Gaussian Accelerated Molecular Dynamics Simulations Combined with NRIMD to Explore the Mechanism of Substrate Selectivity of Cid1 Polymerase for Different Nucleoside Triphosphates
Abstract
1. Introduction
2. Results and Discussion
2.1. Binding of Four Ligand Molecules to Cid1 Polymerase
2.2. Equilibration and Stability of Conventional MD Simulations
2.3. Protein Structural Stability Analysis
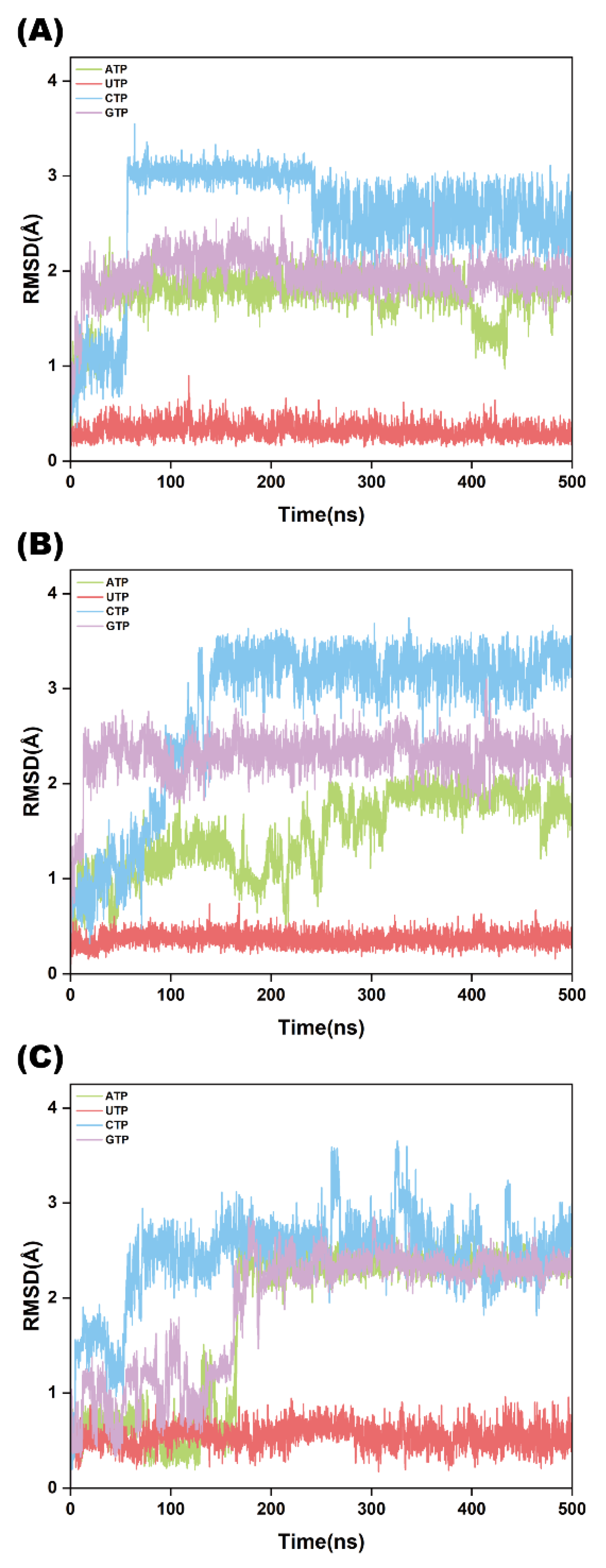
2.4. NTP Structural Stability Analysis
2.5. Binding Free Energy Calculation
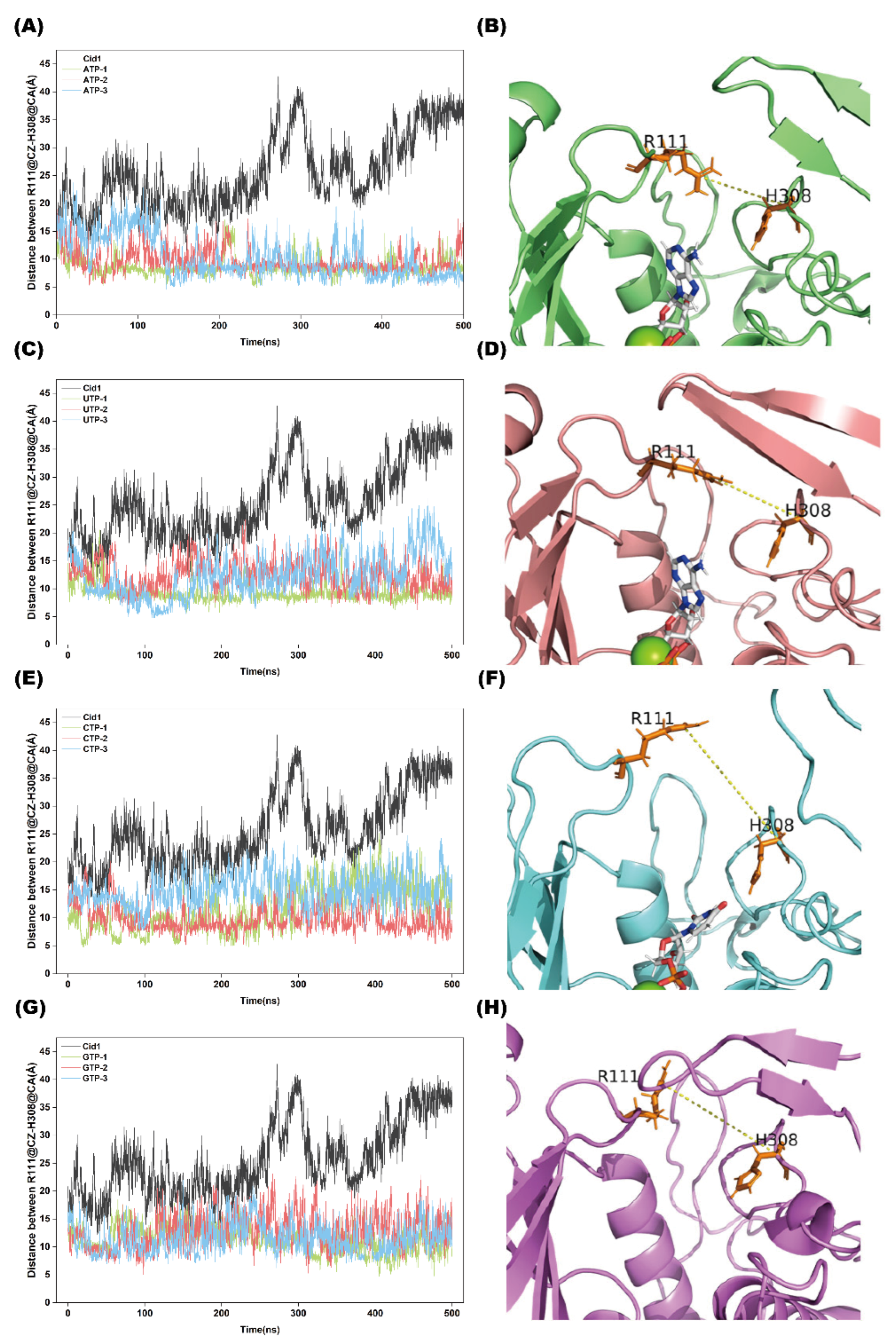
2.6. Distance Analysis
2.7. Salt Bridge Analysis
2.8. Hydrogen Bond Analysis
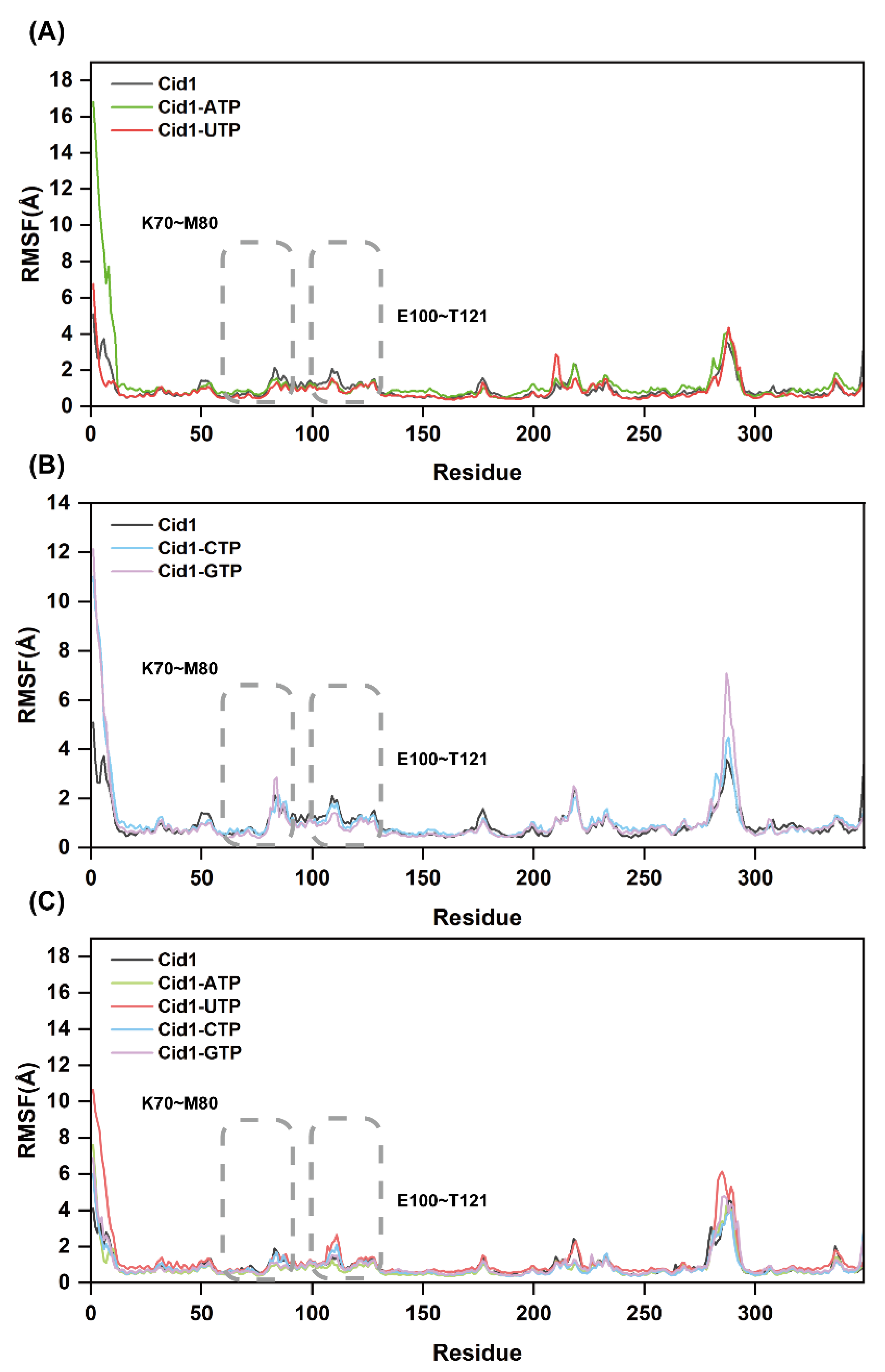
2.9. Protein Residue Flexibility Analysis
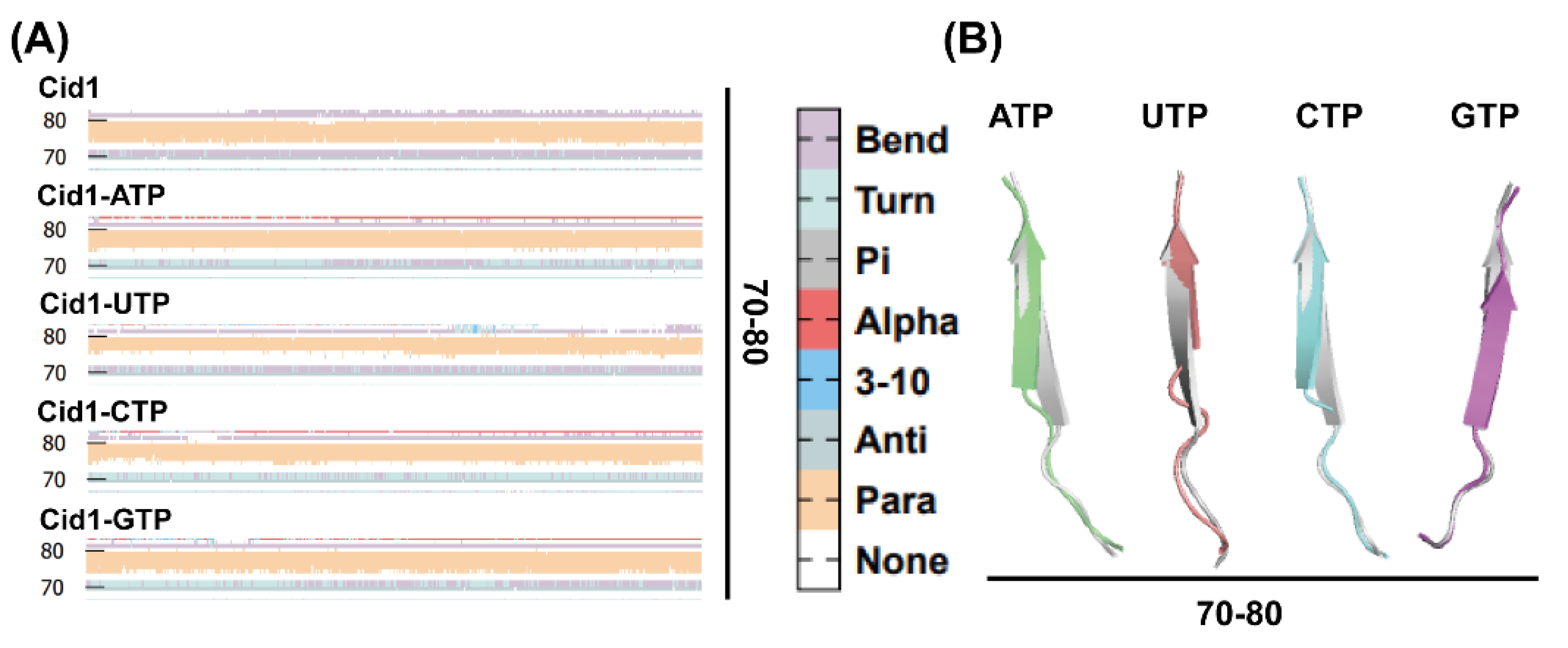
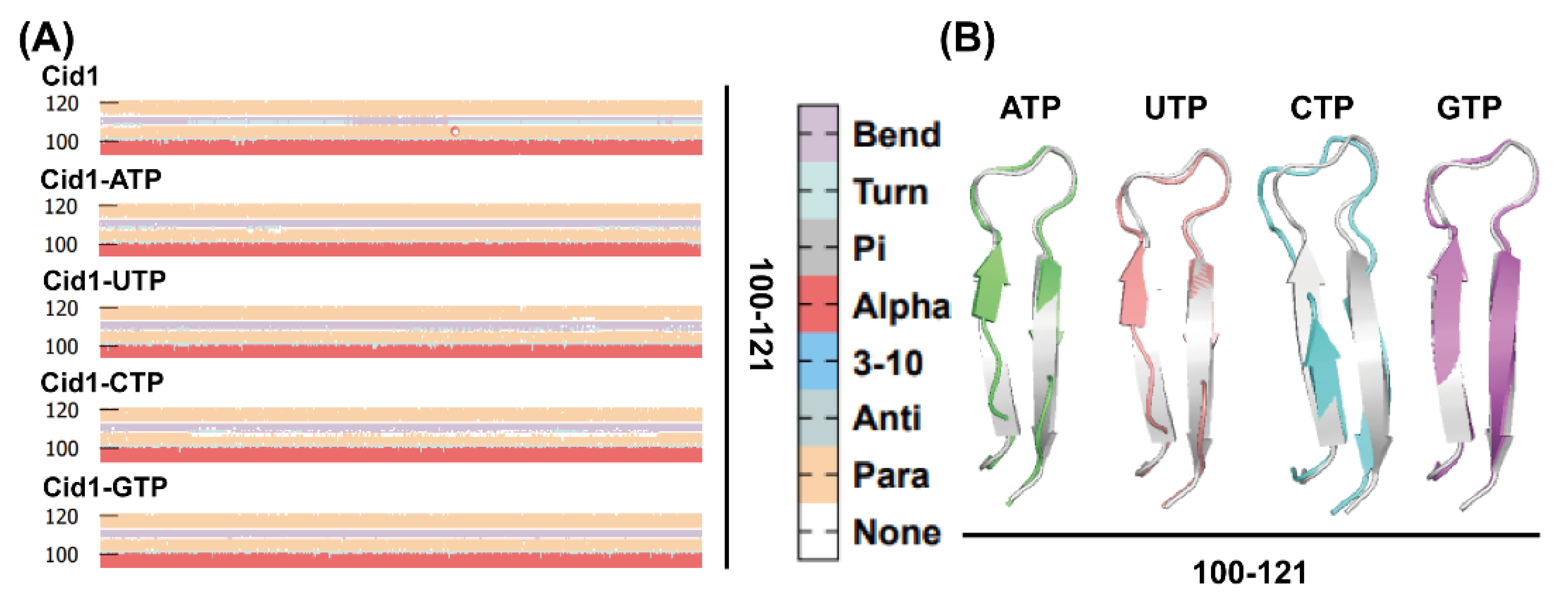
2.10. Secondary Structural Changes During Simulation
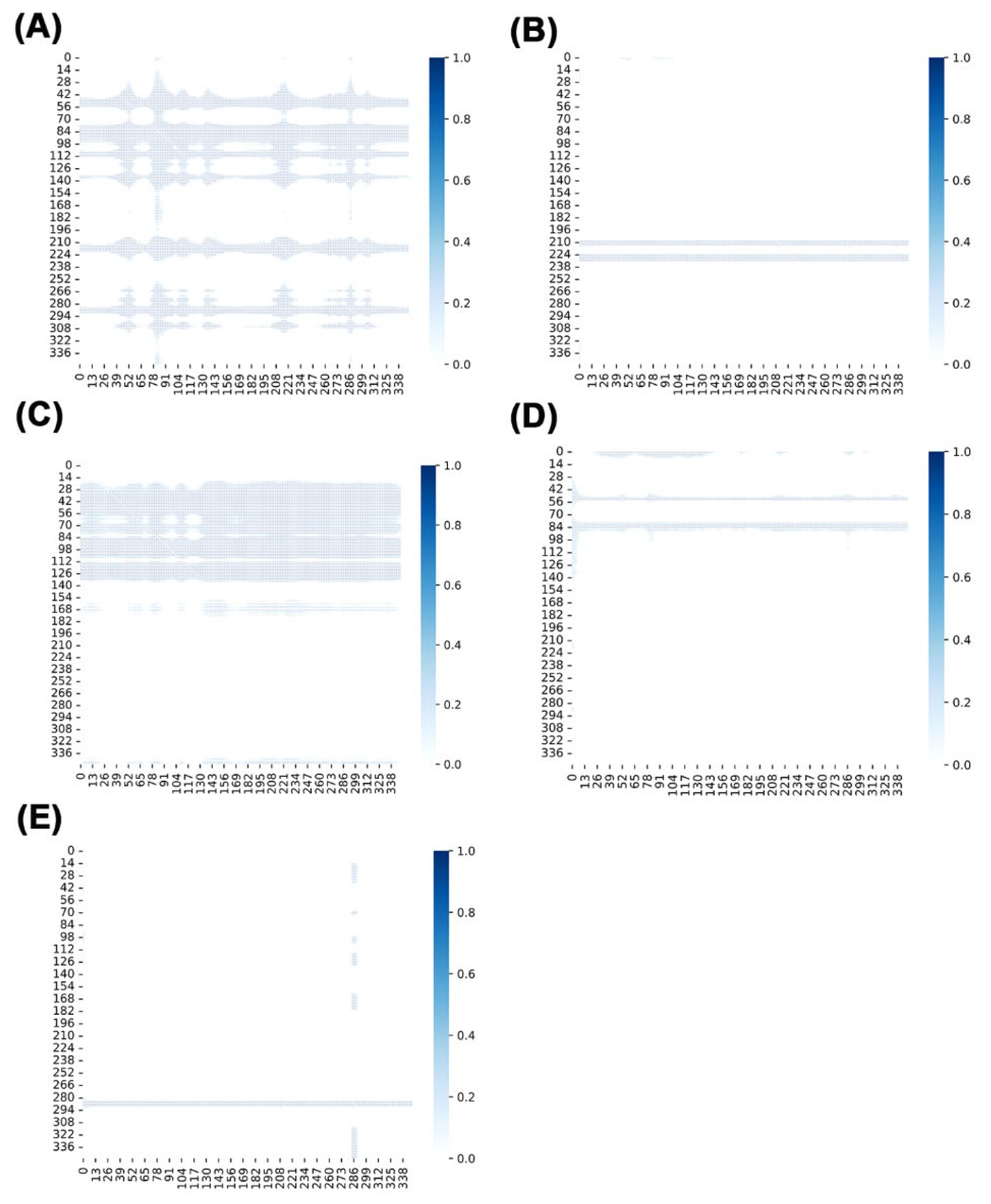
2.11. Analysis via NRIMD Web Server
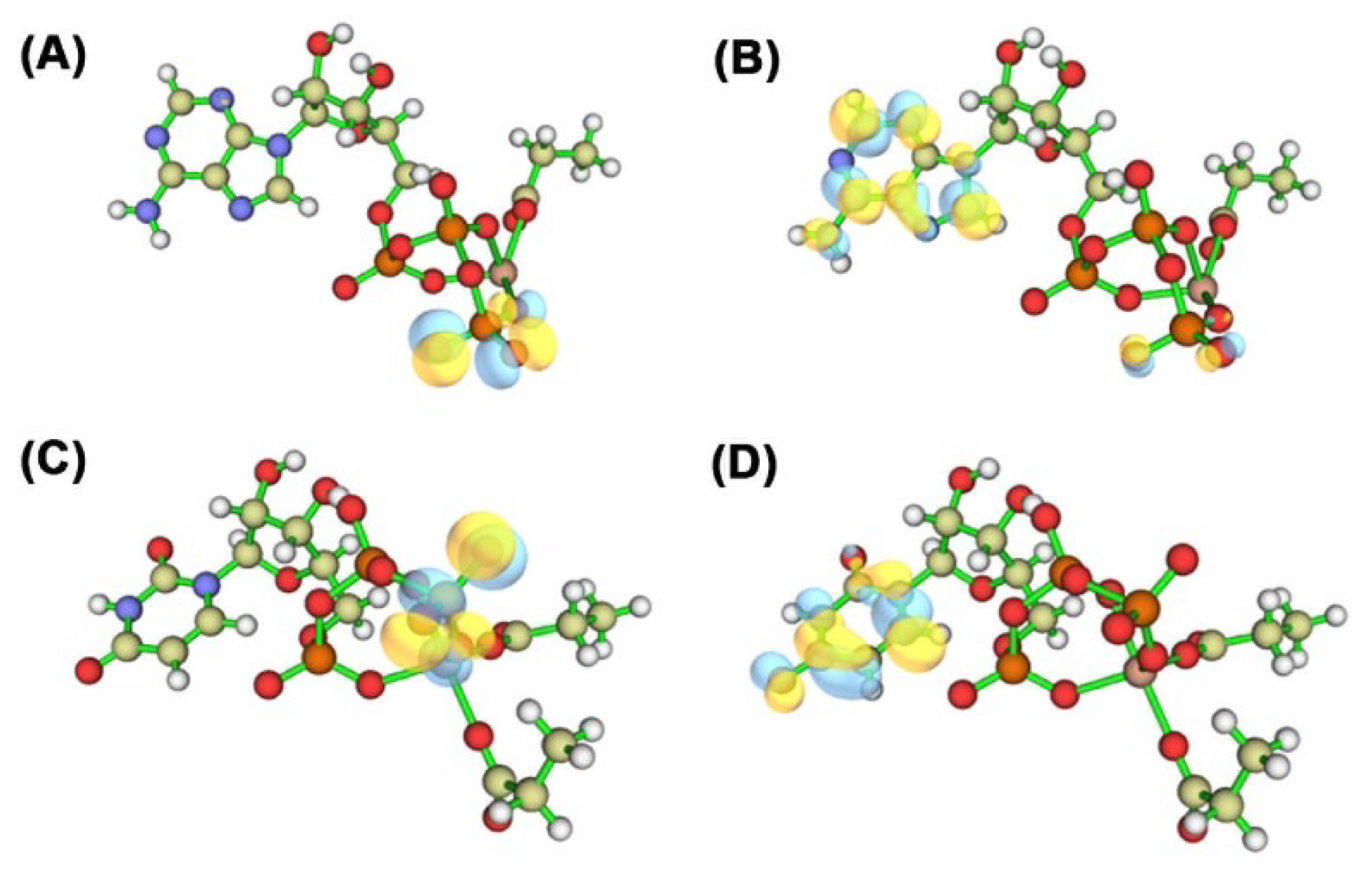
2.12. Quantum Chemical Calculations
3. Materials and Methods
3.1. System Preparation
3.2. Molecular Docking
3.3. Conventional Molecular Dynamics (cMD) Simulation
3.4. Gaussian Accelerated Molecular Dynamics (GaMD) Simulation
3.5. Binding Free Energy Calculation
3.6. Trajectory Analysis
3.7. Salt Bridges Analysis
3.8. NRIMD Web Server
3.9. Quantum Chemical Calculations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brueckner, F.; Ortiz, J.; Cramer, P. A movie of the RNA polymerase nucleotide addition cycle. Curr. Opin. Struct. Biol. 2009, 19, 294–299. [Google Scholar] [CrossRef]
- Steitz, T.A. A mechanism for all polymerases. Nature 1998, 391, 231–232. [Google Scholar] [CrossRef] [PubMed]
- Drygin, D.; Lin, A.; Bliesath, J.; Ho, C.B.; O’Brien, S.E.; Proffitt, C.; Omori, M.; Haddach, M.; Schwaebe, M.K.; Siddiqui-Jain, A.; et al. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 2011, 71, 1418–1430. [Google Scholar] [CrossRef]
- Drygin, D.; Rice, W.G.; Grummt, I. The RNA polymerase I transcription machinery: An emerging target for the treatment of cancer. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 131–156. [Google Scholar] [CrossRef]
- Engel, C.; Sainsbury, S.; Cheung, A.C.; Kostrewa, D.; Cramer, P. RNA polymerase I structure and transcription regulation. Nature 2013, 502, 650–655. [Google Scholar] [CrossRef]
- Fatica, A.; Tollervey, D. Making ribosomes. Curr. Opin. Cell Biol. 2002, 14, 313–318. [Google Scholar] [CrossRef]
- Fernández-Tornero, C.; Moreno-Morcillo, M.; Rashid, U.J.; Taylor, N.M.; Ruiz, F.M.; Gruene, T.; Legrand, P.; Steuerwald, U.; Müller, C.W. Crystal structure of the 14-subunit RNA polymerase I. Nature 2013, 502, 644–649. [Google Scholar] [CrossRef] [PubMed]
- French, S.L.; Osheim, Y.N.; Cioci, F.; Nomura, M.; Beyer, A.L. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell Biol. 2003, 23, 1558–1568. [Google Scholar] [CrossRef] [PubMed]
- Gadal, O.; Labarre, S.; Boschiero, C.; Thuriaux, P. Hmo1, an HMG-box protein, belongs to the yeast ribosomal DNA transcription system. EMBO J. 2002, 21, 5498–5507. [Google Scholar] [CrossRef]
- Turowski, T.W.; Boguta, M. Specific Features of RNA Polymerases I and III: Structure and Assembly. Front. Mol. Biosci. 2021, 8, 680090. [Google Scholar] [CrossRef]
- Ganley, A.R.; Kobayashi, T. Highly efficient concerted evolution in the ribosomal DNA repeats: Total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Res. 2007, 17, 184–191. [Google Scholar] [CrossRef]
- Geiger, S.R.; Kuhn, C.D.; Leidig, C.; Renkawitz, J.; Cramer, P. Crystallization of RNA polymerase I subcomplex A14/A43 by iterative prediction, probing and removal of flexible regions. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2008, 64 Pt 5, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Viktorovskaya, O.V.; Schneider, D.A. Functional divergence of eukaryotic RNA polymerases: Unique properties of RNA polymerase I suit its cellular role. Gene 2015, 556, 19–26. [Google Scholar] [CrossRef]
- Schwank, K.; Schmid, C.; Fremter, T.; Engel, C.; Milkereit, P.; Griesenbeck, J.; Tschochner, H. Features of yeast RNA polymerase I with special consideration of the lobe binding subunits. Biol. Chem. 2023, 404, 979–1002. [Google Scholar] [CrossRef]
- Kwak, J.E.; Wickens, M. A family of poly(U) polymerases. RNA 2007, 13, 860–867. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Malecki, M.; Viegas, S.C.; Carneiro, T.; Golik, P.; Dressaire, C.; Ferreira, M.G.; Arraiano, C.M. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J 2013, 32, 1842–1854. [Google Scholar] [CrossRef]
- Martin, G.; Keller, W. RNA-specific ribonucleotidyl transferases. RNA 2007, 13, 1834–1849. [Google Scholar] [CrossRef]
- Sement, F.M.; Ferrier, E.; Zuber, H.; Merret, R.; Alioua, M.; Deragon, J.M.; Bousquet-Antonelli, C.; Lange, H.; Gagliardi, D. Uridylation prevents 3′ trimming of oligoadenylated mRNAs. Nucleic Acids Res. 2013, 41, 7115–7127. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.J.; West, S.; Norbury, C.J. The human cytoplasmic RNA terminal U-transferase ZCCHC11 targets histone mRNAs for degradation. RNA 2011, 17, 39–44. [Google Scholar] [CrossRef][Green Version]
- Munoz-Tello, P.; Gabus, C.; Thore, S. A critical switch in the enzymatic properties of the Cid1 protein deciphered from its product-bound crystal structure. Nucleic Acids Res. 2014, 42, 3372–3380. [Google Scholar] [CrossRef]
- Rissland, O.S.; Mikulasova, A.; Norbury, C.J. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol. Cell Biol. 2007, 27, 3612–3624. [Google Scholar] [CrossRef] [PubMed]
- Lunde, B.M.; Magler, I.; Meinhart, A. Crystal structures of the Cid1 poly (U) polymerase reveal the mechanism for UTP selectivity. Nucleic Acids Res. 2012, 40, 9815–9824. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, J.; Sui, Y.; Liu, S.; Yang, M. In silico drug repurposing carvedilol and its metabolites against SARS-CoV-2 infection using molecular docking and molecular dynamic simulation approaches. Sci. Rep. 2023, 13, 21404. [Google Scholar] [CrossRef] [PubMed]
- Skariyachan, S.; Gopal, D.; Deshpande, D.; Joshi, A.; Uttarkar, A.; Niranjan, V. Carbon fullerene and nanotube are probable binders to multiple targets of SARS-CoV-2: Insights from computational modeling and molecular dynamic simulation studies. Infect. Genet. Evol. 2021, 96, 105155. [Google Scholar] [CrossRef]
- Parihar, A.; Sonia, Z.F.; Akter, F.; Ali, M.A.; Hakim, F.T.; Hossain, M.S. Phytochemicals-based targeting RdRp and main protease of SARS-CoV-2 using docking and steered molecular dynamic simulation: A promising therapeutic approach for Tackling COVID-19. Comput. Biol. Med. 2022, 145, 105468. [Google Scholar] [CrossRef] [PubMed]
- Kawsar, S.M.A.; Hosen, M.A.; Ahmad, S.; El Bakri, Y.; Laaroussi, H.; Ben Hadda, T.; Almalki, F.A.; Ozeki, Y.; Goumri-Said, S. Potential SARS-CoV-2 RdRp inhibitors of cytidine derivatives: Molecular docking, molecular dynamic simulations, ADMET, and POM analyses for the identification of pharmacophore sites. PLoS ONE 2022, 17, e0273256. [Google Scholar] [CrossRef]
- Azeem, M.; Mustafa, G.; Ahmed, S.; Mushtaq, A.; Arshad, M.; Usama, M.; Farooq, M. Structure based screening and molecular docking with dynamic simulation of natural secondary metabolites to target RNA-dependent RNA polymerase of five different retroviruses. PLoS ONE 2024, 19, e0307615. [Google Scholar] [CrossRef] [PubMed]
- Meng, E.C.; Gschwend, D.A.; Blaney, J.M.; Kuntz, I.D. Orientational sampling and rigid-body minimization in molecular docking. Proteins 1993, 17, 266–278. [Google Scholar] [CrossRef]
- Morris, G.M.; Lim-Wilby, M. Molecular docking. Methods Mol. Biol. 2008, 443, 365–382. [Google Scholar]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Flyvbjerg, H.; Petersen, H.G. Error estimates on averages of correlated data. J. Chem. Phys. 1989, 91, 461–466. [Google Scholar] [CrossRef]
- Grossfield, A.; Patrone, P.N.; Roe, D.R.; Schultz, A.J.; Siderius, D.W.; Zuckerman, D.M. Best Practices for Quantification of Uncertainty and Sampling Quality in Molecular Simulations [Article v1.0]. Living J. Comput. Mol. Sci. 2018, 1, 5067. [Google Scholar] [CrossRef]
- Chodera, J.D. A Simple Method for Automated Equilibration Detection in Molecular Simulations. J. Chem. Theory Comput. 2016, 12, 1799–1805. [Google Scholar] [CrossRef]
- Homeyer, N.; Gohlke, H. Free Energy Calculations by the Molecular Mechanics Poisson-Boltzmann Surface Area Method. Mol. Inform. 2012, 31, 114–122. [Google Scholar] [CrossRef]
- Wang, E.; Sun, H.; Wang, J.; Wang, Z.; Liu, H.; Zhang, J.Z.H.; Hou, T. End-Point Binding Free Energy Calculation with MM/PBSA and MM/GBSA: Strategies and Applications in Drug Design. Chem. Rev. 2019, 119, 9478–9508. [Google Scholar] [CrossRef]
- DeRocco, A.G. Modern Quantum Chemistry. Am. J. Phys. 1967, 35, 1097. [Google Scholar] [CrossRef]
- Eryilmaz, S.; Gul, M.; Inkaya, E. Investigation of global reactivity descriptors of some perillaldehyde derivatives in different solvents by DFT method. Indian J. Chem. Technol. 2019, 26, 235–238. [Google Scholar]
- Dixit, V.; Yadav, R.A. DFT-B3LYP computations of electro and thermo molecular characteristics and mode of action of fungicides (chlorophenols). Int. J. Pharm. 2015, 491, 277–284. [Google Scholar] [CrossRef]
- Wang, C.; Greene, D.; Xiao, L.; Qi, R.; Luo, R. Recent Developments and Applications of the MMPBSA Method. Front. Mol. Biosci. 2017, 4, 87. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Cheatham, T.E., 3rd; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M., Jr.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef]
- Tian, C.; Kasavajhala, K.; Belfon, K.A.A.; Raguette, L.; Huang, H.; Migues, A.N.; Bickel, J.; Wang, Y.; Pincay, J.; Wu, Q.; et al. ff19SB: Amino-Acid-Specific Protein Backbone Parameters Trained against Quantum Mechanics Energy Surfaces in Solution. J. Chem. Theory Comput. 2020, 16, 528–552. [Google Scholar] [CrossRef]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef]
- Izadi, S.; Anandakrishnan, R.; Onufriev, A.V. Building Water Models: A Different Approach. J. Phys. Chem. Lett. 2014, 5, 3863–3871. [Google Scholar] [CrossRef]
- Bullerjahn, J.T.; von Bülow, S.; Heidari, M.; Hénin, J.; Hummer, G. Unwrapping NPT Simulations to Calculate Diffusion Coefficients. J. Chem. Theory Comput. 2023, 19, 3406–3417. [Google Scholar] [CrossRef]
- Erban, R.; Togashi, Y. Asymmetric Periodic Boundary Conditions for All-Atom Molecular Dynamics and Coarse-Grained Simulations of Nucleic Acids. J. Phys. Chem. B 2023, 127, 8257–8267. [Google Scholar] [CrossRef] [PubMed]
- Howard, M.P.; Statt, A.; Stone, H.A.; Truskett, T.M. Stability of force-driven shear flows in nonequilibrium molecular simulations with periodic boundaries. J. Chem. Phys. 2020, 152, 214113. [Google Scholar] [CrossRef] [PubMed]
- Simonnin, P.; Noetinger, B.T.; Nieto-Draghi, C.; Marry, V.; Rotenberg, B. Diffusion under Confinement: Hydrodynamic Finite-Size Effects in Simulation. J. Chem. Theory Comput. 2017, 13, 2881–2889. [Google Scholar] [CrossRef]
- Winkler, R.G.; Wysocki, A.; Gompper, G. Virial pressure in systems of spherical active Brownian particles. Soft Matter 2015, 11, 6680–6691. [Google Scholar] [CrossRef] [PubMed]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Tao, M.; Owhadi, H.; Marsden, J.E. Symplectic, linearly-implicit and stable integrators, with applications to constrained dynamics. arXiv 2011, arXiv:1103.4645. [Google Scholar]
- Harvey, M.J.; De Fabritiis, G. An Implementation of the Smooth Particle Mesh Ewald Method on GPU Hardware. J. Chem. Theory Comput. 2009, 5, 2371–2377. [Google Scholar] [CrossRef]
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E., III. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Miao, Y.; McCammon, J.A. Gaussian Accelerated Molecular Dynamics: Theory, Implementation, and Applications. Annu. Rep. Comput. Chem. 2017, 13, 231–278. [Google Scholar]
- Khan, M.T.; Ali, S.; Zeb, M.T.; Kaushik, A.C.; Malik, S.I.; Wei, D.Q. Gibbs Free Energy Calculation of Mutation in PncA and RpsA Associated with Pyrazinamide Resistance. Front. Mol. Biosci. 2020, 7, 52. [Google Scholar] [CrossRef]
- Massova, I.; Kollman, P.A. Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspect. Drug Discov. Des. 2000, 18, 113–135. [Google Scholar] [CrossRef]
- Case, D.A.B.K.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Ghoreishi, D. AMBER 2022; University of California: San Francisco, CA, USA, 2022. [Google Scholar]
- Roe, D.R.; Cheatham, T.E., 3rd. Parallelization of CPPTRAJ enables large scale analysis of molecular dynamics trajectory data. J. Comput. Chem. 2018, 39, 2110–2117. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Nussinov, R. Close-range electrostatic interactions in proteins. Chembiochem 2002, 3, 604–617. [Google Scholar] [CrossRef]
- Thomas, A.S.; Elcock, A.H. Direct Observation of Salt Effects on Molecular Interactions through Explicit-Solvent Molecular Dynamics Simulations: Differential Effects on Electrostatic and Hydrophobic Interactions and Comparisons to Poisson-Boltzmann Theory. J. Am. Chem. Soc. 2006, 128, 7796–7806. [Google Scholar] [CrossRef]
- Donald, J.E.; Kulp, D.W.; DeGrado, W.F. Salt bridges: Geometrically specific, designable interactions. Proteins 2011, 79, 898–915. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wang, S.; Zeng, S.; Zhu, J.; Xu, D.; Han, W.; Wang, J. NRIMD, a Web Server for Analyzing Protein Allosteric Interactions Based on Molecular Dynamics Simulation. J. Chem. Inf. Model. 2024, 64, 7176–7183. [Google Scholar] [CrossRef]
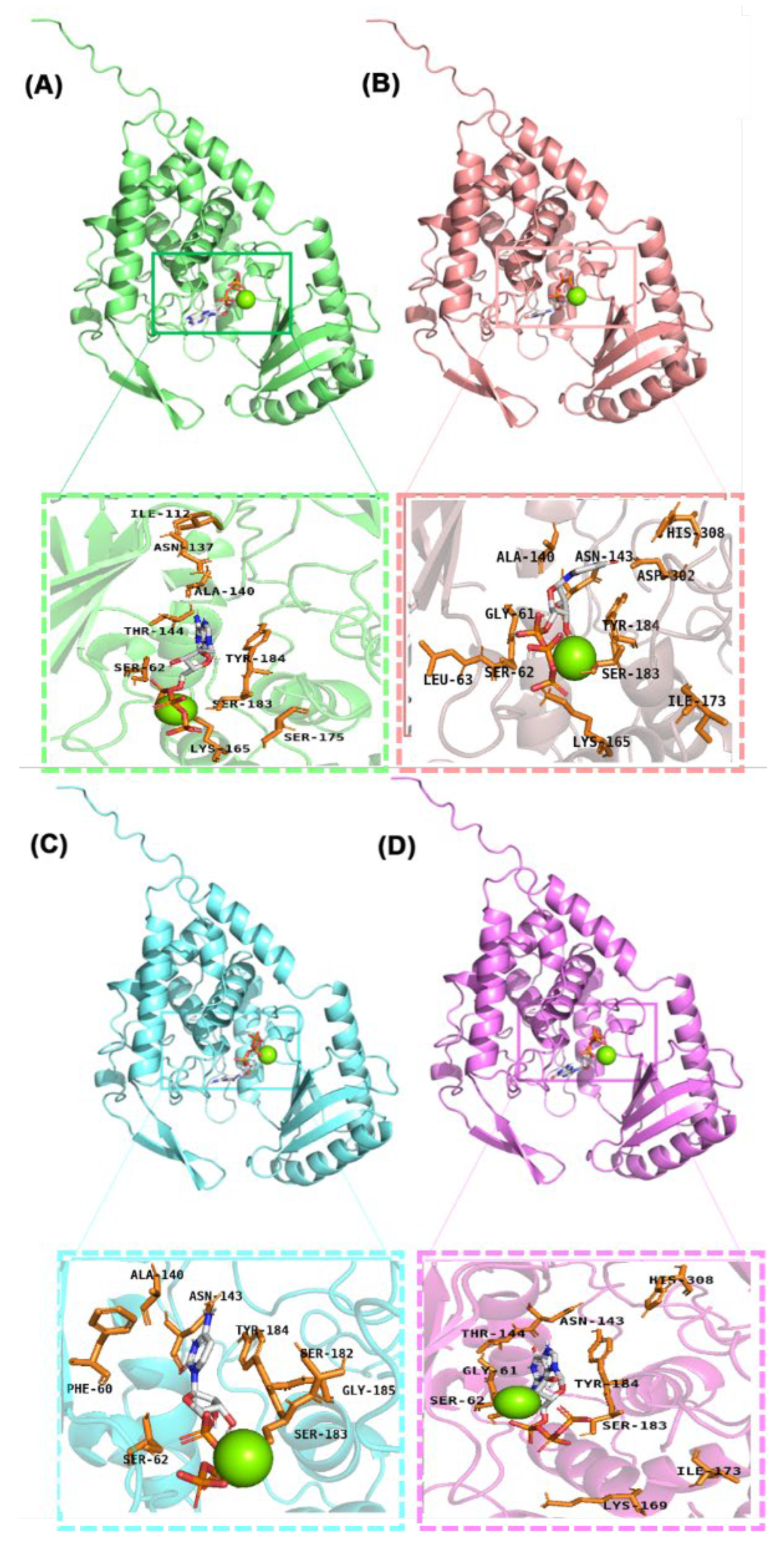
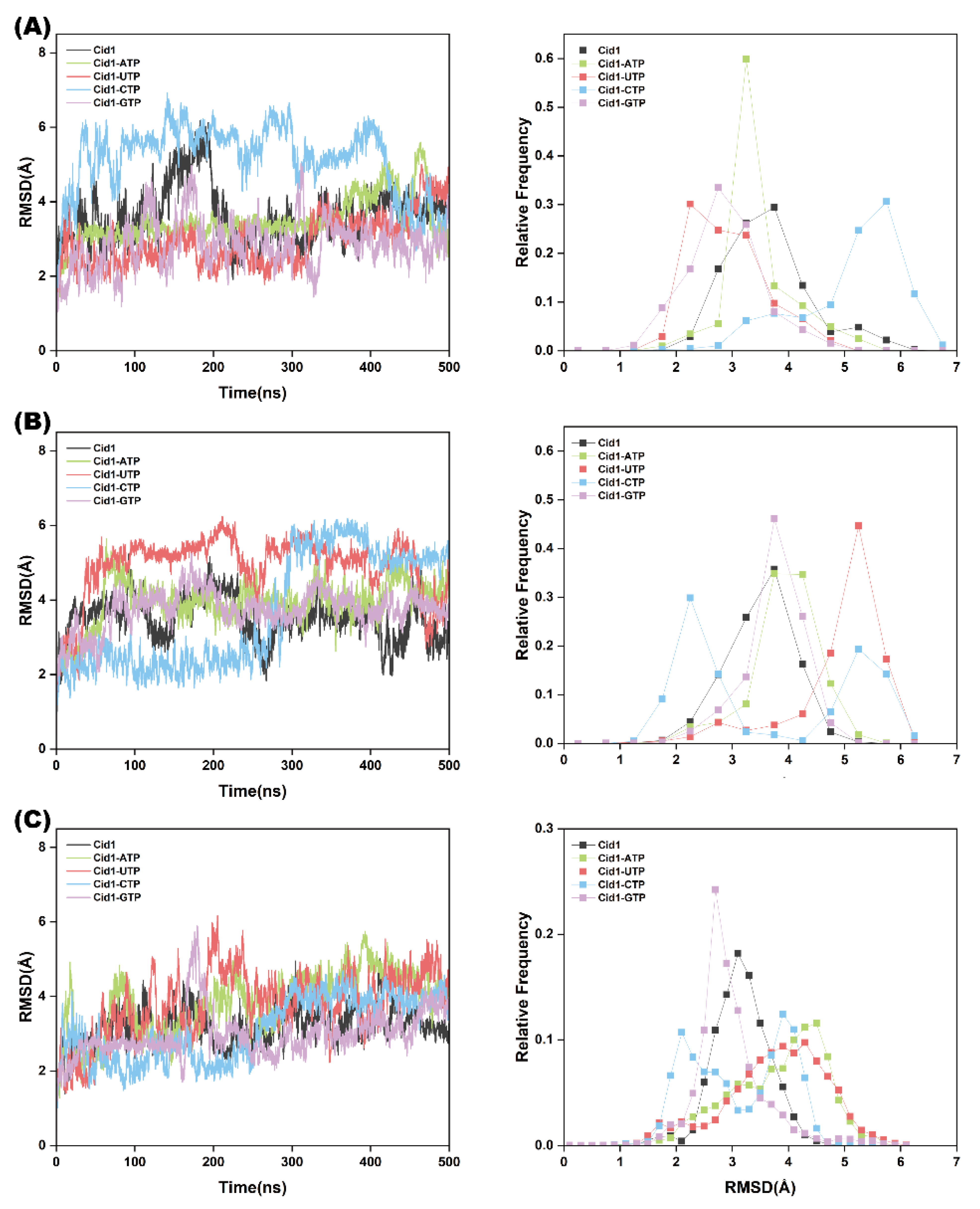
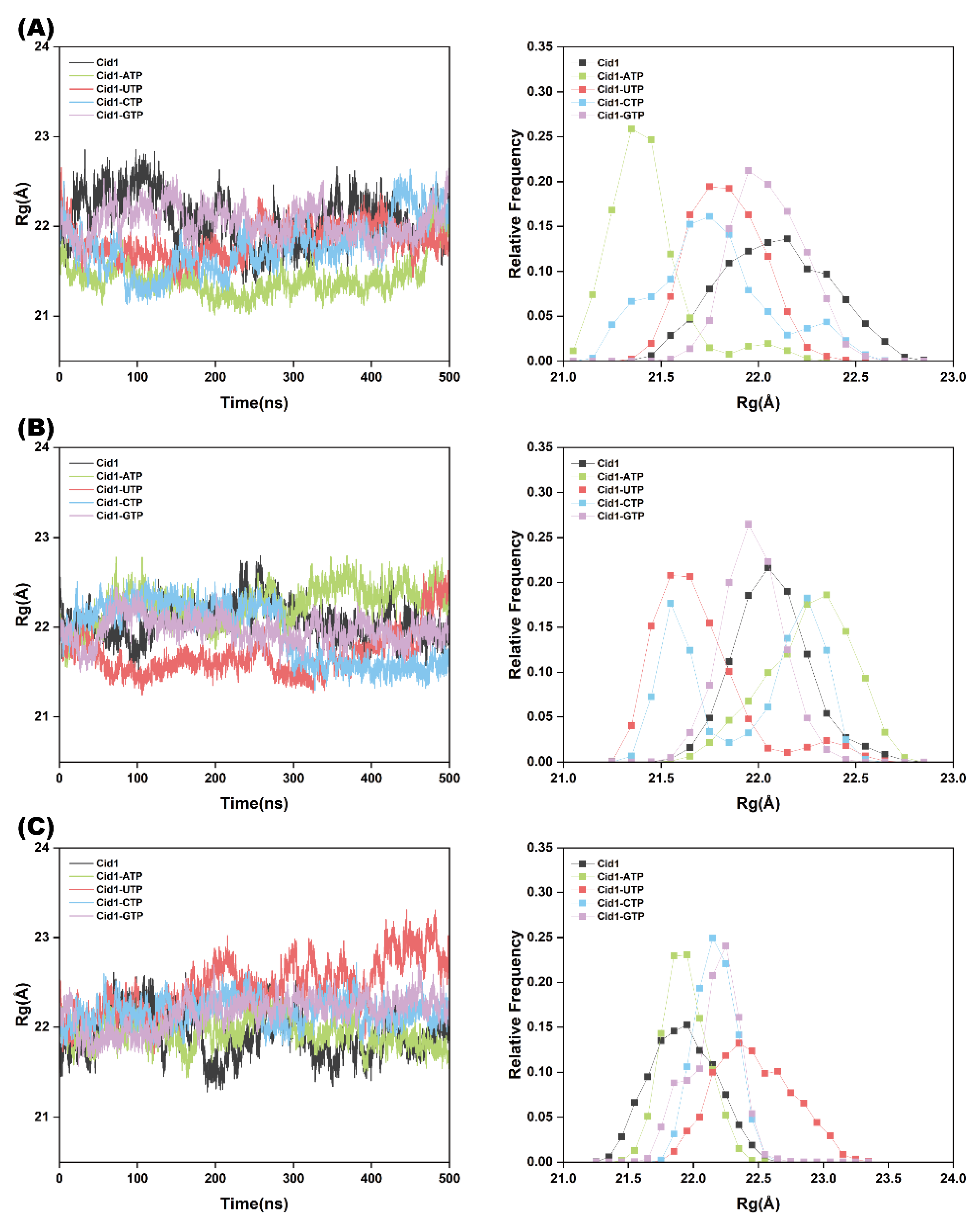
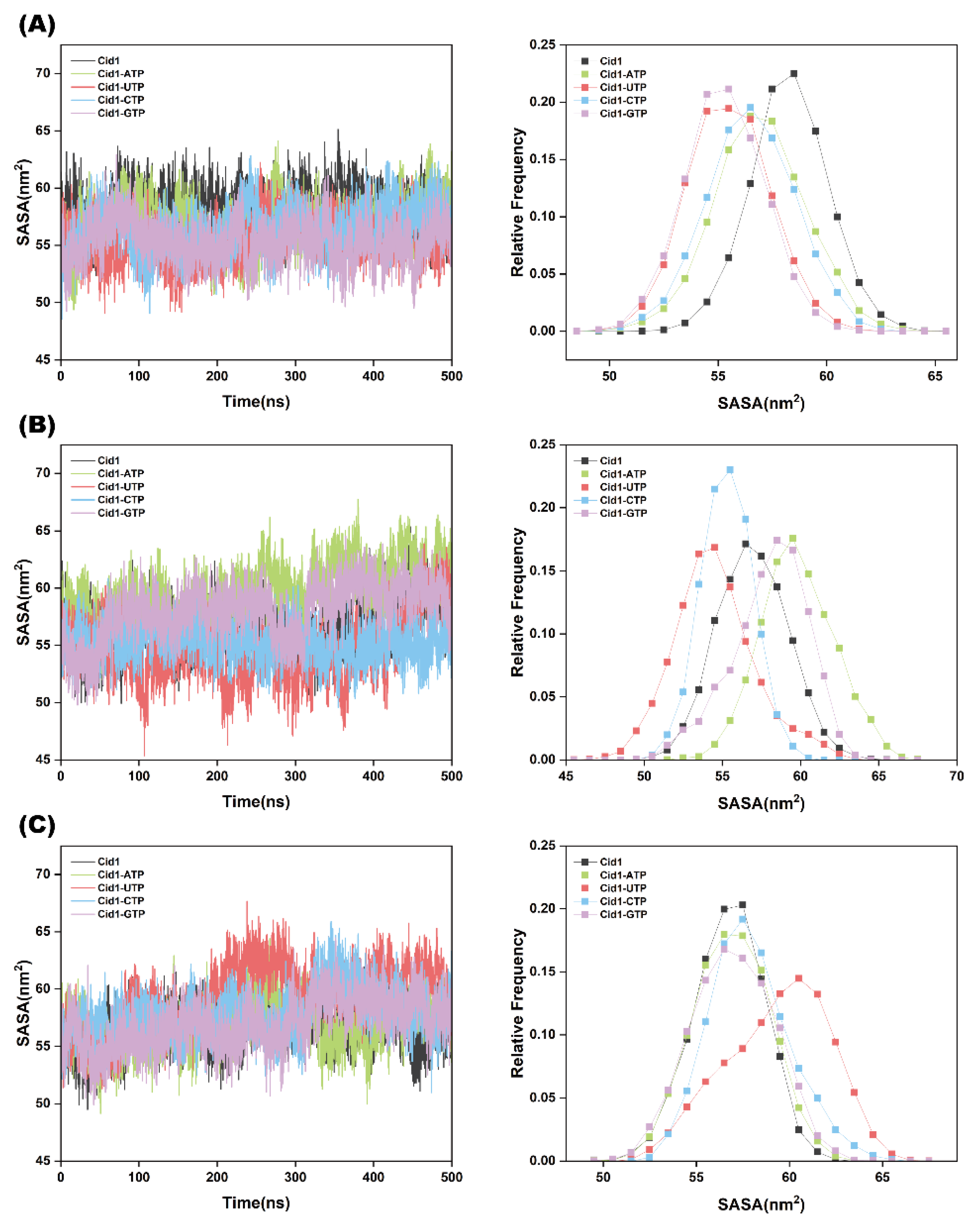

| Systems | Cid1-ATP | Cid1-UTP | Cid1-CTP | Cid1-GTP |
|---|---|---|---|---|
| ∆GvdW | −33.20 ± 0.53 | −41.35 ± 0.64 | −45.77 ± 0.59 | −46.20 ± 0.57 |
| ∆Gele | 123.74 ± 5.48 | −186.07 ± 3.96 | −319.88 ± 3.64 | −2.92 ± 5.00 |
| ∆Gsolv | 203.56 ± 5.14 | 728.83 ± 4.01 | 358.93 ± 3.30 | 406.24 ± 4.23 |
| ∆Ggas | −307.07 ± 5.43 | −862.52 ± 4.25 | −396.36 ± 3.66 | −475.08 ± 4.92 |
| ∆Gtotal | −103.51 ± 0.85 | −133.69 ± 0.78 | −37.43 ± 0.73 | −68.84 ± 1.44 |
| Systems | Saltbridge |
|---|---|
| Cid1-ATP | 0.028 |
| Cid1-UTP | 0.028 |
| Cid1-CTP | 0.016 |
| Cid1-GTP | 0.024 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Zhou, X.; Wang, H.; Cao, F.; Han, W. Gaussian Accelerated Molecular Dynamics Simulations Combined with NRIMD to Explore the Mechanism of Substrate Selectivity of Cid1 Polymerase for Different Nucleoside Triphosphates. Int. J. Mol. Sci. 2025, 26, 9325. https://doi.org/10.3390/ijms26199325
Liu H, Zhou X, Wang H, Cao F, Han W. Gaussian Accelerated Molecular Dynamics Simulations Combined with NRIMD to Explore the Mechanism of Substrate Selectivity of Cid1 Polymerase for Different Nucleoside Triphosphates. International Journal of Molecular Sciences. 2025; 26(19):9325. https://doi.org/10.3390/ijms26199325
Chicago/Turabian StyleLiu, Hanwen, Xue Zhou, Haohao Wang, Fuyan Cao, and Weiwei Han. 2025. "Gaussian Accelerated Molecular Dynamics Simulations Combined with NRIMD to Explore the Mechanism of Substrate Selectivity of Cid1 Polymerase for Different Nucleoside Triphosphates" International Journal of Molecular Sciences 26, no. 19: 9325. https://doi.org/10.3390/ijms26199325
APA StyleLiu, H., Zhou, X., Wang, H., Cao, F., & Han, W. (2025). Gaussian Accelerated Molecular Dynamics Simulations Combined with NRIMD to Explore the Mechanism of Substrate Selectivity of Cid1 Polymerase for Different Nucleoside Triphosphates. International Journal of Molecular Sciences, 26(19), 9325. https://doi.org/10.3390/ijms26199325








