Enteroflow: Automated Pipeline for In Silico Characterization of Enterococcus faecium/faecalis Isolates from Short Reads
Abstract
1. Introduction
2. Results
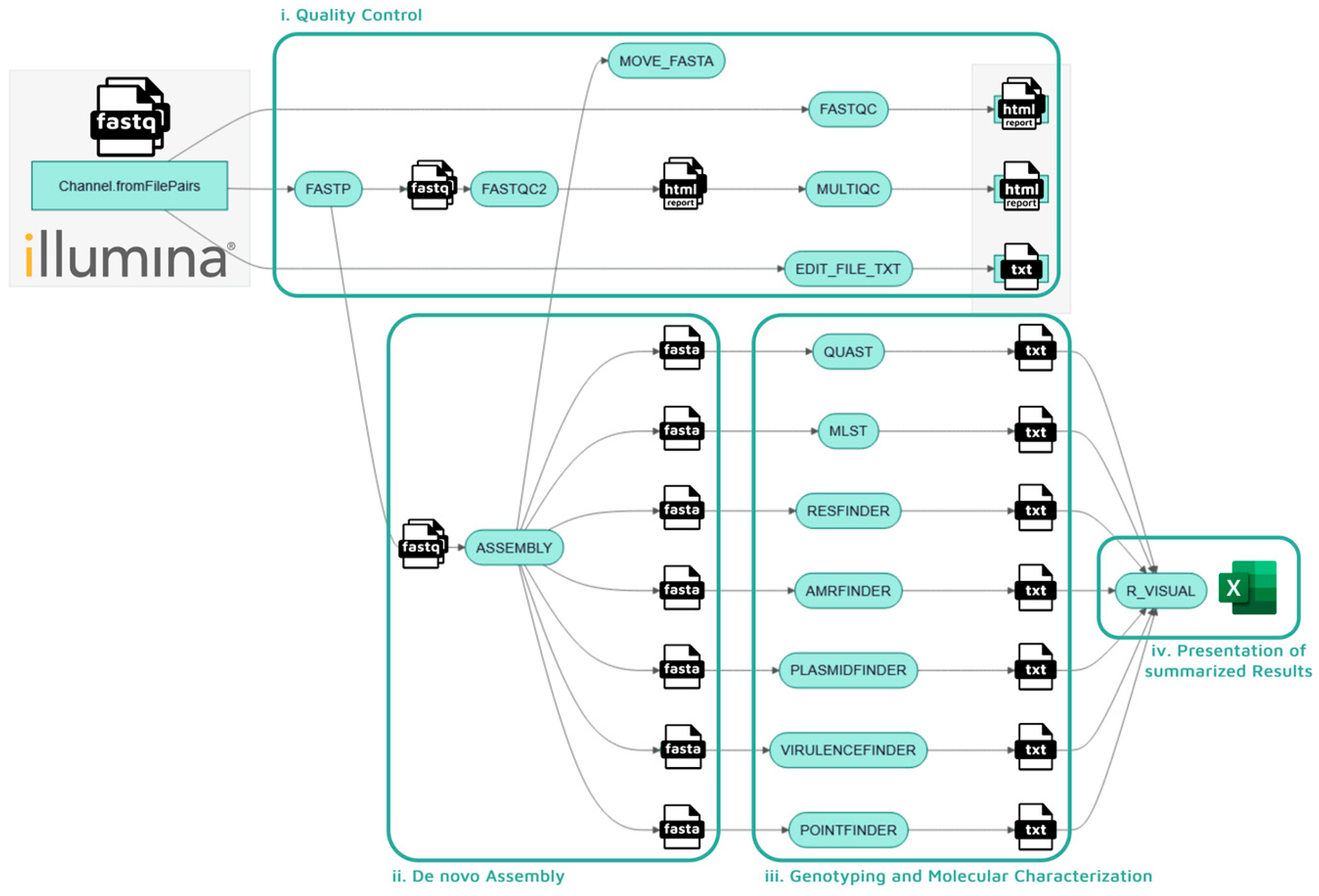
2.1. Pipeline: Enteroflow
2.2. Performance Results
2.2.1. Quality Testing Results
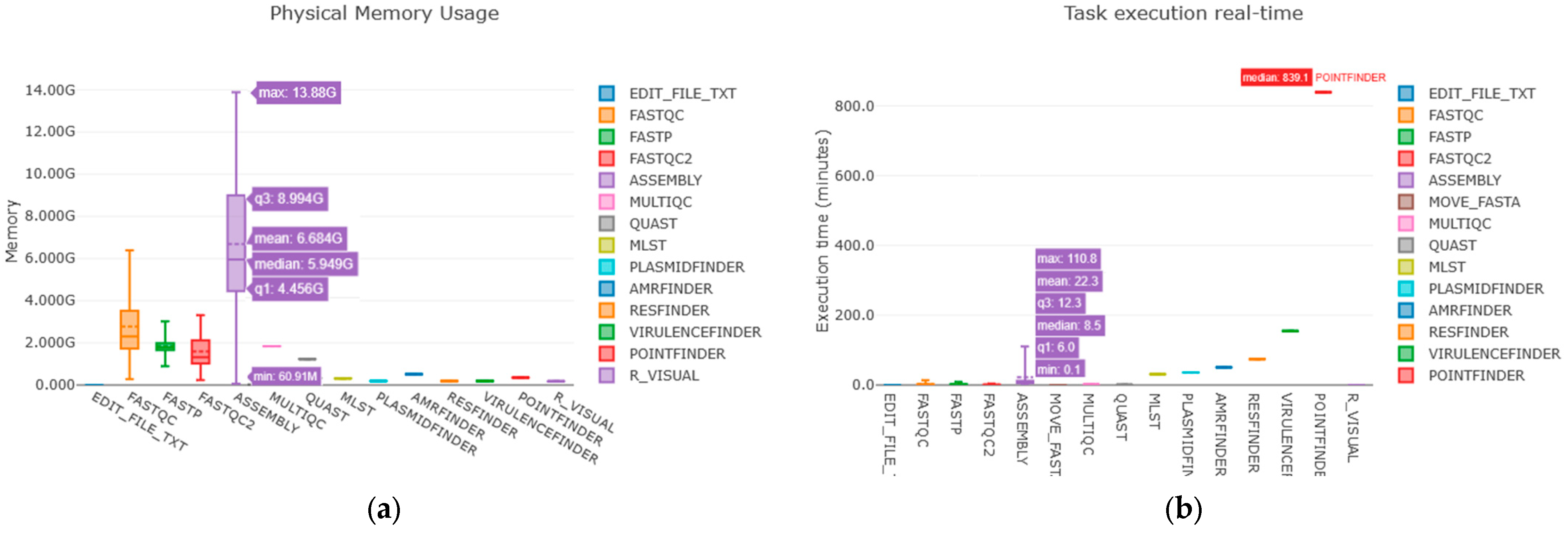
2.2.2. Computational Performance Testing
2.3. Final Output
2.4. Benchmarking Test
3. Discussion
4. Materials and Methods
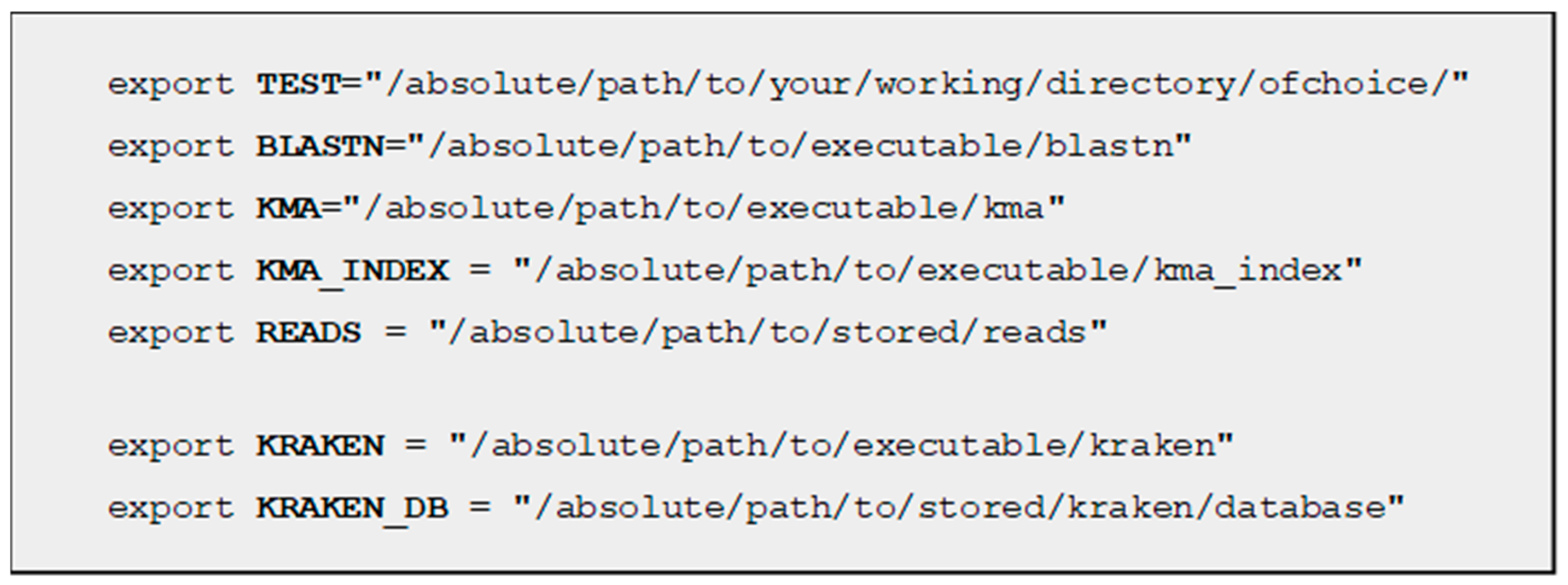
4.1. Installation and Dependencies
4.2. Pipeline Architecture
- i.
- Quality control
- ii.
- De Novo assembly
- iii.
- Genotyping and molecular characterization
- iv.
- Presentation of summarized results.
4.3. Quality Testing
4.4. Benchmarking
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AMR | Antimicrobial Resistance |
| CRN-AR | National Reference Center for Antimicrobial Resistance |
| NRL-AR | National Reference Laboratory for Antimicrobial Resistance |
| IZSLT | Istituto Zooprofilattico Sperimentale del Lazio e della Toscana |
| E. | Enterococcus |
| HTS | High-Throughput Sequencing |
| DSL | Domani Specific Language |
| WHO | World Health Organization |
| EU | European Union |
| HGT | Horizontal Genetic Transfer |
| MGEs | Mobile Genetic Elements |
| VRE | Vancomycin-Resistant Enterococci |
| MLST | Multi-Locus Sequence Typing |
| MDR | Multi-drug Resistant |
| ST | Sequence Type |
References
- 2013/652/EU: Commission Implementing Decision of 12 November 2013 on the Monitoring and Reporting of Antimicrobial Resistance in Zoonotic and Commensal Bacteria (Notified Under Document C(2013) 7145). Official Journal of the European Union, EUR-lex. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32013D0652&from=EN (accessed on 22 September 2025).
- 2020/1729/EU: Commission Implementing Decision (EU) 2020/1729 of 17 November 2020 on the Monitoring and Reporting of Antimicrobial Resistance in Zoonotic and Commensal Bacteria and Repealing Implementing Decision 2013/652/EU. Official Journal of the European Union, EUR-lex. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32020D1729&from=EN (accessed on 22 September 2025).
- Miller, W.R.; Arias, C.A. ESKAPE pathogens: Antimicrobial resistance, epidemiology, clinical impact and therapeutics. Nat. Rev. Microbiol. 2024, 22, 598–616. [Google Scholar] [CrossRef]
- Krawczyk, B.; Wysocka, M.; Kotłowski, R.; Bronk, M.; Michalik, M.; Samet, A. Linezolid-resistant Enterococcus faecium strains isolated from one hospital in Poland -commensals or hospital-adapted pathogens? PLoS ONE 2020, 15, e0233504. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.; Zaheer, R.; Zovoilis, A.; McAllister, T. Enterococci as a One Health indicator of antimicrobial resistance. Can. J. Microbiol. 2024, 70, 303–335. [Google Scholar] [CrossRef] [PubMed]
- Hota, S.; Patil, S.R.; Mane, P.M. Enterococcus: Understanding Their Resistance Mechanisms, Therapeutic Challenges, and Emerging Threats. Cureus 2025, 17, e79628. [Google Scholar] [CrossRef]
- Di Tommaso, P.; Chatzou, M.; Floden, E.W.; Barja, P.P.; Palumbo, E.; Notredame, C. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 2017, 35, 316–319. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, A.; Ripabelli, G.; Sammarco, M.L.; Tamburro, M. Enterococcus faecium as an Emerging Pathogen: Molecular Epidemiology and Antimicrobial Resistance in Clinical Strains. Pathogens 2025, 14, 483. [Google Scholar] [CrossRef]
- Navarra, A.; Cicalini, S.; D’Arezzo, S.; Pica, F.; Selleri, M.; Nisii, C.; Venditti, C.; Cannas, A.; Mazzarelli, A.; Vulcano, A.; et al. Vancomycin-Resistant Enterococci: Screening Efficacy and the Risk of Bloodstream Infections in a Specialized Healthcare Setting. Antibiotics 2025, 14, 304. [Google Scholar] [CrossRef]
- Seemann, T. Nullarbor, Github. Available online: https://github.com/tseemann/nullarbor (accessed on 22 September 2025).
- Petit, R.A., 3rd; Read, T.D. Bactopia: A Flexible Pipeline for Complete Analysis of Bacterial Genomes. mSystems. 2020, 5, e00190-20. [Google Scholar] [CrossRef]
- Schwengers, O.; Hoek, A.; Fritzenwanker, M.; Falgenhauer, L.; Hain, T.; Chakraborty, T.; Goesmann, A. ASA3P: An automatic and scalable pipeline for the assembly, annotation and higher-level analysis of closely related bacterial isolates. PLoS Comput. Biol. 2020, 16, e1007134. [Google Scholar] [CrossRef]
- Sserwadda, I.; Mboowa, G. rMAP: The Rapid Microbial Analysis Pipeline for ESKAPE bacterial group whole-genome sequence data. Microb. Genom. 2021, 7, 000583. [Google Scholar] [CrossRef]
- Gurbich, T.A.; Beracochea, M.; De Silva, N.H.; Finn, R.D. mettannotator: A comprehensive and scalable Nextflow annotation pipeline for prokaryotic assemblies. Bioinformatics 2025, 41, btaf037. [Google Scholar] [CrossRef]
- Conda Contributors. Conda. Available online: https://docs.conda.io/projects/conda/ (accessed on 22 September 2025).
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Clausen, P.T.L.C.; Aarestrup, F.M.; Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 2018, 19, 307. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- Katz, L.S.; Griswold, T.; Morrison, S.S.; Caravas, J.A.; Zhang, S.; den Bakker, H.C.; Deng, X.; Carleton, H.A. Mashtree: A rapid comparison of whole genome sequence files. J. Open Source Softw. 2019, 4, 1762. [Google Scholar] [CrossRef]
- Homan, W.L.; Tribe, D.; Poznanski, S.; Li, M.; Hogg, G.; Spalburg, E.; Van Embden, J.D.; Willems, R.J. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 2002, 40, 1963–1971, Correction in J. Clin. Microbiol. 2002, 40, 3548. [Google Scholar] [CrossRef]
- Ruiz-Garbajosa, P.; Bonten, M.J.; Robinson, D.A.; Top, J.; Nallapareddy, S.R.; Torres, C.; Coque, T.M.; Cantón, R.; Baquero, F.; Murray, B.E.; et al. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol. 2006, 44, 2220–2228. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Galaxy Community. The Galaxy platform for accessible, reproducible, and collaborative data analyses: 2024 update. Nucleic Acids Res. 2024, 52, W83–W94. [Google Scholar] [CrossRef]
- Sørensen, L.H.; Pedersen, S.K.; Jensen, J.D.; Lacy-Roberts, N.; Andrea, A.; Brouwer, M.S.M.; Veldman, K.T.; Lou, Y.; Hoffmann, M.; Hendriksen, R.S. Whole-genome sequencing for antimicrobial surveillance: Species-specific quality thresholds and data evaluation from the network of the European Union Reference Laboratory for Antimicrobial Resistance genomic proficiency tests of 2021 and 2022. mSystems. 2024, 9, e0016024. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 22 September 2025).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. ABRicate. Github. Available online: https://github.com/tseemann/abricate (accessed on 22 September 2025).
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2017, 72, 2764–2768. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef]
- Zhou, S.; Liu, B.; Zheng, D.; Chen, L.; Yang, J. VFDB 2025: An integrated resource for exploring anti-virulence compounds. Nucleic Acids Res. 2025, 53, D871–D877. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Seeman, T. mlst, Github. Available online: https://github.com/tseemann/mlst (accessed on 22 September 2025).
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Lee, R.S.; Gonçalves da Silva, A.; Baines, S.L.; Strachan, J.; Ballard, S.; Carter, G.P.; Kwong, J.C.; Schultz, M.B.; Bulach, D.M.; Seemann, T.; et al. The changing landscape of vancomycin-resistant Enterococcus faecium in Australia: A population-level genomic study. J. Antimicrob. Chemother. 2018, 73, 3268–3278. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
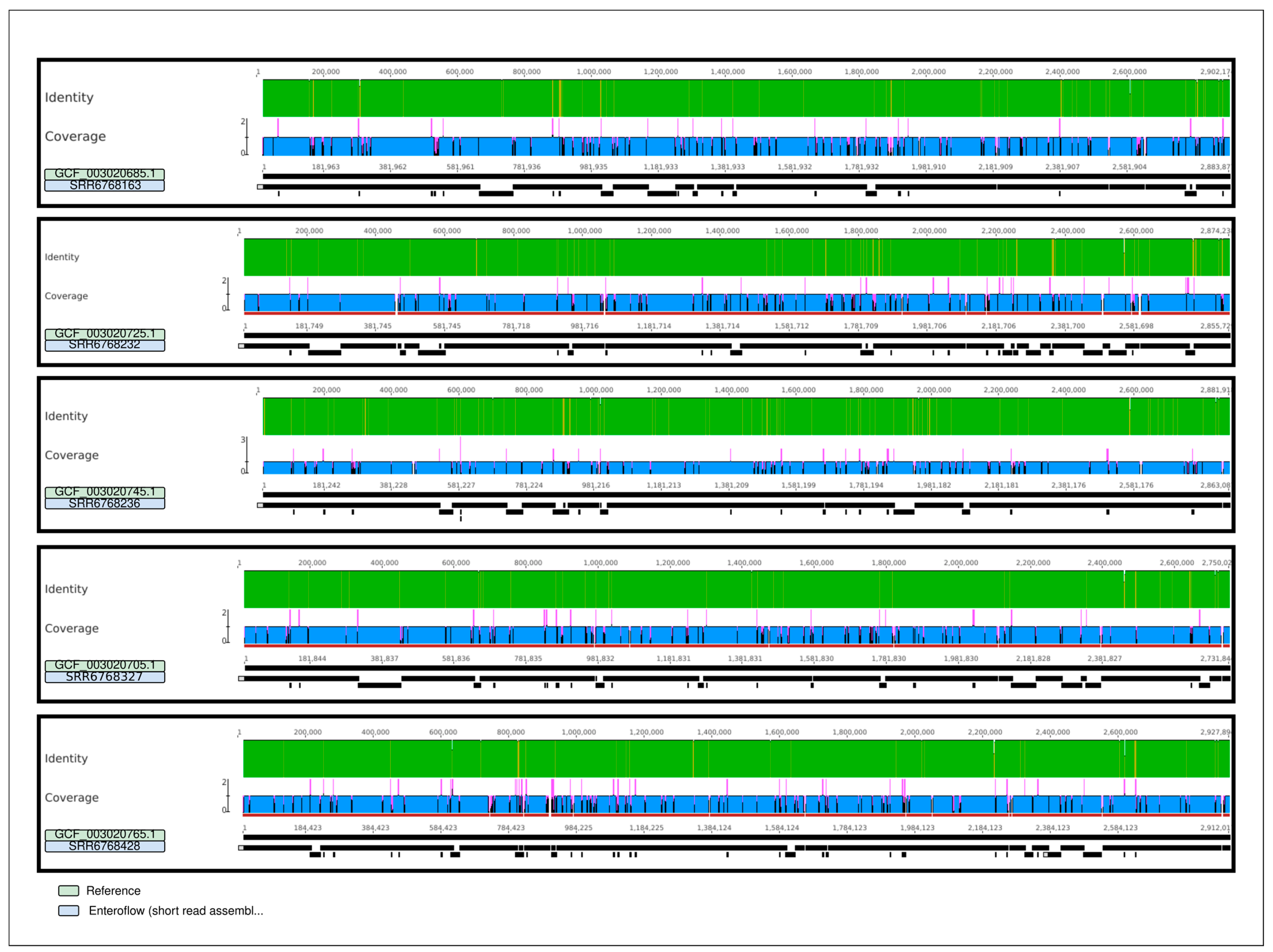

| SRR6768163 | SRR6768232 | SRR6768236 | SRR6768327 | SRR6768428 | ||
|---|---|---|---|---|---|---|
| Assembly | N° contigs (>1000 bp) | 183 | 183 | 157 | 165 | 179 |
| Total length | 2,973,147 bp | 3,019,906 bp | 2,966,970 bp | 2,947,552 bp | 2,911,576 bp | |
| Largest contig | 180,945 bp | 158,984 bp | 124,974 bp | 117,522 bp | 100,544 bp | |
| N50 | 34,377 bp | 38,672 bp | 45,124 bp | 39,545 bp | 36,946 bp | |
| N90 | 6992 bp | 7473 bp | 10,553 bp | 8733 bp | 7339 bp | |
| auN | 49,162.4 | 51,633.3 | 49,986.1 | 47,108.9 | 41,424.1 | |
| L50 | 23 | 22 | 21 | 23 | 25 | |
| L90 | 95 | 90 | 78 | 79 | 92 | |
| N° N’s per 100 kps | 24.39 | 26.34 | 13.74 | 17.53 | 17.06 | |
| Alignment to Reference | N° mapped contigs | 167 | 152 | 140 | 141 | 198 |
| Identical sites | 2,715,830 | 2,710,104 | 2,721,660 | 2,597,602 | 2,724,329 | |
| Identity | 99.9% | 99.9% | 99.9% | 99.9% | 99.8% | |
| Ref. chromosome length | 2,883,877 bp | 2,855,729 bp | 2,863,087 bp | 2,731,844 bp | 2,912,017 bp | |
| Coverage of Ref. | 94.2% (2,717,947) | 95.0% (2,711,659 bp) | 95.1% (2,723,740 bp) | 95.1% (2,599,315 bp) | 93.7% (2,728,020 bp) |
| Execution Time | Max Ram | Output File | |
|---|---|---|---|
| Enteroflow | 52 min 55 s | 9048 GB | One Excel + txt file |
| Nullarbor | 2 h 1 min 57 s | N.A. | Several tsv and txt files |
| Bactopia | 2 h 26 min 36 s (sum of 10 runs) | 8039 GB (mean of 10 runs) | Subdirectories |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smedile, D.; Diaconu, E.L.; Grelloni, M.; Middei, B.; Carfora, V.; Battisti, A.; Alba, P.; Franco, A. Enteroflow: Automated Pipeline for In Silico Characterization of Enterococcus faecium/faecalis Isolates from Short Reads. Int. J. Mol. Sci. 2025, 26, 9441. https://doi.org/10.3390/ijms26199441
Smedile D, Diaconu EL, Grelloni M, Middei B, Carfora V, Battisti A, Alba P, Franco A. Enteroflow: Automated Pipeline for In Silico Characterization of Enterococcus faecium/faecalis Isolates from Short Reads. International Journal of Molecular Sciences. 2025; 26(19):9441. https://doi.org/10.3390/ijms26199441
Chicago/Turabian StyleSmedile, Daniele, Elena L. Diaconu, Matteo Grelloni, Barbara Middei, Virginia Carfora, Antonio Battisti, Patricia Alba, and Alessia Franco. 2025. "Enteroflow: Automated Pipeline for In Silico Characterization of Enterococcus faecium/faecalis Isolates from Short Reads" International Journal of Molecular Sciences 26, no. 19: 9441. https://doi.org/10.3390/ijms26199441
APA StyleSmedile, D., Diaconu, E. L., Grelloni, M., Middei, B., Carfora, V., Battisti, A., Alba, P., & Franco, A. (2025). Enteroflow: Automated Pipeline for In Silico Characterization of Enterococcus faecium/faecalis Isolates from Short Reads. International Journal of Molecular Sciences, 26(19), 9441. https://doi.org/10.3390/ijms26199441








