Cytokine and Chemokine-Associated Signatures Underlying Dermal Invasion and Skin Metastasis in Melanoma
Abstract
1. Introduction
2. Results
2.1. Effect of HDMEC-Conditioned Medium on Melanoma Cell Invasion
2.2. Cytokine and Chemokine Receptor Gene Expression in Melanoma Cell Lines
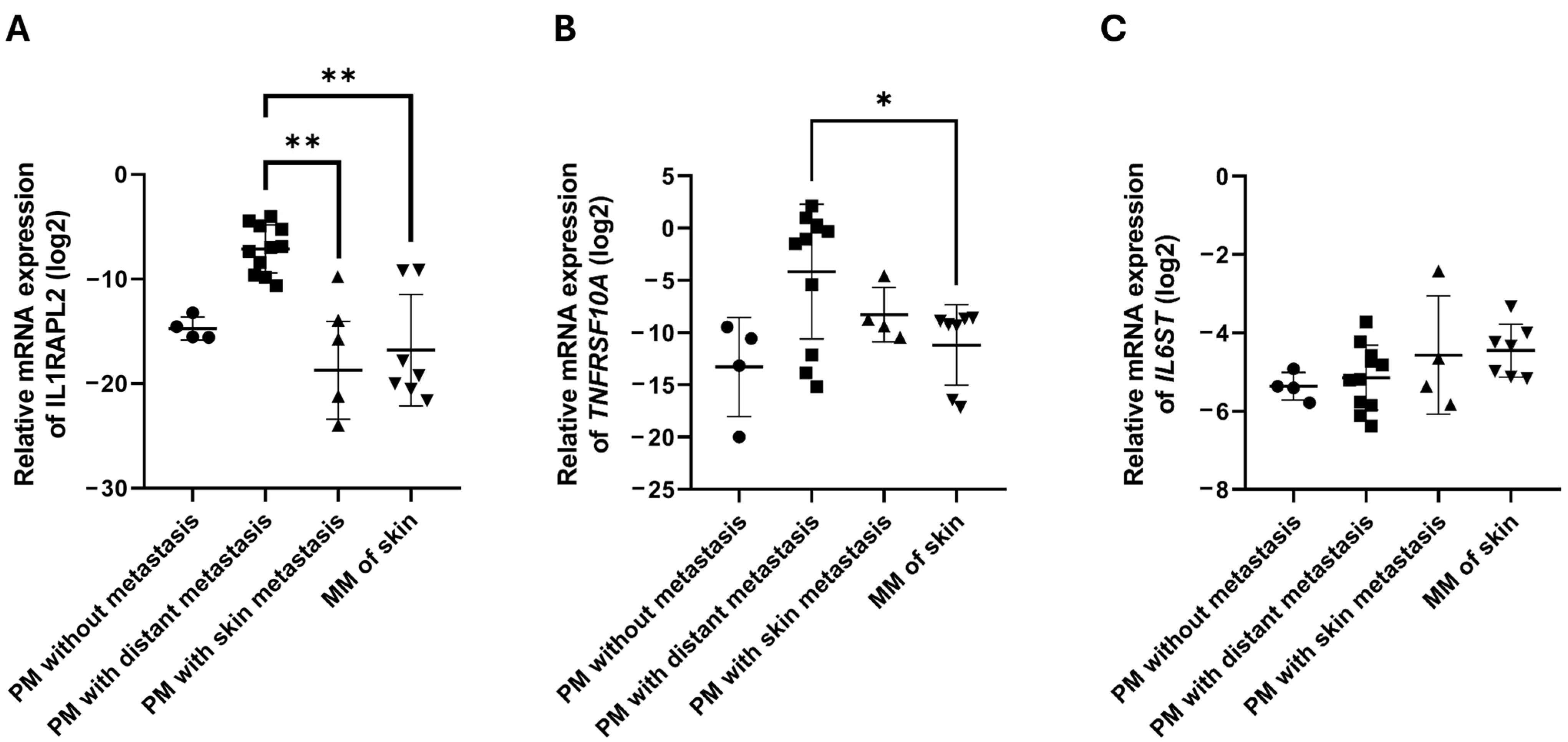
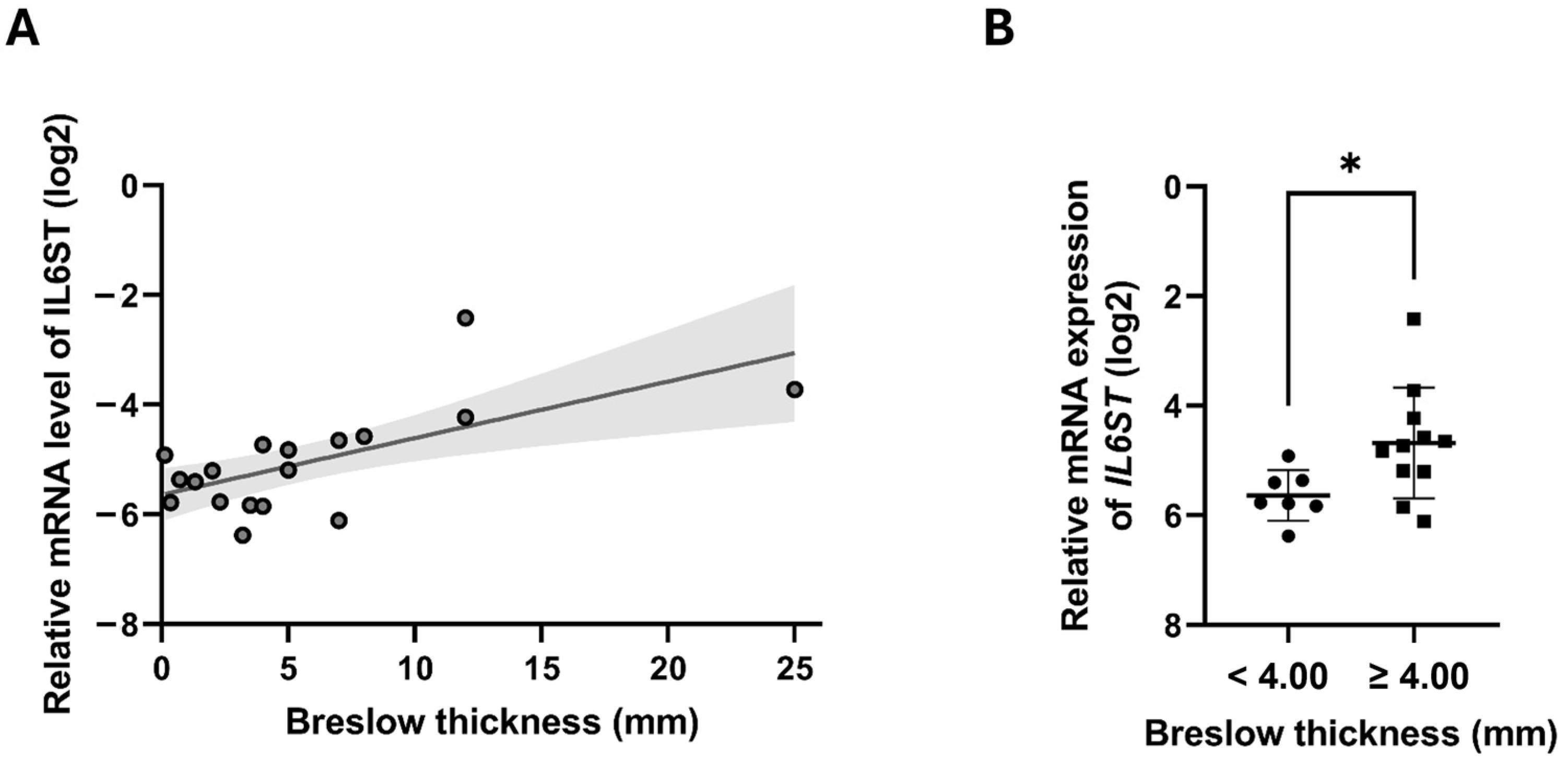
2.3. Cytokine and Chemokine Receptor Gene Expression in Melanoma Tumor Samples
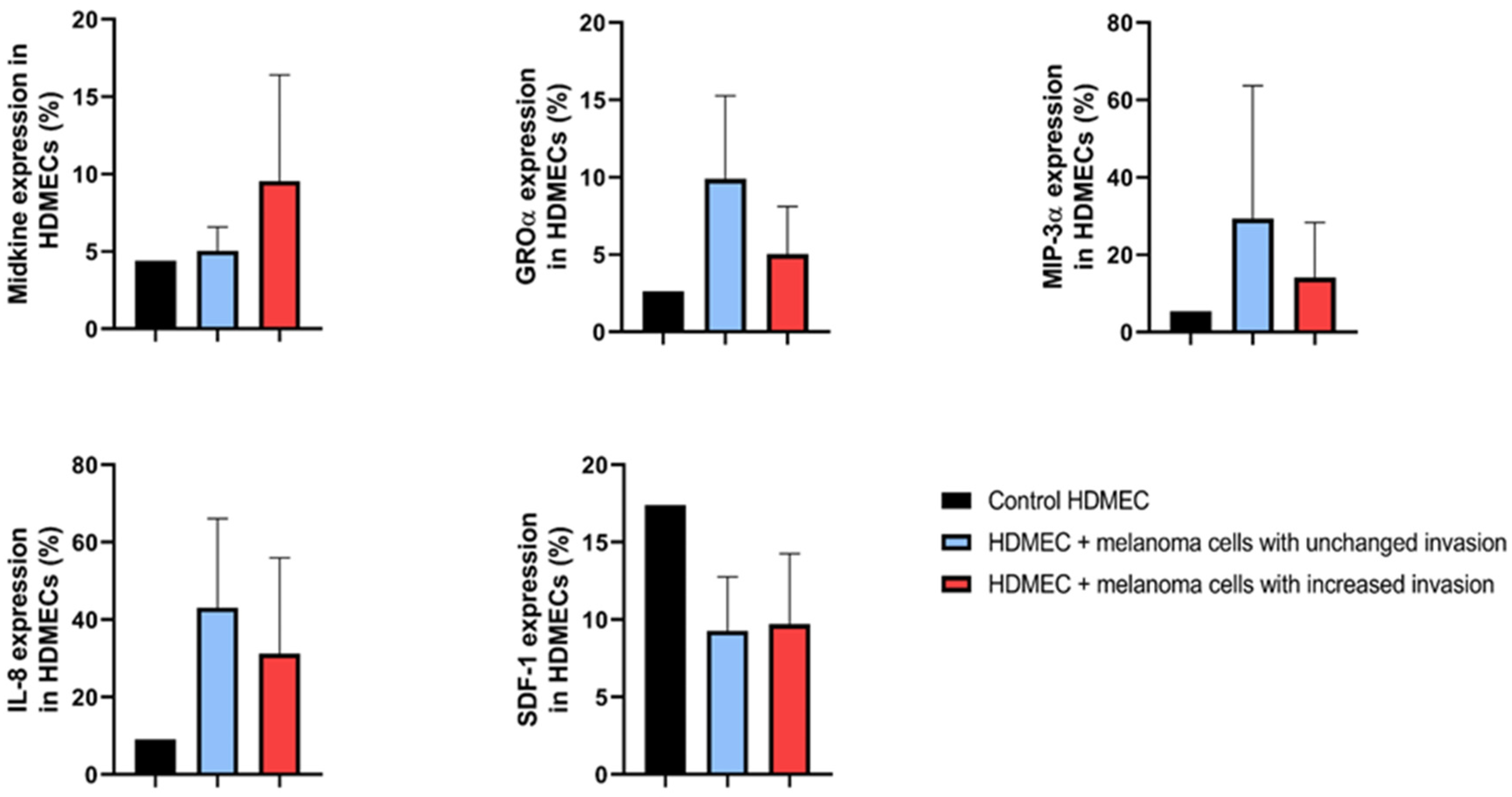
2.4. Proteome Profile of HDMECs
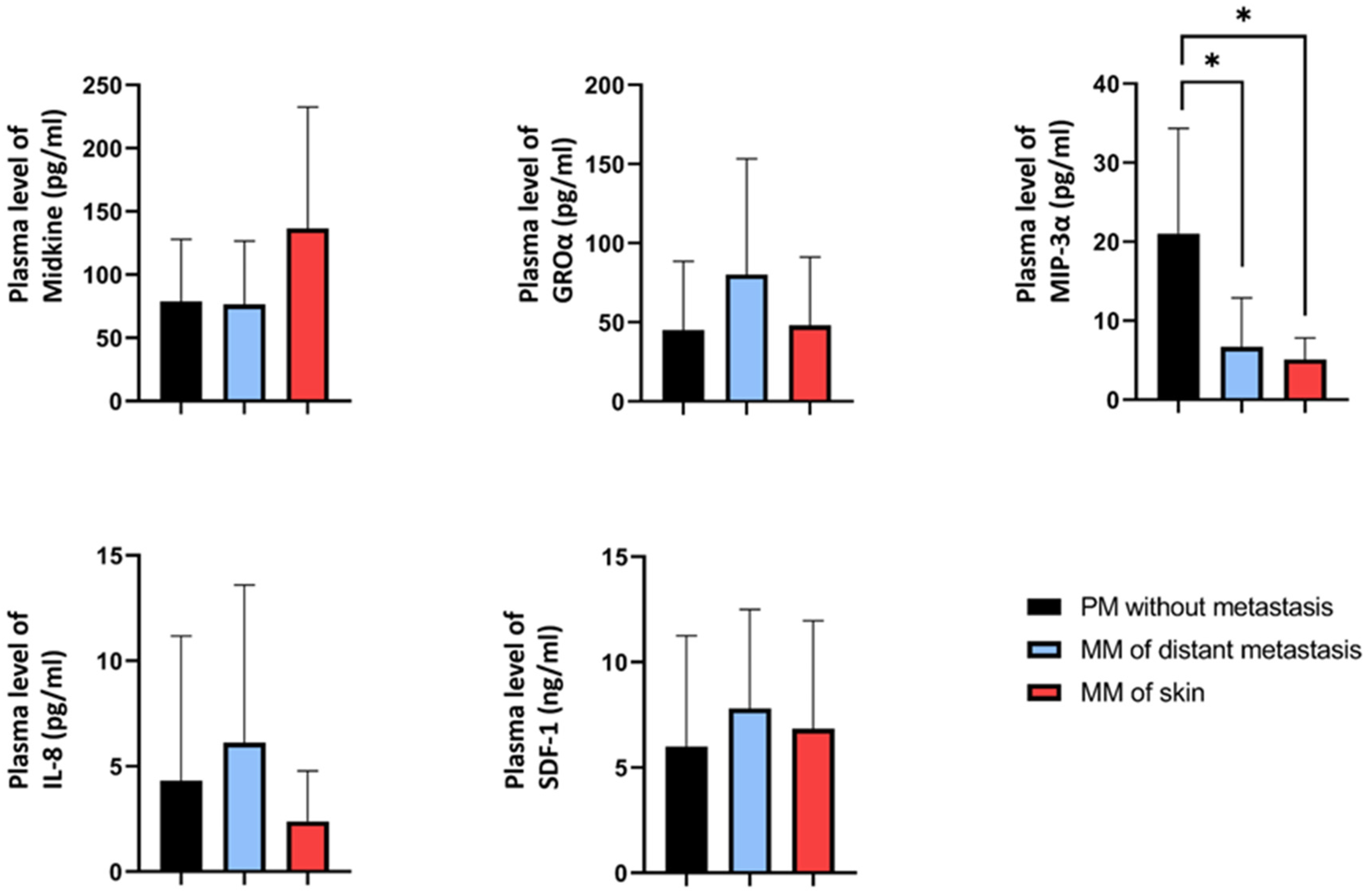
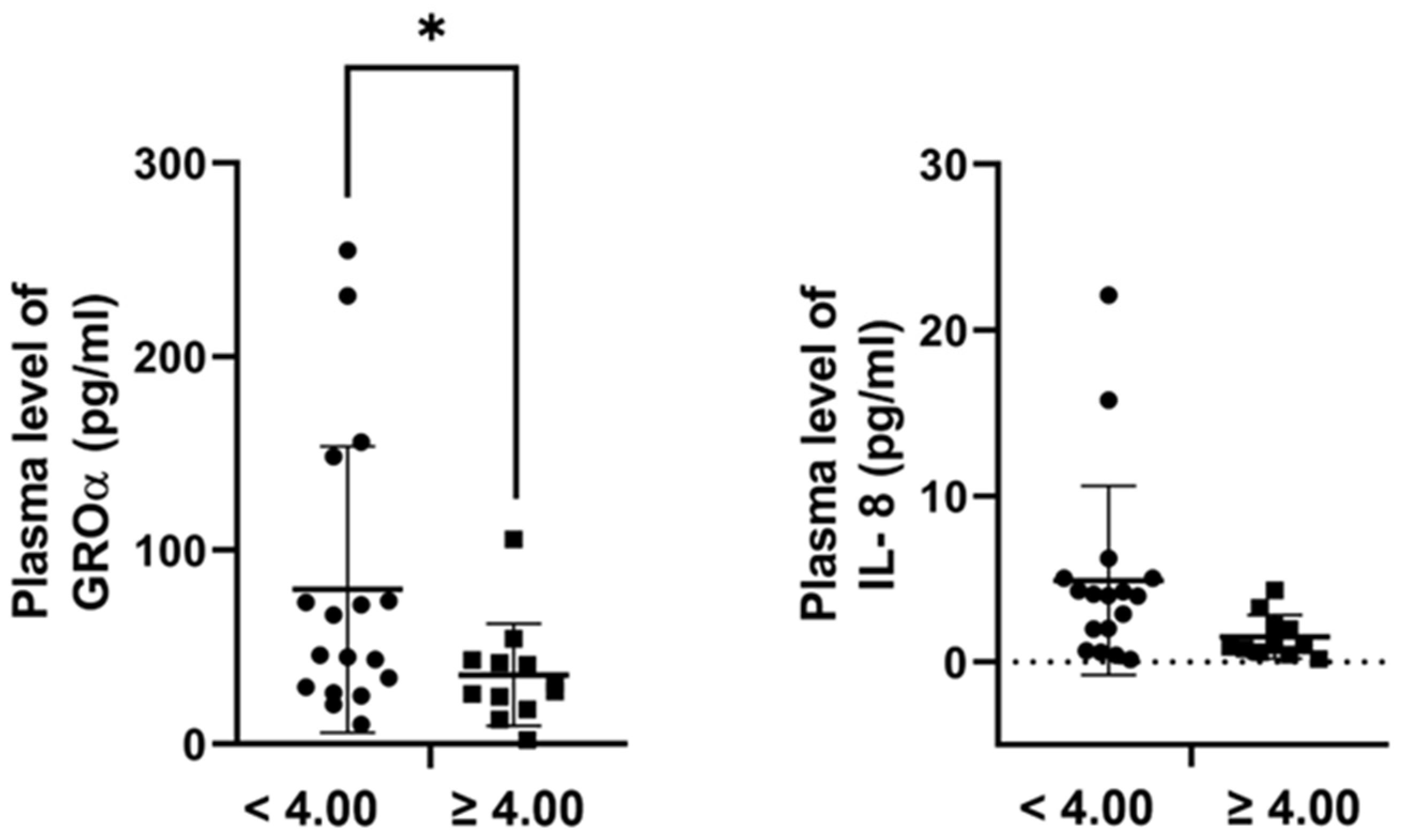
2.5. Plasma Concentrations of the Candidate Proteins in Melanoma Patients
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cell Culture
4.2. Melanoma Tissue and Plasma Samples
4.3. In Vitro Invasion Assay
4.4. Real-Time Quantitative PCR Analysis
4.5. Proteome Profiler Assay
4.6. Enzyme-Linked Immunosorbent Assay (ELISA)
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reed, K.B.; Cook-Norris, R.H.; Brewer, J.D. The cutaneous manifestations of metastatic malignant melanoma. Int. J. Dermatol. 2012, 51, 243–249. [Google Scholar] [CrossRef]
- Rebecca, V.W.; Somasundaram, R.; Herlyn, M. Pre-clinical modeling of cutaneous melanoma. Nat. Commun. 2020, 11, 2858. [Google Scholar] [CrossRef]
- Dhanyamraju, P.K.; Patel, T.N. Melanoma therapeutics: A literature review. J. Biomed. Res. 2022, 36, 77–97. [Google Scholar] [CrossRef] [PubMed]
- Hartman, R.I.; Lin, J.Y. Cutaneous Melanoma-A Review in Detection, Staging, and Management. Hematol. Oncol. Clin. N. Am. 2019, 33, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Di Raimondo, C.; Lozzi, F.; Di Domenico, P.P.; Campione, E.; Bianchi, L. The Diagnosis and Management of Cutaneous Metastases from Melanoma. Int. J. Mol. Sci. 2023, 24, 4535. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, G.C.; Falzone, L.; Salemi, R.; Zanghi, A.; Spandidos, D.A.; McCubrey, J.A.; Candido, S.; Libra, M. Cutaneous melanoma: From pathogenesis to therapy (Review). Int. J. Oncol. 2018, 52, 1071–1080. [Google Scholar] [CrossRef]
- Wong, C.Y.; Helm, M.A.; Helm, T.N.; Zeitouni, N. Patterns of skin metastases: A review of 25 years’ experience at a single cancer center. Int. J. Dermatol. 2014, 53, 56–60. [Google Scholar] [CrossRef]
- Savoia, P.; Fava, P.; Nardo, T.; Osella-Abate, S.; Quaglino, P.; Bernengo, M.G. Skin metastases of malignant melanoma: A clinical and prognostic survey. Melanoma Res. 2009, 19, 321–326. [Google Scholar] [CrossRef]
- Wong, J.Y.; Gilson, N.D.; Bush, R.A.; Brown, W.J. Patterns and perceptions of physical activity and sedentary time in male transport drivers working in regional Australia. Aust. N. Z. J. Public Health 2014, 38, 314–320. [Google Scholar] [CrossRef]
- Plaza, J.A.; Torres-Cabala, C.; Evans, H.; Diwan, H.A.; Suster, S.; Prieto, V.G. Cutaneous metastases of malignant melanoma: A clinicopathologic study of 192 cases with emphasis on the morphologic spectrum. Am. J. Dermatopathol. 2010, 32, 129–136. [Google Scholar] [CrossRef]
- Chambers, A.F.; Groom, A.C.; MacDonald, I.C. Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer 2002, 2, 563–572. [Google Scholar] [CrossRef]
- Orgaz, J.L.; Sanz-Moreno, V. Emerging molecular targets in melanoma invasion and metastasis. Pigment. Cell Melanoma Res. 2013, 26, 39–57. [Google Scholar] [CrossRef] [PubMed]
- Murakami, T.; Cardones, A.R.; Hwang, S.T. Chemokine receptors and melanoma metastasis. J. Dermatol. Sci. 2004, 36, 71–78. [Google Scholar] [CrossRef]
- Guan, X. Cancer metastases: Challenges and opportunities. Acta Pharm. Sin. B 2015, 5, 402–418. [Google Scholar] [CrossRef]
- Hembruff, S.L.; Cheng, N. Chemokine signaling in cancer: Implications on the tumor microenvironment and therapeutic targeting. Cancer Ther. 2009, 7, 254–267. [Google Scholar]
- Cameron, M.J.; Kelvin, D.J. Cytokines and chemokines--their receptors and their genes: An overview. Adv. Exp. Med. Biol. 2003, 520, 8–32. [Google Scholar] [CrossRef]
- Amos, S.E.; Choi, Y.S. The Cancer Microenvironment: Mechanical Challenges of the Metastatic Cascade. Front. Bioeng. Biotechnol. 2021, 9, 625859. [Google Scholar] [CrossRef]
- Muller, A.; Homey, B.; Soto, H.; Ge, N.; Catron, D.; Buchanan, M.E.; McClanahan, T.; Murphy, E.; Yuan, W.; Wagner, S.N.; et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001, 410, 50–56. [Google Scholar] [CrossRef]
- Nguyen, D.X.; Bos, P.D.; Massague, J. Metastasis: From dissemination to organ-specific colonization. Nat. Rev. Cancer 2009, 9, 274–284. [Google Scholar] [CrossRef]
- Li, Q.; Zhu, Z.; Wang, L.; Lin, Y.; Fang, H.; Lei, J.; Cao, T.; Wang, G.; Dang, E. Single-cell transcriptome profiling reveals vascular endothelial cell heterogeneity in human skin. Theranostics 2021, 11, 6461–6476. [Google Scholar] [CrossRef]
- Potente, M.; Makinen, T. Vascular heterogeneity and specialization in development and disease. Nat. Rev. Mol. Cell Biol. 2017, 18, 477–494. [Google Scholar] [CrossRef] [PubMed]
- Ben-Baruch, A. Organ selectivity in metastasis: Regulation by chemokines and their receptors. Clin. Exp. Metastasis 2008, 25, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Habermehl, G.; Ko, J. Cutaneous Metastases: A Review and Diagnostic Approach to Tumors of Unknown Origin. Arch. Pathol. Lab. Med. 2019, 143, 943–957. [Google Scholar] [CrossRef] [PubMed]
- Davis, L.E.; Shalin, S.C.; Tackett, A.J. Current state of melanoma diagnosis and treatment. Cancer Biol. Ther. 2019, 20, 1366–1379. [Google Scholar] [CrossRef]
- Basson, C.; Serem, J.C.; Bipath, P.; Hlophe, Y.N. Chemokines as possible therapeutic targets in metastatic melanoma. Cancer Med. 2023, 12, 14387–14402. [Google Scholar] [CrossRef]
- Floros, T.; Tarhini, A.A. Anticancer Cytokines: Biology and Clinical Effects of Interferon-alpha2, Interleukin (IL)-2, IL-15, IL-21, and IL-12. Semin. Oncol. 2015, 42, 539–548. [Google Scholar] [CrossRef]
- Mortezaee, K.; Majidpoor, J. Checkpoint inhibitor/interleukin-based combination therapy of cancer. Cancer Med. 2022, 11, 2934–2943. [Google Scholar] [CrossRef]
- Bhave, P.; Haydon, A. Treatment of High Risk Resected Melanoma in Australia: Current Landscape and Practises. Australas J. Dermatol. 2020, 61, 203–209. [Google Scholar] [CrossRef]
- Gellrich, F.F.; Schmitz, M.; Beissert, S.; Meier, F. Anti-PD-1 and Novel Combinations in the Treatment of Melanoma-An Update. J. Clin. Med. 2020, 9, 223. [Google Scholar] [CrossRef]
- Lowe, L. Metastatic melanoma and rare melanoma variants: A review. Pathology 2023, 55, 236–244. [Google Scholar] [CrossRef]
- Richmond, A.; Yang, J.; Su, Y. The good and the bad of chemokines/chemokine receptors in melanoma. Pigment. Cell Melanoma Res. 2009, 22, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Simonetti, O.; Goteri, G.; Lucarini, G.; Filosa, A.; Pieramici, T.; Rubini, C.; Biagini, G.; Offidani, A. Potential role of CCL27 and CCR10 expression in melanoma progression and immune escape. Eur. J. Cancer 2006, 42, 1181–1187. [Google Scholar] [CrossRef] [PubMed]
- Homey, B.; Wang, W.; Soto, H.; Buchanan, M.E.; Wiesenborn, A.; Catron, D.; Muller, A.; McClanahan, T.K.; Dieu-Nosjean, M.C.; Orozco, R.; et al. Cutting edge: The orphan chemokine receptor G protein-coupled receptor-2 (GPR-2, CCR10) binds the skin-associated chemokine CCL27 (CTACK/ALP/ILC). J. Immunol. 2000, 164, 3465–3470. [Google Scholar] [CrossRef] [PubMed]
- Mergia Terefe, E.; Catalan Opulencia, M.J.; Rakhshani, A.; Ansari, M.J.; Sergeevna, S.E.; Awadh, S.A.; Polatova, D.S.; Abdulkadhim, A.H.; Mustafa, Y.F.; Kzar, H.H.; et al. Roles of CCR10/CCL27-CCL28 axis in tumour development: Mechanisms, diagnostic and therapeutic approaches, and perspectives. Expert Rev. Mol. Med. 2022, 24, e37. [Google Scholar] [CrossRef]
- Ferrante, M.I.; Ghiani, M.; Bulfone, A.; Franco, B. IL1RAPL2 maps to Xq22 and is specifically expressed in the central nervous system. Gene 2001, 275, 217–221. [Google Scholar] [CrossRef]
- Niu, R.Z.; Feng, W.Q.; Yu, Q.S.; Shi, L.L.; Qin, Q.M.; Liu, J. Integrated analysis of plasma proteome and cortex single-cell transcriptome reveals the novel biomarkers during cortical aging. Front. Aging Neurosci. 2023, 15, 1063861. [Google Scholar] [CrossRef]
- Boraschi, D.; Italiani, P.; Weil, S.; Martin, M.U. The family of the interleukin-1 receptors. Immunol. Rev. 2018, 281, 197–232. [Google Scholar] [CrossRef]
- Singh, D.; Tewari, M.; Singh, S.; Narayan, G. Revisiting the role of TRAIL/TRAIL-R in cancer biology and therapy. Future Oncol. 2021, 17, 581–596. [Google Scholar] [CrossRef]
- Deng, D.; Shah, K. TRAIL of Hope Meeting Resistance in Cancer. Trends Cancer 2020, 6, 989–1001. [Google Scholar] [CrossRef]
- Zhu, J.; Zhou, Q.; Tan, S. Targeting miRNAs associated with surface expression of death receptors to modulate TRAIL resistance in breast cancer. Cancer Lett. 2016, 383, 154–160. [Google Scholar] [CrossRef]
- Park, S.A.; Kim, L.K.; Park, H.M.; Kim, H.J.; Heo, T.H. Inhibition of GP130/STAT3 and EMT by combined bazedoxifene and paclitaxel treatment in ovarian cancer. Oncol. Rep. 2022, 47, 52. [Google Scholar] [CrossRef] [PubMed]
- Rose-John, S. Interleukin-6 Family Cytokines. Cold Spring Harb. Perspect. Biol. 2018, 10, a028415. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; He, Y.; Zhang, Z.; Zhu, Y.; Ren, H.; Zhang, L.; Li, L.; Li, W.; Zhang, W.; Xiao, T.; et al. miR-224-5p Suppresses Non-Small Cell Lung Cancer via IL6ST-Mediated Regulation of the JAK2/STAT3 Pathway. Thorac. Cancer 2025, 16, e15516. [Google Scholar] [CrossRef]
- Yue, Y.; Zhang, Q.; Wu, S.; Wang, S.; Cui, C.; Yu, M.; Sun, Z. Identification of key genes involved in JAK/STAT pathway in colorectal cancer. Mol. Immunol. 2020, 128, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Kadomatsu, K.; Kishida, S.; Tsubota, S. The heparin-binding growth factor midkine: The biological activities and candidate receptors. J. Biochem. 2013, 153, 511–521. [Google Scholar] [CrossRef]
- Karg, M.M.; John, L.; Refaian, N.; Buettner, C.; Rottmar, T.; Sommer, J.; Bock, B.; Resheq, Y.J.; Ksander, B.R.; Heindl, L.M.; et al. Midkine Promotes Metastasis and Therapeutic Resistance via mTOR/RPS6 in Uveal Melanoma. Mol. Cancer Res. 2022, 20, 1320–1336. [Google Scholar] [CrossRef]
- Gungor, C.; Zander, H.; Effenberger, K.E.; Vashist, Y.K.; Kalinina, T.; Izbicki, J.R.; Yekebas, E.; Bockhorn, M. Notch signaling activated by replication stress-induced expression of midkine drives epithelial-mesenchymal transition and chemoresistance in pancreatic cancer. Cancer Res. 2011, 71, 5009–5019. [Google Scholar] [CrossRef]
- Saikia, M.; Cheung, N.; Singh, A.K.; Kapoor, V. Role of Midkine in Cancer Drug Resistance: Regulators of Its Expression and Its Molecular Targeting. Int. J. Mol. Sci. 2023, 24, 8739. [Google Scholar] [CrossRef]
- Olmeda, D.; Cerezo-Wallis, D.; Riveiro-Falkenbach, E.; Pennacchi, P.C.; Contreras-Alcalde, M.; Ibarz, N.; Cifdaloz, M.; Catena, X.; Calvo, T.G.; Canon, E.; et al. Whole-body imaging of lymphovascular niches identifies pre-metastatic roles of midkine. Nature 2017, 546, 676–680. [Google Scholar] [CrossRef]
- Karaman, S.; Alitalo, K. Midkine and Melanoma Metastasis: A Malevolent Mix. Dev. Cell 2017, 42, 205–207. [Google Scholar] [CrossRef]
- Damsky, W.E.; Rosenbaum, L.E.; Bosenberg, M. Decoding melanoma metastasis. Cancers 2010, 3, 126–163. [Google Scholar] [CrossRef] [PubMed]
- Vandyck, H.H.; Hillen, L.M.; Bosisio, F.M.; van den Oord, J.; Zur Hausen, A.; Winnepenninckx, V. Rethinking the biology of metastatic melanoma: A holistic approach. Cancer Metastasis Rev. 2021, 40, 603–624. [Google Scholar] [CrossRef] [PubMed]
- Kadomoto, S.; Izumi, K.; Mizokami, A. The CCL20-CCR6 Axis in Cancer Progression. Int. J. Mol. Sci. 2020, 21, 5186. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Park, K.K.; Kim, H.J.; Park, J.; Son, S.H.; Kim, K.R.; Chung, W.Y. Human antigen R-regulated CCL20 contributes to osteolytic breast cancer bone metastasis. Sci. Rep. 2017, 7, 9610. [Google Scholar] [CrossRef]
- Han, G.; Wu, D.; Yang, Y.; Li, Z.; Zhang, J.; Li, C. CrkL meditates CCL20/CCR6-induced EMT in gastric cancer. Cytokine 2015, 76, 163–169. [Google Scholar] [CrossRef]
- Hughes, C.E.; Nibbs, R.J.B. A guide to chemokines and their receptors. FEBS J. 2018, 285, 2944–2971. [Google Scholar] [CrossRef]
- Korbecki, J.; Bosiacki, M.; Szatkowska, I.; Kupnicka, P.; Chlubek, D.; Baranowska-Bosiacka, I. The Clinical Significance and Involvement in Molecular Cancer Processes of Chemokine CXCL1 in Selected Tumors. Int. J. Mol. Sci. 2024, 25, 4365. [Google Scholar] [CrossRef]
- Fimmel, S.; Devermann, L.; Herrmann, A.; Zouboulis, C. GRO-alpha: A potential marker for cancer and aging silenced by RNA interference. Ann. N. Y. Acad. Sci. 2007, 1119, 176–189. [Google Scholar] [CrossRef]
- Hatano, T.; Yashiro, M.; Fujikawa, H.; Motomura, H. C-X-C Motif Ligand 1 (CXCL1) from melanoma cells down-regulates the invasion of their metastatic melanoma cells. Oncotarget 2018, 9, 31090–31097. [Google Scholar] [CrossRef][Green Version]
- Mendt, M.; Cardier, J.E. Activation of the CXCR4 chemokine receptor enhances biological functions associated with B16 melanoma liver metastasis. Melanoma Res. 2017, 27, 300–308. [Google Scholar] [CrossRef]
- Koroknai, V.; Szasz, I.; Balazs, M. Gene Expression Changes in Cytokine and Chemokine Receptors in Association with Melanoma Liver Metastasis. Int. J. Mol. Sci. 2023, 24, 8901. [Google Scholar] [CrossRef]

| Cell Line | Growth Phase † | Subtype ‡ | BRAF Mutation Status § | NRAS Mutation Status ¶ |
|---|---|---|---|---|
| WM793B | RGP/VGP | SSM | V600E | wt |
| WM1361 | VGP | SSM | wt | Q61L |
| WM278 | VGP | NM | V600E | wt |
| WM983A | VGP | n.a. | V600E | wt |
| WM1366 | VGP | n.a. | wt | Q61L |
| WM3248 | VGP | n.a. | V600E | wt |
| Variables | N | Variables | N |
|---|---|---|---|
| Primary melanoma tumor samples | 19 | Primary melanoma plasma samples | 10 |
| Gender | Gender | ||
| Male | 14 | Male | 6 |
| Female | 5 | Female | 4 |
| Age (years) | Age (years) | ||
| 20–50 | 3 | 20–50 | 2 |
| >50 | 16 | >50 | 8 |
| Breslow thickness | Breslow thickness | ||
| <4.00 | 7 | <4.00 | 8 |
| ≥4.00 | 11 | ≥4.00 | 2 |
| No data | 1 | ||
| Subtype † | Subtype † | ||
| SSM | 11 | SSM | 6 |
| NM | 7 | NM | 4 |
| LM | 1 | LM | 0 |
| Metastasis formation ‡ | Metastasis formation ‡ | ||
| Non-metastatic | 4 | Non-metastatic | 10 |
| Distant organ metastasis | 11 | Distant organ metastasis | 0 |
| Skin metastasis | 4 | Skin metastasis | 0 |
| Ulceration | Ulceration | ||
| Present | 13 | Present | 4 |
| Absent | 6 | Absent | 6 |
| Metastatic melanoma tumor samples | 7 | Metastatic melanoma plasma samples | 30 |
| Gender | Gender | ||
| Male | 4 | Male | 18 |
| Female | 3 | Female | 12 |
| Age (years) | Age (years) | ||
| 20–50 | 2 | 20–50 | 7 |
| >50 | 5 | >50 | 23 |
| Localization | Localization | ||
| Skin | 7 | Skin | 10 |
| Distant organ | 0 | Distant organ | 20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koroknai, V.; Szász, I.; Várvölgyi, T.; Emri, G.; Fodor, Á.; Balázs, M. Cytokine and Chemokine-Associated Signatures Underlying Dermal Invasion and Skin Metastasis in Melanoma. Int. J. Mol. Sci. 2025, 26, 9334. https://doi.org/10.3390/ijms26199334
Koroknai V, Szász I, Várvölgyi T, Emri G, Fodor Á, Balázs M. Cytokine and Chemokine-Associated Signatures Underlying Dermal Invasion and Skin Metastasis in Melanoma. International Journal of Molecular Sciences. 2025; 26(19):9334. https://doi.org/10.3390/ijms26199334
Chicago/Turabian StyleKoroknai, Viktória, István Szász, Tünde Várvölgyi, Gabriella Emri, Ádám Fodor, and Margit Balázs. 2025. "Cytokine and Chemokine-Associated Signatures Underlying Dermal Invasion and Skin Metastasis in Melanoma" International Journal of Molecular Sciences 26, no. 19: 9334. https://doi.org/10.3390/ijms26199334
APA StyleKoroknai, V., Szász, I., Várvölgyi, T., Emri, G., Fodor, Á., & Balázs, M. (2025). Cytokine and Chemokine-Associated Signatures Underlying Dermal Invasion and Skin Metastasis in Melanoma. International Journal of Molecular Sciences, 26(19), 9334. https://doi.org/10.3390/ijms26199334








