Silver(I)-NHC Complexes as Dual-Action Agents Against Pathogenic Acanthamoeba Trophozoites: Anti-Amoebic and Anti-Adhesion Activities
Abstract
1. Introduction
2. Results and Discussion
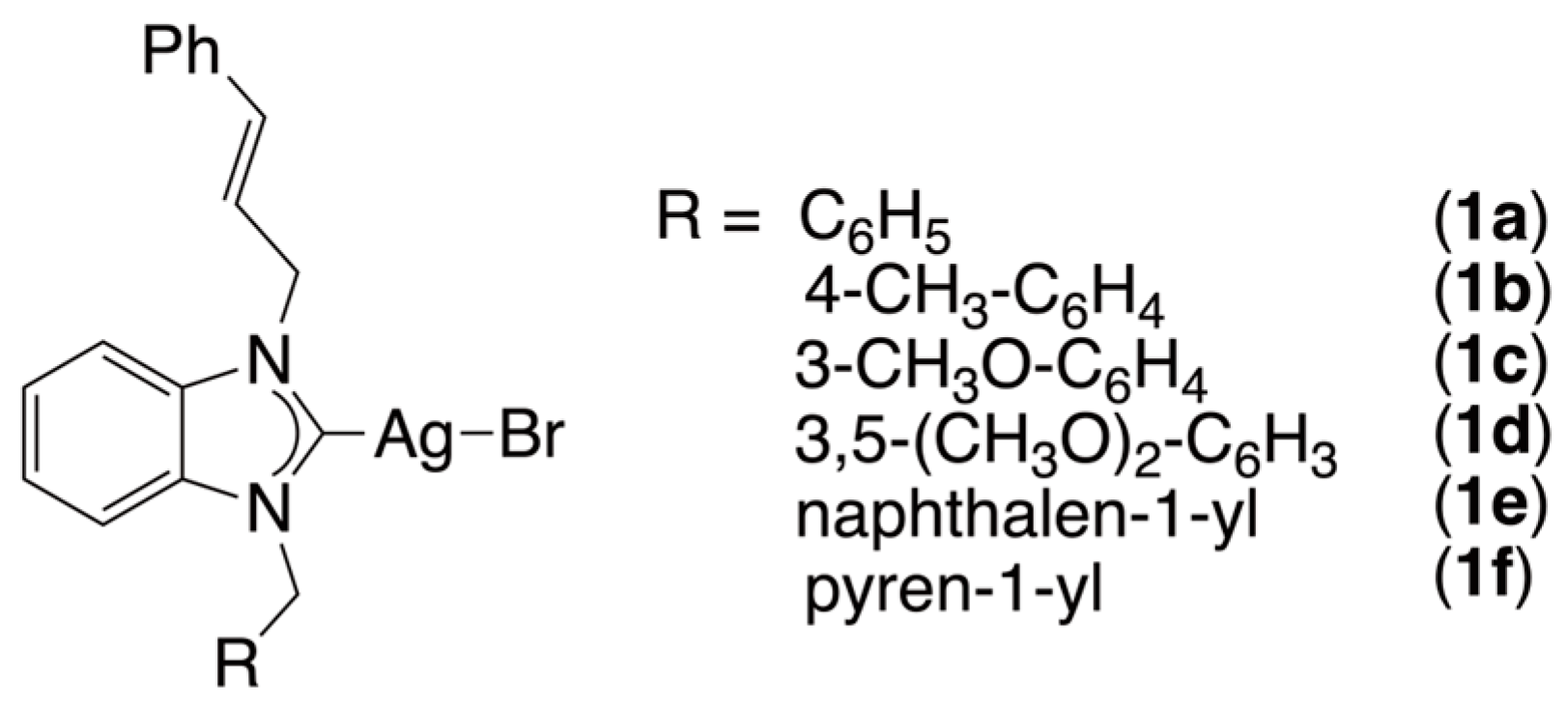
2.1. Synthesis and Characterization of the Silver(I) Complexes
2.2. In Vitro Effects on Acanthamoeba
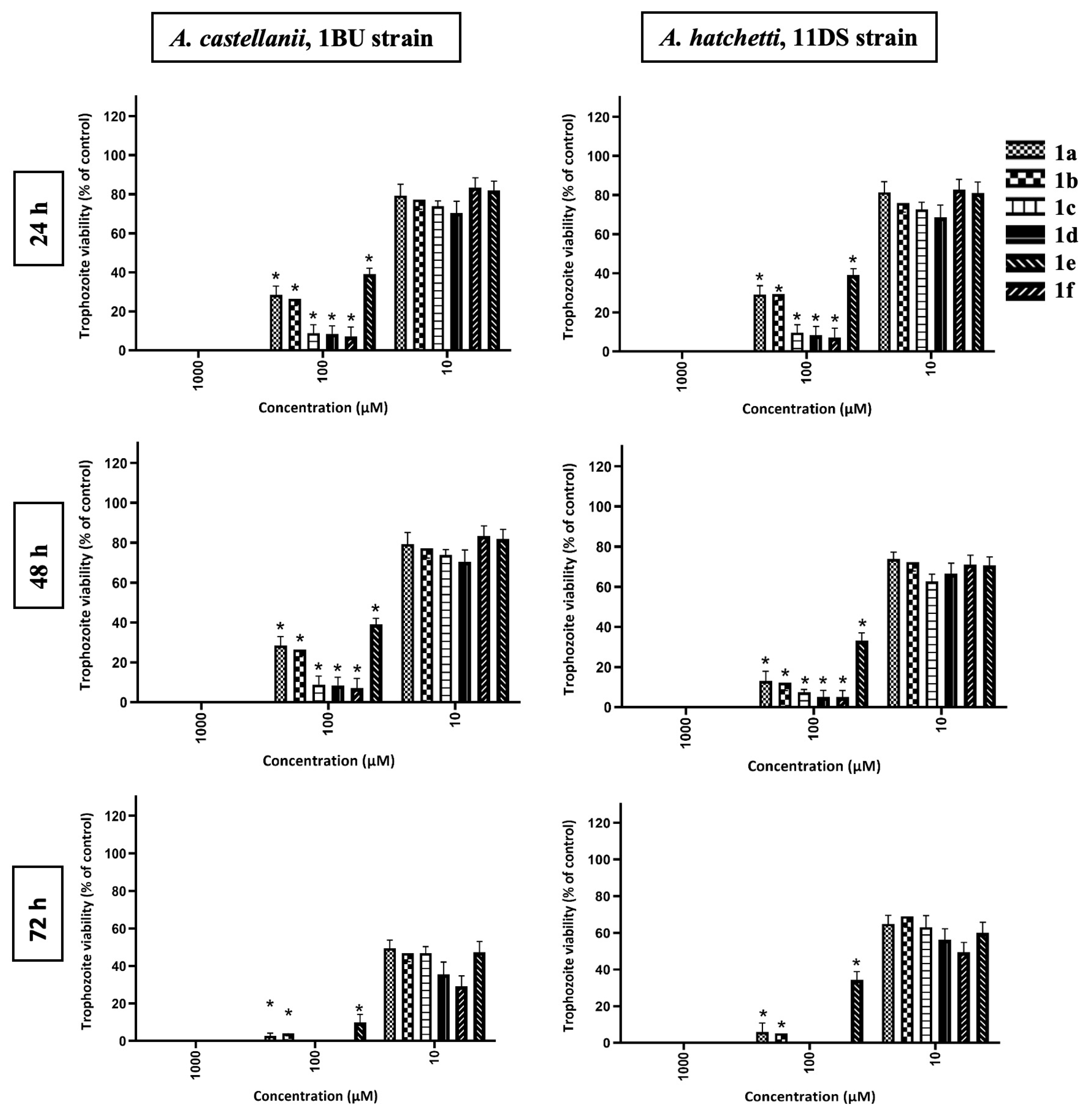
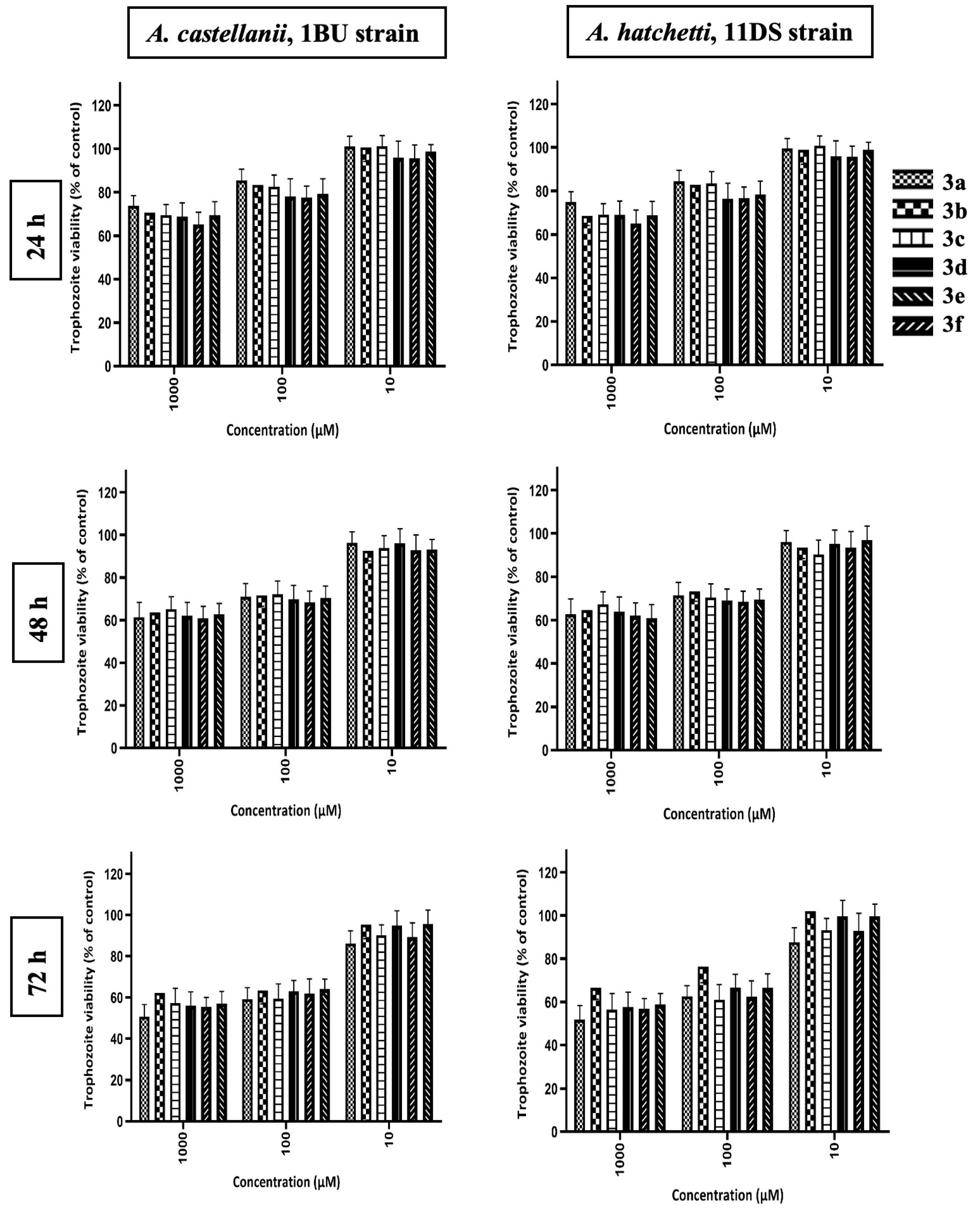
2.2.1. Effect of Benzimidazolium Salts and Silver(I) Complexes on Acanthamoeba Trophozoites
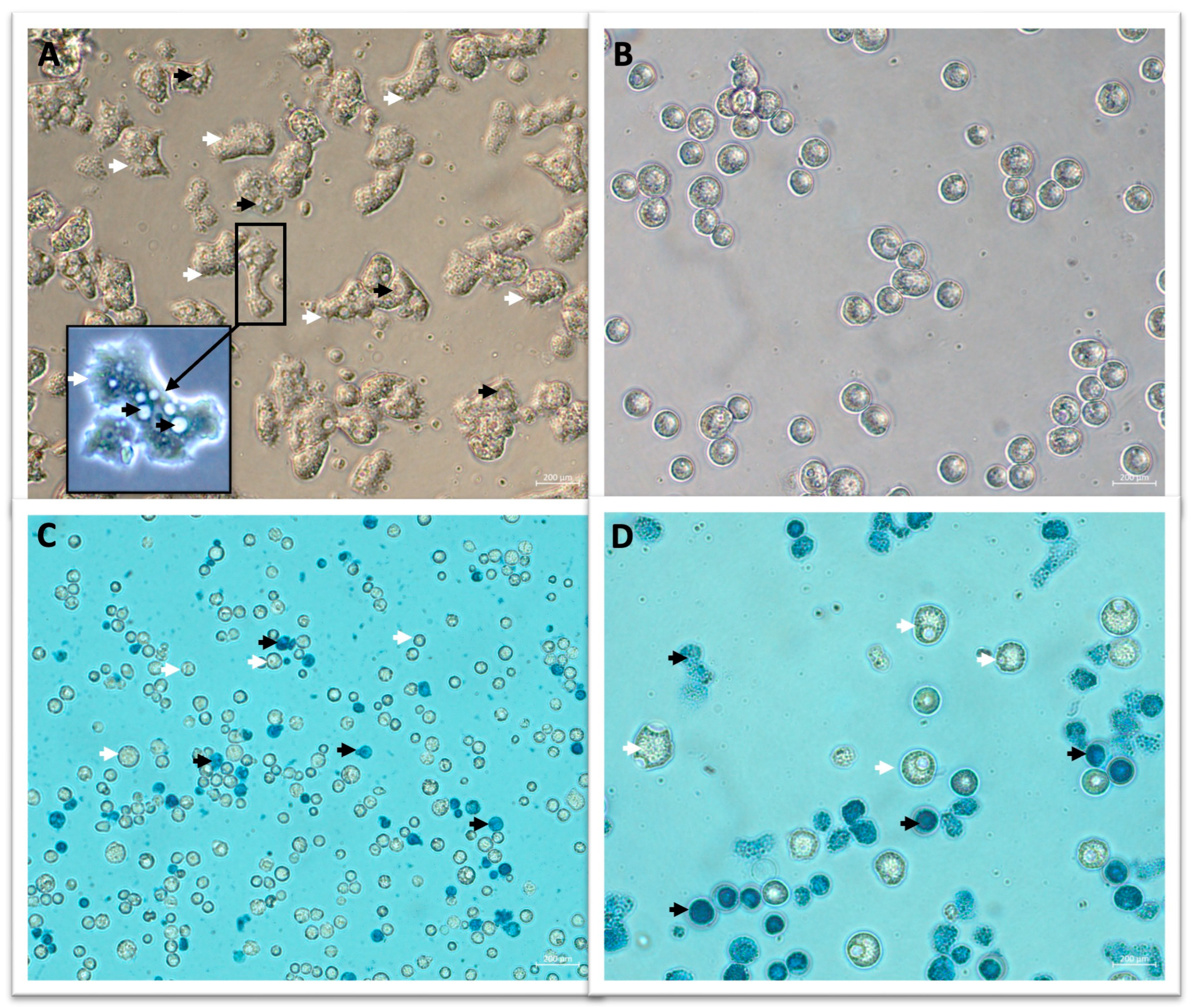
2.2.2. Effect of Silver(I)-NHC Complexes on Trophozoite Morphology and Adhesion Capacity
2.3. Molecular Docking
2.4. Consideration of Effectiveness Against Bacteria A. castellanii
3. Materials and Methods
3.1. General
3.2. General Procedure for the Synthesis of Benzimidazolium Salts
3.2.1. 1-Benzyl-3-cinnamyl-benzimidazolium Bromide (3a)
3.2.2. 1-(4-Methylbenzyl)-3-cinnamyl-benzimidazolium Bromide (3b)
3.2.3. 1-(3-Methoxylbenzyl)-3-cinnamyl-benzimidazolium Bromide (3c)
3.2.4. 1-(3,5-Dimethoxy-benzyl)-3-cinnamyl-benzimidazolium Bromide (3d)
3.2.5. 1-(Naphthalen-1-ylmethyl)-3-cinnamyl-benzimidazolium Bromide (3e)
3.2.6. 1-(Pyren-1-ylmethyl)-3-cinnamyl-benzimidazolium Bromide (3f)
3.3. General Procedure for the Synthesis of Silver Complexes
3.3.1. Bromo(1-benzyl-3-cinnamyl-benzimidazol-2-ylidene)silver (I) (1a)
3.3.2. Bromo[1-(4-methylbenzyl)-3-cinnamyl-benzimidazol-2-yliden]silver(I) (1b)
3.3.3. Bromo[1-(3-methoxylbenzyl)-3-cinnamyl-benzimidazol-2-yliden]silver(I) (1c)
3.3.4. Bromo[1-(3,5-dimethoxy-benzyl)-3-cinnamyl-benzimidazol-2-ylidene]silver(I) (1d)
3.3.5. Bromo[1-(naphthalen-1-ylmethyl)-3-cinnamyl-benzimidazol-2 ylidene]silver(I) (1e)
3.3.6. Bromo[1-(pyren-1-ylmethyl)-3-cinnamyl-benzimidazol-2-yliden]silver(I) (1f)
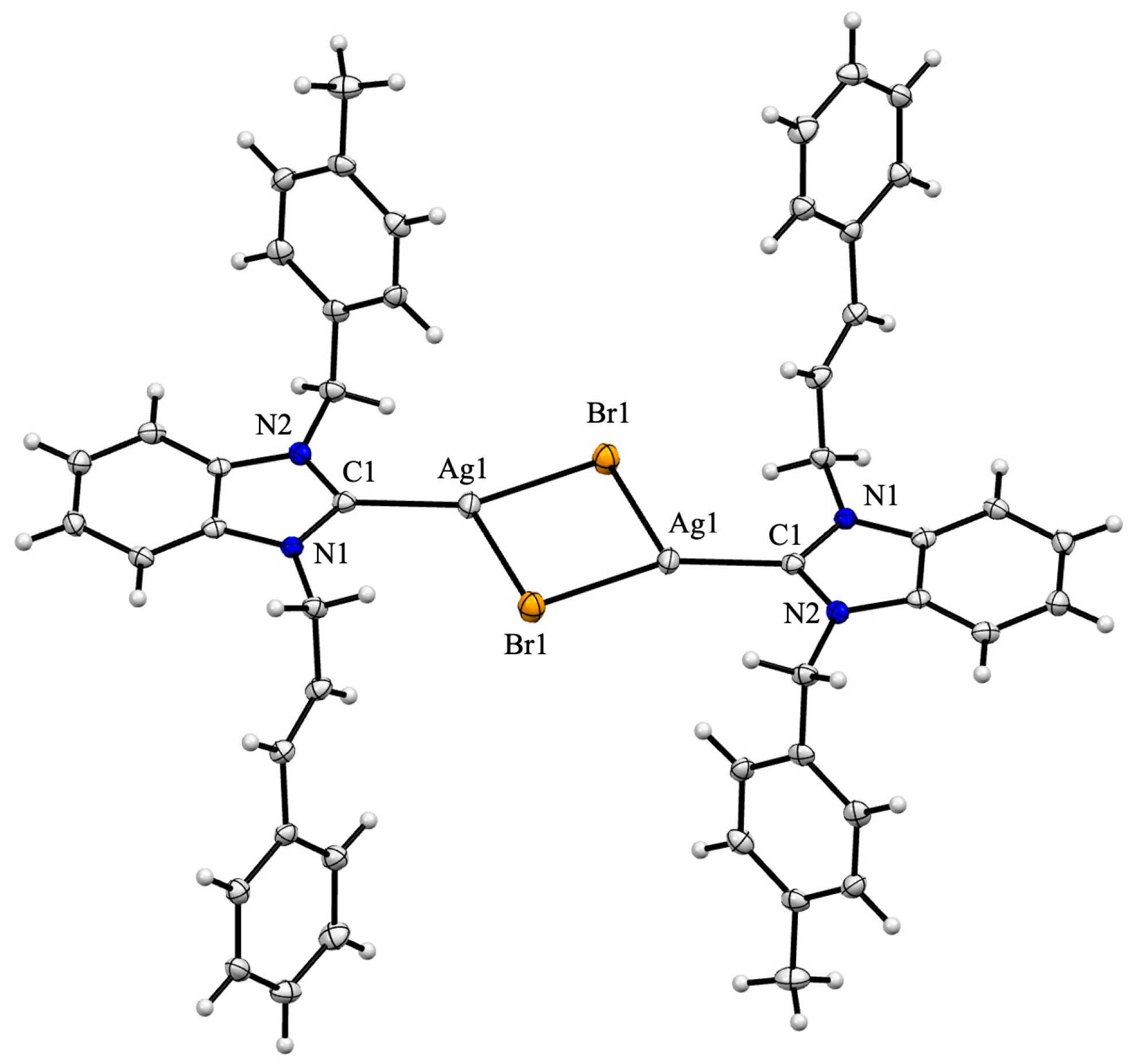
3.4. X-Ray Crystallography
3.5. In Vitro Effects on Acanthamoeba Species
3.5.1. Acanthamoeba Strains and Culture Conditions
3.5.2. Effect on Acanthamoeba Trophozoites
3.5.3. Microscopic Observation of Trophozoite Morphology
3.6. Statistical Analysis
3.7. Molecular Docking Method
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marciano-Cabral, F.; Cabral, G. Acanthamoeba spp. as agents of disease in humans. Clin. Microbiol. Rev. 2003, 16, 273–307. [Google Scholar] [CrossRef]
- Siddiqui, R.; Khan, N.A. Biology and pathogenesis of Acanthamoeba. Parasit. Vectors 2012, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Varacalli, G.; Di Zazzo, A.; Mori, T.; Dohlman, T.H.; Spelta, S.; Coassin, M.; Bonini, S. Challenges in Acanthamoeba Keratitis: A review. J. Clin. Med. 2021, 10, 942. [Google Scholar] [CrossRef]
- Bahrami, S.; Darvishi, M.; Zarei, M.; Sabaeian, M.; Henriquez, F.L. Sublethal exposure to plasma-activated water influences the morphological characteristics, phagocytic ability, and virulence of Acanthamoeba castellanii. Acta Parasitol. 2023, 68, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Iovieno, A.; Miller, D.; Ledee, D.R.; Alfonso, E.C. Cysticidal activity of antifungals against different genotypes of Acanthamoeba. Antimicrob. Agents Chemother. 2014, 58, 5626–5628. [Google Scholar] [CrossRef]
- Khan, N.A. Acanthamoeba: Biology and increasing importance in human health. FEMS Microbiol. Rev. 2006, 30, 564–595. [Google Scholar] [CrossRef]
- Khan, N.A.; Paget, T.A. Molecular tools for speciation and epidemiological studies of Acanthamoeba. Curr. Microbiol. 2002, 44, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Illingworth, C.D.; Cook, S.D. Acanthamoeba Keratitis. Surv. Ophthalmol. 1998, 42, 493–508. [Google Scholar] [CrossRef]
- Padhi, T.R.; Das, S.; Sharma, S.; Rath, S.; Rath, S.; Tripathy, D.; Panda, K.G.; Basu, S.; Besirli, C.G. Ocular parasitoses: A comprehensive review. Surv. Ophthalmol. 2017, 62, 161–189. [Google Scholar] [CrossRef]
- Jones, D.B.; Visvesvara, G.S.; Robinson, N.M. Acanthamoeba polyphaga keratitis and Acanthamoeba uveitis associated with fatal meningoencephalitis. Trans. Ophthalmol. Soc. UK 1975, 95, 221–232. [Google Scholar]
- Martinez, A.J.; Visvesvara, G.S. Free-living, Amphizoic and Opportunistic amebas. Brain Pathol. 1997, 7, 583–598. [Google Scholar] [CrossRef] [PubMed]
- Fanselow, N.; Sirajuddin, N.; Yin, X.-T.; Huang, A.J.W.; Stuart, P.M. Acanthamoeba keratitis, pathology, diagnosis and treatment. Pathogens 2021, 10, 323. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, X.; Wei, Z.; Cao, K.; Zhang, Z.; Liang, Q. The global epidemiology and clinical diagnosis of Acanthamoeba keratitis. J. Infect. Public Health 2023, 16, 841–852. [Google Scholar] [CrossRef]
- Petrillo, F.; Tortori, A.; Vallino, V.; Galdiero, M.; Fea, A.M.; De Sanctis, U.; Reibaldi, M. Understanding Acanthamoeba keratitis: An in-depth review of a sight-threatening eye infection. Microorganisms 2024, 12, 758. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, L.; Zhao, Y.; Ju, X.; Wang, L.; Jin, L.; Fine, R.D.; Li, M. Biological characteristics and pathogenicity of Acanthamoeba. Front. Microbiol. 2023, 14, 1147077. [Google Scholar] [CrossRef]
- Cabrera-Aguas, M.; Khoo, P.; Watson, S.L. Infectious keratitis: A review. Clin. Experiment Ophthalmol. 2022, 50, 543–562. [Google Scholar] [CrossRef]
- Gokhale, N.S. Medical management approach to infectious keratitis. Indian J. Ophthalmol. 2008, 56, 215–220. [Google Scholar] [CrossRef]
- Shareef, O.; Shareef, S.; Saeed, H.N. New frontiers in Acanthamoeba keratitis diagnosis and management. Biology 2023, 12, 1489. [Google Scholar] [CrossRef]
- Durand, M.L.; Barshak, M.B.; Chodosh, J. Infectious keratitis in 2021. JAMA 2021, 326, 1319–1320. [Google Scholar] [CrossRef]
- Goh, J.W.; Harrison, R.; Hau, S.; Alexander, C.L.; Tole, D.M.; Avadhanam, V.S. Comparison of in vivo confocal microscopy, PCR and culture of corneal scrapes in the diagnosis of Acanthamoeba keratitis. Cornea 2018, 37, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Hopkinson, M.N.; Richter, C.; Schedler, M.; Glorius, F. An overview of N-heterocyclic carbenes. Nature 2014, 510, 485–496. [Google Scholar] [CrossRef]
- Öfele, K. 1, 3-Dimethyl-4-imidazolinyliden-(2)-pentacarbonylchrom ein neuer Übergangsmetall-carben-komplex. J. Organomet. Chem. 1968, 12, P42–P43. [Google Scholar]
- Wanzlick, H.W.; Schönherr, H.J. Direct synthesis of a mercury salt-carbene complex. Angew. Chem. Int. Ed. 1968, 7, 141–142. [Google Scholar] [CrossRef]
- Arduengo, A.J., III; Harlow, R.L.; Kline, M. A stable crystalline carbene. J. Am. Chem. Soc. 1991, 113, 361–363. [Google Scholar] [CrossRef]
- Kühl, O. The chemistry of functionalised N-heterocyclic carbenes. Chem. Soc. Rev. 2007, 36, 592–607. [Google Scholar] [CrossRef] [PubMed]
- Normand, A.T.; Cavell, K.J. Donor-functionalised N-heterocyclic carbene complexes of group 9 and 10 metals in catalysis: Trends and directions. Eur. J. Inorg. Chem. 2008, 2781–2800. [Google Scholar] [CrossRef]
- Kascatan-Nebioglu, A.; Panzner, M.J.; Tessier, C.A.; Cannon, C.L.; Youngs, W.J. N-Heterocyclic carbene-silver complexes: A new class of antibiotics. Coord. Chem. Rev. 2007, 251, 884–895. [Google Scholar] [CrossRef]
- Gasser, G.; Ott, I.; Metzler-Nolte, N. Organometallic anticancer compounds. J. Med. Chem. 2011, 54, 3–25. [Google Scholar] [CrossRef]
- Budagumpi, S.; Haque, R.A.; Salman, A.W. Stereochemical and structural characteristics of single-and double-site Pd(II)-N-heterocyclic carbene complexes: Promising catalysts in organic syntheses ranging from CC coupling to olefin polymerizations. Coord. Chem. Rev. 2012, 256, 1787–1830. [Google Scholar] [CrossRef]
- Ceramella, J.; Catalano, A.; Mariconda, A.; D’Amato, A.; Aquila, S.; Saturnino, C.; Rosano, C.; Sinicropi, M.S.; Longo, P. Silver N-heterocyclic carbene (NHC) complexes as antimicrobial and/or anticancer agents. Pharmaceuticals 2025, 18, 9. [Google Scholar] [CrossRef] [PubMed]
- Raju, S.K.; Karunakaran, A.; Kumar, S.; Sekar, P.; Murugesan, M.; Karthikeyan, M. Silver complexes as anticancer agents: A perspective review. Ger. J. Pharm. Biomater. 2022, 1, 6–28. [Google Scholar] [CrossRef]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular docking and structure-based drug design strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef]
- Singh, A.; Singh, K.; Sharma, A.; Kaur, K.; Chadha, R.; Bedi, P.M.S. Recent advances in antifungal drug development targeting lanosterol 14α-demethylase (CYP51): A comprehensive review with structural and molecular insights. Chem. Biol. Drug Des. 2023, 102, 606–639. [Google Scholar] [CrossRef] [PubMed]
- Bubb, M.R.; Baines, I.C.; Korn, E.D. Localization of actobindin, profilin I, profilin II, and phosphatidylinositol-4, 5-bisphosphate (PIP2) in Acanthamoeba castellanii. Cell Motil. Cytoskelet. 1998, 39, 134–146. [Google Scholar] [CrossRef]
- Akın-Polat, Z.; Şahin, N.; Hkiri, S.; Ly, B.M.T.; Özdemir, İ.; Sémeril, D. In vitro evaluation of silver-NHC complexes against a clinical isolate of Acanthamoeba castellanii: Time-and dose-dependent effects. Inorganics 2025, 13, 204. [Google Scholar] [CrossRef]
- Dos Santos, D.L.; Chaúque, B.J.M.; Berté, F.K.; de Miranda Ribeiro, L.; Matiazo, F.F.; Rott, M.B.; Schrekker, H.S.; Sekine, L. Imidazolium salt as potent amoebicide for rapid inactivation of Acanthamoeba spp. trophozoites and cysts. Exp. Parasitol. 2025, 271, 108921. [Google Scholar] [CrossRef]
- Tutar, U.; Çelik, C.; Üstün, E.; Özdemir, N.; ¸Sahin, N.; Sémeril, D.; Gürbüz, N.; Özdemir, İ. Benzimidazol-2-ylidene silver complexes: Synthesis, characterization, antimicrobial and antibiofilm activities, molecular docking and theoretical investigations. Inorganics 2023, 11, 385. [Google Scholar] [CrossRef]
- Garrison, J.C.; Youngs, W.J. Ag(I) N-heterocyclic carbene complexes: synthesis, structure, and application. Chem. Rev. 2005, 105, 3978–4008. [Google Scholar] [CrossRef]
- Tulloch, A.A.D.; Danopoulos, A.A.; Winston, S.; Kleinhenz, S.; Eastham, G. N-Functionalised heterocyclic carbene complexes of silver. J. Chem. Soc. Dalton Trans. 2000, 24, 4499–4506. [Google Scholar] [CrossRef]
- Lin, I.J.B.; Vasam, C.S. Silver(I) N-heterocyclic carbenes. Comments Inorg. Chem. 2004, 25, 75–129. [Google Scholar] [CrossRef]
- Chen, W.; Liu, F. Synthesis and characterization of oligomeric and polymeric silver-imidazol-2-ylidene iodide complexes. J. Organomet. Chem. 2003, 673, 5–12. [Google Scholar] [CrossRef]
- Guarra, F.; Busto, N.; Guerri, A.; Marchetti, L.; Marzo, T.; García, B.; Biver, T.; Gabbiani, C. Cytotoxic Ag(I) and Au(I) NHC-carbenes bind DNA and show TrxR inhibition. J. Inorg. Biochem. 2020, 205, 110998. [Google Scholar] [CrossRef]
- de Lacerda, A.G.; Lira, M. Acanthamoeba keratitis: A review of biology, pathophysiology and epidemiology. Ophthalmic Physiol. Opt. 2020, 41, 116–135. [Google Scholar] [CrossRef] [PubMed]
- Ilyas, M.; Stapleton, F.; Willcox, M.D.P.; Henriquez, F.; Peguda, H.K.; Rayamajhee, B.; Zahid, T.; Petsoglou, C.; Carnt, N.A. Epidemiology of and genetic factors associated with Acanthamoeba keratitis. Pathogens 2024, 13, 142. [Google Scholar] [CrossRef]
- Reyes-Batlle, M.; Sifaoui, I.; Rodríguez-Expósito, R.L.; Piñero, J.E.; Lorenzo-Morales, J. New insights in Acanthamoeba. Pathogens 2022, 11, 609. [Google Scholar] [CrossRef]
- Lee, S.M.; Lee, J.E.; Lee, D.I.; Yu, H.S. Adhesion of Acanthamoeba on cosmetic contact lenses. J. Korean Med. Sci. 2018, 33, e26. [Google Scholar] [CrossRef] [PubMed]
- Hendiger, E.B.; Padzik, M.; Sifaoui, I.; Reyes-Batlle, M.; López-Arencibia, A.; Rizo-Liendo, A.; Bethencourt-Estrella, C.J.; Nicolás-Hernández, D.S.; Chiboub, O.; Rodríguez-Expósito, R.L.; et al. Silver nanoparticles as a novel potential preventive agent against Acanthamoeba keratitis. Pathogens 2020, 9, 350. [Google Scholar] [CrossRef] [PubMed]
- Debeljak, N.; Fink, M.; Rozman, D. Many facets of mammalian lanosterol 14α-demethylase from the evolutionarily conserved cytochrome P450 family CYP51. Arch. Biochem. Biophys. 2003, 409, 159–171. [Google Scholar] [CrossRef]
- Hargrove, T.Y.; Lamb, D.C.; Wawrzak, Z.; Hull, M.; Kelly, S.L.; Guengerich, F.P.; Lepesheva, G.I. Identification of potent and selective inhibitors of Acanthamoeba: Structural insights into sterol 14α-demethylase as a key drug target. J. Med. Chem. 2024, 67, 7443–7457. [Google Scholar] [CrossRef]
- Abdel-Hakeem, S.S.; Hassan, F.A.M.; Hifney, A.F.; Salem, S.H. Combating the causative agent of amoebic keratitis, Acanthamoeba castellanii, using Padina pavonica alcoholic extract: Toxicokinetic and molecular docking approaches. Sci. Rep. 2024, 14, 13610. [Google Scholar] [CrossRef]
- Saeed, B.Q.; Hamdy, R.; Akbar, N.; Sajeevan, S.E.; Khan, N.A.; Soliman, S.S. Azole-based compounds as potential anti-Acanthamoeba agents. RSC Med. Chem. 2024, 15, 1578–1588. [Google Scholar] [CrossRef] [PubMed]
- Kurian, T.; Sebastian, R. Molecular docking study of heterocyclic compounds for antifungal activity against Granulomatous Amoebic Encephalitis. J. Pharm. Res. 2024, 23, 175. [Google Scholar] [CrossRef]
- Aksu, A.; Çelik, M.S.; Polat, Z.A.; Yenidünya, A.F.; Çetinkaya, S.; Tüzün, B. Experimental and theoretical evidence on the amoebicidal activity of synthesized tRNA-palmitic acid esters. Iran. J. Chem. Chem. Eng. 2024, 43, 695–706. [Google Scholar]
- Witke, W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 2004, 14, 461–469. [Google Scholar] [CrossRef]
- Bubb, M.R.; Korn, E.D. [11] Purification of actobindin from Acanthamoeba castellanii. Methods Enzymol. 1991, 196, 119–125. [Google Scholar]
- Khairul, W.M.; Hashim, F.; Rahamathullah, R.; Mohammed, M.; Razali, S.A.; Johari, S.A.T.T.; Azizan, S. Exploring ethynyl-based chalcones as green semiconductor materials for optical limiting interests. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2024, 308, 123776. [Google Scholar] [CrossRef]
- Zamli, K.M.; Hashim, F.; Razali, S.A.; Yusoff, H.M.; Mohamad, H.; Abdullah, F.; Asari, A. Synthesis, anti-amoebic activity and molecular docking simulation of eugenol derivatives against Acanthamoeba sp. Saudi Pharm. J. 2023, 31, 101703. [Google Scholar] [CrossRef] [PubMed]
- Hermosaningtyas, A.A.; Budzianowska, A.; Kruszka, D.; Derda, M.; Długaszewska, J.; Kikowska, M. Can in vitro cell cultures of Eryngium planum, Lychnis flos-cuculi, and Kickxia elatine be an alternative source of plant biomass with biological antimicrobial and anti-Acanthamoeba activities? Appl. Sci. 2025, 15, 8292. [Google Scholar] [CrossRef]
- Meng, G.; Kakalis, L.; Nolan, S.P.; Szostak, M. A simple 1H NMR method for determining the σ-donor properties of N-heterocyclic carbenes. Tetrahedron Lett. 2019, 60, 378–781. [Google Scholar] [CrossRef]
- Asekunowo, P.O.; Haque, R.A.; Razali, M.R. A comparative insight into the bioactivity of mono- and binuclear silver(I)-N-heterocyclic carbene complexes: Synthesis, lipophilicity and substituent effect. Rev. Inorg. Chem. 2017, 37, 29–50. [Google Scholar] [CrossRef]
- Saczewski, J.; Popenda, Ł.; Fedorowicz, J. In silico SwissADME analysis of antibacterial NHC-Silver acetates and halides complexes. Appl. Sci. 2024, 14, 8865. [Google Scholar] [CrossRef]
- Ronga, L.; Varcamonti, M.; Tesauro, D. Structure-activity relationships in NHC-silver complexes as antimicrobial agents. Molecules 2023, 28, 4435. [Google Scholar] [CrossRef] [PubMed]
- Admetlab. Available online: https://admetlab3.scbdd.com (accessed on 26 August 2025).
- Tan, K.L.; Vasudevan, A.; Bergman, R.G.; Ellman, J.A.; Souers, A.J. Microwave-assisted C-H bond activation: A rapid entry into functionalized heterocycles. Org. Lett. 2003, 5, 2131–2134. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT-Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Walochnik, J.; Obwaller, A.; Aspöck, H. Correlations between morphological, molecular biological, and physiological characteristics in clinical and nonclinical isolates of Acanthamoeba spp. Appl. Environ. Microbiol. 2000, 66, 4408–4413. [Google Scholar] [CrossRef]
- Neese, F. The ORCA program system. WIREs Comput. Mol. Sci. 2012, 2, 73–78. [Google Scholar] [CrossRef]
- Neese, F. Software update: The ORCA program system, version 4.0. WIREs Comput. Mol. Sci. 2018, 8, e1327. [Google Scholar] [CrossRef]
- Cosconati, S.; Forli, S.; Perryman, A.L.; Harris, R.; Goodsell, D.S.; Olson, A.J. Virtual screening with AutoDock: Theory and practice. Expert Opin. Drug Discov. 2010, 5, 597–607. [Google Scholar] [CrossRef]
- Sharma, V.; Shing, B.; Hernandez-Alvarez, L.; Debnath, A.; Podust, L.M. Domain-swap dimerization of Acanthamoeba castellanii CYP51 and a unique mechanism of inactivation by isavuconazole. Mol. Pharmacol. 2020, 98, 770–780. [Google Scholar] [CrossRef]
- Liu, S.; Fedorov, A.A.; Pollard, T.D.; Lattman, E.E.; Almo, S.C.; Magnus, K.A. Crystal packing induces a conformational change in profilin-I from Acanthamoeba castellanii. J. Struct. Biol. 1998, 123, 22–29. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fedorov, A.A.; Magnus, K.A.; Graupe, M.H.; Lattman, E.E.; Pollard, T.D.; Almo, S.C. X-ray structures of isoforms of the actin-binding protein profilin that differ in their affinity for phosphatidylinositol phosphates. Proc. Natl. Acad. Sci. USA 1994, 91, 8636–8640. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Hkiri, S.; Coşkun, K.A.; Üstün, E.; Samarat, A.; Tutar, Y.; Şahin, N.; Sémeril, D. Silver(I) complexes based on oxadiazole-functionalized α-aminophosphonate: Synthesis, structural study, and biological activities. Molecules 2022, 27, 8131. [Google Scholar] [CrossRef]


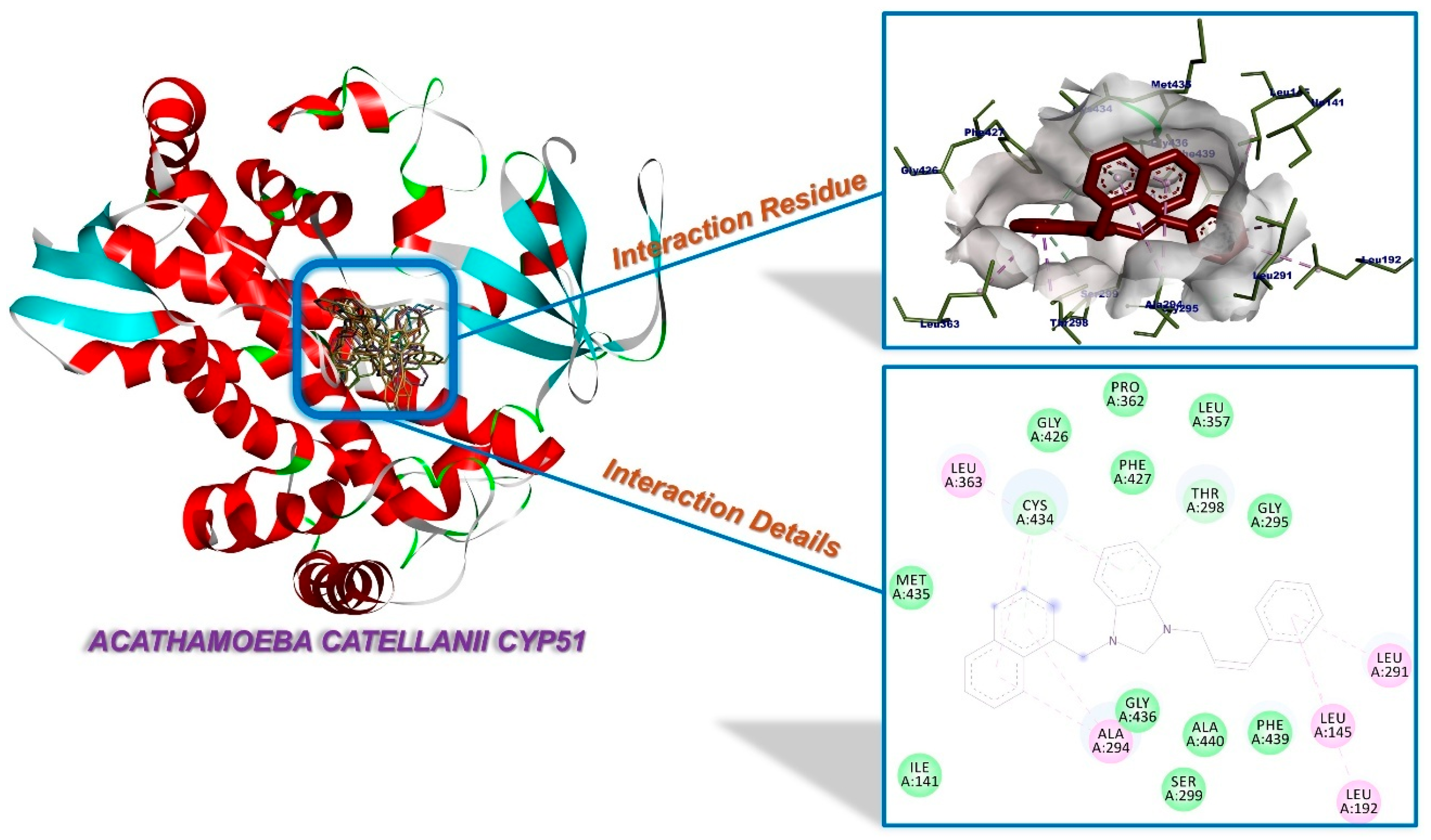
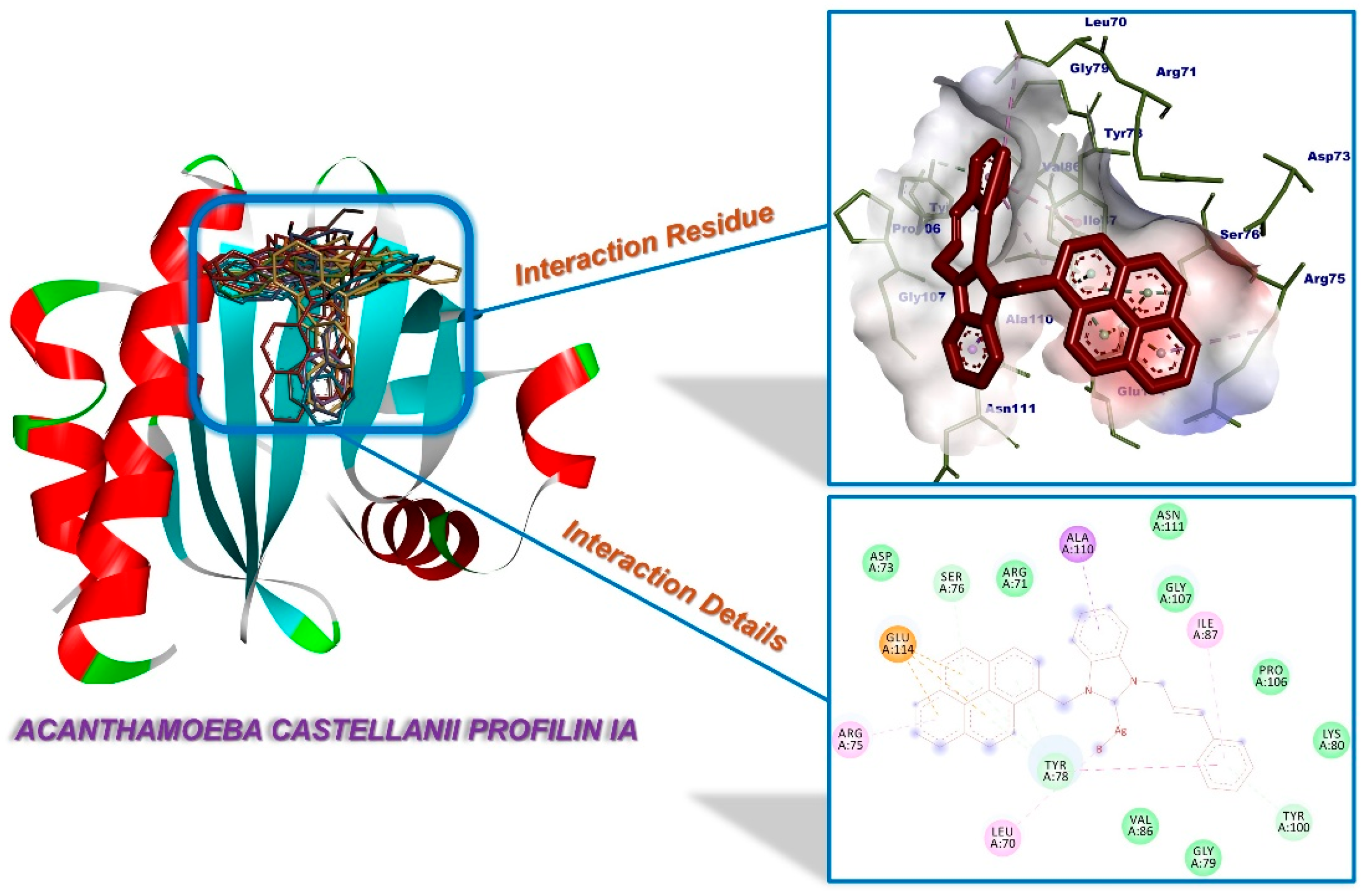

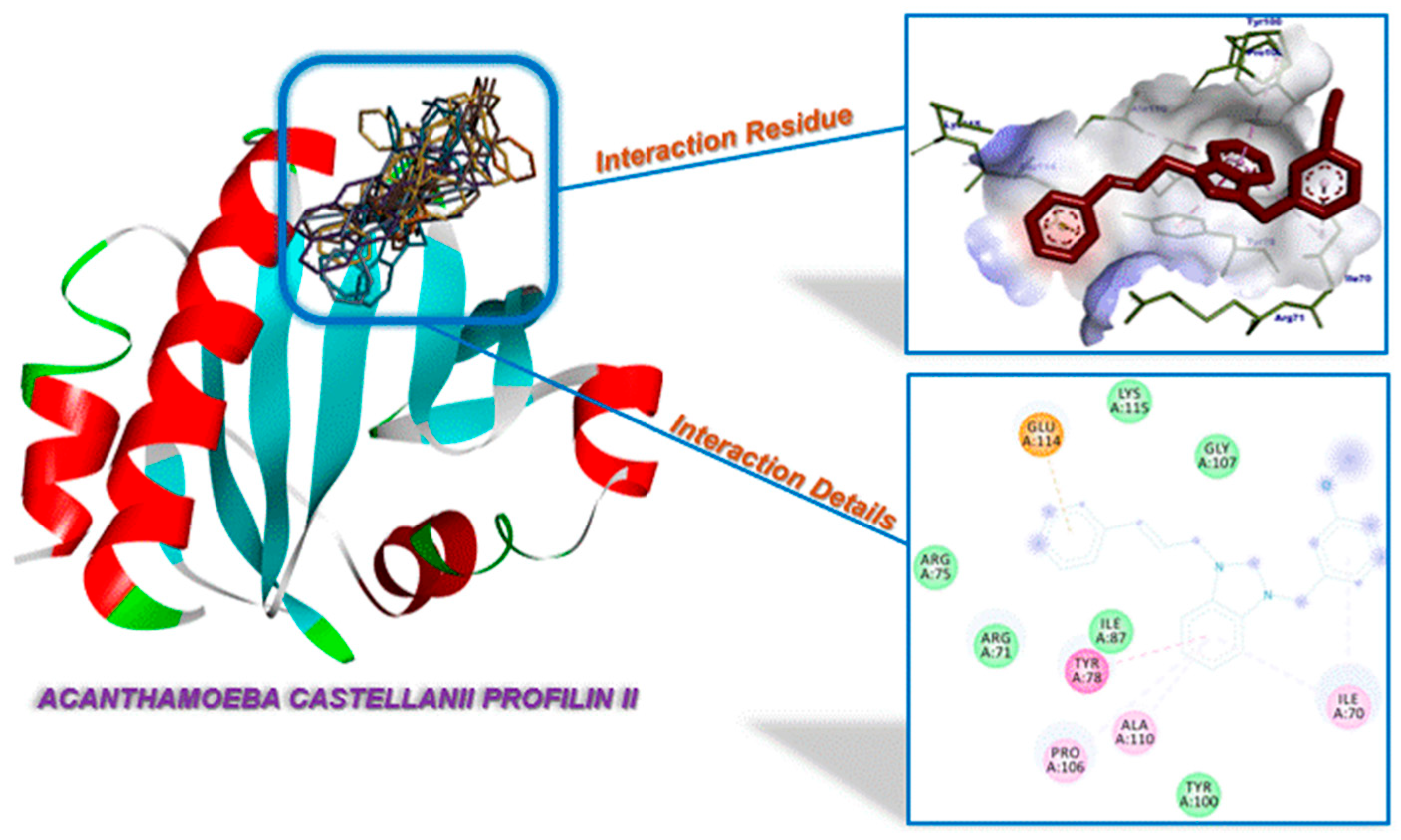
| Compounds | Binding Affinity (kcal/mol) | Amino Acids Residue A. castellanii CYP51 |
|---|---|---|
| 1a | −8.36 | Thr298 (H-bond), Phe427, Cys434, Met435 (π-interactions), Ile141, Leu291, Ala294, Pro362, Ala440 (alkylic interactions), Leu145, Ala290, Gly295, Ser299, Thr302, Leu357, Leu363, His432, Gly436, Phe439 (van der Waals interactions) |
| 1b | −8.37 | Thr298, Ser299 (H-bonds), Ala294 (π-interaction), Leu145, Leu291, Leu357, Pro362, Leu363, Ala440 (alkylic interactions), Phe121, Leu192, Phe293, Gly295, Thr302, Gly426, Phe427, Cys434, Gly436, Phe439 (van der Waals interactions) |
| 1c | −8.68 | Ala294, Cys434, Met435 (π-interactions), Ile141, Ala290, Leu291, Ala440 (alkylic interactions), Leu139, Leu145, Gly295, Thr298, Ser299, Leu363, Phe427, Gly428, His432, Gly433, Gly436, Phe439 (van der Waals interactions) |
| 1d | −8.91 | Gly433, Cys434, Met435, Gly436 (H-bonds), Phe121, Ala294, Leu363, Ala440 (alkylic interactions), Leu138, Leu291, Phe293, Gly295, Thr298, Ser299, His426, Phe427, Gly428, His432, Phe439 (van der Waals interactions) |
| 1e | −9.85 | Ala294, Thr298, Cys434, Met435 (H-bonds), Leu145, Leu291 (alkylic interactions), Leu138, Ile141, Leu192, Gly295, Leu357, Pro362, Leu363, Gly426, Phe427, His432, Gly436, Phe439 (van der Waals interactions) |
| 1f | −11.26 | Ala294, Thr298 (π-interactions), Leu145, Ala290, Leu291, Leu357, Pro362, Leu363, Cys434 (alkylic interactions), Phe121, Val126, Leu192, Phe293, Gly295, His297, Ser299, Gly426, Phe427, Gly436, Phe439, Ala440 (van der Waals interactions) |
| 3a | −8.25 | Cys434 (π-interaction), Leu145, Leu291, Leu357, Pro362 (alkylic interactions), Leu192, Ala294, Gly295, Thr298, Ser299, Leu363, Gly426, Phe427, Gly428, His432, Gly433, Gly436, Phe439, Ala440 (van der Waals interactions) |
| 3b | −8.27 | Phe427, Cys434 (π-interactions), Leu145, Leu291, Leu357, Pro362 (alkylic interactions), Leu192, Ala294, Gly295, Thr298, Ser299, Leu363, Gly426, Gly428, Ala429, His532, Gly433, Gly436, Phe439, Ala440 (van der Waals interactions) |
| 3c | −8.63 | His432, Gly433, Cys434, Met435 (H-bonds), Thr298 (π-interactions), Leu145, Leu291, Pro362 (alkylic interactions), Leu192, Ala294, Gly295, Ser299, Leu357, Leu363, Gly426, Phe427, Gly436, Phe439, Ala440 (van der Waals interactions) |
| 3d | −8.67 | Thr298, His432, Cys434 (H-bonds), Phe427 (π-interaction), Leu145, Leu291, Leu357, Pro362, Leu363, Ala440 (alkylic interactions), Leu192, Ala294, Gly295, Ser299, Thr302, Gly426, Gly428, Gly433, Gly436 Glu437, Phe439, Ile444 (van der Waals interactions) |
| 3e | −9.30 | Thr298, Cys434 (H-bonds), Leu145, Leu192, Leu291, Ala294, Leu363 (alkylic interactions), Ile141, Gly295, Ser299, Leu357, Pro362, Gly426, Phe427, Met435, Gly436, Phe439, Ala440 (van der Waals interactions) |
| 3f | −10.56 | Thr298, Cys434 (H-bonds), Phe427 (π-interaction), Leu145, Leu291, Ala294, Leu357, Pro362, Leu363, Ala440 (alkylic interactions), Leu192, Gly295, Ser299, Thr302, Gly426, Gly428, His432, Gly433, Met435, Gly436, Glu437, Phe439, Ile444 (van de Waals interactions) |
| Compounds | Binding Affinity (kcal/mol) | Amino Acids Residue A. castellanii Profilin IA |
|---|---|---|
| 1a | −6.49 | Tyr100 (H-bond), Tyr78 (π-interaction), Arg71, Ile87, Pro106, Ala110 (alkylic interactions), Leu70, Asp73, Ser76, Gly79, Lys80, Gly85, Val86, Gly107, Asn111, Glu114 (van der Waals interactions) |
| 1b | −6.32 | Arg71, Tyr78, Ala110 (π-interactions), Leu70, Ile87, Tyr100, Pro106 (alkylic interactions), Asp73, Arg75, Ser76, Gly79, Gly85, Val86, Gly107, Asn111, Glu114 (van der Waals interactions) |
| 1c | −6.74 | Ser76, Tyr78, Tyr100 (H-bonds), Glu114 (π-interaction), Leu70, Arg71, Arg75, Ile87, Pro106, Ala110 (alkylic interactions), Asp73, Gly79, Lys80, Gly85, Val86, Gly107, Ala109 (van der Waals interactions) |
| 1d | −6.12 | Ser76, Gly79, Val86, Tyr100 (H-bonds), Glu114 (π-interaction), Leu70, Arg71, Tyr78, Pro106, Ala110 (alkylic interactions), Asp73, Arg75, Gly85, Ile87, Gly107 (van der Waals interactions) |
| 1e | −7.47 | Tyr100 (H-bond), Tyr78 (π-interaction), Arg71, Ile87, Ala110 (alkylic interactions), Leu70, Gly79, Lys79, Lys80, Gly85, Val86, Pro106, Gly107, Glu114 (van der Waals interactions) |
| 1f | −7.48 | Ser76, Tyr78, Tyr100 (H-bonds), Ala110, Glu114 (π-interactions), Leu70, Arg75, Ile87 (alkylic interactions), Arg71, Asp73, Gly79, Lys80, Gly85, Val86, Pro106, Gly107, Asn111 (van der Waals interactions) |
| 3a | −6.24 | Ser76, Tyr78, Tyr100 (H-bonds), Glu114 (π-interaction), Ile87, Pro106, Ala110 (alkylic interactions), Leu70, Arg71, Asp73, Arg75, Gly79, Lys80, Gly85, Val86, Gly107 (van der Waals interactions) |
| 3b | −6.30 | Tyr (H-bond), Tyr78 (π-interactions), Leu70, Arg71, Ile87, Pro106, Ala110 (alkylic interactions), Gly79, Lys80, Gly85, Val86, Gly107, Asn111 (van der Waals interactions) |
| 3c | −6.20 | Gly79, Val86, Tyr100 (H-bonds), Tyr78, Ala110 (π-interactions), Leu70, Arg71, Pro106 (alkylic interactions), Gly85, Ile87, Gly107, Asn111 (van der Waals interactions) |
| 3d | −5.59 | Val86, Tyr100 (H-bonds), Ala110 (π-interaction), Leu70 (alkylic interaction), Arg71, Tyr78, Gly79, Lys80, Gly85, Ile87, Gly107, Asn111, Glu114 (van der Waals interactions) |
| 3e | −7.24 | Ser76, Tyr78 (H-bonds), Ala110, Glu114 (π-interactions), Arg75, Ile87, Pro106 (alkylic interactions), Leu70, Arg71, Asp73, Tyr100, Gly107, Ala109 (van der Waals interactions) |
| 3f | −6.85 | Tyr100 (H-bond), Arg71, Tyr78, Ala110 (π-interactions), Ile87, Pro106 (alkylic interactions), Leu70, Gly79, Lys80, Gly85, Val86, Gly107, Asn111, Glu114 (van der Waals interactions) |
| Compounds | Binding Affinity (kcal/mol) | Amino Acids Residue A. castellanii Profilin IB |
|---|---|---|
| 1a | −5.32 | Tyr78, Tyr100 (H-bonds), Leu70, Arg71, Ile87, Ala110 (alkylic interactions), Val86, Pro106, Gly107, Ala109, Glu114 (van der Waals interactions) |
| 1b | −5.44 | Tyr100 (H-bond), Tyr78, Ala110 (π-interactions), Leu70, Arg71, Ile87 (alkylic interactions), Gly79, Gly85, Val86, Pro106, Gly107, Asn111 (van der Waaals interactions) |
| 1c | −5.80 | Gly79, Val86, Tyr100 (H-bonds), Tyr78, Pro106, Glu114 (π-interaction), Arg71, Arg75, Ala110 (alkylic interactions), Leu70, Arg75, Gly85, Ile87, Gly107, Asn111, Lys115 (van der Waals interactions) |
| 1d | −5.60 | Leu70, Arg71, Tyr78, Tyr100 (H-bonds), Glu114 (π-interaction), Ile87, Ala110 (alkylic interactions), Gly79, Val86, Pro106, Gly107, Asn111 (van der Waals interactions) |
| 1e | −6.24 | Leu70, Tyr78, Glu114 (π-interactions), Arg71, Ile87, Pro106, Ala110 (alkylic interactions), Gly79, Lys80, Gly107, Asn111, Lys115 (van der Waals interactions) |
| 1f | −6.31 | Tyr78 (H-bond), Leu70, Lys80, Ile87, Tyr100, Pro106, Ala110 (alkylic interactions), Arg71, Gly79, Gly82, Ser83, Ser84, Gly85, Val86, Gly107, Asn111, Lys115 (van der Waals interactions) |
| 3a | −6.08 | Tyr78, Tyr100, Pro106, Gly107 (H-bonds), Glu114 (π-interaction), Ile87, Ala110 (alkylic interactions), Leu70, Arg71, Val86, Ala109, Asn111, Lys115 (van der waals interactions) |
| 3b | −6.37 | Tyr78 (H-bond), Ala110 (π-interaction), Leu70, Arg71, Tyr78, Ile87 (alkylic interactions), Gly79, Gly85, Val86, Pro106, Gly107, Asn111 (van der Waals interactions) |
| 3c | −5.80 | Gly79, Val86, Tyr100 (H-bonds), Ala110 (π-interaction), Leu70, Arg71, Pro106 (alkylic interactions), Tyr78, Gly85, Ile87, Gly107, Asn111, Glu114 (van der Waals interactions) |
| 3d | −5.70 | Gly79, Val86 (H-bonds), Tyr78, Ala110 (π-interactions), Leu70, Arg71, Pro106 (alkylic interactions), Gly85, Ile87, Tyr100, Gly107, Asn111 (van der Waals interactions) |
| 3e | −6.14 | Tyr78, Ala110 (π-interactions), Leu70, Arg71, Ile87, Pro106 (alkylic interactions), Tyr100, Gly107, Asn111, Glu114 (van der Waals interactions) |
| 3f | −6.11 | Tyr78, Gly107 (H-bonds), Leu70, Arg71, Pro106, Ala110 (π-interactions), Ile87, Tyr100, Asn111, Glu114 (van der Waals interactions) |
| Compounds | Binding Affinity (kcal/mol) | Amino Acids Residue A. castellanii Profilin II |
|---|---|---|
| 1a | −3.37 | Tyr78, Pro106 (π-interactions), İle70, Ala110 (alkylic interactions), Arg71, Gly79, Lys80, Gly85, Val86, Tyr100, Gly107, Glu114 (van der Waals interactions) |
| 1b | −3.67 | Tyr100 (H-bond), Tyr78 (π-interaction), Ile70, Pro106 (alkylic interactions), Arg71, Gly79, Lys80, Gly85, Val86, Ile87, Gly107, Ala110 (van der Waals interactions) |
| 1c | −3.46 | Tyr78 (H-bond), Glu114 (π-interaction), Ile70, Pro106, Ala110 (alkylic interactions), Arg71, Arg78, Gly79, Lys80, Ile87, Tyr100, Gly107, Asn111, Lys115 (van der Waals interactions) |
| 1d | −3.05 | Ile87 (π-interaction), Ile70, Arg71 (alkylic interactions), Gly107, Glu114 (van der Waal interactions), Tyr78, Tyr100, Pro106, Ala109, Ala110 (Unfavorable Bumb) |
| 1e | −3.08 | Tyr78 (H-bond), Ile70, Ala110 (π-interactions), Pro106 (alkylic interaction), Arg71, Lys80, Tyr100, Gly107, Asn111, Glu114, Lys115 (van der Waal interactions) |
| 1f | −3.31 | Tyr100 (H-bond), Ile70 (alkylic interaction), Val77, Lys80, Thr88, Gly107 (van der Waals interactions), Tyr78, Gly79, Val86, Ile87, Pro106, Ala110 (Unfavorable Bump) |
| 3a | −4.38 | Tyr78 (H-bond), Glu114 (π-interaction), Ile70, Lys80, Pro106, Ala110 (alkylic interactions), Arg71, Tyr100, Gly107, Asn111, Lys115 (van der Waals interactions) |
| 3b | −4.41 | Tyr78, Pro106, Ala110 (alkylic interactions), Ile70, Arg71, Lys80, Ser83, Ala84, Tyr100, Glu102, Ile104, Gln105, Gly107 (van der Waals interactions) |
| 3c | −5.22 | Tyr78, Glu114 (π-interactions), Ile70, Pro106, Ala110 (alkylic interactions), Arg71, Arg75, Ile87, Tyr100, Gly107, Lys115 (van der Waals interactions) |
| 3d | −4.04 | Tyr100, Pro106 (H-bonds), Arg71 (π-interaction), Ile70, Lys80 (alkylic interactions), Tyr78, Gly79, Gly85, Ile87, Gly107, Ala110 (van der Waals interactions) |
| 3e | −4.11 | Ile70 (π-interaction), Lys80, Pro106 (alkylic interaction), Arg71, Tyr78, Tyr100, Ala110 (van der Waals interactions) |
| 3f | −3.52 | Ala110, Glu114 (π-interactions), Ile70, Lys80, Pro106 (alkylic interactions), Arg71, Tyr78, Tyr100, Gly107, Asn111, Lys115 (van der Waals interactions) |
| Silver Complex | Calculated logP | Silver Complex | Calculated logP |
| 1a | 4.60 | 1d | 4.52 |
| 1b | 5.01 | 1e | 5.14 |
| 1c | 4.52 | 1f | 6.69 |
| CCDC depository | 2,482,191 | Chemical formula | C48H44Ag2Br2N4 |
| Molar mass (g·mol−1) | 1052.43 | Temperature (K) | 120(2) |
| Crystal system | Triclinic | Space group | |
| a (Å) | 8.7251(4) | α (°) | 69.405(2) |
| b (Å) | 10.7811(6) | β (°) | 78.683(2) |
| c (Å) | 11.7674(6) | γ (°) | 86.031(3) |
| Volume (Å3) | 1016.03(9) | Z | 1 |
| ρcalc. (g·cm−3) | 1.720 | μ (mm−1) | 2.970 |
| F000 | 524 | Crystal size (mm) | 0.140 × 0.130 × 0.120 |
| θ range for data collection (°) | 1.881 ≤ θ ≤ 27.877 | Rint | 0.0538 |
| Reflections collected | 34781 | Goodness-of-fit on F2 | 1.018 |
| R1, wR2 (all data) | R1 = 0.0473 wR2 = 0.0894 | R1, wR2 [I > 2σ(I)] | R1 = 0.0359 wR2 = 0.0819 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hkiri, S.; Şahin, N.; Akın-Polat, Z.; Üstün, E.; Ly, B.M.T.; Özdemir, İ.; Sémeril, D. Silver(I)-NHC Complexes as Dual-Action Agents Against Pathogenic Acanthamoeba Trophozoites: Anti-Amoebic and Anti-Adhesion Activities. Int. J. Mol. Sci. 2025, 26, 9393. https://doi.org/10.3390/ijms26199393
Hkiri S, Şahin N, Akın-Polat Z, Üstün E, Ly BMT, Özdemir İ, Sémeril D. Silver(I)-NHC Complexes as Dual-Action Agents Against Pathogenic Acanthamoeba Trophozoites: Anti-Amoebic and Anti-Adhesion Activities. International Journal of Molecular Sciences. 2025; 26(19):9393. https://doi.org/10.3390/ijms26199393
Chicago/Turabian StyleHkiri, Shaima, Neslihan Şahin, Zübeyda Akın-Polat, Elvan Üstün, Bui Minh Thu Ly, İsmail Özdemir, and David Sémeril. 2025. "Silver(I)-NHC Complexes as Dual-Action Agents Against Pathogenic Acanthamoeba Trophozoites: Anti-Amoebic and Anti-Adhesion Activities" International Journal of Molecular Sciences 26, no. 19: 9393. https://doi.org/10.3390/ijms26199393
APA StyleHkiri, S., Şahin, N., Akın-Polat, Z., Üstün, E., Ly, B. M. T., Özdemir, İ., & Sémeril, D. (2025). Silver(I)-NHC Complexes as Dual-Action Agents Against Pathogenic Acanthamoeba Trophozoites: Anti-Amoebic and Anti-Adhesion Activities. International Journal of Molecular Sciences, 26(19), 9393. https://doi.org/10.3390/ijms26199393





