Codon Composition in Human Oocytes Reveals Age-Associated Defects in mRNA Decay
Abstract
1. Introduction
2. Results
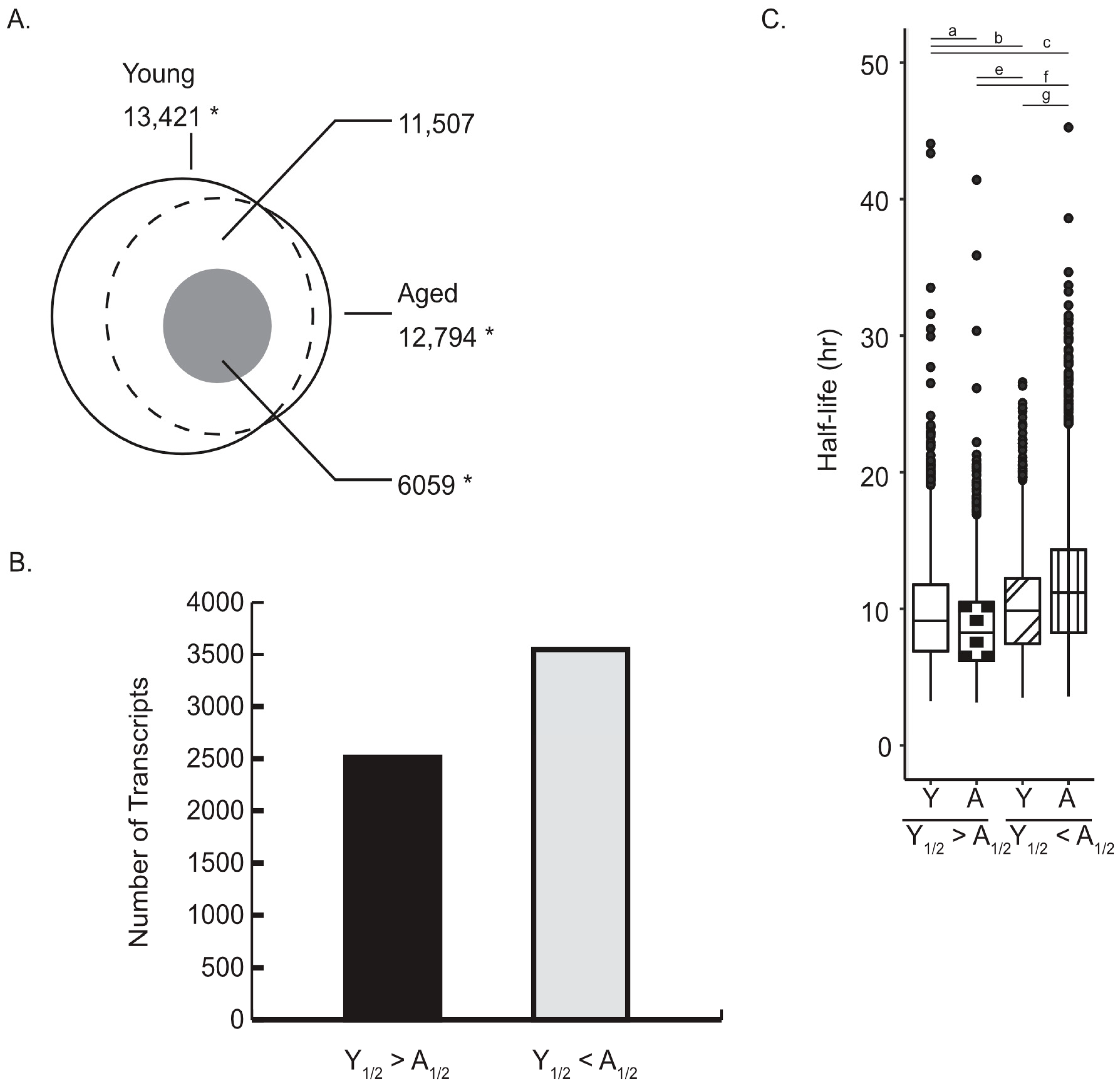
2.1. Age-Associated Changes in mRNA Half-Life
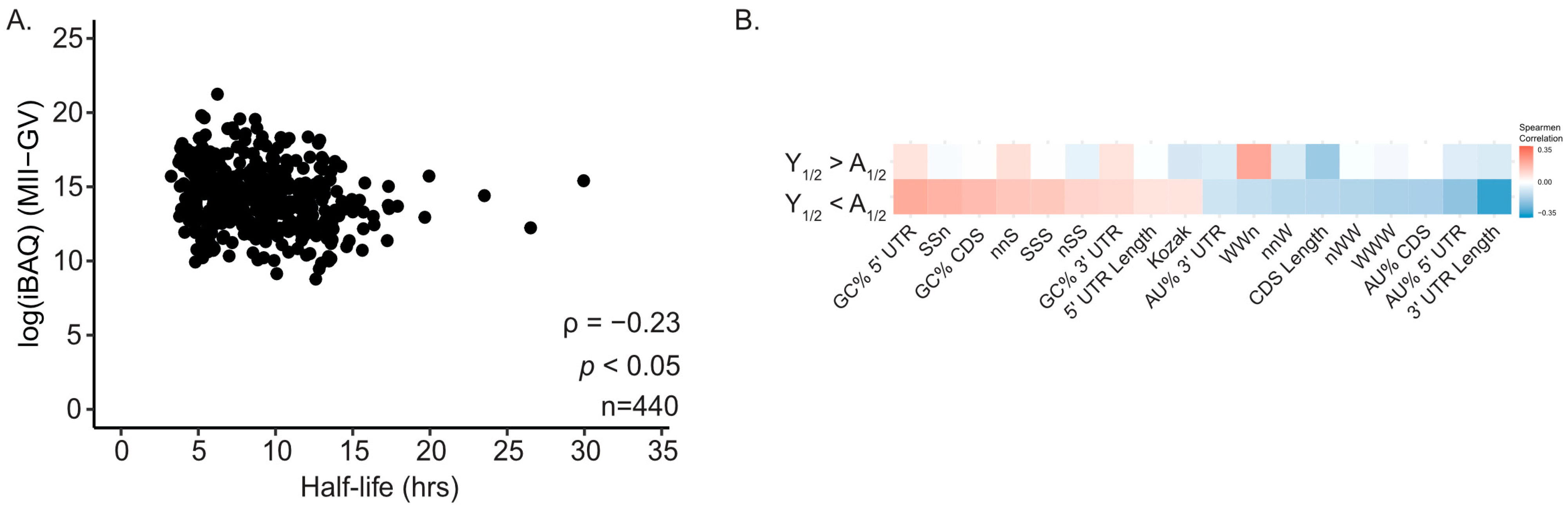
2.2. GC Content of mRNA Positively Correlates with Half-Life During Reproductive Aging
2.3. GC Content Reveals a Difference Between mRNA Half-Life and Protein Abundance
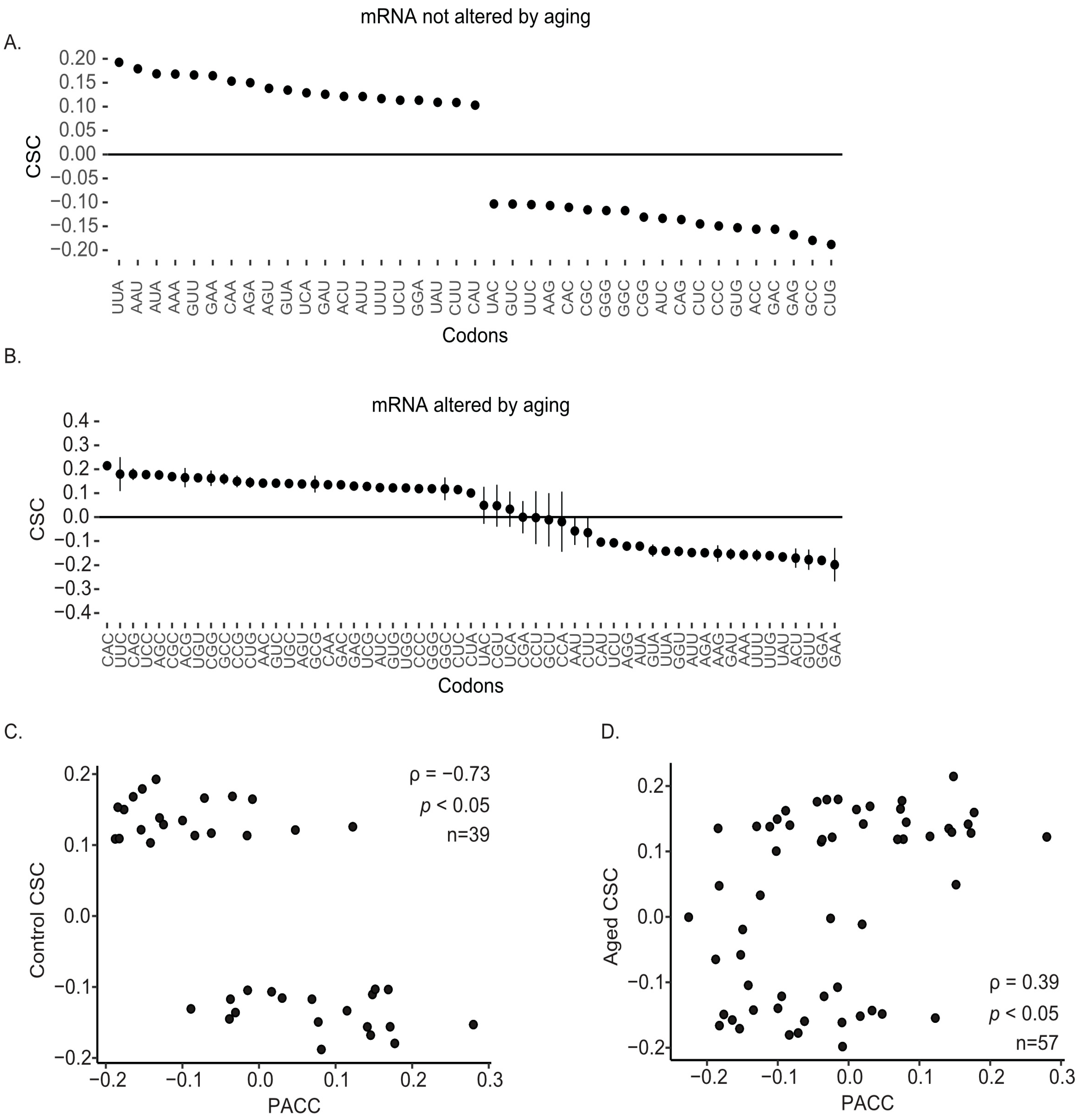
2.4. Age-Associated Changes in Codon Optimality
3. Discussion
4. Materials and Methods
4.1. Alignment and Half-Life Calculation
4.2. mRNA Feature, CSC, and Pathway Analysis
4.3. Mass Spectrometry Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ART | Assisted Reproductive Technology |
| CDS | Coding Sequence |
| COMD | Codon-Optimality-Mediated mRNA Decay |
| CSC | Codon Stability Coefficient |
| FACS | Fluorescence-Activated Cell Sorting |
| FDR | False Discovery Rate |
| GC | Guanine-Cytosine |
| GV | Germinal Vesicle |
| hCG | Human Chorionic Gonadotropin |
| HR | Hour |
| iBAQ | intensity-Based Absolute Quantification |
| IVF | In Vitro Fertilization |
| IVM | In Vitro Maturation |
| KS test | Kolmogorov-Smirnov test |
| MII | Metaphase II |
| MS | Mass Spectrometry |
| PACC | Protein Abundance Codon Correlation |
| Poly(A) | Polyadenylated |
| ppm | Parts Per Million |
| RNA | Ribonucleic Acid |
| RNA-seq | RNA Sequencing |
| TPM | Transcripts Per Million |
| UTR | Untranslated Region |
| ZGA | Zygotic Genome Activation |
References
- Navot, D.; Bergh, R.A.; Williams, M.A.; Garrisi, G.J.; Guzman, I.; Sandler, B.; Grunfeld, L. Poor oocyte quality rather than implantation failure as a cause of age-related decline in female fertility. Lancet 1991, 337, 1375–1377. [Google Scholar] [CrossRef]
- Sandalinas, M.; Márquez, C.; Munné, S. Spectral karyotyping of fresh, non-inseminated oocytes. Mol. Hum. Reprod. 2002, 8, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Mai, C.T.; Kucik, J.E.; Isenburg, J.; Feldkamp, M.L.; Marengo, L.K.; Bugenske, E.M.; Thorpe, P.G.; Jackson, J.M.; Correa, A.; Rickard, R.; et al. Selected birth defects data from population-based birth defects surveillance programs in the United States, 2006 to 2010: Featuring trisomy conditions. Birth Defects Res. A Clin. Mol. Teratol. 2013, 97, 709–725. [Google Scholar] [CrossRef] [PubMed]
- Treff, N.R.; Krisher, R.L.; Tao, X.; Garnsey, H.; Bohrer, C.; Silva, E.; Landis, J.; Taylor, D.; Scott, R.T.; Woodruff, T.K.; et al. Next Generation Sequencing-Based Comprehensive Chromosome Screening in Mouse Polar Bodies, Oocytes, and Embryos. Biol. Reprod. 2016, 94, 76. [Google Scholar] [CrossRef]
- Duncan, F.E.; Jasti, S.; Paulson, A.; Kelsh, J.M.; Fegley, B.; Gerton, J.L. Age-associated dysregulation of protein metabolism in the mammalian oocyte. Aging Cell 2017, 16, 1381–1393. [Google Scholar] [CrossRef] [PubMed]
- Del Llano, E.; Masek, T.; Gahurova, L.; Pospisek, M.; Koncicka, M.; Jindrova, A.; Jansova, D.; Iyyappan, R.; Roucova, K.; Bruce, A.W.; et al. Age-related differences in the translational landscape of mammalian oocytes. Aging Cell 2020, 19, 1547. [Google Scholar] [CrossRef]
- Wu, Y.-W.; Li, S.; Zheng, W.; Li, Y.-C.; Chen, L.; Zhou, Y.; Deng, Z.-Q.; Lin, G.; Fan, H.-Y.; Sha, Q.-Q. Dynamic mRNA degradome analyses indicate a role of histone H3K4 trimethylation in association with meiosis-coupled mRNA decay in oocyte aging. Nat. Commun. 2022, 13, 3191. [Google Scholar] [CrossRef]
- Liu, Y.; Tao, W.; Wu, S.; Zhang, Y.; Nie, H.; Hou, Z.; Zhang, J.; Yang, Z.; Chen, Z.-J.; Wang, J.; et al. Maternal mRNA deadenylation is defective in in vitro matured mouse and human oocytes. Nat. Commun. 2024, 15, 5550. [Google Scholar] [CrossRef]
- Soeda, S.; Oyama, M.; Kozuka-Hata, H.; Yamamoto, T. The CCR4-NOT complex suppresses untimely translational activation of maternal mRNAs. Development 2023, 150, dev201773. [Google Scholar] [CrossRef]
- Piko, L.; Clegg, K.B. Quantitative changes in total RNA, total poly(A), and ribosomes in early mouse embryos. Dev. Biol. 1982, 89, 362–378. [Google Scholar] [CrossRef]
- Bachvarova, R.; De Leon, V.; Johnson, A.; Kaplan, G.; Paynton, B.V. Changes in total RNA, polyadenylated RNA, and actin mRNA during meiotic maturation of mouse oocytes. Dev. Biol. 1985, 108, 325–331. [Google Scholar] [CrossRef]
- De La Fuente, R.; Eppig, J.J. Transcriptional activity of the mouse oocyte genome: Companion granulosa cells modulate transcription and chromatin remodeling. Dev. Biol. 2001, 229, 224–236. [Google Scholar] [CrossRef]
- Yu, C.; Ji, S.-Y.; Sha, Q.-Q.; Dang, Y.; Zhou, J.-J.; Zhang, Y.-L.; Liu, Y.; Wang, Z.-W.; Hu, B.; Sun, Q.-Y.; et al. BTG4 is a meiotic cell cycle-coupled maternal-zygotic-transition licensing factor in oocytes. Nat. Struct. Mol. Biol. 2016, 23, 387–394. [Google Scholar] [CrossRef]
- Sha, Q.; Yu, J.; Guo, J.; Dai, X.; Jiang, J.; Zhang, Y.; Yu, C.; Ji, S.; Jiang, Y.; Zhang, S.; et al. CNOT6L couples the selective degradation of maternal transcripts to meiotic cell cycle progression in mouse oocyte. EMBO J. 2018, 37, e99333. [Google Scholar] [CrossRef]
- Medina-Muñoz, S.G.; Kushawah, G.; Castellano, L.A.; Diez, M.; DeVore, M.L.; Salazar, M.J.B.; Bazzini, A.A. Crosstalk between codon optimality and cis-regulatory elements dictates mRNA stability. Genome Biol. 2021, 22, 14. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Cheng, Y.; Zhu, Y.; Xu, C.; Li, Q.; Xing, X.; Li, W.; Zou, J.; Meng, L.; Azhar, M.; et al. Maternal NAT10 orchestrates oocyte meiotic cell-cycle progression and maturation in mice. Nat. Commun. 2023, 14, 3729. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.J.; Zavolan, M. Alternative cleavage and polyadenylation in health and disease. Nat. Rev. Genet. 2019, 20, 599–614. [Google Scholar] [CrossRef]
- Mayya, V.K.; Duchaine, T.F. Ciphers and Executioners: How 3′-Untranslated Regions Determine the Fate of Messenger RNAs. Front. Genet. 2019, 10, 6. [Google Scholar] [CrossRef] [PubMed]
- Schall, P.Z.; Latham, K.E. Predictive modeling of oocyte maternal mRNA features for five mammalian species reveals potential shared and species-restricted regulators during maturation. Physiol. Genom. 2024, 56, 9–31. [Google Scholar] [CrossRef]
- Harvey, R.F.; Smith, T.S.; Mulroney, T.; Queiroz, R.M.; Pizzinga, M.; Dezi, V.; Villenueva, E.; Ramakrishna, M.; Lilley, K.S.; Willis, A.E. Trans-acting translational regulatory RNA binding proteins. Wiley Interdiscip. Rev. RNA 2018, 9, e1465. [Google Scholar] [CrossRef]
- Mendez, R.; Richter, J.D. Translational control by CPEB: A means to the end. Nat. Rev. Mol. Cell Biol. 2001, 2, 521–529. [Google Scholar] [CrossRef]
- Roy, B.; Jacobson, A. The intimate relationships of mRNA decay and translation. Trends Genet. 2013, 29, 691–699. [Google Scholar] [CrossRef]
- Shan, L.-Y.; Tian, Y.; Liu, W.-X.; Fan, H.-T.; Li, F.-G.; Liu, W.-J.; Li, A.; Shen, W.; Sun, Q.-Y.; Liu, Y.-B.; et al. LSM14B controls oocyte mRNA storage and stability to ensure female fertility. Cell. Mol. Life Sci. 2023, 80, 247. [Google Scholar] [CrossRef]
- Guo, J.; Zhao, H.; Zhang, J.; Lv, X.; Zhang, S.; Su, R.; Zheng, W.; Dai, J.; Meng, F.; Gong, F.; et al. Selective translation of maternal mRNA by eIF4E1B controls oocyte to embryo transition. Adv. Sci. 2023, 10, e2205500. [Google Scholar] [CrossRef]
- Presnyak, V.; Alhusaini, N.; Chen, Y.-H.; Martin, S.; Morris, N.; Kline, N.; Olson, S.; Weinberg, D.; Baker, K.E.; Graveley, B.R.; et al. Codon optimality is a major determinant of mRNA stability. Cell 2015, 160, 1111–1124. [Google Scholar] [CrossRef]
- Bazzini, A.A.; Del Viso, F.; Moreno-Mateos, M.A.; Johnstone, T.G.; Vejnar, C.E.; Qin, Y.; Yao, J.; Khokha, M.K.; Giraldez, A.J. Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J. 2016, 35, 2087–2103. [Google Scholar] [CrossRef]
- Carneiro, R.L.; Requião, R.D.; Rossetto, S.; Domitrovic, T.; Palhano, F.L. Codon stabilization coefficient as a metric to gain insights into mRNA stability and codon bias and their relationships with translation. Nucleic Acids Res. 2019, 47, 2216–2228. [Google Scholar] [CrossRef]
- Horstick, E.J.; Jordan, D.C.; Bergeron, S.A.; Tabor, K.M.; Serpe, M.; Feldman, B.; Burgess, H.A. Increased functional protein expression using nucleotide sequence features enriched in highly expressed genes in zebrafish. Nucleic Acids Res. 2015, 43, e48. [Google Scholar] [CrossRef] [PubMed]
- Pop, C.; Rouskin, S.; Ingolia, N.T.; Han, L.; Phizicky, E.M.; Weissman, J.S.; Koller, D. Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol. Syst. Biol. 2014, 10, 770. [Google Scholar] [CrossRef] [PubMed]
- Bergman, S.; Tuller, T. Widespread non-modular overlapping codes in the coding regions. Phys Biol. 2020, 17, 031002. [Google Scholar] [CrossRef] [PubMed]
- Mishima, Y.; Tomari, Y. Codon Usage and 3′ UTR Length Determine Maternal mRNA Stability in Zebrafish. Mol. Cell 2016, 61, 874–885. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, A.; Chen, Y.-H.; Martin, S.; Alhusaini, N.; Green, R.; Coller, J. The DEAD-Box Protein Dhh1p Couples mRNA Decay and Translation by Monitoring Codon Optimality. Cell 2016, 167, 122–132.e9. [Google Scholar] [CrossRef]
- Webster, M.W.; Chen, Y.-H.; Stowell, J.A.; Alhusaini, N.; Sweet, T.; Graveley, B.R.; Coller, J.; Passmore, L.A. mRNA Deadenylation Is Coupled to Translation Rates by the Differential Activities of Ccr4-Not Nucleases. Mol. Cell 2018, 70, 1089–1100.e8. [Google Scholar] [CrossRef]
- Bae, H.; Coller, J. Codon optimality-mediated mRNA degradation: Linking translational elongation to mRNA stability. Mol. Cell 2022, 82, 1467–1476. [Google Scholar] [CrossRef]
- Barrington, C.L.; Galindo, G.; Koch, A.L.; Horton, E.R.; Morrison, E.J.; Tisa, S.; Stasevich, T.J.; Rissland, O.S. Synonymous codon usage regulates translation initiation. Cell Rep. 2023, 42, 113413. [Google Scholar] [CrossRef] [PubMed]
- Nedialkova, D.D.; Leidel, S.A. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell 2015, 161, 1606–1618. [Google Scholar] [CrossRef] [PubMed]
- Boël, G.; Letso, R.; Neely, H.; Price, W.N.; Wong, K.-H.; Su, M.; Luff, J.D.; Valecha, M.; Everett, J.K.; Acton, T.B.; et al. Codon influence on protein expression in E. coli correlates with mRNA levels. Nature 2016, 529, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Medina, S.G.; Kushawah, G.; DeVore, M.L.; ACastellano, L.; Hand, J.M.; Wright, M.; Bazzini, A.A. Translation affects mRNA stability in a codon-dependent manner in human cells. eLife 2019, 8, e45396. [Google Scholar] [CrossRef]
- Narula, A.; Ellis, J.; Taliaferro, J.M.; Rissland, O.S. Coding regions affect mRNA stability in human cells. RNA 2019, 25, 1751–1764. [Google Scholar] [CrossRef]
- Brachova, P.; Alvarez, N.S.; Hong, X.; Gunewardena, S.; Vincent, A.K.; ELatham, K.; Christenson, L.K. Inosine RNA modifications are enriched at the codon wobble position in mouse oocytes and eggs. Biol. Reprod. 2019, 101, 938–949. [Google Scholar] [CrossRef]
- Brachova, P.; Alvarez, N.S.; Christenson, L.K. Loss of Cnot6l Impairs Inosine RNA Modifications in Mouse Oocytes. Int. J. Mol. Sci. 2021, 22, 1191. [Google Scholar] [CrossRef]
- Xiong, Z.; Xu, K.; Lin, Z.; Kong, F.; Wang, Q.; Quan, Y.; Sha, Q.-Q.; Li, F.; Zou, Z.; Liu, L.; et al. Ultrasensitive Ribo-seq reveals translational landscapes during mammalian oocyte-to-embryo transition and pre-implantation development. Nat. Cell Biol. 2022, 24, 968–980. [Google Scholar] [CrossRef]
- Zou, Z.; Zhang, C.; Wang, Q.; Hou, Z.; Xiong, Z.; Kong, F.; Wang, Q.; Song, J.; Liu, B.; Liu, B.; et al. Translatome and transcriptome co-profiling reveals a role of TPRXs in human zygotic genome activation. Science 2022, 378, abo7923. [Google Scholar] [CrossRef]
- Yang, J.; Bu, J.; Liu, B.; Liu, Y.; Zhang, Z.; Li, Z.; Lu, F.; Zhu, B.; Li, Y. MARTRE family proteins negatively regulate CCR4-NOT activity to protect poly(A) tail length and promote translation of maternal mRNA. Nat. Commun. 2025, 16, 248. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.M.; Silva, E.; Chitwood, J.L.; Schoolcraft, W.B.; Krisher, R.L.; Ross, P.J. Differing molecular response of young and advanced maternal age human oocytes to IVM. Hum. Reprod. 2017, 32, 2199–2208. [Google Scholar] [CrossRef] [PubMed]
- Friedel, C.C.; Dölken, L.; Ruzsics, Z.; Koszinowski, U.H.; Zimmer, R. Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res. 2009, 37, e115. [Google Scholar] [CrossRef] [PubMed]
- Combelles, C.M.H.; Cekleniak, N.A.; Racowsky, C.; Albertini, D.F. Assessment of nuclear and cytoplasmic maturation in in-vitro matured human oocytes. Hum. Reprod. 2002, 17, 1006–1016. [Google Scholar] [CrossRef]
- Escrich, L.; Grau, N.; de los Santos, M.J.; Romero, J.-L.; Pellicer, A.; Escribá, M.-J. The dynamics of in vitro maturation of germinal vesicle oocytes. Fertil. Steril. 2012, 98, 1147–1151. [Google Scholar] [CrossRef]
- Virant-Klun, I.; Leicht, S.; Hughes, C.; Krijgsveld, J. Identification of Maturation-Specific Proteins by Single-Cell Proteomics of Human Oocytes. Mol. Cell. Proteom. 2016, 15, 2616–2627. [Google Scholar] [CrossRef]
- Galatidou, S.; Petelski, A.A.; Pujol, A.; Lattes, K.; Latorraca, L.B.; Fair, T.; Popovic, M.; Vassena, R.; Slavov, N.; Barragán, M. Single-cell proteomics reveals decreased abundance of proteostasis and meiosis proteins in advanced maternal age oocytes. Mol. Hum. Reprod. 2024, 30, gaae023. [Google Scholar] [CrossRef]
- Courel, M.; Clément, Y.; Bossevain, C.; Foretek, D.; Cruchez, O.V.; Yi, Z.; Bénard, M.; Benassy, M.-N.; Kress, M.; Vindry, C.; et al. GC content shapes mRNA storage and decay in human cells. eLife 2019, 8, e49708. [Google Scholar] [CrossRef]
- Svoboda, P.; Franke, V.; Schultz, R.M. Sculpting the Transcriptome During the Oocyte-to-Embryo Transition in Mouse. Curr. Top. Dev. Biol. 2015, 113, 305–349. [Google Scholar]
- Sha, Q.-Q.; Zhang, J.; Fan, H.-Y. A story of birth and death: mRNA translation and clearance at the onset of Maternal-to-Zygotic transition in mammals. Biol. Reprod. 2019, 101, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner; Lawrence Berkeley National Lab. (LBNL): Berkeley, CA, USA, 2014. Available online: https://www.osti.gov/biblio/1241166 (accessed on 18 February 2020).
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. 2013. Available online: http://www.R-project.org/ (accessed on 21 March 2020).
- Sorenson, R.S.; Deshotel, M.J.; Johnson, K.; Adler, F.R.; Sieburth, L.E. Arabidopsis mRNA decay landscape arises from specialized RNA decay substrates, decapping-mediated feedback, and redundancy. Proc. Natl. Acad. Sci. USA 2018, 115, E1485–E1494. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Noderer, W.L.; Flockhart, R.J.; Bhaduri, A.; de Arce, A.J.D.; Zhang, J.; AKhavari, P.; Wang, C.L. Quantitative analysis of mam-malian translation initiation sites by FACS-seq. Mol. Syst. Biol. 2014, 10, 748. [Google Scholar]
- Tremblay, B.J.-M. Universalmotif: An R Package for Biological Motif Analysis. J. Open Source Softw. 2024, 9, 7012. [Google Scholar]
- Yu, G.; He, Q.-Y. ReactomePA: An R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 2016, 12, 477–479. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef]
- Schwanhäusser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brachova, P.; Christenson, L.K.; Alvarez, N.S. Codon Composition in Human Oocytes Reveals Age-Associated Defects in mRNA Decay. Int. J. Mol. Sci. 2025, 26, 9395. https://doi.org/10.3390/ijms26199395
Brachova P, Christenson LK, Alvarez NS. Codon Composition in Human Oocytes Reveals Age-Associated Defects in mRNA Decay. International Journal of Molecular Sciences. 2025; 26(19):9395. https://doi.org/10.3390/ijms26199395
Chicago/Turabian StyleBrachova, Pavla, Lane K. Christenson, and Nehemiah S. Alvarez. 2025. "Codon Composition in Human Oocytes Reveals Age-Associated Defects in mRNA Decay" International Journal of Molecular Sciences 26, no. 19: 9395. https://doi.org/10.3390/ijms26199395
APA StyleBrachova, P., Christenson, L. K., & Alvarez, N. S. (2025). Codon Composition in Human Oocytes Reveals Age-Associated Defects in mRNA Decay. International Journal of Molecular Sciences, 26(19), 9395. https://doi.org/10.3390/ijms26199395






