Meiotic Recombination May Be Initiated by Copy Choice During DNA Synthesis Rather than Break/Join Mechanism
Abstract
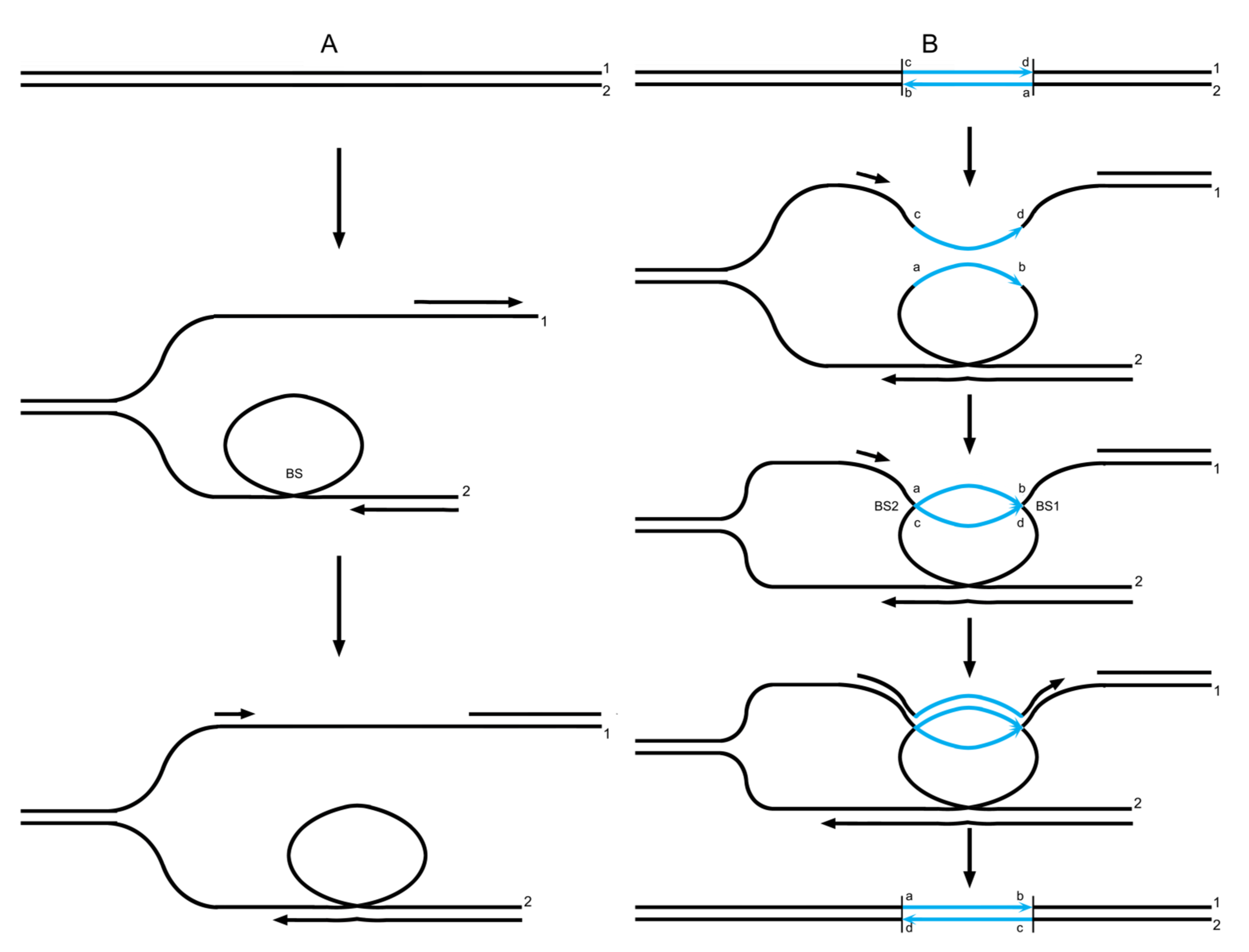
1. Introduction
2. Materials and Methods
2.1. Literature Search
2.2. Selection Criteria
2.3. Data Collection
2.4. Study Quality
2.5. Model Revisions and Upgrades
2.6. Data Reanalysis
3. Results
3.1. Search Results
3.2. Data Collection Summary
3.3. Model Revisions and Upgrades Results
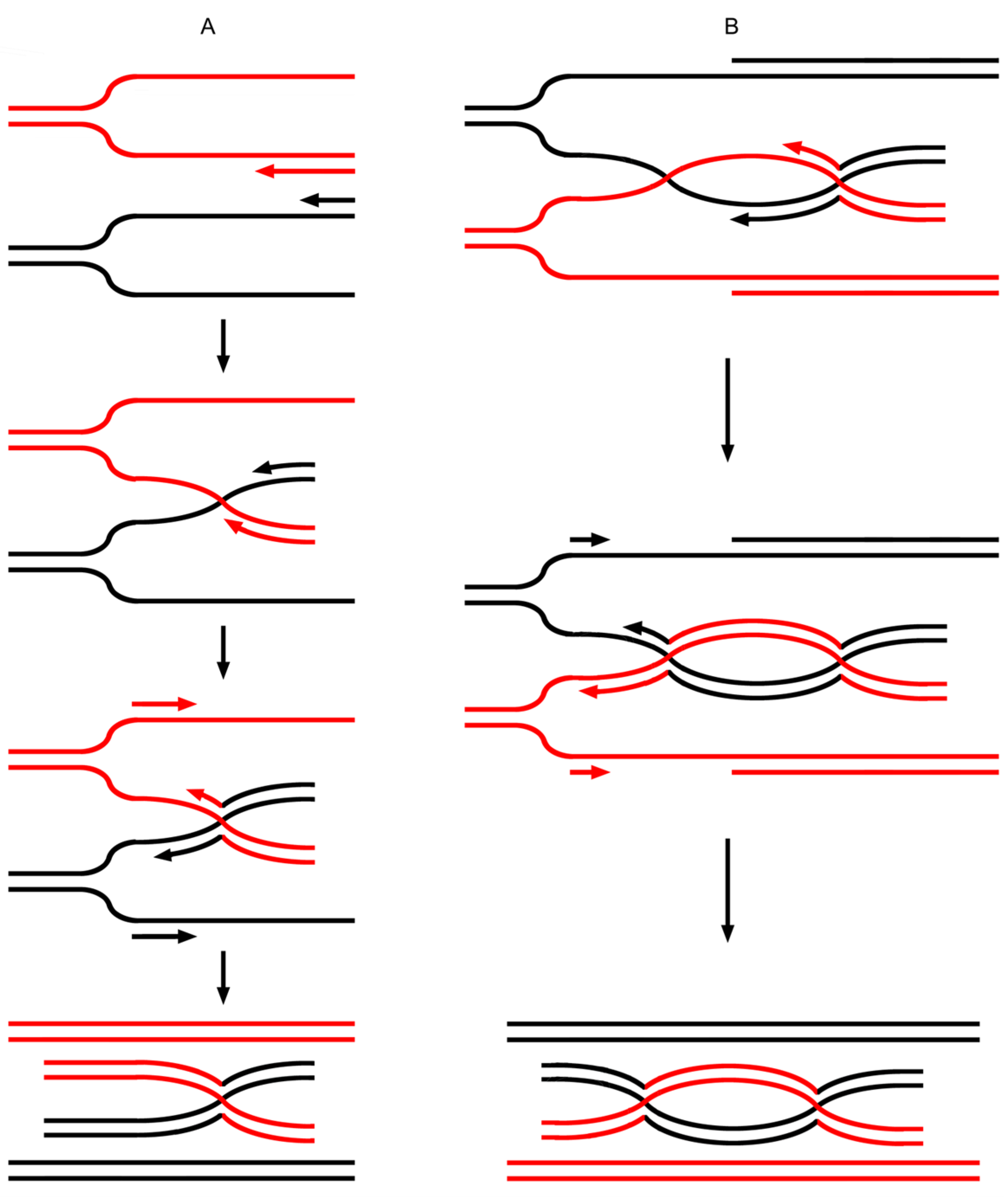
3.4. Data Reanalysis: The Intertwinement Model Embodies the Particular Characteristics of the SDSA Model
3.5. Data Reanalysis: hHDNA Can Also Arise from JM Resolution Rather than Being Followed by a JM
3.6. Data Reanalysis: Strand Specificity of hDNA Mismatch Repair Seems to Be an Illusion and Copy Choice Is More Likely to Be the Actual State
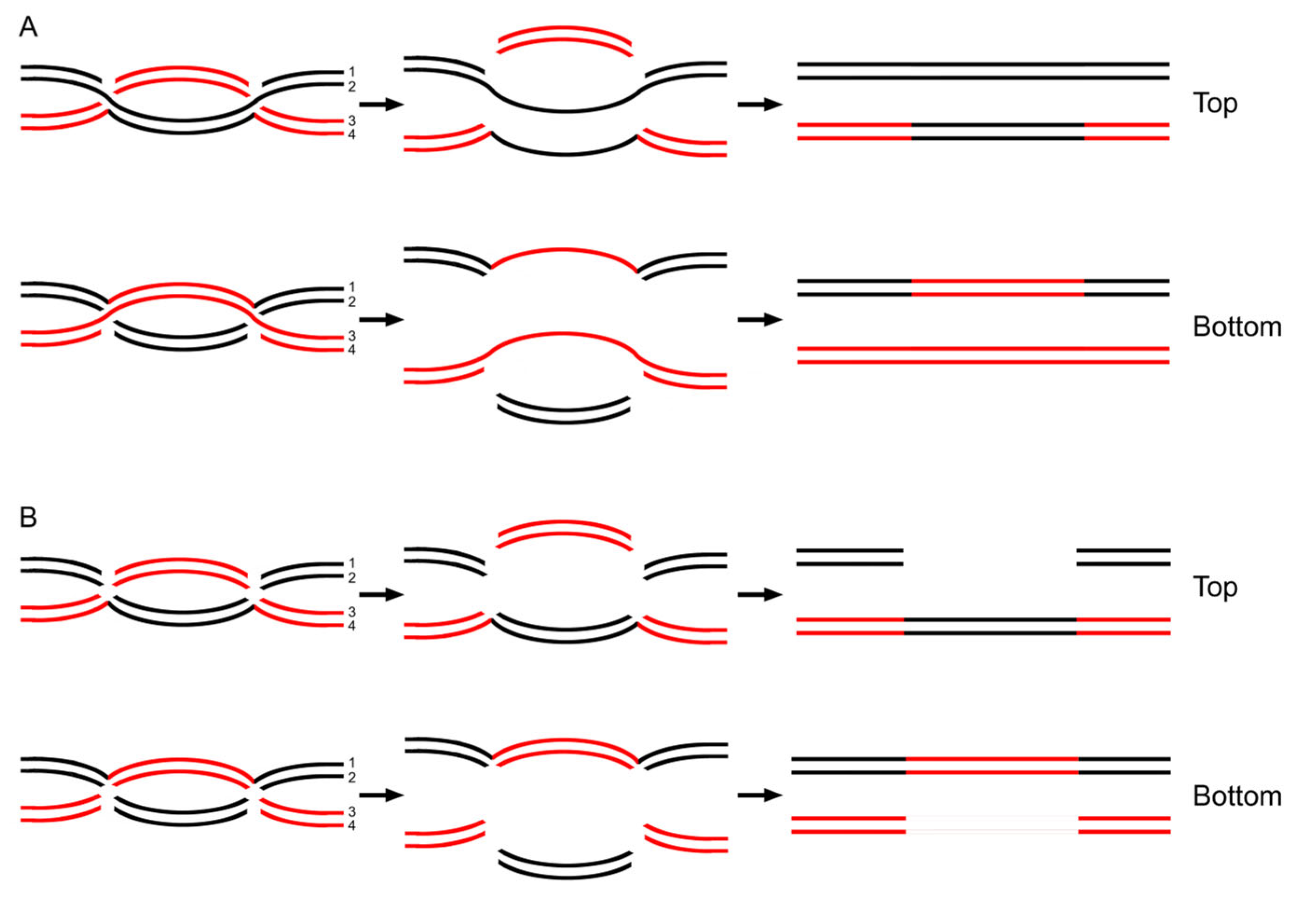
3.7. Data Reanalysis: Parity in Resolution Patterns of a dHJ Would Lead to Parity of Gene Conversion
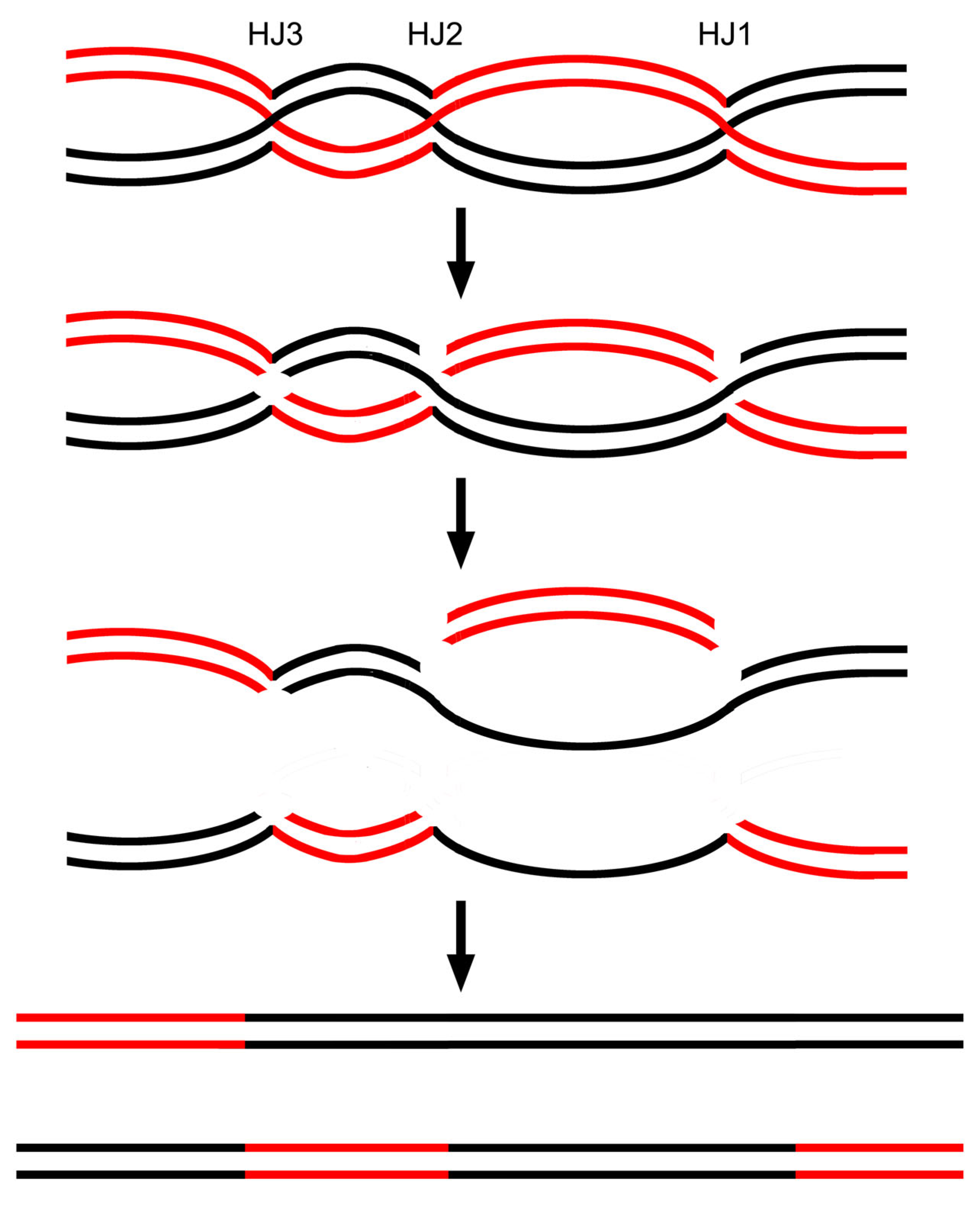
3.8. Data Reanalysis: The Cooperation of Multiple HJs Would Readily Generate High Correlation Between Gene Conversion and Crossover
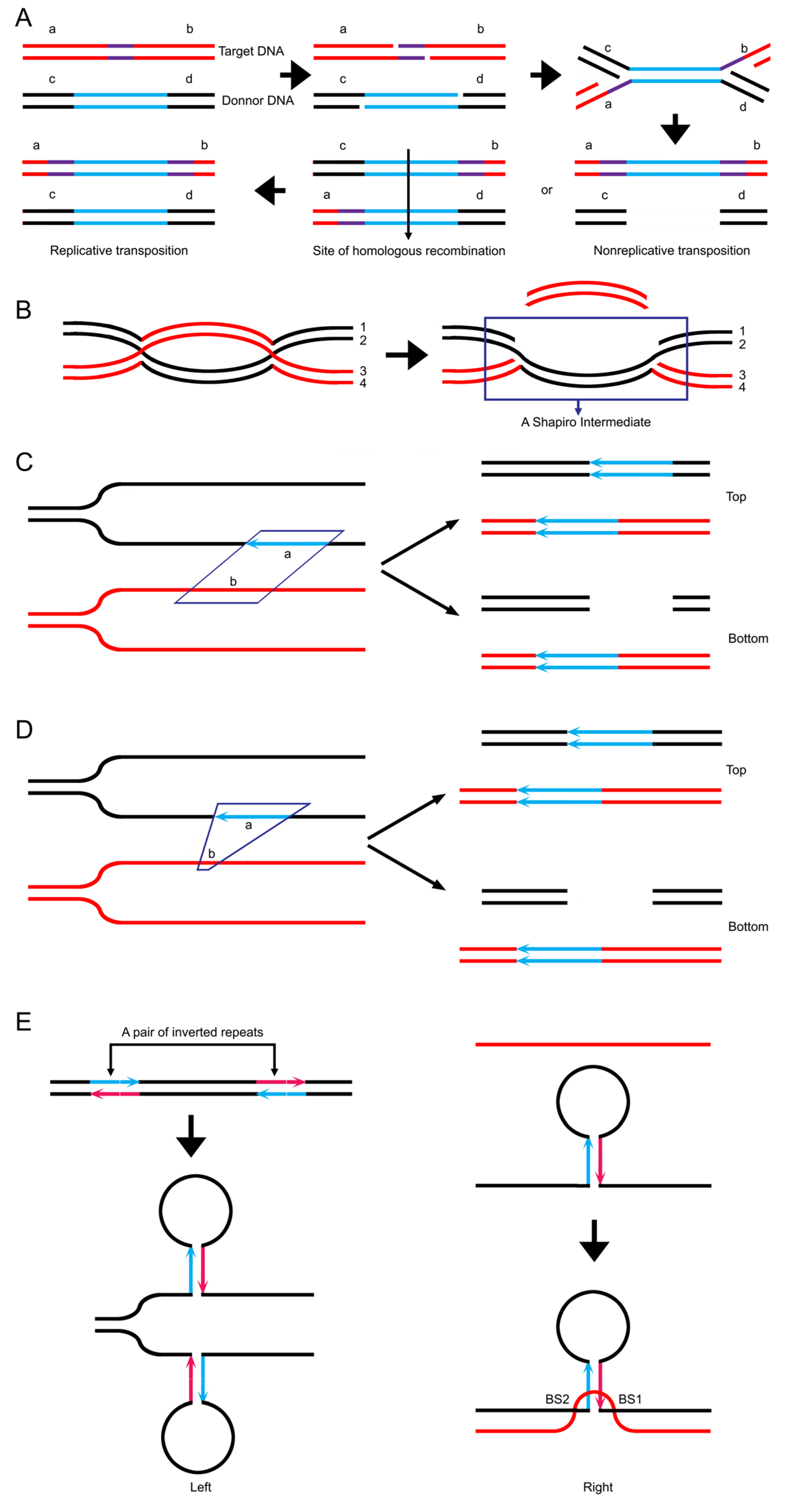
3.9. Data Reanalysis: Transpositional Recombination and Site-Specific Recombination May Have a Common Pathway to Meiotic Recombination
| Reference(s) | Result(s)/Data | Representative Viewpoint(s) |
|---|---|---|
| [1] | hDNA forms early during strand invasion and is followed by a JM. Finally, crossover products are generated via JM resolution. | hDNA > JM > products |
| [12], See summary in [13] | JMs have been detected in advance of hDNA. Experimentally detectable hDNA emerges just prior to, or concomitantly with, the appearance of mature recombination products | JM > hDNA ≈ products |
| [1] | A specific asymmetric configuration of hDNA regions—relative to the site where the initiating DSB occurs—is what the DSBR model predicts (Figure S2A). | |
| [14,15,16] | The bi-directional configuration of heteroduplex segments, which was predicted by the DSBR model, was rarely observed. | |
| [1,6] | hDNA has an equal probability of being corrected in either direction (to wild-type or mutant). | No strand specificity during mismatch repair. |
| See summary in [1] | Experiments by Savage and Hastings with the his1 locus and by Fogel et al. with arg4 locus suggest that when carrying out repair, the invading strand is the only template that mismatch correction is permitted to use. | There is an apparent strand specificity during mismatch repair. |
| See summary in [4] | Excision originates from exchange between sites in a head-to-tail orientation, while inversion arises from recombination between inverted (head-to-head) sites. | The outcome is determined by a break/join mechanism. |
| [31,39] | Recombination of short direct repeats might result from DNA replication errors—specifically, the tip of a growing DNA chain could slip between repeats and be utilized as a primer to continue DNA synthesis. | Strongly support that recombination between short direct repeats operates via a copy-choice mechanism, rather than a break/join mechanism. |
| [29] | Nearly precise excision was significantly stimulated by rolling circle replication, and this process did not involve the transfer of DNA from the parental molecule to the recombinant molecule. | The same as above. |
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Transparency Statement
References
- Szostak, J.W.; Orr-Weaver, T.L.; Rothstein, R.J.; Stahl, F.W. The double-strand-break repair model for recombination. Cell 1983, 33, 25–35. [Google Scholar] [CrossRef]
- Pâques, F.; Haber, J.E. Multiple Pathways of Recombination Induced by Double-Strand Breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999, 63, 349–404. [Google Scholar] [CrossRef]
- Shapiro, J.A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc. Natl. Acad. Sci. USA 1979, 76, 1933–1937. [Google Scholar] [CrossRef]
- Grindley, N.D.; Whiteson, K.L.; Rice, P.A. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006, 75, 567–605. [Google Scholar] [CrossRef]
- Mcmahill, M.S.; Sham, C.W.; Bishop, D.K. Synthesis-dependent strand annealing in meiosis. PLoS Biol. 2007, 5, e299. [Google Scholar] [CrossRef] [PubMed]
- Holliday, R. A mechanism for gene conversion in fungi. Genet. Res. 1964, 5, 282–304. [Google Scholar] [CrossRef]
- Meselson, M.S.; Radding, C.M. A general model for genetic recombination. Proc. Natl. Acad. Sci. USA 1975, 72, 358–361. [Google Scholar] [CrossRef]
- Allers, T.; Lichten, M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 2001, 106, 47–57. [Google Scholar] [CrossRef]
- Potter, H.; Dressler, D. On the mechanism of genetic recombination: Electron microscopic observation of recombination intermediates. Proc. Natl. Acad. Sci. USA 1976, 73, 3000–3004. [Google Scholar] [CrossRef]
- Bell, L.R.; Byers, B. Homologous association of chromosomal DNA during yeast meiosis. Cold Spring Harb. Symp. Quant. Biol. 1983, 47 Pt 2, 829–840. [Google Scholar] [CrossRef] [PubMed]
- Collins, I.; Newlon, C.S. Meiosis-specific formation of joint DNA molecules containing sequences from homologous chromosomes. Cell 1994, 76, 65–75. [Google Scholar] [CrossRef]
- Schwacha, A.; Kleckner, N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell 1994, 76, 51–63. [Google Scholar] [CrossRef]
- Schwacha, A.; Kleckner, N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 1995, 83, 783–791. [Google Scholar] [CrossRef]
- Jessop, L.; Allers, T.; Lichten, M. Infrequent co-conversion of markers flanking a meiotic recombination initiation site in Saccharomyces cerevisiae. Genetics 2005, 169, 1353–1367. [Google Scholar] [CrossRef]
- Merker, J.D.; Dominska, M.; Petes, T.D. Patterns of heteroduplex formation associated with the initiation of meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics 2003, 165, 47–63. [Google Scholar] [CrossRef]
- Hillers, K.J.; Stahl, F.W. The conversion gradient at HIS4 of Saccharomyces cerevisiae. I. Heteroduplex rejection and restoration of Mendelian segregation. Genetics 1999, 153, 555–572. [Google Scholar] [CrossRef]
- Jia, L.; Li, L.; Gui, T.; Liu, S.; Li, H.; Han, J.; Guo, W.; Liu, Y.; Li, J. Analysis of HIV-1 intersubtype recombination breakpoints suggests region with high pairing probability may be a more fundamental factor than sequence similarity affecting HIV-1 recombination. Virol. J. 2016, 13, 156. [Google Scholar] [CrossRef] [PubMed]
- Andraszek, K.; Smalec, E.; Czyzewska, D. The structure of the synaptonemal complexes in meiocytes of European domestic goose (Anser anser). Folia Biol. 2008, 56, 139–147. [Google Scholar] [CrossRef]
- Hochwagen, A. Meiosis. Curr. Biol. 2008, 18, R641–R645. [Google Scholar] [CrossRef] [PubMed]
- Mackey, Z.B.; Ramos, W.; Levin, D.S.; Walter, C.A.; McCarrey, J.R.; Tomkinson, A.E. An alternative splicing event which occurs in mouse pachytene spermatocytes generates a form of DNA ligase III with distinct biochemical properties that may function in meiotic recombination. Mol. Cell Biol. 1997, 17, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.M.; Plug, A.W.; Prolla, T.A.; Bronner, C.E.; Harris, A.C.; Yao, X.; Christie, D.-M.; Monell, C.; Arnheim, N.; Bradley, A.; et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 1996, 13, 336–342. [Google Scholar] [CrossRef]
- Nassif, N.; Penney, J.; Pal, S.; Engels, W.R.; Gloor, G.B. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell Biol. 1994, 14, 1613–1625. [Google Scholar]
- Pâques, F.; Leung, W.Y.; Haber, J.E. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol. Cell Biol. 1998, 18, 2045–2054. [Google Scholar] [CrossRef]
- Gilbertson, L.A.; Stahl, F.W. A test of the double-strand break repair model for meiotic recombination in Saccharomyces cerevisiae. Genetics 1996, 144, 27–41. [Google Scholar] [CrossRef]
- Hurst, D.D.; Fogel, S.; Mortimer, R.K. Conversion-associated recombination in yeast (hybrids-meiosis-tetrads-marker loci-models). Proc. Natl. Acad. Sci. USA 1972, 69, 101–105. [Google Scholar] [CrossRef]
- Fogel, S.; Mortimer, R.; Lusnak, K.; Tavares, F. Meiotic gene conversion: A signal of the basic recombination event in yeast. Cold Spring Harb. Symp. Quant. Biol. 1979, 43 Pt 2, 1325–1341. [Google Scholar] [CrossRef]
- Fink, G.R.; Styles, C.A. Gene conversion of deletions in the his4 region of yeast. Genetics 1974, 77, 231–244. [Google Scholar] [CrossRef]
- Lawrence, C.W.; Sherman, F.; Jackson, M.; Gilmore, R.A. Mapping and gene conversion studies with the structural gene for iso-1-cytochrome C in yeast. Genetics 1975, 81, 615–629. [Google Scholar] [CrossRef] [PubMed]
- D’alençon, E.; Petranovic, M.; Michel, B.; Noirot, P.; Aucouturier, A.; Uzest, M.; Ehrlich, S. Copy-choice illegitimate DNA recombination revisited. EMBO J. 1994, 13, 2725–2734. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P. Anecdotal, historical and critical commentaries on genetics twenty years of illegitimate recombination. Genetics 1987, 115, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Albertini, A.M.; Hofer, M.; Calos, M.P.; Miller, J.H. On the formation of spontaneous deletions: The importance of short sequence homologies in the generation of large deletions. Cell 1982, 29, 319–328. [Google Scholar] [CrossRef]
- Lopez, P.; Espinosa, M.; Greenberg, B.; Lacks, S.A. Generation of deletions in pneumococcal mal genes cloned in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 1984, 81, 5189–5193. [Google Scholar] [CrossRef]
- Conrad, B.; Bashkirov, V.I.; Hofemeister, J. Imprecise excision of plasmid pE194 from the chromosomes of Bacillus subtilis pE194 insertion strains. J. Bacteriol. 1992, 174, 6997–7002. [Google Scholar] [CrossRef]
- Efstratiadis, A.; Posakony, J.W.; Maniatis, T.; Lawn, R.M.; O’Connell, C.; Spritz, R.A.; Deriel, J.K.; Forget, B.G.; Weissman, S.M.; Slightom, J.L.; et al. The structure and evolution of the human beta-globin gene family. Cell 1980, 21, 653–668. [Google Scholar] [CrossRef] [PubMed]
- De Zamaroczy, M.; Faugeron-Fonty, G.; Bernardi, G. Excision sequences in the mitochondrial genome of yeast. Gene 1983, 21, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Nalbantoglu, J.; Hartley, D.; Phear, G.; Tear, G.; Meuth, M. Spontaneous deletion formation at the aprt locus of hamster cells: The presence of short sequence homologies and dyad symmetries at deletion termini. EMBO J. 1986, 5, 1199–1204. [Google Scholar] [CrossRef]
- Wallace, D.C. Diseases of the Mitochondrial DNA. Annu. Rev. Biochem. 1992, 61, 1175–1212. [Google Scholar] [CrossRef] [PubMed]
- Morris, T.; Thacker, J. Formation of large deletions by illegitimate recombination in the HPRT gene of primary human fibroblasts. Proc. Natl. Acad. Sci. USA 1993, 90, 1392–1396. [Google Scholar] [CrossRef]
- Egner, C.; Berg, D.E. Excision of transposon Tn5 is dependent on the inverted repeats but not on the transposase function of Tn5. Proc. Natl. Acad. Sci. USA 1981, 78, 459–463. [Google Scholar] [CrossRef]
- Streisinger, G.; Okada, Y.; Emrich, J.; Newton, J.; Tsugita, A.; Terzaghi, E.; Inouye, M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb. Symp. Quant. Biol. 1966, 31, 77–84. [Google Scholar] [CrossRef]
- Lederberg, J. Recombination mechanisms in bacteria. J. Cell. Physiol. 1955, 45 (Suppl. S2), 75–107. [Google Scholar] [CrossRef]
- Chen, M.; Jia, L.; Zheng, X.; Han, M.; Li, L.; Zhang, L. Ancient human endogenous retroviruses contribute to genetic evolution and regulate cancer cell type-specific gene expression. Cancer Res. 2022, 82, 3457–3473. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Huang, X.; Wang, C.; Wang, S.; Jia, L.; Li, L. Endogenous retroviral solo-LTRs in human genome. Front. Genet. 2024, 15, 1358078. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, M.; Guo, X.; Zhang, B.; Li, H.; Liu, Y.; Han, J.; Jia, L.; Li, L. Endogenous Retrovirus Elements Are Co-Expressed with IFN Stimulation Genes in the JAK–STAT Pathway. Viruses 2023, 15, 60. [Google Scholar] [CrossRef]
- Liu, M.; Jia, L.; Li, H.; Liu, Y.; Han, J.; Wang, X.; Li, T.; Li, J.; Zhang, B.; Zhai, X.; et al. p53 Binding Sites in Long Terminal Repeat 5Hs (LTR5Hs) of Human Endogenous Retrovirus K Family (HML-2 Subgroup) Play Important Roles in the Regulation of LTR5Hs Transcriptional Activity. Microbiol. Spectr. 2022, 10, e00485-22. [Google Scholar] [CrossRef]
- Jia, L.; Liu, M.; Yang, C.; Li, H.; Liu, Y.; Han, J.; Zhai, X.; Wang, X.; Li, T.; Li, J.; et al. Comprehensive identification and characterization of the HERV-K (HML-9) group in the human genome. Retrovirology 2022, 19, 11. [Google Scholar] [CrossRef]
- Beggs, J.D. Transformation of yeast by a replicating hybrid plasmid. Nature 1978, 275, 104–109. [Google Scholar] [CrossRef]
- Stinchcomb, D.T.; Struhl, K.; Davis, R.W. Isolation and characterisation of a yeast chromosomal replicator. Nature 1979, 282, 39–43. [Google Scholar] [CrossRef]
- Thaler, D.S.; Stahl, M.M.; Stahl, F.W. Tests of the double-strand-break repair model for red-mediated recombination of phage lambda and plasmid lambda dv. Genetics 1987, 116, 501–511. [Google Scholar] [CrossRef]
- Moffat, A.S. Triplex DNA finally comes of age. Science 1991, 252, 1374–1375. [Google Scholar] [CrossRef] [PubMed]
- Mirkin, S.M.; Lyamichev, V.I.; Drushlyak, K.N.; Dobrynin, V.N.; Filippov, S.A.; Frank-Kamenetskii, M.D. DNA H form requires a homopurine-homopyrimidine mirror repeat. Nature 1987, 330, 495–497. [Google Scholar] [CrossRef]
- Moser, H.E.; Dervan, P.B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science 1987, 238, 645–650. [Google Scholar] [CrossRef]
- Meselson, M.; Stahl, F.W. The Replication of DNA in Escherichia Coli. Proc. Natl. Acad. Sci. USA 1958, 44, 671–682. [Google Scholar] [CrossRef]
- Paigen, K.; Petkov, P.M. PRDM9 and Its Role in Genetic Recombination. Trends Genet. 2018, 34, 291–300. [Google Scholar] [CrossRef]
- Baudat, F.; Buard, J.; Grey, C.; Fledel-Alon, A.; Ober, C.; Przeworski, M.; Coop, G.; de Massy, B. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 2010, 327, 836–840. [Google Scholar] [CrossRef]
- Ladias, P.; Markopoulos, G.S.; Kostoulas, C.; Bouba, I.; Markoula, S.; Georgiou, I. Cancer Associated PRDM9: Implications for Linking Genomic Instability and Meiotic Recombination. Int. J. Mol. Sci. 2023, 24, 16522. [Google Scholar] [CrossRef] [PubMed]
- Ladias, P.; Markopoulos, G.S.; Kostoulas, C.; Bouba, I.; Georgiou, A.; Markoula, S.; Georgiou, I. Sequence Motif Analysis of PRDM9 and Short Inverted Repeats Suggests Their Contribution to Human Microdeletion and Microduplication Syndromes. BioMedInformatics 2023, 3, 267–279. [Google Scholar] [CrossRef]
- Ladias, P.; Markopoulos, G.; Lazaros, L.; Markoula, S.; Tzavaras, T.; Georgiou, I. Holliday Junctions Are Associated with Transposable Element Sequences in the Human Genome. J. Mol. Biol. 2016, 428, 658–667. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, L.; Yin, N.; Wang, X.; Li, J.; Li, L. Meiotic Recombination May Be Initiated by Copy Choice During DNA Synthesis Rather than Break/Join Mechanism. Int. J. Mol. Sci. 2025, 26, 9464. https://doi.org/10.3390/ijms26199464
Jia L, Yin N, Wang X, Li J, Li L. Meiotic Recombination May Be Initiated by Copy Choice During DNA Synthesis Rather than Break/Join Mechanism. International Journal of Molecular Sciences. 2025; 26(19):9464. https://doi.org/10.3390/ijms26199464
Chicago/Turabian StyleJia, Lei, Na Yin, Xiaolin Wang, Jingyun Li, and Lin Li. 2025. "Meiotic Recombination May Be Initiated by Copy Choice During DNA Synthesis Rather than Break/Join Mechanism" International Journal of Molecular Sciences 26, no. 19: 9464. https://doi.org/10.3390/ijms26199464
APA StyleJia, L., Yin, N., Wang, X., Li, J., & Li, L. (2025). Meiotic Recombination May Be Initiated by Copy Choice During DNA Synthesis Rather than Break/Join Mechanism. International Journal of Molecular Sciences, 26(19), 9464. https://doi.org/10.3390/ijms26199464






