Gut as a Target of Ochratoxin A: Toxicological Insights and the Role of Microbiota
Abstract
1. Introduction
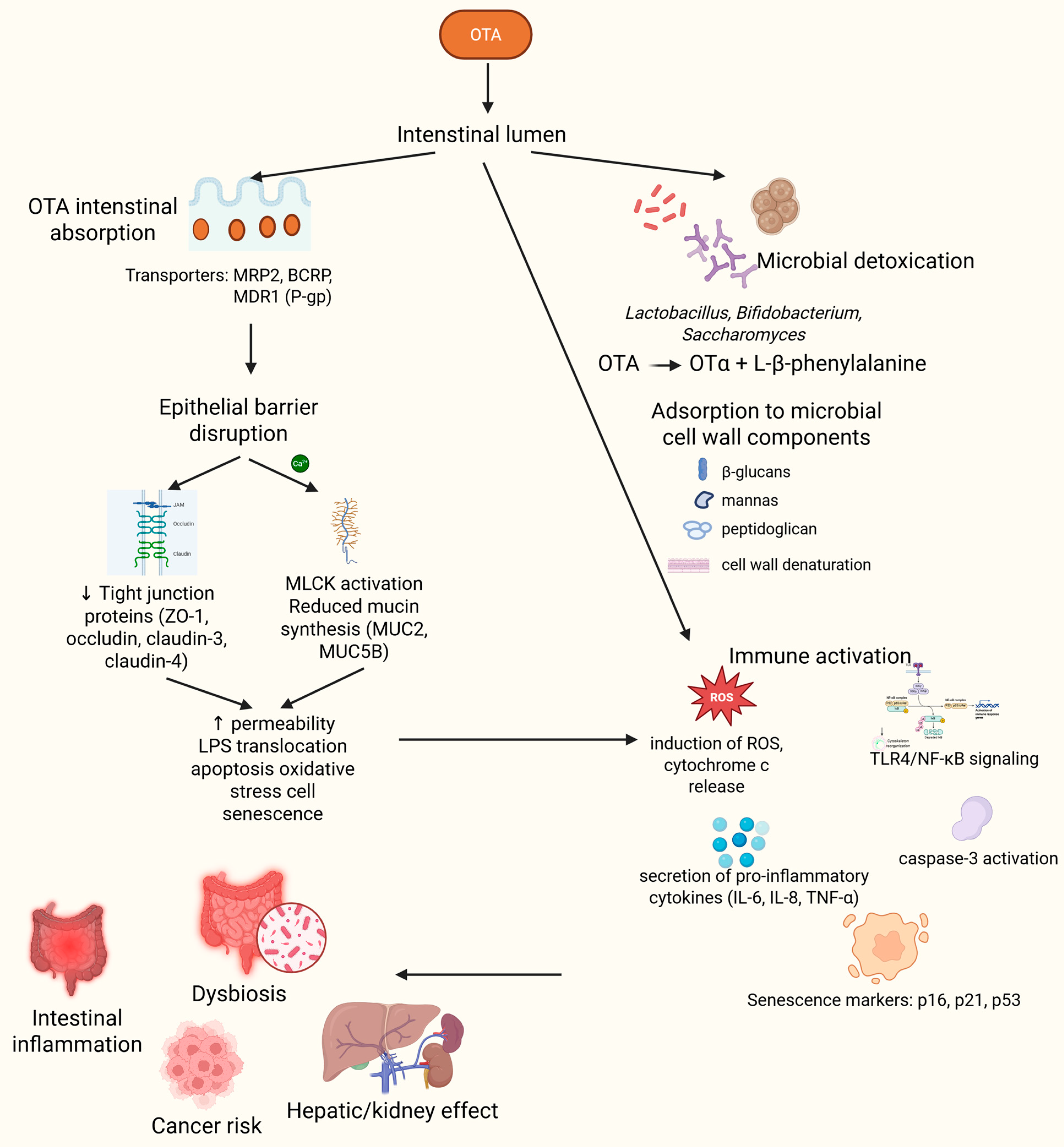
2. Intestines and Ochratoxin A Pharmacokinetics
3. Biodetoxification of OTA
| Microorganism | Reaction Conditions | Medium | OTA Concentration | The Percentage of OTA Degradation | Reference |
|---|---|---|---|---|---|
| Yarrowia lipolytica Y-2 | Rotatory shaker 180 rpm, 28 °C, 20 h | PM Broth | 1 µg/mL | 97.2% | [54] |
| Alcaligens faecalis 0D-1 | 30 °C, 48 h | LB Medium | 1 µg/mL 2 µg/mL 5 µg/mL | Up to 100% 22–64% 23–68% | [55] |
| Lysobacter sp. CW239 | With agitation at 180 rpm, 30 °C, 24 h | LB Broth | 30 µg/L | 86.2% | [43] |
| Bacillus amyloliquefaciens ASAG1 | 31 °C, 10 h | No 4. Medium | 1 µg/mL | 98.5% | [50] |
| Bifidobacterium bifidum CECT 870T | 37 °C, 24 h | MRS Medium | 0.6 µg/mL | pH 3.5/pH 6.5 80.4/74.1 | [56] |
| Bf. breve CECT 4839T | 87.2/94.1 | ||||
| Lactobacillus bulgaricus CECT 4005 | 73.9/96.4 | ||||
| Lb. casei CECT 4040 | 81.4/88.5 | ||||
| Lb. casei CECT 4045 | 87.1/88.5 | ||||
| Lb. johnsonii CECT 289 | 76.4/93.1 | ||||
| Lb. paracasei CECT 4022 | 64.2/89.9 | ||||
| Lb. plantarum CECT 220 | 31.1/64.6 | ||||
| Lb. plantarum CECT 221 | 29.6/64.4 | ||||
| Lb. plantarum CECT 222 | 72.6/64.8 | ||||
| Lb. plantarum CECT 223 | 63.7/66.3 | ||||
| Lb. plantarum CECT 748 | 30.1/58.4 | ||||
| Lb. plantarum CECT 749 | 90.5/97.1 | ||||
| Lb. rhamnosus CECT 278T | 92.1/90.3 | ||||
| Lb. rhamnosus CECT 288 | 86/95 | ||||
| Lb. salivarius CECT 4062 | 71.4/87.7 | ||||
| Leuconostoc mesenteroides CECT 215 | -/52.4 | ||||
| Acinetobacter sp. Neg1 ITEM 17016 | Rotatory shaker 120 rpm, 28 °C, 6 days | MMP Medium | 1 µg/mL | >70% | [51] |
| Brevidobacterium casei (RM101, DSM 20657, DSM 9657, DSM 20658), B. linens DSM 20425, B. iodinum DSM 20626, B. epidermidis DSM 20660 | Rotatory shaker 150 rpm, 30 °C, 10 days | BSM Medium | 40 µg/L | 100 | [52] |
| Pediococcus parvulus UTAD 111B, | 30 °C, 7 days | MRS broth | 1 µg/mL | 72% | [53] |
| UTAD 168, | 89% | ||||
| UTAD 333, | 97% | ||||
| UTAD 334, | 94% | ||||
| UTAD 335, | 98% | ||||
| UTAD 473 | 100% | ||||
| Aspergillus strains M100120 | With agitation at 3.8× g, 30 °C, 6 days | MEA Medium | 10 µg/mL | 99% | [57] |
| M30011 | 81% | ||||
| M10012 | 76% | ||||
| M4001 | 71% | ||||
| X6121 | 44.3% | ||||
| X1011 | 30% | ||||
| Aspergillus oryzae | 30 °C, 72 h | PDA Medium | 10 µg/mL | 94% | [58] |
| Mechanism | Animal | Source/Conditions | OTA Concentration | The Percentage of OTA Degradation | Reference |
|---|---|---|---|---|---|
| OTA ⟶ OTα | Sprague-Dawley male albino rats | Digesta from cecum and large intestine. Incubated at 37 °C, 6 h in a shaking water bath | 20 µg OTA addition into 1 g of digesta sample | Cecum: 52.5% Large intestine: 54.5% | [60] |
| OTA ⟶ OTα | Sheep | Rumen fluid | 2 ppm and 5 ppm. OTA was added to the diet for 4 days. | 99–100% | [61] |
| OTA ⟶ OTα | Brown Swiss Cow | Rumen fluid. 39 °C, 8 h in a shaking water bath, in the dark with constant CO2 administration. | Pure OTA was added at zero time (equivalent to 200 µg/L rumen fluid) | ~100% | [62] |
| OTA ⟶ OTα | Suffolk sheep | Urine | i.v.: 0.2 mg/kg of bw i.r.: 0.5 mg/kg of bw | i.v.: 2–4% i.r.: 90–99% | [64] |
| OTA ⟶ OTα after OAH supplementation | Weaning piglets | Plasma, DBS, kidney, liver, muscle, GIT (digesta content of stomach, jejunum, cecum, and colon) | OTA: 50 or 500 µg/kg of bw OAH: 50 or 500 µg/kg of bw | Plasma: 54–59% DBS: 50–53% Kidney: 52% (OAH500) Liver: 67% (OAH500) Muscle: 59% (OAH500) GIT (OAH500): Stomach: 67% Jejunum: 68% Cecum: 86% Colon: 93% | [59] |
4. Understanding the Interplay Between Ochratoxin A, the Gut, and the Microbiome
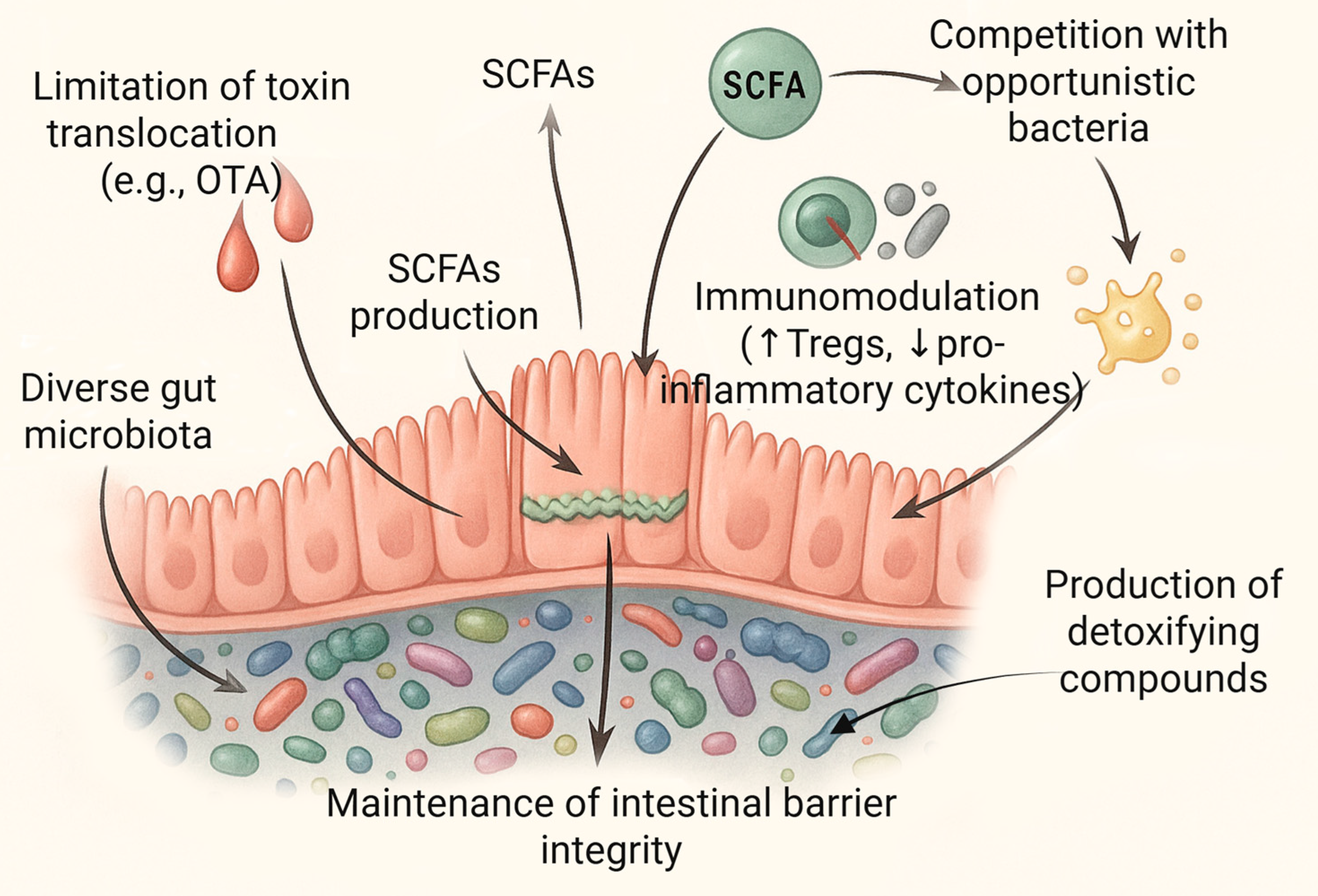
5. The Microbiome as a Determinant of Resistance to Toxins
5.1. Dysbiosis and Its Implications for Susceptibility to OTA
5.2. Individual Microbiome Profiles and Variability in Toxin Susceptibility
6. Conclusions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABC | ATP-binding cassette |
| ABCB1 | ATP binding cassette subfamily B member 1 |
| ABCC2 | ATP binding cassette subfamily C member 2 |
| AC | acid-treated cells |
| AFB1 | aflatoxin B1 |
| AHR | aryl hydrocarbon receptor |
| AMPK | AMP-activated protein kinase |
| Anxa3 | Annexin A3 |
| ATP | adenosine triphosphate |
| BCRP | breast cancer resistance protein |
| BEN | Balkan Endemic Nephropathy |
| BSM | basal salts medium |
| CAR | nuclear receptor subfamily 1 group I member 3 |
| CASP3 | caspase 3 |
| CD | Cluster of Differentiation |
| Cdkn1a | Cyclin-dependent kinase inhibitor 1A |
| COX-2 | Cyclooxygenase-2 |
| CP | carboxypeptidase |
| CRC | colorectal cancer |
| CYP450 | cytochrome P450 |
| cyt-c | cytochrome c |
| DOX | deoxynivalenol |
| DUSP9 | Dual-Specificity Phosphatase 9 |
| EFSA | European Food Safety Authority |
| FBs | fumonisins |
| FMT | Fecal microbiota transplantation |
| Gen1 | GEN1 Holliday Junction 5′ Flap Endonuclease |
| GIT | gastrointestinal tract |
| HAT | histone acetyltransferase |
| HC | heat-treated cells |
| HCC | hepatocellular carcinoma |
| HCT116 | human colon cancer cell line 116 |
| HDACs | histone deacetylases |
| IL | interleukin |
| IPEC-J2 | intestinal porcine epithelial cells-jejunum 2 |
| LAB | lactic acid bacteria |
| LB | Lauria-Bertani |
| LOX-5 | arachidonate 5-lipoxygenase |
| LPS | lipopolysaccharide |
| MAPK | mitogen-activated protein kinase |
| MDM2 | murine double minute 2 |
| MDR1 | multidrug resistance protein 1 |
| MEA | malt extract agar |
| MLCK | myosin light chain kinase |
| MMP | minimal medium peptone |
| mPTP | mitochondrial permeability transition pore |
| MRP2 | multidrug resistance-associated protein 2 |
| MRS | De Man, Rogosa and Sharpe |
| mTOR | mechanistic target of rapamycin |
| MUC2 | mucin 2 |
| MUC5B | mucin 5B |
| NAD+ | oxidized nicotinamide adenine dinucleotide |
| NAT2 | N-acetyltransferase 2 |
| NF-κB | nuclear factor kappa-light-chain enhancer of activated B cells |
| Nrf2 | nuclear factor erythroid 2-related factor 2 |
| OAH | ochratoxin amidohydrolase |
| Osm | Oncostatin M |
| OTA | ochratoxin A |
| OTα | ochratoxin α |
| PAK6 | Serine/threonine-protein kinase PAK 6 |
| PC | phosphatidylcholine |
| PCNA | proliferating cell nuclear antigen |
| PDA | potato dextrose agar |
| PE | phosphatidylethanolamine |
| P-gp | glycoprotein P |
| PLA2G2D | Phospholipase A2 group IID |
| PM | polytoma medium |
| PPAR-γ | peroxisome proliferator-activated receptor gamma |
| PXR | nuclear receptor subfamily 1 group I member 2 |
| Ras | rat sarcoma |
| ROS | reactive oxygen species |
| SA-β-gal | senescence-associated β-galactosidase |
| SCFAs | short-chain fatty acids |
| SGM | synthetic grape juice medium |
| TAMs | tumor-associated macrophages |
| TCR | T cell receptor |
| TEER | transepithelial electrical resistance |
| TET | Ten-Eleven Translocation dioxygenases |
| TJ | tight junction |
| TLR4 | toll-like receptor 4 |
| TNF-α | tumor necrosis factor α |
| Tregs | regulatory T cells |
| VC | viable cells |
| YMB | yeast mold broth |
| YPG | yeast peptone glucose |
| ZO-1 | zonula occludens-1 |
References
- Ringot, D.; Chango, A.; Schneider, Y.J.; Larondelle, Y. Toxicokinetics and toxicodynamics of ochratoxin A, an update. Chem. Biol. Interact. 2006, 159, 18–46. [Google Scholar] [CrossRef]
- Więckowska, M.; Szelenberger, R.; Niemcewicz, M.; Harmata, P.; Poplawski, T.; Bijak, M. Ochratoxin A—The Current Knowledge Concerning Hepatotoxicity, Mode of Action and Possible Prevention. Molecules 2023, 28, 6617. [Google Scholar] [CrossRef]
- Klarić, M.Š.; Rašić, D.; Peraica, M. Deleterious Effects of Mycotoxin Combinations Involving Ochratoxin A. Toxins 2013, 5, 1965–1987. [Google Scholar] [CrossRef]
- Ostry, V.; Malir, F.; Toman, J.; Grosse, Y. Mycotoxins as human carcinogens-the IARC Monographs classification. Mycotoxin Res. 2017, 33, 65–73. [Google Scholar] [CrossRef]
- Iqbal, S.Z.; Mumtaz, A.; Mahmood, Z.; Waqas, M.; Ghaffar, A.; Ismail, A.; Pervaiz, W. Assessment of aflatoxins and ochratoxin a in chili sauce samples and estimation of dietary intake. Food Control 2021, 121, 107621. [Google Scholar] [CrossRef]
- Iqbal, S.Z.; Mehmood, Z.; Asi, M.R.; Shahid, M.; Sehar, M.; Malik, N. Co-occurrence of aflatoxins and ochratoxin A in nuts, dry fruits, and nuty products. J. Food Saf. 2018, 38, e12462. [Google Scholar] [CrossRef]
- Iqbal, S.Z.; Asi, M.R.; Hanif, U.; Zuber, M.; Jinap, S. The presence of aflatoxins and ochratoxin A in rice and rice products; and evaluation of dietary intake. Food Chem. 2016, 210, 135–140. [Google Scholar] [CrossRef] [PubMed]
- El Darra, N.; Gambacorta, L.; Solfrizzo, M. Multimycotoxins occurrence in spices and herbs commercialized in Lebanon. Food Control 2019, 95, 63–70. [Google Scholar] [CrossRef]
- Hassan, H.F.; Koaik, L.; Khoury, A.E.; Atoui, A.; El Obeid, T.; Karam, L. Dietary Exposure and Risk Assessment of Mycotoxins in Thyme and Thyme-Based Products Marketed in Lebanon. Toxins 2022, 14, 331. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.; Fungaro, M.H.P.; Silva, J.J.; Martins, L.M.; Taniwaki, M.H.; Iamanaka, B.T. Ochratoxin A and related fungi in Brazilian black pepper (Piper nigrum L.). Food Res. Int. 2021, 142, 110207. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.J.G.; Teixeira, A.C.; Pereira, A.; Pena, A.; Lino, C.M. Ochratoxin A in Beers Marketed in Portugal: Occurrence and Human Risk Assessment. Toxins 2020, 12, 249. [Google Scholar] [CrossRef]
- Remiro, R.; Irigoyen, A.; González-Peñas, E.; Lizarraga, E.; López de Cerain, A. Levels of ochratoxins in Mediterranean red wines. Food Control 2013, 32, 63–68. [Google Scholar] [CrossRef]
- Iqbal, S.Z.; Nisar, S.; Asi, M.R.; Jinap, S. Natural incidence of aflatoxins, ochratoxin A and zearalenone in chicken meat and eggs. Food Control 2014, 43, 98–103. [Google Scholar] [CrossRef]
- Breitholtz-Emanuelsson, A.; Olsen, M.; Oskarsson, A.; Palminger, I.; Hult, K. Ochratoxin A in Cow’s Milk and in Human Milk with Corresponding Human Blood Samples. J. AOAC Int. 1993, 76, 842–846. [Google Scholar] [CrossRef] [PubMed]
- Altafini, A.; Roncada, P.; Guerrini, A.; Minkoumba Sonfack, G.; Fedrizzi, G.; Caprai, E. Occurrence of Ochratoxin A in Different Types of Cheese Offered for Sale in Italy. Toxins 2021, 13, 540. [Google Scholar] [CrossRef] [PubMed]
- Altafini, A.; Fedrizzi, G.; Roncada, P. Occurrence of ochratoxin A in typical salami produced in different regions of Italy. Mycotoxin Res. 2019, 35, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Armorini, S.; Altafini, A.; Zaghini, A.; Roncada, P. Ochratoxin A in artisan salami produced in Veneto (Italy). Food Addit. Contam. Part B Surveill. 2016, 9, 9–14. [Google Scholar] [CrossRef]
- Vlachou, M.; Pexara, A.; Solomakos, N.; Govaris, A. Ochratoxin A in Slaughtered Pigs and Pork Products. Toxins 2022, 14, 67. [Google Scholar] [CrossRef]
- Battacone, G.; Nudda, A.; Pulina, G. Effects of ochratoxin a on livestock production. Toxins 2010, 2, 1796–1824. [Google Scholar] [CrossRef]
- EFSA Panel on Contaminants in the Food Chain (CONTAM); Schrenk, D.; Bignami, M.; Bodin, L.; Chipman, J.K.; del Mazo, J.; Grasl-Kraupp, B.; Hogstrand, C.; Hoogenboom, L.; Leblanc, J.-C.; et al. Risks for animal health related to the presence of ochratoxin A (OTA) in feed. EFSA J. 2023, 21, e08375. [Google Scholar] [CrossRef]
- Gümüş, R.; Ercan, N.; Imik, H. Determination of Ochratoxin A Levels in Mixed Feed and Feed Stuffs Used in Some Laying Hens and Ruminant Enterprises of Sivas City. Rev. Bras. Ciência Avícola 2018, 20, 85–90. [Google Scholar] [CrossRef]
- Pietruszka, K.; Piątkowska, M.; Jedziniak, P. Occurrence of Ochratoxin a in Animal Tissues and Feeds in Poland in 2014–2016. J. Vet. Res. 2017, 61, 483–487. [Google Scholar] [CrossRef]
- Ganesan, A.R.; Balasubramanian, B.; Park, S.; Jha, R.; Andretta, I.; Bakare, A.G.; Kim, I.H. Ochratoxin A: Carryover from animal feed into livestock and the mitigation strategies. Anim. Nutr. 2021, 7, 56–63. [Google Scholar] [CrossRef]
- Tolosa, J.; Rodríguez-Carrasco, Y.; Ruiz, M.J.; Vila-Donat, P. Multi-mycotoxin occurrence in feed, metabolism and carry-over to animal-derived food products: A review. Food Chem. Toxicol. 2021, 158, 112661. [Google Scholar] [CrossRef]
- Damiano, S.; Longobardi, C.; De Marchi, L.; Piscopo, N.; Meucci, V.; Lenzi, A.; Ciarcia, R. Detection of Ochratoxin A in Tissues of Wild Boars (Sus scrofa) from Southern Italy. Toxins 2025, 17, 74. [Google Scholar] [CrossRef]
- Zhai, S.; Zhu, Y.; Feng, P.; Li, M.; Wang, W.; Yang, L.; Yang, Y. Ochratoxin A: Its impact on poultry gut health and microbiota, an overview. Poult. Sci. 2021, 100, 101037. [Google Scholar] [CrossRef]
- Liu, X.; Yan, C.; Chang, C.; Meng, F.; Shen, W.; Wang, S.; Zhang, Y. Ochratoxin A promotes chronic enteritis and early colorectal cancer progression by targeting Rinck signaling. Phytomedicine 2024, 122, 155095. [Google Scholar] [CrossRef]
- Wang, H.; Chen, Y.; Zhai, N.; Chen, X.; Gan, F.; Li, H.; Huang, K. Ochratoxin A-Induced Apoptosis of IPEC-J2 Cells through ROS-Mediated Mitochondrial Permeability Transition Pore Opening Pathway. J. Agric. Food Chem. 2017, 65, 10630–10637. [Google Scholar] [CrossRef]
- Aggarwal, N.; Kitano, S.; Puah, G.R.Y.; Kittelmann, S.; Hwang, I.Y.; Chang, M.W. Microbiome and Human Health: Current Understanding, Engineering, and Enabling Technologies. Chem. Rev. 2023, 123, 31–72. [Google Scholar] [CrossRef] [PubMed]
- Ursell, L.K.; Metcalf, J.L.; Parfrey, L.W.; Knight, R. Defining the human microbiome. Nutr. Rev. 2012, 70 (Suppl. S1), S38–S44. [Google Scholar] [CrossRef] [PubMed]
- Assefa, S.; Köhler, G. Intestinal Microbiome and Metal Toxicity. Curr. Opin. Toxicol. 2020, 19, 21–27. [Google Scholar] [CrossRef]
- El-Sayed, A.; Aleya, L.; Kamel, M. Microbiota’s role in health and diseases. Environ. Sci. Pollut. Res. Int. 2021, 28, 36967–36983. [Google Scholar] [CrossRef]
- Vila-Donat, P.; Sánchez, D.; Manyes, L.; Cimbalo, A. Impact of Bioactive Ingredients on the Fecal Excretion of Aflatoxin B1 and Ochratoxin A in Wistar Rats. Molecules 2025, 30, 647. [Google Scholar] [CrossRef]
- Zhu, Q.; Qu, H.; Kang, R.; Zheng, Y.; Guo, Q.; Huang, S.; Zhao, L.; Ma, Q. The Toxicokinetics, Excretion Patterns, and Milk Transmission of Ochratoxin A in Lactating Sows. Toxins 2024, 16, 128. [Google Scholar] [CrossRef]
- Skaug, M.A.; Helland, I.; Solvoll, K.; Saugstad, O.D. Presence of ochratoxin A in human milk in relation to dietary intake. Food Addit. Contam. 2001, 18, 321–327. [Google Scholar] [CrossRef]
- Tuntiteerawit, P.; Jarukamjorn, K.; Porasuphatana, S. The effect of green tea catechins on breast cancer resistance protein activity and intestinal efflux of aflatoxin B1 via breast cancer resistance protein in Caco-2 cells. Toxicol. Res. 2020, 36, 293–300. [Google Scholar] [CrossRef]
- Antonissen, G.; Devreese, M.; De Baere, S.; Martel, A.; Van Immerseel, F.; Croubels, S. Impact of Fusarium mycotoxins on hepatic and intestinal mRNA expression of cytochrome P450 enzymes and drug transporters, and on the pharmacokinetics of oral enrofloxacin in broiler chickens. Food Chem. Toxicol. 2017, 101, 75–83. [Google Scholar] [CrossRef]
- Schrickx, J.; Lektarau, Y.; Fink-Gremmels, J. Ochratoxin A secretion by ATP-dependent membrane transporters in Caco-2 cells. Arch. Toxicol. 2006, 80, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M.; Mahbub, N.U.; Islam, M.A. Gut Microorganism-Mediated Neutralization of Mycotoxins: A Promising Approach to Combat Fungal Toxicity. Adv. Gut Microbiome Res. 2024, 2024, 8448547. [Google Scholar] [CrossRef]
- Shekhar, R.; Raghavendra, V.B.; Rachitha, P. A comprehensive review of mycotoxins, their toxicity, and innovative detoxification methods. Toxicol. Rep. 2025, 14, 101952. [Google Scholar] [CrossRef] [PubMed]
- Shanakhat, H.; Sorrentino, A.; Raiola, A.; Romano, A.; Masi, P.; Cavella, S. Current methods for mycotoxins analysis and innovative strategies for their reduction in cereals: An overview. J. Sci. Food Agric. 2018, 98, 4003–4013. [Google Scholar] [CrossRef]
- Nahle, S.; El Khoury, A.; Savvaidis, I.; Chokr, A.; Louka, N.; Atoui, A. Detoxification approaches of mycotoxins: By microorganisms, biofilms and enzymes. Int. J. Food Contam. 2022, 9, 3. [Google Scholar] [CrossRef]
- Wei, W.; Qian, Y.; Wu, Y.; Chen, Y.; Peng, C.; Luo, M.; Xu, J.; Zhou, Y. Detoxification of ochratoxin A by Lysobacter sp. CW239 and characteristics of a novel degrading gene carboxypeptidase cp4. Environ. Pollut. 2020, 258, 113677. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Q.; Wang, S.; Cai, R.; Yuan, Y.; Yue, T.; Wang, Z. Bio-control on the contamination of Ochratoxin A in food: Current research and future prospects. Curr. Res. Food Sci. 2022, 5, 1539–1549. [Google Scholar] [CrossRef]
- Chen, W.; Li, C.; Zhang, B.; Zhou, Z.; Shen, Y.; Liao, X.; Yang, J.; Wang, Y.; Li, X.; Li, Y.; et al. Advances in Biodetoxification of Ochratoxin A-A Review of the Past Five Decades. Front. Microbiol. 2018, 9, 1386. [Google Scholar] [CrossRef]
- Pitout, M.J.; Nel, W. The inhibitory effect of ochratoxin a on bovine carboxypeptidase a in vitro. Biochem. Pharmacol. 1969, 18, 1837–1843. [Google Scholar] [CrossRef] [PubMed]
- Stander, M.A.; Bornscheuer, U.T.; Henke, E.; Steyn, P.S. Screening of Commercial Hydrolases for the Degradation of Ochratoxin A. J. Agric. Food Chem. 2000, 48, 5736–5739. [Google Scholar] [CrossRef]
- Abrunhosa, L.; Santos, L.; Venâncio, A. Degradation of Ochratoxin A by Proteases and by a Crude Enzyme of Aspergillus niger. Food Biotechnol. 2006, 20, 231–242. [Google Scholar] [CrossRef]
- Dobritzsch, D.; Wang, H.; Schneider, G.; Yu, S. Structural and functional characterization of ochratoxinase, a novel mycotoxin-degrading enzyme. Biochem. J. 2014, 462, 441–452. [Google Scholar] [CrossRef]
- Chang, X.; Wu, Z.; Wu, S.; Dai, Y.; Sun, C. Degradation of ochratoxin A by Bacillus amyloliquefaciens ASAG1. Food Addit. Contam. Part A 2015, 32, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Liuzzi, V.C.; Fanelli, F.; Tristezza, M.; Haidukowski, M.; Picardi, E.; Manzari, C.; Lionetti, C.; Grieco, F.; Logrieco, A.F.; Thon, M.R.; et al. Transcriptional Analysis of Acinetobacter sp. neg1 Capable of Degrading Ochratoxin A. Front. Microbiol. 2017, 7, 2162. [Google Scholar] [CrossRef]
- Rodriguez, H.; Reveron, I.; Doria, F.; Costantini, A.; De Las Rivas, B.; Muňoz, R.; Garcia-Moruno, E. Degradation of Ochratoxin A by Brevibacterium Species. J. Agric. Food Chem. 2011, 59, 10755–10760. [Google Scholar] [CrossRef]
- Abrunhosa, L.; Inês, A.; Rodrigues, A.I.; Guimarães, A.; Pereira, V.L.; Parpot, P.; Mendes-Faia, A.; Venâncio, A. Biodegradation of ochratoxin A by Pediococcus parvulus isolated from Douro wines. Int. J. Food Microbiol. 2014, 188, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yang, H.; Apaliya, M.T.; Zhao, L.; Gu, X.; Zheng, X.; Hu, W.; Zhang, H. The mechanisms involved in ochratoxin A elimination by Yarrowia lipolytica Y-2. Ann. Appl. Biol. 2018, 173, 164–174. [Google Scholar] [CrossRef]
- Zhang, H.H.; Wang, Y.; Zhao, C.; Wang, J.; Zhang, X.L. Biodegradation of ochratoxin A by Alcaligenes faecalis isolated from soil. J. Appl. Microbiol. 2017, 123, 661–668. [Google Scholar] [CrossRef]
- Luz, C.; Ferrer, J.; Mañes, J.; Meca, G. Toxicity reduction of ochratoxin A by lactic acid bacteria. Food Chem. Toxicol. 2018, 112, 60–66. [Google Scholar] [CrossRef]
- Xiong, K.; Wang, X.L.; Zhi, H.W.; Sun, B.G.; Li, X.T. Identification and safety evaluation of a product from the biodegradation of ochratoxin A by an Aspergillus strain. J. Sci. Food Agric. 2017, 97, 434–443. [Google Scholar] [CrossRef]
- Xiong, K.; Zhi, H.W.; Liu, J.Y.; Wang, X.Y.; Zhao, Z.Y.; Pei, P.G.; Deng, L.; Xiong, S.Y. Detoxification of Ochratoxin A by a novel Aspergillus oryzae strain and optimization of its biodegradation. Rev. Argent. Microbiol. 2021, 53, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Prasad, S.; Streit, B.; Gruber, C.; Gonaus, C. Enzymatic degradation of ochratoxin A in the gastrointestinal tract of piglets. J. Anim. Sci. 2023, 101, skad171. [Google Scholar] [CrossRef]
- Madhyastha, M.S.; Marquardt, R.R.; Frohlich, A.A. Hydrolysis of ochratoxin A by the microbial activity of digesta in the gastrointestinal tract of rats. Arch. Environ. Contam. Toxicol. 1992, 23, 468–472. [Google Scholar] [CrossRef]
- Kiessling, K.H.; Pettersson, H.; Sandholm, K.; Olsen, M. Metabolism of Aflatoxin, Ochratoxin, Zearalenone and Three Trichothecenes by Intact Rumen Fluid, Rumen Protozoa and Rumen Bacteria. Appl. Envieon. Microbiol. 1984, 47, 1070–1073. [Google Scholar] [CrossRef]
- Müller, H.M.; Lerch, C.; Müller, K.; Eggert, W. Kinetic profiles of ochratoxin A and ochratoxin alpha during in vitro incubation in buffered forestomach and abomasal contents from cows. Nat. Toxins 1998, 6, 251–258. [Google Scholar] [CrossRef]
- Xiao, H.; Marquardt, R.R.; Frohlich, A.A.; Phillips, G.D.; Vitti, T.G. Effect of a hay and a grain diet on the bioavailability of ochratoxin A in the rumen of sheep. J. Anim. Sci. 1991, 69, 3715–3723. [Google Scholar] [CrossRef]
- Xiao, H.; Marquardt, R.R.; Frohlich, A.A.; Phillips, G.D.; Vitti, T.G. Effect of a hay and a grain diet on the rate of hydrolysis of ochratoxin A in the rumen of sheep. J. Anim. Sci. 1991, 69, 3706–3714. [Google Scholar] [CrossRef]
- Zoghi, A.; Khosravi-Darani, K.; Sohrabvandi, S. Surface binding of toxins and heavy metals by probiotics. Mini Rev. Med. Chem. 2014, 14, 84–98. [Google Scholar] [CrossRef]
- Niderkorn, V.; Morgavi, D.P.; Aboab, B.; Lemaire, M.; Boudra, H. Cell wall component and mycotoxin moieties involved in the binding of fumonisin B1 and B2 by lactic acid bacteria. J. Appl. Microbiol. 2009, 106, 977–985. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xie, M.; Wei, D. Biological Detoxification of Mycotoxins: Current Status and Future Advances. Int. J. Mol. Sci. 2022, 23, 1064. [Google Scholar] [CrossRef]
- Piotrowska, M. The adsorption of ochratoxin a by lactobacillus species. Toxins 2014, 6, 2826–2839. [Google Scholar] [CrossRef] [PubMed]
- Møller, C.O.A.; Freire, L.; Rosim, R.E.; Margalho, L.P.; Balthazar, C.F.; Franco, L.T.; Sant’Ana, A.S.; Corassin, C.H.; Rattray, F.P.; de Oliveira, C.A.F. Effect of Lactic Acid Bacteria Strains on the Growth and Aflatoxin Production Potential of Aspergillus parasiticus, and Their Ability to Bind Aflatoxin B(1), Ochratoxin A, and Zearalenone in vitro. Front. Microbiol. 2021, 12, 655386. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.; Sontag, G.; Stidl, R.; Ehrlich, V.; Kundi, M.; Knasmüller, S. Detoxification of patulin and ochratoxin A, two abundant mycotoxins, by lactic acid bacteria. Food Chem. Toxicol. 2008, 46, 1398–1407. [Google Scholar] [CrossRef]
- Piotrowska, M.; Zakowska, Z. The elimination of ochratoxin A by lactic acid bacteria strains. Pol. J. Microbiol. 2005, 54, 279–286. [Google Scholar] [PubMed]
- Khoury, R.E.; Mathieu, F.; Atoui, A.; Kawtharani, H.; Khoury, A.E.; Afif, C.; Maroun, R.G. Ability of Soil Isolated Actinobacterial Strains to Prevent, Bind and Biodegrade Ochratoxin A. Toxins 2017, 9, 222. [Google Scholar] [CrossRef]
- Böhm, J.; Grajewski, J.; Asperger, H.; Cecon, B.; Rabus, B.; Razzazi, E. Study on biodegradation of some A- and B-trichothecenes and ochratoxin A by use of probiotic microorganisms. Mycotoxin Res. 2000, 16, 70–74. [Google Scholar] [CrossRef]
- Petchkongkaew, A.; Taillandier, P.; Gasaluck, P.; Lebrihi, A. Isolation of Bacillus spp. from Thai fermented soybean (Thua-nao): Screening for aflatoxin B1 and ochratoxin A detoxification. J. Appl. Microbiol. 2008, 104, 1495–1502. [Google Scholar] [CrossRef]
- Škrinjar, M.; Rašić, J.L.; Stojičić, V. Lowering of ochratoxin A level in milk by yoghurt bacteria and bifidobacteria. Folia Microbiol. 1996, 41, 26–28. [Google Scholar] [CrossRef]
- Jouany, J.-P.; Yiannikouris, A.; Bertin, G. The chemical bonds between mycotoxins and cell wall components of Saccharomyces cerevisiae have been identified. Arch. Zootech. 2005, 8, 4. [Google Scholar]
- Armando, M.; Pizzolitto, R.; Dogi, C.; Cristofolini, A.; Merkis, C.; Poloni, V.; Dalcero, A.; Cavaglieri, L. Adsorption of ochratoxin A and zearalenone by potential probiotic Saccharomyces cerevisiae strains and its relation with cell wall thickness. J. Appl. Microbiol. 2012, 113, 256–264. [Google Scholar] [CrossRef]
- Bejaoui, H.; Mathieu, F.; Taillandier, P.; Lebrihi, A. Ochratoxin A removal in synthetic and natural grape juices by selected oenological Saccharomyces strains. J. Appl. Microbiol. 2004, 97, 1038–1044. [Google Scholar] [CrossRef]
- Piotrowska, M.; Masek, A. Saccharomyces cerevisiae cell wall components as tools for ochratoxin a decontamination. Toxins 2015, 7, 1151–1162. [Google Scholar] [CrossRef] [PubMed]
- Piotrowska, M.; Nowak, A.; Czyzowska, A. Removal of ochratoxin A by wine Saccharomyces cerevisiae strains. Eur. Food Res. Technol. 2013, 236, 441–447. [Google Scholar] [CrossRef]
- Farbo, M.G.; Urgeghe, P.P.; Fiori, S.; Marceddu, S.; Jaoua, S.; Migheli, Q. Adsorption of ochratoxin A from grape juice by yeast cells immobilised in calcium alginate beads. Int. J. Food Microbiol. 2016, 217, 29–34. [Google Scholar] [CrossRef] [PubMed]
- G Gil-Serna, J.; Patiño, B.; Cortés, L.; González-Jaén, M.T.; Vázquez, C. Mechanisms involved in reduction of ochratoxin A produced by Aspergillus westerdijkiae using Debaryomyces hansenii CYC 1244. Int. J. Food Microbiol. 2011, 151, 113–118. [Google Scholar] [CrossRef]
- Csutoras, C.; Rácz, L.; Racz, K.; Fűtő, P.; Forgo, P.; Attila, K. Monitoring of ochratoxin A during the fermentation of different wines by applying high toxin concentrations. Microchem. J. 2013, 107, 182–184. [Google Scholar] [CrossRef]
- Taheur, F.B.; Fedhila, K.; Chaieb, K.; Kouidhi, B.; Bakhrouf, A.; Abrunhosa, L. Adsorption of aflatoxin B1, zearalenone and ochratoxin A by microorganisms isolated from Kefir grains. Int. J. Food Microbiol. 2017, 251, 1–7. [Google Scholar] [CrossRef]
- Maresca, M.; Mahfoud, R.; Pfohl-Leszkowicz, A.; Fantini, J. The mycotoxin ochratoxin A alters intestinal barrier and absorption functions but has no effect on chloride secretion. Toxicol. Appl. Pharmacol. 2001, 176, 54–63. [Google Scholar] [CrossRef]
- Abassi, H.; Ayed-Boussema, I.; Shirley, S.; Abid, S.; Bacha, H. Ochratoxin A and T-2 Toxin Induce Clonogenicity and Cell Migration in Human Colon Carcinoma and Fetal Lung Fibroblast Cell Lines. J. Biochem. Mol. Toxicol. 2016, 30, 128–135. [Google Scholar] [CrossRef]
- Huang, X.; Gao, Y.; Li, S.; Wu, C.; Wang, J.; Zheng, N. Modulation of Mucin (MUC2, MUC5AC and MUC5B) mRNA Expression and Protein Production and Secretion in Caco-2/HT29-MTX Co-Cultures Following Exposure to Individual and Combined Aflatoxin M1 and Ochratoxin A. Toxins 2019, 11, 132. [Google Scholar] [CrossRef]
- González-Arias, C.A.; Crespo-Sempere, A.; Marín, S.; Sanchis, V.; Ramos, A.J. Modulation of the xenobiotic transformation system and inflammatory response by ochratoxin A exposure using a co-culture system of Caco-2 and HepG2 cells. Food Chem. Toxicol. 2015, 86, 245–252. [Google Scholar] [CrossRef]
- Yang, X.; Gao, Y.; Yan, Q.; Bao, X.; Zhao, S.; Wang, J.; Zheng, N. Transcriptome Analysis of Ochratoxin A-Induced Apoptosis in Differentiated Caco-2 Cells. Toxins 2019, 12, 23. [Google Scholar] [CrossRef]
- McLaughlin, J.; Padfield, P.J.; Burt, J.P.H.; O’Neill, C.A. Ochratoxin A increases permeability through tight junctions by removal of specific claudin isoforms. Am. J. Physiol.—Cell Physiol. 2004, 287, C1412–C1417. [Google Scholar] [CrossRef] [PubMed]
- Romero, A.; Ares, I.; Ramos, E.; Castellano, V.; Martínez, M.; Martínez-Larrañaga, M.-R.; Anadón, A.; Martínez, M.-A. Mycotoxins modify the barrier function of Caco-2 cells through differential gene expression of specific claudin isoforms: Protective effect of illite mineral clay. Toxicology 2016, 353–354, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Li, S.; Bao, X.; Luo, C.; Yang, H.; Wang, J.; Zhao, S.; Zheng, N. Transcriptional and Proteomic Analysis Revealed a Synergistic Effect of Aflatoxin M1 and Ochratoxin A Mycotoxins on the Intestinal Epithelial Integrity of Differentiated Human Caco-2 Cells. J. Proteome Res. 2018, 17, 3128–3142. [Google Scholar] [CrossRef]
- Wang, H.; Zhai, N.; Chen, Y.; Fu, C.; Huang, K. OTA induces intestinal epithelial barrier dysfunction and tight junction disruption in IPEC-J2 cells through ROS/Ca(2+)-mediated MLCK activation. Environ. Pollut. 2018, 242, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.W.; Lee, S.I. Gene expression profiling after ochratoxin A treatment in small intestinal epithelial cells from pigs. J. Anim. Sci. Technol. 2022, 64, 842–853. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, S.; Lan, H.; Zheng, X. Ochratoxin A (OTA) causes intestinal aging damage through the NLRP3 signaling pathway mediated by calcium overload and oxidative stress. Environ. Sci. Pollut. Res. 2024, 31, 27864–27882. [Google Scholar] [CrossRef]
- Nejdfors, P.; Ekelund, M.; Jeppsson, B.; Weström, B.R. Mucosal in vitro permeability in the intestinal tract of the pig, the rat, and man: Species- and region-related differences. Scand. J. Gastroenterol. 2000, 35, 501–507. [Google Scholar] [CrossRef]
- Szczech, G.M.; Carlton, W.W.; Tuite, J.; Caldwell, R. Ochratoxin A Toxicosis in Swine. Vet. Pathol. 1973, 10, 347–364. [Google Scholar] [CrossRef]
- Solcan, C.; Pavel, G.; Floristean, V.; Chiriac, I.; Slencu, B.; Solcan, G. Effect of ochratoxin A on the intestinal mucosa and mucosa-associated lymphoid tissues in broiler chickens. Acta Vet. Hung. 2015, 63, 30–40. [Google Scholar] [CrossRef]
- Manafi, M.; Mohan, K.; Ali, M.N. Effect of ochratoxin A on coccidiosis-challenged broiler chicks. World Mycotoxin J. 2011, 4, 177–181. [Google Scholar] [CrossRef]
- El Cafsi, I.; Bjeoui, S.; Rabeh, I.; Nechi, S.; Chelbi, E.; El Cafsi, M.; Ghram, A. Effects of ochratoxin A on membrane phospholipids of the intestine of broiler chickens, practical consequences. Animal 2020, 14, 933–941. [Google Scholar] [CrossRef]
- Gao, Y.N.; Min, L.; Yang, X.; Wang, J.Q.; Zheng, N. The coexistence of aflatoxin M1 and ochratoxin A induced intestinal barrier disruption via the regulation of key differentially expressed microRNAs and long non-coding RNAs in BALB/c mice. Ecotoxicol. Environ. Saf. 2023, 264, 115428. [Google Scholar] [CrossRef]
- Peng, X.; Fan, H.; Liu, J.; Jiang, X.; Liu, C.; Yang, Y.; Zhai, S. Embryo injected with Ochratoxin A induced jejunum injury in ducklings by activating the TLR4 signaling pathway: Involvement of intestinal microbiota. Ecotoxicol. Environ. Saf. 2024, 281, 116666. [Google Scholar] [CrossRef]
- Izco, M.; Vettorazzi, A.; de Toro, M.; Sáenz, Y.; Alvarez-Erviti, L. Oral Sub-chronic Ochratoxin A Exposure Induces Gut Microbiota Alterations in Mice. Toxins 2021, 13, 106. [Google Scholar] [CrossRef]
- Guo, M.; Huang, K.; Chen, S.; Qi, X.; He, X.; Cheng, W.H.; Luo, Y.; Xia, K.; Xu, W. Combination of metagenomics and culture-based methods to study the interaction between ochratoxin a and gut microbiota. Toxicol. Sci. 2014, 141, 314–323. [Google Scholar] [CrossRef]
- Ansari, I.; Raddatz, G.; Gutekunst, J.; Ridnik, M.; Cohen, D.; Abu-Remaileh, M.; Tuganbaev, T.; Shapiro, H.; Pikarsky, E.; Elinav, E.; et al. The microbiota programs DNA methylation to control intestinal homeostasis and inflammation. Nat. Microbiol. 2020, 5, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Demirkan, A.; van Dongen, J.; Finnicum, C.T.; Westra, H.-J.; Jankipersadsing, S.; Willemsen, G.; Ijzerman, R.G.; Boomsma, D.I.; Ehli, E.A.; Bonder, M.J.; et al. Linking the gut microbiome to host DNA methylation by a discovery and replication epigenome-wide association study. BMC Genom. 2024, 25, 1224. [Google Scholar] [CrossRef]
- Ozawa, S.; Ojiro, R.; Tang, Q.; Zou, X.; Jin, M.; Yoshida, T.; Shibutani, M. Involvement of multiple epigenetic mechanisms by altered DNA methylation from the early stage of renal carcinogenesis before proliferative lesion formation upon repeated administration of ochratoxin A. Toxicology 2024, 506, 153875. [Google Scholar] [CrossRef]
- Fellows, R.; Denizot, J.; Stellato, C.; Cuomo, A.; Jain, P.; Stoyanova, E.; Balázsi, S.; Hajnády, Z.; Liebert, A.; Kazakevych, J.; et al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat. Commun. 2018, 9, 105. [Google Scholar] [CrossRef] [PubMed]
- Czakai, K.; Müller, K.; Mosesso, P.; Pepe, G.; Schulze, M.; Gohla, A.; Patnaik, D.; Dekant, W.; Higgins, J.M.; Mally, A. Perturbation of mitosis through inhibition of histone acetyltransferases: The key to ochratoxin a toxicity and carcinogenicity? Toxicol. Sci. 2011, 122, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Albillos, A.; de Gottardi, A.; Rescigno, M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J. Hepatol. 2020, 72, 558–577. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Chen, D.Q.; Chen, L.; Liu, J.R.; Vaziri, N.D.; Guo, Y.; Zhao, Y.Y. Microbiome-metabolome reveals the contribution of gut-kidney axis on kidney disease. J. Transl. Med. 2019, 17, 5. [Google Scholar] [CrossRef]
- Quigley, E.M.M. Microbiota-Brain-Gut Axis and Neurodegenerative Diseases. Curr. Neurol. Neurosci. Rep. 2017, 17, 94. [Google Scholar] [CrossRef]
- Wang, W.; Zhai, S.; Xia, Y.; Wang, H.; Ruan, D.; Zhou, T.; Zhu, Y.; Zhang, H.; Zhang, M.; Ye, H.; et al. Ochratoxin A induces liver inflammation: Involvement of intestinal microbiota. Microbiome 2019, 7, 151. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Fu, Y.; Zhu, S.; Xia, D.; Zhai, S.; Xiao, D.; Zhu, Y.; Dione, M.; Ben, L.; Yang, L.; et al. Ochratoxin A induces abnormal tryptophan metabolism in the intestine and liver to activate AMPK signaling pathway. J. Anim. Sci. Biotechnol. 2023, 14, 125. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, M.; Li, X.; Ye, Y.; Xie, Y.; Wu, T.; Chen, Y.; Yu, H.; Wu, H.; Yang, Z.; et al. Inulin Supplementation Alleviates Ochratoxin A-Induced Kidney Injury through Modulating Intestinal Microbiota. J. Agric. Food Chem. 2024, 72, 18682–18696. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, D.; Lv, L.; Zhao, K.; Wang, D.; Yang, Y. Neuroprotective effects of Walnut kernel polysaccharides against ochratoxin A-induced hippocampal neurotoxicity by modulating the gut-brain axis. Food Biosci. 2015, 71, 107177. [Google Scholar] [CrossRef]
- Godley, F.A.; Shogan, B.D.; Hyman, N.H. Role of the Microbiome in Malignancy. Surg. Infect. 2023, 24, 271–275. [Google Scholar] [CrossRef]
- Liew, W.P.; Mohd-Redzwan, S. Mycotoxin: Its Impact on Gut Health and Microbiota. Front. Cell Infect. Microbiol. 2018, 8, 60. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Lin, Y.; Cao, C.; Chen, D.; Huang, X.; Li, C.; Xu, H.; Lai, H.; Chen, H.; et al. Roles of the gut microbiota in hepatocellular carcinoma: From the gut dysbiosis to the intratumoral microbiota. Cell Death Discov. 2025, 11, 140. [Google Scholar] [CrossRef]
- Xu, Y.; Gao, Z.; Liu, J.; Yang, Q.; Xu, S. Role of gut microbiome in suppression of cancers. Gut Microbes 2025, 17, 2495183. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.H.; Yu, J. Gut microbiota in colorectal cancer: Mechanisms of action and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 690–704. [Google Scholar] [CrossRef]
- Gaines, S.; van Praagh, J.B.; Williamson, A.J.; Jacobson, R.A.; Hyoju, S.; Zaborin, A.; Mao, J.; Koo, H.Y.; Alpert, L.; Bissonnette, M.; et al. Western Diet Promotes Intestinal Colonization by Collagenolytic Microbes and Promotes Tumor Formation After Colorectal Surgery. Gastroenterology 2020, 158, 958–970.e952. [Google Scholar] [CrossRef]
- Li, R.; Zhou, R.; Wang, H.; Li, W.; Pan, M.; Yao, X.; Zhan, W.; Yang, S.; Xu, L.; Ding, Y.; et al. Gut microbiota-stimulated cathepsin K secretion mediates TLR4-dependent M2 macrophage polarization and promotes tumor metastasis in colorectal cancer. Cell Death Differ. 2019, 26, 2447–2463. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef]
- Peng, L.; Li, Z.R.; Green, R.S.; Holzman, I.R.; Lin, J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J. Nutr. 2009, 139, 1619–1625. [Google Scholar] [CrossRef] [PubMed]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Núñez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef]
- Zhao, Q.; Yu, J.; Hao, Y.; Zhou, H.; Hu, Y.; Zhang, C.; Zheng, H.; Wang, X.; Zeng, F.; Hu, J.; et al. Akkermansia muciniphila plays critical roles in host health. Crit. Rev. Microbiol. 2023, 49, 82–100. [Google Scholar] [CrossRef] [PubMed]
- Markowiak, P.; Śliżewska, K.; Nowak, A.; Chlebicz, A.; Żbikowski, A.; Pawłowski, K.; Szeleszczuk, P. Probiotic microorganisms detoxify ochratoxin A in both a chicken liver cell line and chickens. J. Sci. Food Agric. 2019, 99, 4309–4318. [Google Scholar] [CrossRef]
- Kalyanaraman, B.; Cheng, G.; Hardy, M. Gut microbiome, short-chain fatty acids, alpha-synuclein, neuroinflammation, and ROS/RNS: Relevance to Parkinson’s disease and therapeutic implications. Redox Biol. 2024, 71, 103092. [Google Scholar] [CrossRef]
- van de Wouw, M.; Boehme, M.; Lyte, J.M.; Wiley, N.; Strain, C.; O’Sullivan, O.; Clarke, G.; Stanton, C.; Dinan, T.G.; Cryan, J.F. Short-chain fatty acids: Microbial metabolites that alleviate stress-induced brain-gut axis alterations. J. Physiol. 2018, 596, 4923–4944. [Google Scholar] [CrossRef] [PubMed]
- Stecher, B.; Hardt, W.D. Mechanisms controlling pathogen colonization of the gut. Curr. Opin. Microbiol. 2011, 14, 82–91. [Google Scholar] [CrossRef]
- Kostic, A.D.; Howitt, M.R.; Garrett, W.S. Exploring host-microbiota interactions in animal models and humans. Genes. Dev. 2013, 27, 701–718. [Google Scholar] [CrossRef] [PubMed]
- Azcárate-Peril, M.A.; Sikes, M.; Bruno-Bárcena, J.M. The intestinal microbiota, gastrointestinal environment and colorectal cancer: A putative role for probiotics in prevention of colorectal cancer? Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 301, G401–G424. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, F.; Ding, X.; Wu, G.; Lam, Y.Y.; Wang, X.; Fu, H.; Xue, X.; Lu, C.; Ma, J.; et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 2018, 359, 1151–1156. [Google Scholar] [CrossRef]
- De Angelis, M.; Siragusa, S.; Berloco, M.; Caputo, L.; Settanni, L.; Alfonsi, G.; Amerio, M.; Grandi, A.; Ragni, A.; Gobbetti, M. Selection of potential probiotic lactobacilli from pig feces to be used as additives in pelleted feeding. Res. Microbiol. 2006, 157, 792–801. [Google Scholar] [CrossRef]
- Tap, J.; Mondot, S.; Levenez, F.; Pelletier, E.; Caron, C.; Furet, J.P.; Ugarte, E.; Muñoz-Tamayo, R.; Paslier, D.L.; Nalin, R.; et al. Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 2009, 11, 2574–2584. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef]
- Zmora, N.; Suez, J.; Elinav, E. You are what you eat: Diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 35–56. [Google Scholar] [CrossRef]
- Marco, M.L.; Heeney, D.; Binda, S.; Cifelli, C.J.; Cotter, P.D.; Foligné, B.; Gänzle, M.; Kort, R.; Pasin, G.; Pihlanto, A.; et al. Health benefits of fermented foods: Microbiota and beyond. Curr. Opin. Biotechnol. 2017, 44, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Holscher, H.D. Dietary fiber and prebiotics and the gastrointestinal microbiota. Gut Microbes 2017, 8, 172–184. [Google Scholar] [CrossRef]
- Haran, J.P.; McCormick, B.A. Aging, Frailty, and the Microbiome-How Dysbiosis Influences Human Aging and Disease. Gastroenterology 2021, 160, 507–523. [Google Scholar] [CrossRef]
- Wallen, Z.D.; Appah, M.; Dean, M.N.; Sesler, C.L.; Factor, S.A.; Molho, E.; Zabetian, C.P.; Standaert, D.G.; Payami, H. Characterizing dysbiosis of gut microbiome in PD: Evidence for overabundance of opportunistic pathogens. NPJ Park. Dis. 2020, 6, 11. [Google Scholar] [CrossRef]
- Ghosh, S.; Whitley, C.S.; Haribabu, B.; Jala, V.R. Regulation of Intestinal Barrier Function by Microbial Metabolites. Cell Mol. Gastroenterol. Hepatol. 2021, 11, 1463–1482. [Google Scholar] [CrossRef]
- Bischoff, S.C.; Barbara, G.; Buurman, W.; Ockhuizen, T.; Schulzke, J.D.; Serino, M.; Tilg, H.; Watson, A.; Wells, J.M. Intestinal permeability--a new target for disease prevention and therapy. BMC Gastroenterol. 2014, 14, 189. [Google Scholar] [CrossRef]
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C.; et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007, 56, 1761–1772. [Google Scholar] [CrossRef]
- Khosravi, A.; Mazmanian, S.K. Disruption of the gut microbiome as a risk factor for microbial infections. Curr. Opin. Microbiol. 2013, 16, 221–227. [Google Scholar] [CrossRef]
- Wang, Y.; Cui, J.; Zheng, G.; Zhao, M.; Hao, Z.; Lian, H.; Li, Y.; Wu, W.; Zhang, X.; Wang, J. Ochratoxin A induces cytotoxicity through ROS-mediated endoplasmic reticulum stress pathway in human gastric epithelium cells. Toxicology 2022, 479, 153309. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, S.; Jiao, D.; Yao, B.; Yang, S.; Li, P.; Long, M. Astaxanthin Alleviates Ochratoxin A-Induced Cecum Injury and Inflammation in Mice by Regulating the Diversity of Cecal Microbiota and TLR4/MyD88/NF-κB Signaling Pathway. Oxid. Med. Cell Longev. 2021, 2021, 8894491. [Google Scholar] [CrossRef]
- Shin, N.R.; Whon, T.W.; Bae, J.W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Levy, M.; Kolodziejczyk, A.A.; Thaiss, C.A.; Elinav, E. Dysbiosis and the immune system. Nat. Rev. Immunol. 2017, 17, 219–232. [Google Scholar] [CrossRef]
- Belkaid, Y.; Harrison, O.J. Homeostatic Immunity and the Microbiota. Immunity 2017, 46, 562–576. [Google Scholar] [CrossRef]
- Bonerba, E.; Manfredi, A.; Dimuccio, M.M.; Lorusso, P.; Pandiscia, A.; Terio, V.; Di Pinto, A.; Panseri, S.; Ceci, E.; Bozzo, G. Ochratoxin A in Poultry Supply Chain: Overview of Feed Occurrence, Carry-Over, and Pathognomonic Lesions in Target Organs to Promote Food Safety. Toxins 2024, 16, 487. [Google Scholar] [CrossRef]
- Clarke, G.; Stilling, R.M.; Kennedy, P.J.; Stanton, C.; Cryan, J.F.; Dinan, T.G. Minireview: Gut microbiota: The neglected endocrine organ. Mol. Endocrinol. 2014, 28, 1221–1238. [Google Scholar] [CrossRef]
- Rao, J.; Qiu, P.; Zhang, Y.; Wang, X. Gut microbiota trigger host liver immune responses that affect drug-metabolising enzymes. Front. Immunol. 2024, 15, 1511229. [Google Scholar] [CrossRef]
- Li, M.; Wang, B.; Zhang, M.; Rantalainen, M.; Wang, S.; Zhou, H.; Zhang, Y.; Shen, J.; Pang, X.; Wei, H.; et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc. Natl. Acad. Sci. USA 2008, 105, 2117–2122. [Google Scholar] [CrossRef]
- Wilson, A.; Urquhart, B.L.; Ponich, T.; Chande, N.; Gregor, J.C.; Beaton, M.; Kim, R.B. Crohn’s Disease Is Associated with Decreased CYP3A4 and P-Glycoprotein Protein Expression. Mol. Pharm. 2019, 16, 4059–4064. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 2009, 294, 1–8. [Google Scholar] [CrossRef]
- Boronat, A.; Rodriguez-Morató, J.; Serreli, G.; Fitó, M.; Tyndale, R.F.; Deiana, M.; de la Torre, R. Contribution of Biotransformations Carried Out by the Microbiota, Drug-Metabolizing Enzymes, and Transport Proteins to the Biological Activities of Phytochemicals Found in the Diet. Adv. Nutr. 2021, 12, 2172–2189. [Google Scholar] [CrossRef]
- Zhai, S.S.; Ruan, D.; Zhu, Y.W.; Li, M.C.; Ye, H.; Wang, W.C.; Yang, L. Protective effect of curcumin on ochratoxin A-induced liver oxidative injury in duck is mediated by modulating lipid metabolism and the intestinal microbiota. Poult. Sci. 2020, 99, 1124–1134. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Abeltino, A.; Hatem, D.; Serantoni, C.; Riente, A.; De Giulio, M.M.; De Spirito, M.; De Maio, F.; Maulucci, G. Unraveling the Gut Microbiota: Implications for Precision Nutrition and Personalized Medicine. Nutrients 2024, 16, 3806. [Google Scholar] [CrossRef]
- Kumar, B.; Lorusso, E.; Fosso, B.; Pesole, G. A comprehensive overview of microbiome data in the light of machine learning applications: Categorization, accessibility, and future directions. Front. Microbiol. 2024, 15, 1343572. [Google Scholar] [CrossRef]
- Johnson, K.V. Gut microbiome composition and diversity are related to human personality traits. Hum. Microb. J. 2020, 15, 100069. [Google Scholar] [CrossRef]
- Larzul, C.; Estellé, J.; Borey, M.; Blanc, F.; Lemonnier, G.; Billon, Y.; Thiam, M.G.; Quinquis, B.; Galleron, N.; Jardet, D.; et al. Driving gut microbiota enterotypes through host genetics. Microbiome 2024, 12, 116. [Google Scholar] [CrossRef]
- Fan, C.; Zhang, L.; Fu, H.; Liu, C.; Li, W.; Cheng, Q.; Zhang, H.; Jia, S.; Zhang, Y. Enterotypes of the Gut Microbial Community and Their Response to Plant Secondary Compounds in Plateau Pikas. Microorganisms 2020, 8, 1311. [Google Scholar] [CrossRef]
- Gorvitovskaia, A.; Holmes, S.P.; Huse, S.M. Interpreting Prevotella and Bacteroides as biomarkers of diet and lifestyle. Microbiome 2016, 4, 15. [Google Scholar] [CrossRef]
- Tremaroli, V.; Bäckhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef]
- Marin, D.E.; Pistol, G.C.; Gras, M.A.; Palade, M.L.; Taranu, I. Comparative effect of ochratoxin A on inflammation and oxidative stress parameters in gut and kidney of piglets. Regul. Toxicol. Pharmacol. 2017, 89, 224–231. [Google Scholar] [CrossRef]
- Claus, S.P.; Guillou, H.; Ellero-Simatos, S. The gut microbiota: A major player in the toxicity of environmental pollutants? NPJ Biofilms Microbiomes 2016, 2, 16003. [Google Scholar] [CrossRef]
- Abdelsalam, N.A.; Ramadan, A.T.; ElRakaiby, M.T.; Aziz, R.K. Toxicomicrobiomics: The Human Microbiome vs. Pharmaceutical, Dietary, and Environmental Xenobiotics. Front. Pharmacol. 2020, 11, 390. [Google Scholar] [CrossRef]
- Petrosino, J.F. The microbiome in precision medicine: The way forward. Genome Med. 2018, 10, 12. [Google Scholar] [CrossRef]
- Kashyap, P.C.; Chia, N.; Nelson, H.; Segal, E.; Elinav, E. Microbiome at the Frontier of Personalized Medicine. Mayo Clin. Proc. 2017, 92, 1855–1864. [Google Scholar] [CrossRef]
- Schupack, D.A.; Mars, R.A.T.; Voelker, D.H.; Abeykoon, J.P.; Kashyap, P.C. The promise of the gut microbiome as part of individualized treatment strategies. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 7–25. [Google Scholar] [CrossRef]
- Song, E.J.; Shin, J.H. Personalized Diets based on the Gut Microbiome as a Target for Health Maintenance: From Current Evidence to Future Possibilities. J. Microbiol. Biotechnol. 2022, 32, 1497–1505. [Google Scholar] [CrossRef]
- Simon, B.O.; Nnaji, N.D.; Anumudu, C.K.; Aleke, J.C.; Ekwueme, C.T.; Uhegwu, C.C.; Ihenetu, F.C.; Obioha, P.; Ifedinezi, O.V.; Ezechukwu, P.S.; et al. Microbiome-Based Interventions for Food Safety and Environmental Health. Appl. Sci. 2025, 15, 5219. [Google Scholar] [CrossRef]
- Singh, A.V.; Bhardwaj, P.; Laux, P.; Pradeep, P.; Busse, M.; Luch, A.; Hirose, A.; Osgood, C.J.; Stacey, M.W. AI and ML-based risk assessment of chemicals: Predicting carcinogenic risk from chemical-induced genomic instability. Front. Toxicol. 2024, 6, 1461587. [Google Scholar] [CrossRef]
- Marcos-Zambrano, L.J.; Karaduzovic-Hadziabdic, K.; Loncar Turukalo, T.; Przymus, P.; Trajkovik, V.; Aasmets, O.; Berland, M.; Gruca, A.; Hasic, J.; Hron, K.; et al. Applications of Machine Learning in Human Microbiome Studies: A Review on Feature Selection, Biomarker Identification, Disease Prediction and Treatment. Front. Microbiol. 2021, 12, 634511. [Google Scholar] [CrossRef]
- Ali, N.; Muñoz, K.; Degen, G.H. Ochratoxin A and its metabolites in urines of German adults-An assessment of variables in biomarker analysis. Toxicol. Lett. 2017, 275, 19–26. [Google Scholar] [CrossRef]
- Aslam, M.; Beg, A.E.; Blaszkewicz, M.; Degen, G.H.; Golka, K. Ochratoxin A blood concentration in healthy subjects and bladder cancer cases from Pakistan. Mycotoxin Res. 2005, 21, 164–167. [Google Scholar] [CrossRef]
- Coronel, M.B.; Sanchis, V.; Ramos, A.J.; Marin, S. Review. Ochratoxin A: Presence in Human Plasma and Intake Estimation. Food Sci. Technol. Int. 2010, 16, 5–18. [Google Scholar] [CrossRef]
- Märtlbauer, E.; Usleber, E.; Dietrich, R.; Schneider, E. Ochratoxin A in human blood serum—Retrospective long-term data. Mycotoxin Res. 2009, 25, 175–186. [Google Scholar] [CrossRef]
- Coronel, M.B.; Marin, S.; Tarragó, M.; Cano-Sancho, G.; Ramos, A.J.; Sanchis, V. Ochratoxin A and its metabolite ochratoxin alpha in urine and assessment of the exposure of inhabitants of Lleida, Spain. Food Chem. Toxicol. 2011, 49, 1436–1442. [Google Scholar] [CrossRef]
- Carballo, D.; Pallarés, N.; Ferrer, E.; Barba, F.J.; Berrada, H. Assessment of Human Exposure to Deoxynivalenol, Ochratoxin A, Zearalenone and Their Metabolites Biomarker in Urine Samples Using LC-ESI-qTOF. Toxins 2021, 13, 530. [Google Scholar] [CrossRef]
- Arce-López, B.; Lizarraga, E.; Irigoyen, Á.; González-Peñas, E. Presence of 19 Mycotoxins in Human Plasma in a Region of Northern Spain. Toxins 2020, 12, 750. [Google Scholar] [CrossRef]
- Das Trisha, A.; Hafsa, J.M.; Hasan, A.; Habib, A.; Tuba, H.R.; Degen, G.H.; Ali, N. Occurrence of ochratoxin A in breast milk and urine samples of nursing mothers in Bangladesh. Mycotoxin Res. 2024, 40, 135–146. [Google Scholar] [CrossRef]
- European Food Safety Authority. Opinion of the Scientific Panel on contaminants in the food chain on a request from the commission related to ochratoxin A in food. EFSA J. 2006, 4, 1–56. [Google Scholar] [CrossRef]
- Bui-Klimke, T.R.; Wu, F. Ochratoxin A and human health risk: A review of the evidence. Crit. Rev. Food Sci. Nutr. 2015, 55, 1860–1869. [Google Scholar] [CrossRef]
- Mally, A.; Zepnik, H.; Wanek, P.; Eder, E.; Dingley, K.; Ihmels, H.; Völkel, W.; Dekant, W. Ochratoxin A: Lack of formation of covalent DNA adducts. Chem. Res. Toxicol. 2004, 17, 234–242. [Google Scholar] [CrossRef]
- Gross-Steinmeyer, K.; Weymann, J.; Hege, H.G.; Metzler, M. Metabolism and lack of DNA reactivity of the mycotoxin ochratoxin a in cultured rat and human primary hepatocytes. J. Agric. Food Chem. 2002, 50, 938–945. [Google Scholar] [CrossRef]
- Stiborová, M.; Arlt, V.M.; Schmeiser, H.H. Balkan endemic nephropathy: An update on its aetiology. Arch. Toxicol. 2016, 90, 2595–2615. [Google Scholar] [CrossRef]
- Więckowska, M.; Cichon, N.; Szelenberger, R.; Gorniak, L.; Bijak, M. Ochratoxin A and Its Role in Cancer Development: A Comprehensive Review. Cancers 2024, 16, 3473. [Google Scholar] [CrossRef]
- Śliżewska, K.; Piotrowska, M. Reduction of ochratoxin A in chicken feed using probiotic. Ann. Agric. Environ. Med. 2014, 21, 676–680. [Google Scholar] [CrossRef]
- Śliżewska, K.; Markowiak-Kopeć, P.; Sip, A.; Lipiński, K.; Mazur-Kuśnirek, M. The Effect of Using New Synbiotics on the Turkey Performance, the Intestinal Microbiota and the Fecal Enzymes Activity in Turkeys Fed Ochratoxin A Contaminated Feed. Toxins 2020, 12, 578. [Google Scholar] [CrossRef]
- Śliżewska, K.; Markowiak, P.; Żbikowski, A.; Szeleszczuk, P. Effects of synbiotics on the gut microbiota, blood and rearing parameters of chickens. FEMS Microbiol. Lett. 2019, 366, fnz116. [Google Scholar] [CrossRef]
- Śliżewska, K.; Markowiak-Kopeć, P.; Żbikowski, A.; Szeleszczuk, P. The effect of synbiotic preparations on the intestinal microbiota and her metabolism in broiler chickens. Sci. Rep. 2020, 10, 4281. [Google Scholar] [CrossRef]
- Jiang, S.; Du, L.; Zhao, Q.; Su, S.; Huang, S.; Zhang, J. Tropical postbiotics alleviate the disorders in the gut microbiota and kidney damage induced by ochratoxin A exposure. Food Funct. 2024, 15, 3980–3992. [Google Scholar] [CrossRef]
- Sahle, Z.; Engidaye, G.; Shenkute Gebreyes, D.; Adenew, B.; Abebe, T.A. Fecal microbiota transplantation and next-generation therapies: A review on targeting dysbiosis in metabolic disorders and beyond. SAGE Open Med. 2024, 12, 20503121241257486. [Google Scholar] [CrossRef]
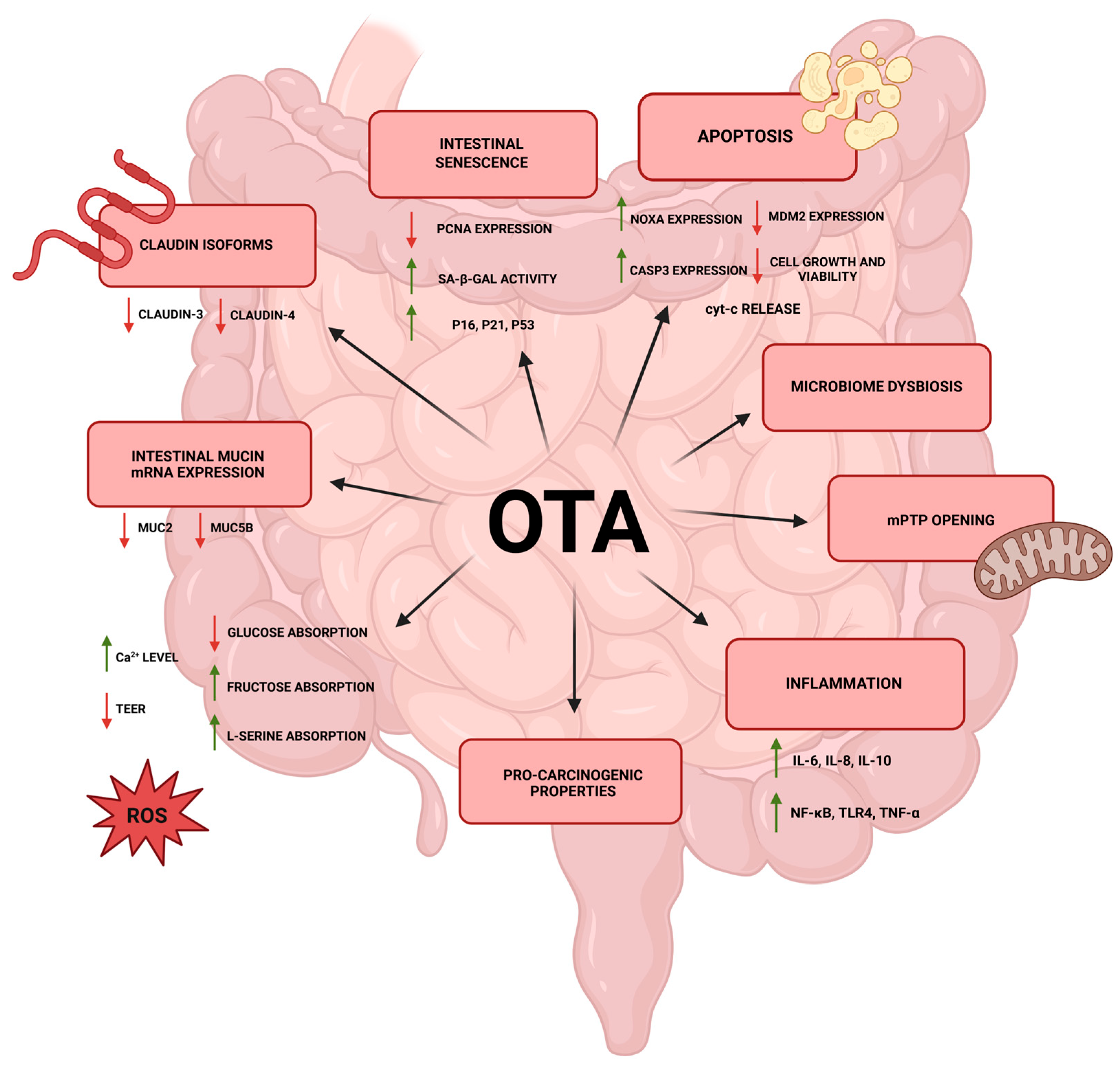
| Microorganism | Reaction Conditions | Medium | OTA Concentration | The Percentage of OTA Degradation | Reference |
|---|---|---|---|---|---|
| Saccharomyces cerevisiae: Malaga LOCK 0173 | 30 °C, 24 h, thermally inactivated cells | YPG medium White grape juice medium Blackcurrant juice medium | 1 µg/mL | YPG medium: 35.4% White grape juice medium: 82.8% Blackcurrant juice medium: 10.7% | [80] |
| Syrena ŁOCK 0201 | YPG medium: 21% White grape juice medium: 85.1% Blackcurrant juice medium: 65.2% | ||||
| Bakery BS | YPG Medium: 54.1% White grape juice medium: 64.4% Blackcurrant juice medium: 62.4% | ||||
| Candida intermedia | Rotary shaker 100 rpm, 25 °C, 48 h, in dark, immobilized yeast cells on calcium alginate beads | Grape juice | 20 µg/kg | >80% | [81] |
| Debaryomyces hansenii | Rotary shaker 300 rpm, 28 °C, 24 h, pH 3 | YMB medium | 7 µg/mL | >98% After 5 min | [82] |
| Saccharomyces cerevisiae | 12 °C, 90 days, viable cells | Red, rose, and white wine musts | 0.01–4 µg/mL | Red: 88–90% Rose: 83–86% White: 73–76% | [83] |
| Lactobacillus kefiri KFLM3, | Aerobically, 25 °C, 24 h, viable cells | Milk | 1 µg/mL | 81% | [84] |
| Kazachstania servazzi KFGY7, | 62% | ||||
| Acetobacter syzygii KFGM 1 | 50% | ||||
| Lactobacillus plantarum LOCK 0862 | 30 °C, 24 h, viable cells | MRS medium | (A) 1 mg s.m./mL (B) 5 s.m./mL | (A) 21.23% (B) 35.01% | [68] |
| Lactobacillus brevis LOCK 0845 | (A) 14.64% (B) 20.53% | ||||
| Lactobacillus sanfranciscensis LOCK 0866 | (A) 16.91% (B) 32% | ||||
| Actinobacterial strains: | 1 h, viable cells | PBS solution | 45.12 ng/mL | [72] | |
| AT10 | 25.62% | ||||
| AT8 | 16.07% | ||||
| SN7 | 33.93% | ||||
| MS1 | 4.33% | ||||
| ML5 | 9.46% | ||||
| G10 | 16.28% | ||||
| PT1 | 24.85% | ||||
| Saccharomyces cerevisiae: RC008 | 37 °C, 24 h | YPD medium | (A) 1 µg/mL (B) 5 µg/mL (C) 10 µg/mL (D) 40 µg/mL (E) 100 µg/mL | (A) 46% (B) 16% (C) 14.5% (D) 17.9% (E) 56.7% | [77] |
| RC009 | (A) 43% (B) 16% (C) 49.4% (D) 37.3% (E) 67.2% | ||||
| RC012 | (A) 63% (B) 39.2% (C) 56.4% (D) 39.2% (E) 71.2% | ||||
| RC016 | (A) 74% (B) 30.4% (C) 58% (D) 39.2% (E) 74.2% | ||||
| Saccharomyces cerevisiae LALVIN BM45 LALVIN Rhône 2226 UVAFERM 43 LALVIN Rhône 2323 LALVIN Rhône 2056 S. bayanus LALVIN QA23 | 30 °C, 2 h, viable cells | YPD medium SGM medium | 2 µg/mL | YPG medium: 11–45% SGM medium: 1–35% | [78] |
| Bifidobacterium bifidum CECT 870T | 37 °C, 24 h | MRS Medium | 0.6 µg/mL | pH 3.5/pH 6.5 9.1/3.1% | [56] |
| Bf. breve CECT 4839T | 4.2/1.4% | ||||
| Lactobacillus bulgaricus CECT 4005 | 16/1.6% | ||||
| Lb. casei CECT 4040 | 5.3/3.5% | ||||
| Lb. casei CECT 4045 | 6.2/2.2% | ||||
| Lb. johnsonii CECT 289 | 16.3/4% | ||||
| Lb. paracasei CECT 4022 | 9.4/3.3% | ||||
| Lb. plantarum CECT 220 | 2.1/7% | ||||
| Lb. plantarum CECT 221 | 1.1/4.8% | ||||
| Lb. plantarum CECT 222 | 4.6/5.4% | ||||
| Lb. plantarum CECT 223 | 1.2/1.2% | ||||
| Lb. plantarum CECT 748 | 3/5.6% | ||||
| Lb. plantarum CECT 749 | 5.3/1.7% | ||||
| Lb. rhamnosus CECT 278T | 5.6/3.3% | ||||
| Lb. rhamnosus CECT 288 | 5.1/2.1% | ||||
| Lb. salivarius CECT 4062 | 16.1/4.4% | ||||
| Leuconostoc mesenteroides CECT 215 | -/- |
| Mechanism | Description | Reference |
|---|---|---|
| Increase in intestinal permeability | Loss of tight junction integrity facilitates OTA translocation | [145,146,147,149] |
| Decrease in SCFA production | Depletion of butyrate producers impairs barrier function and immune control | [103,139,158] |
| Increase in inflammation | Expansion of Proteobacteria, ↑ IL-6, TNF-α, leads to tissue sensitization | [150,151,152,153] |
| Decrease in detoxication enzymes | Disrupted signaling to CYP450s (e.g., via PXR) lowers OTA metabolism | [154,155,159] |
| Decrease in transporter activity | Decreased P-gp/MRP2 reduces OTA excretion | [156,157] |
| Increase in oxidative stress | Lower antioxidant capacity in dysbiotic microbiota increases damage | [159,160] |
| Microbiotype | Dominant Genus | Functional Features | Possible Effect on OTA Toxicity | Level of Evidence | References |
|---|---|---|---|---|---|
| Bacteroides | Bacteroides | Low SCFA, bile acid metabolism | Possibly higher risk | Moderate (indirect) | [167,168,169] |
| Prevotella | Prevotella | High SCFA, fiber fermentation | Possibly protective | Moderate (indirect) | [161,167,168] |
| Dysbiosis/increse of Proteobacteria level | Proteobacteria | Inflammation, decrease in detox enzymes | Higher risk | Strong (preclinical) | [150,151,153] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Więckowska, M.; Szelenberger, R.; Poplawski, T.; Bijak, M.; Gorniak, L.; Stela, M.; Cichon, N. Gut as a Target of Ochratoxin A: Toxicological Insights and the Role of Microbiota. Int. J. Mol. Sci. 2025, 26, 9438. https://doi.org/10.3390/ijms26199438
Więckowska M, Szelenberger R, Poplawski T, Bijak M, Gorniak L, Stela M, Cichon N. Gut as a Target of Ochratoxin A: Toxicological Insights and the Role of Microbiota. International Journal of Molecular Sciences. 2025; 26(19):9438. https://doi.org/10.3390/ijms26199438
Chicago/Turabian StyleWięckowska, Magdalena, Rafał Szelenberger, Tomasz Poplawski, Michal Bijak, Leslaw Gorniak, Maksymilian Stela, and Natalia Cichon. 2025. "Gut as a Target of Ochratoxin A: Toxicological Insights and the Role of Microbiota" International Journal of Molecular Sciences 26, no. 19: 9438. https://doi.org/10.3390/ijms26199438
APA StyleWięckowska, M., Szelenberger, R., Poplawski, T., Bijak, M., Gorniak, L., Stela, M., & Cichon, N. (2025). Gut as a Target of Ochratoxin A: Toxicological Insights and the Role of Microbiota. International Journal of Molecular Sciences, 26(19), 9438. https://doi.org/10.3390/ijms26199438






