radiol. 2000, 30, 432–433. [Google Scholar] [CrossRef] [PubMed]
- Merugu, S.; Chen, L.; Gavens, E.; Gabra, H.; Brougham, M.; Makin, G.; Ng, A.; Murphy, D.; Gabriel, A.S.; Robinson, M.L.; et al. Detection of Circulating and Disseminated Neuroblastoma Cells Using the ImageStream Flow Cytometer for Use as Predictive and Pharmacodynamic Biomarkers. Clin. Cancer Res. 2020, 26, 122–134. [Google Scholar] [CrossRef]
- Amoroso, L.; Ognibene, M.; Morini, M.; Conte, M.; Di Cataldo, A.; Tondo, A.; D’Angelo, P.; Castellano, A.; Garaventa, A.; Lasorsa, V.A.; et al. Genomic coamplification of CDK4/MDM2/FRS2 is associated with very poor prognosis and atypical clinical features in neuroblastoma patients. Genes Chromosomes Cancer 2020, 59, 277–285. [Google Scholar] [CrossRef]
- Rihani, A.; Vandesompele, J.; Speleman, F.; Van Maerken, T. Inhibition of CDK4/6 as a novel therapeutic option for neuroblastoma. Cancer Cell Int. 2015, 15, 76. [Google Scholar] [CrossRef] [PubMed]
- Tucker, E.R.; Jiménez, I.; Chen, L.; Bellini, A.; Gorrini, C.; Calton, E.; Gao, Q.; Che, H.; Poon, E.; Jamin, Y.; et al. Combination Therapies Targeting ALK-aberrant Neuroblastoma in Preclinical Models. Clin. Cancer Res. 2023, 29, 1317–1331. [Google Scholar] [CrossRef] [PubMed]
- Bagley, S.J.; Nabavizadeh, S.A.; Mays, J.J.; Till, J.E.; Ware, J.B.; Levy, S.; Sarchiapone, W.; Hussain, J.; Prior, T.; Guiry, S.; et al. Clinical Utility of Plasma Cell-Free DNA in Adult Patients with Newly Diagnosed Glioblastoma: A Pilot Prospective Study. Clin. Cancer Res. 2020, 26, 397–407. [Google Scholar] [CrossRef]
- Fleischhacker, M.; Schmidt, B.; Weickmann, S.; Fersching, D.M.; Leszinski, G.S.; Siegele, B.; Stötzer, O.J.; Nagel, D.; Holdenrieder, S. Methods for isolation of cell-free plasma DNA strongly affect DNA yield. Clin. Chim. Acta 2011, 412, 2085–2088. [Google Scholar] [CrossRef]
- Page, K.; Guttery, D.S.; Zahra, N.; Primrose, L.; Elshaw, S.R.; Pringle, J.H.; Blighe, K.; Marchese, S.D.; Hills, A.; Woodley, L.; et al. Influence of plasma processing on recovery and analysis of circulating nucleic acids. PLoS ONE 2013, 8, e77963. [Google Scholar] [CrossRef]
- Bosse, K.R.; Giudice, A.M.; Lane, M.V.; McIntyre, B.; Schürch, P.M.; Pascual-Pasto, G.; Buongervino, S.N.; Suresh, S.; Fitzsimmons, A.; Hyman, A.; et al. Serial Profiling of Circulating Tumor DNA Identifies Dynamic Evolution of Clinically Actionable Genomic Alterations in High-Risk Neuroblastoma. Cancer Discov. 2022, 12, 2800–2819. [Google Scholar] [CrossRef] [PubMed]
- Chicard, M.; Colmet-Daage, L.; Clement, N.; Danzon, A.; Bohec, M.; Bernard, V.; Baulande, S.; Bellini, A.; Deveau, P.; Pierron, G.; et al. Whole-Exome Sequencing of Cell-Free DNA Reveals Temporo-spatial Heterogeneity and Identifies Treatment-Resistant Clones in Neuroblastoma. Clin. Cancer Res. 2018, 24, 939–949. [Google Scholar] [CrossRef]
- Bobin, C.; Iddir, Y.; Butterworth, C.; Masliah-Planchon, J.; Saint-Charles, A.; Bellini, A.; Bhalshankar, J.; Pierron, G.; Combaret, V.; Attignon, V.; et al. Sequential Analysis of cfDNA Reveals Clonal Evolution in Patients with Neuroblastoma Receiving ALK-Targeted Therapy. Clin. Cancer Res. 2024, 30, 3316–3328. [Google Scholar] [CrossRef]
- Berko, E.R.; Witek, G.M.; Matkar, S.; Petrova, Z.O.; Wu, M.A.; Smith, C.M.; Daniels, A.; Kalna, J.; Kennedy, A.; Gostuski, I.; et al. Circulating tumor DNA reveals mechanisms of lorlatinib resistance in patients with relapsed/refractory ALK-driven neuroblastoma. Nat. Commun. 2023, 14, 2601. [Google Scholar] [CrossRef]
- Mossé, Y.P.; Laudenslager, M.; Longo, L.; Cole, K.A.; Wood, A.; Attiyeh, E.F.; Laquaglia, M.J.; Sennett, R.; Lynch, J.E.; Perri, P.; et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 2008, 455, 930–935. [Google Scholar] [CrossRef]
- Bresler, S.C.; Weiser, D.A.; Huwe, P.J.; Park, J.H.; Krytska, K.; Ryles, H.; Laudenslager, M.; Rappaport, E.F.; Wood, A.C.; McGrady, P.W.; et al. ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma. Cancer Cell 2014, 26, 682–694. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, S.; Cartolano, M.; Hero, B.; Welte, A.; Kahlert, Y.; Roderwieser, A.; Bartenhagen, C.; Walter, E.; Gecht, J.; Kerschke, L.; et al. A mechanistic classification of clinical phenotypes in neuroblastoma. Science 2018, 362, 1165–1170. [Google Scholar] [CrossRef]
- Rahmqvist, I.; Engström, E.; Mellström, E.; Ibrahim, R.R.; Pujol-Calderón, F.; Dahlstrand Rudin, A.; Ordqvist Redfors, A.; Rostamzadeh, N.; Di Rienzo, R.; Franssila, W.; et al. Personalized circulating tumor DNA analysis for sensitive disease monitoring and detection of relapse in neuroblastoma. Biomark. Res. 2024, 12, 148. [Google Scholar] [CrossRef]
- Ek, T.; Ibrahim, R.R.; Vogt, H.; Georgantzi, K.; Träger, C.; Gaarder, J.; Djos, A.; Rahmqvist, I.; Mellström, E.; Pujol-Calderón, F.; et al. Long-Lasting Response to Lorlatinib in Patients with ALK-Driven Relapsed or Refractory Neuroblastoma Monitored with Circulating Tumor DNA Analysis. Cancer Res. Commun. 2024, 4, 2553–2564. [Google Scholar] [CrossRef]
- Chen, L.; Humphreys, A.; Turnbull, L.; Bellini, A.; Schleiermacher, G.; Salwen, H.; Cohn, S.L.; Bown, N.; Tweddle, D.A. Identification of different ALK mutations in a pair of neuroblastoma cell lines established at diagnosis and relapse. Oncotarget 2016, 7, 87301–87311. [Google Scholar] [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443, Erratum in Nature 2021, 590, E53. [Google Scholar]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 76, 7.20. 1–7.20. 41. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef] [PubMed]
- Eijkelenboom, A.; Tops, B.B.J.; van den Berg, A.; van den Brule, A.J.C.; Dinjens, W.N.M.; Dubbink, H.J.; Ter Elst, A.; Geurts-Giele, W.R.R.; Groenen, P.; Groenendijk, F.H.; et al. Recommendations for the clinical interpretation and reporting of copy number gains using gene panel NGS analysis in routine diagnostics. Virchows Arch. 2019, 474, 673–680. [Google Scholar] [CrossRef] [PubMed]
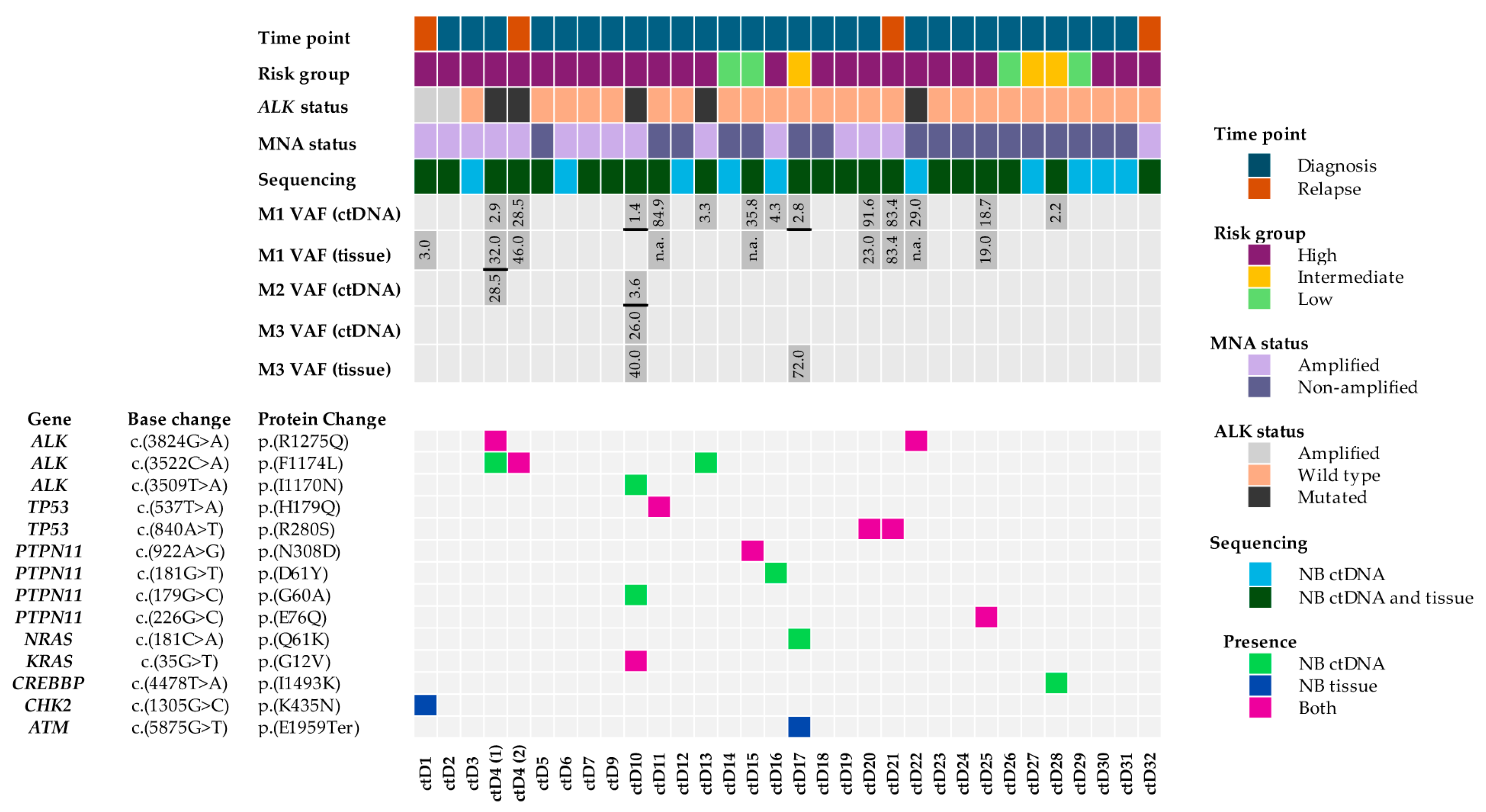


| Sample | VAF | Variant Gene | Chr. Location | Base Change | Protein Change | COSMIC ID | SIFT Score | Polyphen | REVEL |
|---|---|---|---|---|---|---|---|---|---|
| Score | Score | ||||||||
| ctD4 | 2.85 | ALK | chr2 (p23.2) | c.(3824G>A) | p.(R1275Q) | COSV66555567 | Del (0) | PRD (1) | 0.885 |
| ctD4 | 28.5 | ALK | chr2 (p23.2) | c.(3522C>A) | p.(F1174L) | COSV66556325 | Del (0.03) | POD (0.641) | 0.752 |
| ctD10 | 1.38 | ALK | chr2 (p23.2) | c.(3509T>A) | p.(I1170N) | COSV66589132 | Del (0) | PRD (1) | 0.906 |
| ctD10 | 26.16 | KRAS | chr12 (p12.1) | c.(35G>T) | p.(G12V) | COSV55497419 | Del (0) | PRD (0.972) | 0.91 |
| ctD10 | 3.61 | PTPN11 | chr12 (q24.13) | c.(179G>C) | p.(G60A) | COSV61006397 | Del (0.01) | PRD (0.953) | 0.907 |
| a ctD11 | 84.88 | TP53 | chr17 (p13.1) | c.(537T>A) | p.(H179G) | COSV52673406 | Del (0) | PRD (0.98) | 0.79 |
| a gD1 | 1.4 | TP53 | chr17 (p13.1) | c.(537T>A) | p.(H179Q) | COSV52669519 | Del (0) | PRD (0.98) | 0.79 |
| ctD13 | 3.33 | ALK | chr2 (p23.2) | c.(3522C>A) | p.(F1174L) | COSV66556325 | Del (0.03) | POD (0.641) | 0.752 |
| b ctD15 | 35.8 | PTPN11 | chr12 (q24.13) | c.(922A>G) | p.(N308D) | COSV61006575 | Del (0.03) | B (0.134) | 0.838 |
| b gD2 | 43.9 | PTPN11 | chr12 (q24.13) | c.(922A>G) | p.(N308D) | COSV61006575 | Del (0.03) | B (0.134) | 0.838 |
| ctD16 | 4.34 | PTPN11 | chr12 (q24.13) | c.(181G>T) | p.(D61Y) | COSV61004841 | Del (0) | PRD (0.997) | 0.933 |
| ctD17 | 2.26 | NRAS | chr1 (p13.2) | c.(181C>A) | p.(Q61K) | COSV54736310 | Del (0.01) | POD (0.709) | N/A |
| c ctD20 | 91.64 | TP53 | chr17 (p13.1) | c.(840A>T) | p.(R280S) | COSV52782181 | Del (0.03) | POD (0.843) | 0.878 |
| c ctD21 | 83.4 | TP53 | chr17 (p13.1) | c.(840A>T) | p.(R280S) | COSV52801834 | Del (0.03) | POD (0.843) | 0.878 |
| ctD22 | 29.03 | ALK | chr2 (p23.2) | c.(3824G>A) | p.(R1275Q) | COSV66555567 | Del (0) | PRD (1) | 0.885 |
| ctD25 | 18.7 | PTPN11 | chr12 (q24.13) | c.(226G>C) | p.(E76Q) | COSV61004751 | Del (0.01) | PRD (0.979) | 0.733 |
| ctD28 | 2.2 | CREBBP | chr16 (p13.3) | c.(4478T>A) | p.(I1493K) | COSV52129182 | Del (0) | PRD (0.991) | 0.929 |
| 1 | 44.4 | KRAS | chr12 (p12.1) | c.(35G>T) | p.(G12V) | COSV55497419 | Del (0) | PRD (0.972) | 0.91 |
| 1 | 50.9 | SMARCA4 | chr19 (p13.2) | c.(2917C>T) | p.(R973W) | COSV60787034 | Del (0) | PRD (1) | 0.86 |
| 1 | 49.6 | ALK | chr2 (p23.2) | c.(3522C>A) | p.(F1174L) | COSV66555460 | Del (0.03) | POD (0.641) | 0.752 |
| 2 | 99.1 | TP53 | chr17 (p13.1) | c.(404G>T) | p.(C135F) | COSV52680475 | Del (0) | PRD (1) | 0.96 |
| 2 | 100 | NF1 | chr17 (q11.2) | c.(1989_2001del) | p.(G663fs) | N/A | N/A | N/A | N/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Danilenko, M.; Nath, S.; Baines, J.; Gordon, F.; Merugu, S.; Allinson, L.M.; Potts, A.; Collins, B.; Goodman, A.; Kidman, S.E.; et al. Detection of Targetable Genetic Abnormalities in Neuroblastoma Circulating Tumour DNA. Int. J. Mol. Sci. 2025, 26, 9466. https://doi.org/10.3390/ijms26199466
Danilenko M, Nath S, Baines J, Gordon F, Merugu S, Allinson LM, Potts A, Collins B, Goodman A, Kidman SE, et al. Detection of Targetable Genetic Abnormalities in Neuroblastoma Circulating Tumour DNA. International Journal of Molecular Sciences. 2025; 26(19):9466. https://doi.org/10.3390/ijms26199466
Chicago/Turabian StyleDanilenko, Marina, Sharanya Nath, Jack Baines, Freya Gordon, Swathi Merugu, Lisa M. Allinson, Aaron Potts, Bethany Collins, Angharad Goodman, Samuel E. Kidman, and et al. 2025. "Detection of Targetable Genetic Abnormalities in Neuroblastoma Circulating Tumour DNA" International Journal of Molecular Sciences 26, no. 19: 9466. https://doi.org/10.3390/ijms26199466
APA StyleDanilenko, M., Nath, S., Baines, J., Gordon, F., Merugu, S., Allinson, L. M., Potts, A., Collins, B., Goodman, A., Kidman, S. E., McAnulty, C., Jamieson, D., & Tweddle, D. A. (2025). Detection of Targetable Genetic Abnormalities in Neuroblastoma Circulating Tumour DNA. International Journal of Molecular Sciences, 26(19), 9466. https://doi.org/10.3390/ijms26199466







