Cerebrospinal Fluid-Derived Small Extracellular Vesicles May Better Reflect Medulloblastoma Proteomes than Those from Blood Plasma
Abstract
1. Introduction
2. Results
2.1. sEVs Elute in Different Fractions Depending on the Body Fluid
2.2. Analysis of BP and CSF sEV Proteome Reveals Body-Fluid-Specific Protein Ontology Enrichments
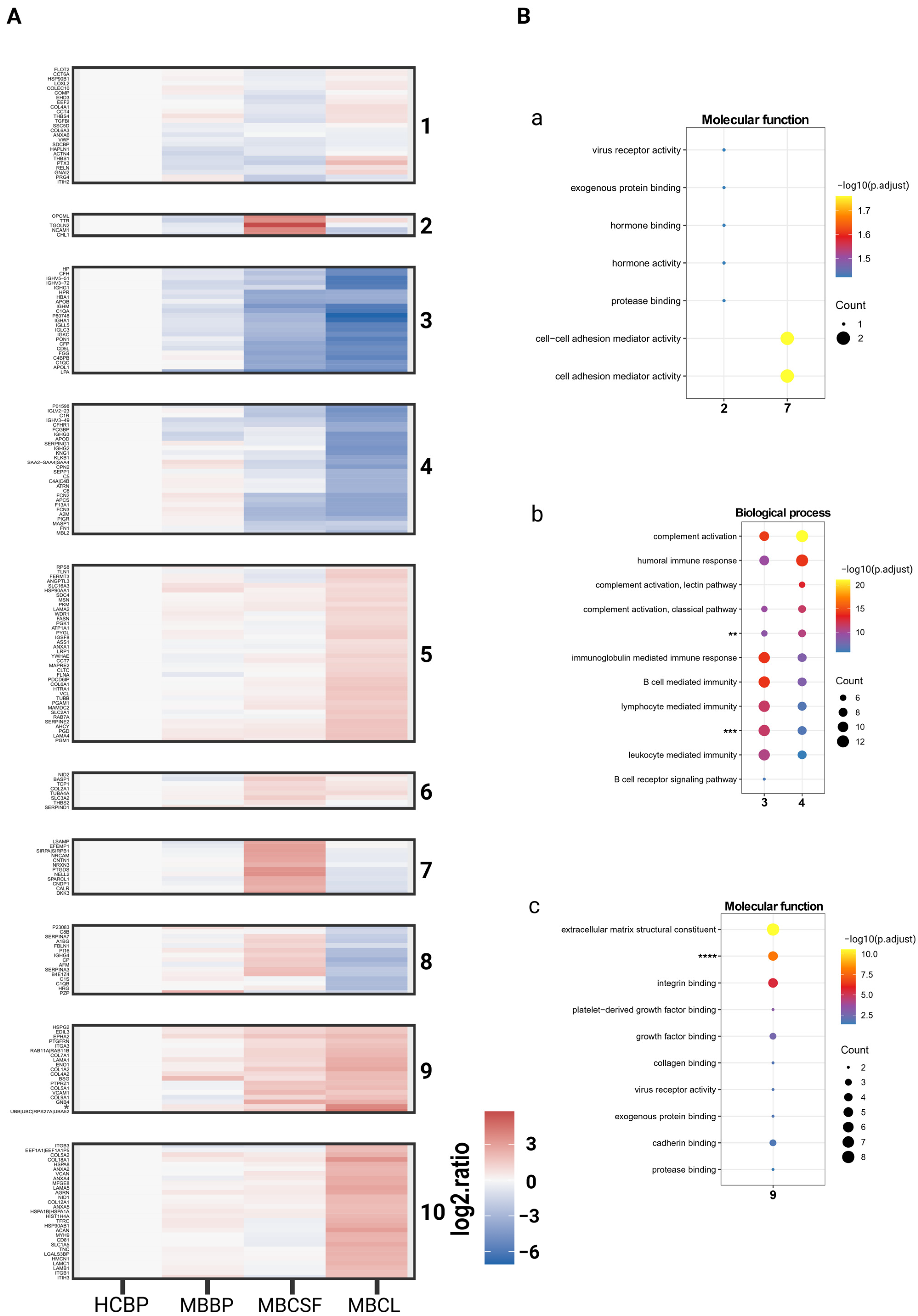
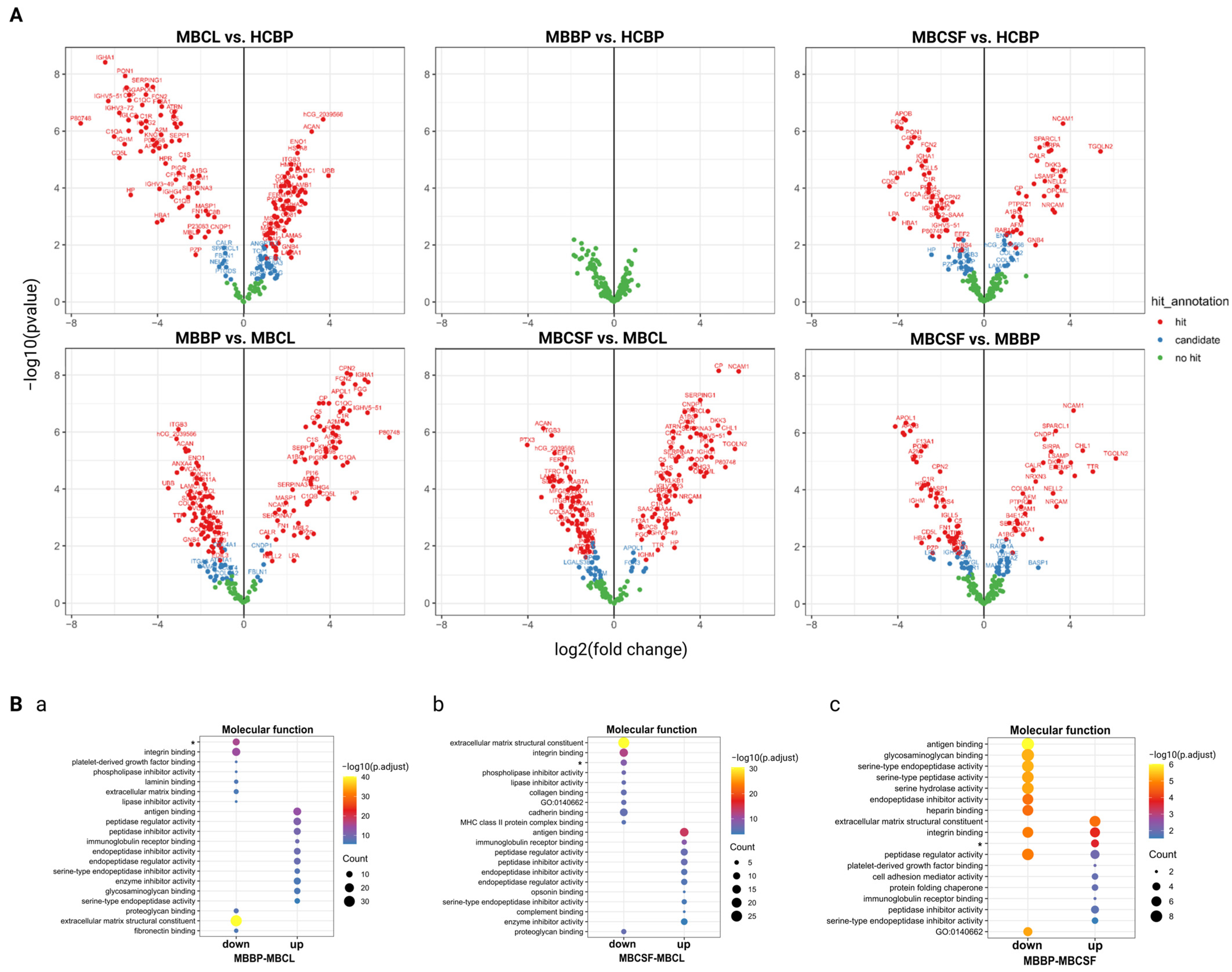
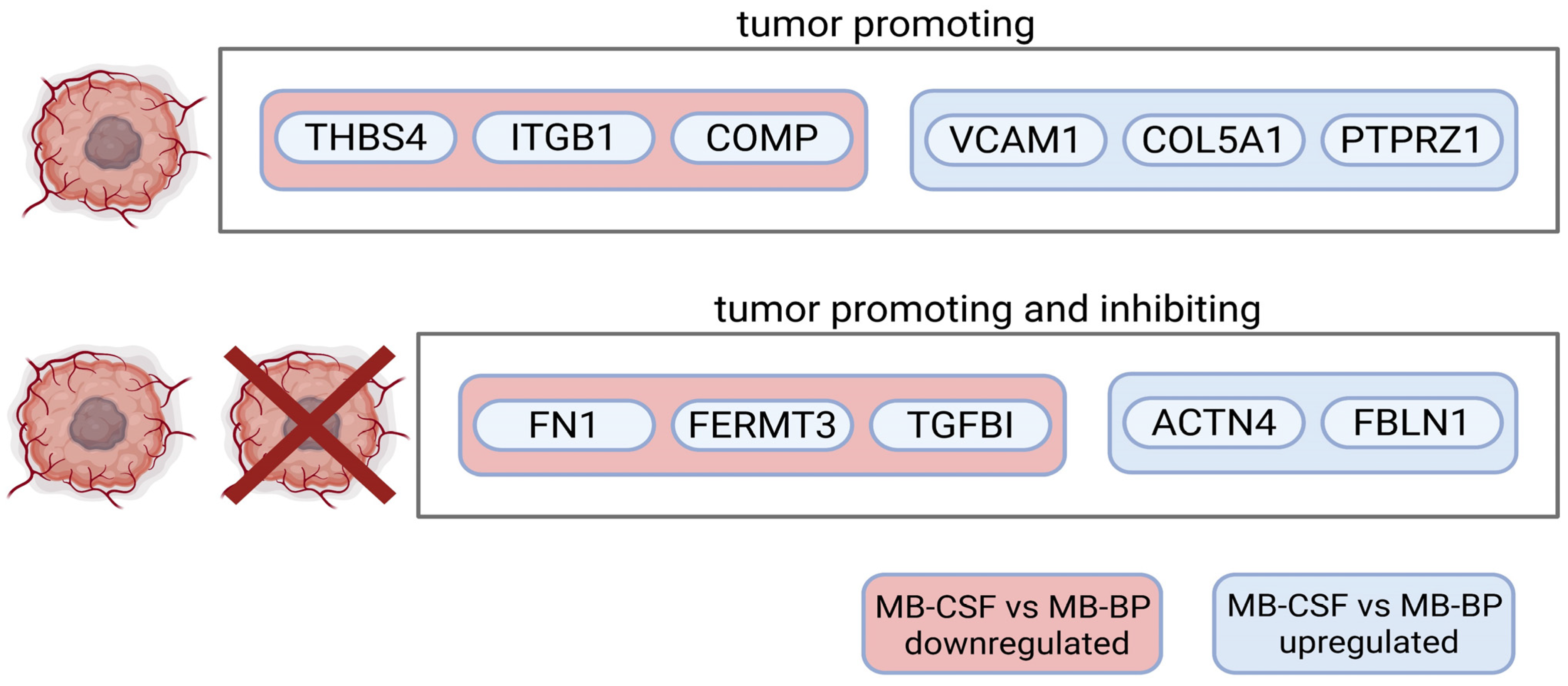
2.3. Extracellular Matrix and Integrin Binding Are Differently Expressed Between MBCL, MBCSF, and MBBP
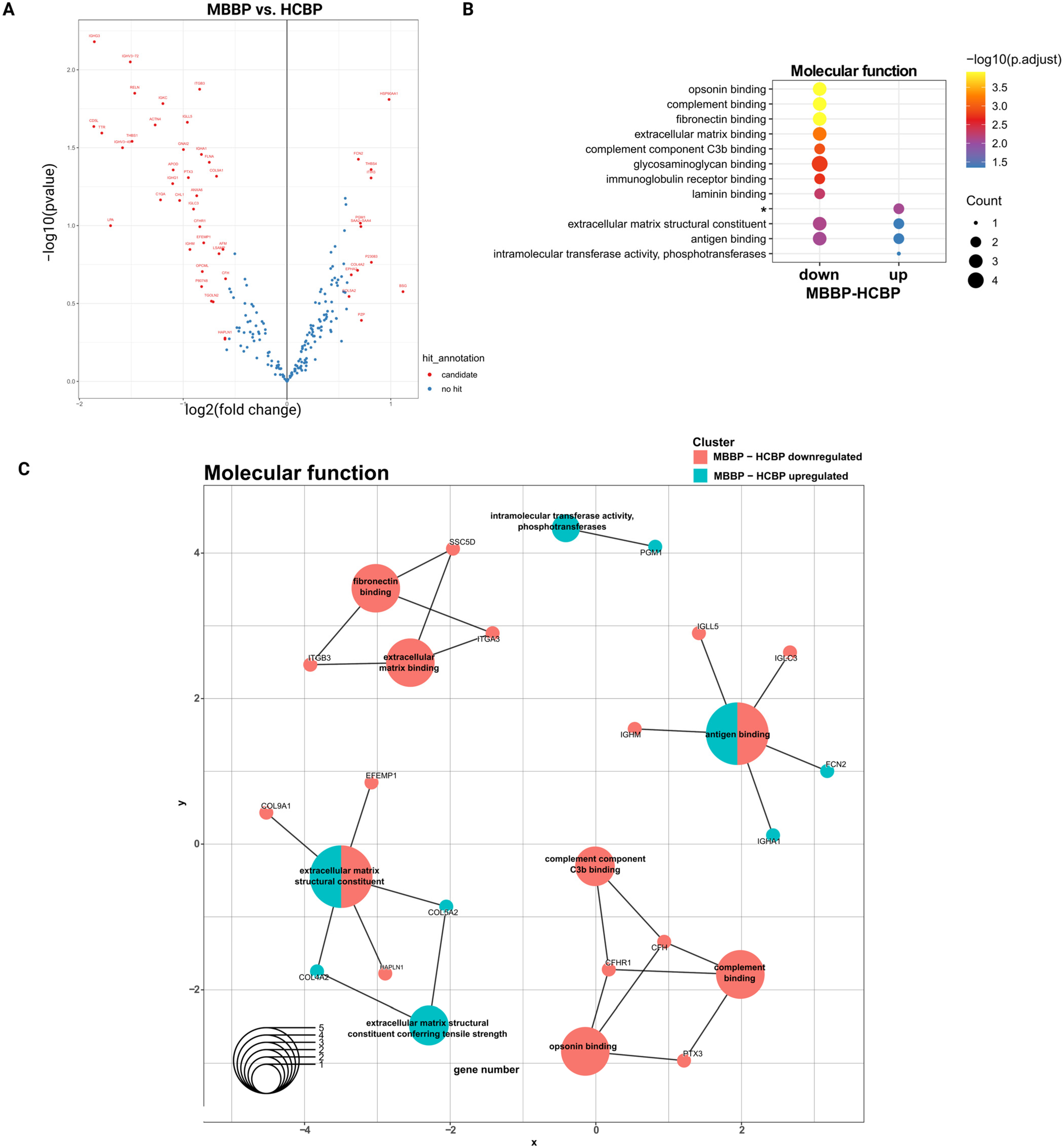
2.4. MBBP and HCBP Differ in ECM- and Complement-Related Proteins Based on logFC Values
2.5. Complement System and Integrin Cell Surface Interactions Represent Central Pathways in MB-sEVs
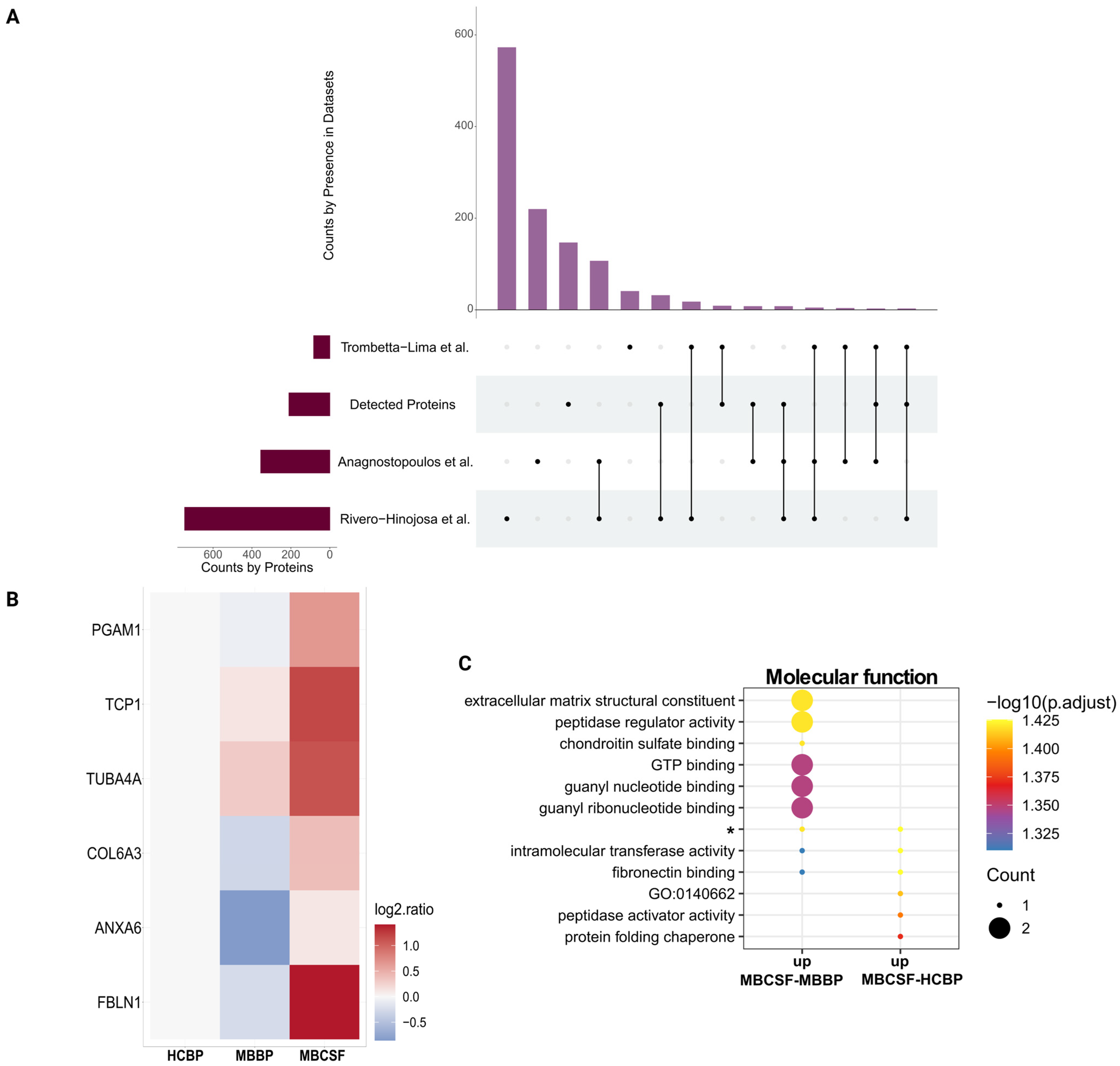
2.6. CSF-sEVs Surpass BP-sEVs in Recovering MB Proteins
3. Discussion
4. Materials and Methods
4.1. Collection and Processing of Patient Samples
4.2. Isolation of Small Extracellular Vesicles from Blood Plasma and Cerebrospinal Fluid (UF-SEC)
4.3. Isolation of Small Extracellular Vesicles from Medulloblastoma Cell Lines
4.4. Characterization of Small Extracellular Vesicles
4.4.1. Particle Concentration
4.4.2. Protein Concentration
4.4.3. Bead-Assisted Flow Cytometry
4.4.4. Transmission Electron Microscopy (TEM) and Measurement of sEV Size
4.5. Mass Spectrometry and Data Analysis
4.5.1. Sample Preparation
4.5.2. LC-MS/MS P2258
4.5.3. Data Processing, MaxQuant
4.5.4. Data Analysis
4.6. Selection of MB, CSF and sEV Datasets and Comparison of Identified Proteins
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| EV | Extracellular vesicle |
| sEV | Small extracellular vesicle |
| CSF | Cerebrospinal fluid |
| BP | Blood plasma |
| CNS | Central nervous system |
| MB | Medulloblastoma |
| UF-SEC | Ultrafiltration–size exclusion chromatography |
| GO | Gene Ontology |
| ECM | Extracellular matrix |
| cfDNA | Cell-free DNA |
| BBB | Blood–brain barrier |
| RT | Room temperature |
| DPBS | Dulbecco’s Phosphate-Buffered Saline |
| NTA | Nanoparticle Tracking Analysis |
| BCA | Bicinchoninic acid assay |
| BSA | Bovine Serum Albumine |
| FITC | Fluorescein-isothiocyanat |
| FACS | Fluorescence-activated cell sorting |
| MFI | Mean fluorescence intensity |
| LC-MS/MS | Liquid Chromatography–Tandem Mass Spectrometry |
| FDR | False discovery rate |
| MBBP | Blood plasma from medulloblastoma patient(s) |
| MBCSF | Cerebrospinal fluid from medulloblastoma patient(s) |
| HCBP | Blood plasma from healthy control(s) |
| MBCL | Medulloblastoma cell lines |
| TGOLN2 | Trans-golgi network protein 2 |
| FC | Fold change |
| ANXA6 | Annexin A6 |
| ITGB1 | Integrin β-1 |
| TME | Tumor microenvironment |
| COL5A1 | Collagen type V alpha 1 chain |
| COL9A1 | Collagen type IX alpha 1 chain |
| COL4A1 | Collagen type IV alpha 1 chain |
| BTB | Brain-tumor-barrier |
| WNT | Wingless-related integration site |
References
- Serra, R.; Mangraviti, A. A systematic view of pediatric medulloblastoma proteomics-current state of the field and future directions. Child’s Nerv. Syst. 2021, 37, 779–788. [Google Scholar] [CrossRef]
- Schwalbe, E.C.; Lindsey, J.C.; Nakjang, S.; Crosier, S.; Smith, A.J.; Hicks, D.; Rafiee, G.; Hill, R.M.; Iliasova, A.; Stone, T.; et al. Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: A cohort study. Lancet Oncol. 2017, 18, 958–971. [Google Scholar] [CrossRef]
- Miller, A.M.; Karajannis, M.A. Current Role and Future Potential of CSF ctDNA for the Diagnosis and Clinical Management of Pediatric Central Nervous System Tumors. J. Natl. Compr. Canc Netw. 2022, 20, 1363–1369. [Google Scholar] [PubMed]
- Ghanam, J.; Chetty, V.K.; Barthel, L.; Reinhardt, D.; Hoyer, P.F.; Thakur, B.K. DNA in extracellular vesicles: From evolution to its current application in health and disease. Cell Biosci. 2022, 12, 37. [Google Scholar] [CrossRef]
- Chetty, V.K.; Ghanam, J.; Anchan, S.; Reinhardt, K.; Brenzel, A.; Gelleri, M.; Cremer, C.; Grueso-Navarro, E.; Schneider, M.; von Neuhoff, N.; et al. Efficient Small Extracellular Vesicles (EV) Isolation Method and Evaluation of EV-Associated DNA Role in Cell-Cell Communication in Cancer. Cancers 2022, 14, 2068. [Google Scholar] [CrossRef] [PubMed]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrugger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404, Correction in J. Extracell. Vesicles 2024, 13, e12451. [Google Scholar] [CrossRef]
- Marar, C.; Starich, B.; Wirtz, D. Extracellular vesicles in immunomodulation and tumor progression. Nat. Immunol. 2021, 22, 560–570. [Google Scholar] [CrossRef]
- Morad, G.; Carman, C.V.; Hagedorn, E.J.; Perlin, J.R.; Zon, L.I.; Mustafaoglu, N.; Park, T.E.; Ingber, D.E.; Daisy, C.C.; Moses, M.A. Tumor-Derived Extracellular Vesicles Breach the Intact Blood-Brain Barrier via Transcytosis. ACS Nano 2019, 13, 13853–13865. [Google Scholar] [CrossRef] [PubMed]
- Kadry, H.; Noorani, B.; Cucullo, L. A blood–brain barrier overview on structure, function, impairment, and biomarkers of integrity. Fluids Barriers CNS 2020, 17, 69. [Google Scholar] [CrossRef]
- Arvanitis, C.D.; Ferraro, G.B.; Jain, R.K. The blood-brain barrier and blood-tumour barrier in brain tumours and metastases. Nat. Rev. Cancer 2020, 20, 26–41. [Google Scholar] [CrossRef]
- Barthel, L.; Hadamitzky, M.; Dammann, P.; Schedlowski, M.; Sure, U.; Thakur, B.K.; Hetze, S. Glioma: Molecular signature and crossroads with tumor microenvironment. Cancer Metastasis Rev. 2022, 41, 53–75. [Google Scholar] [CrossRef]
- Garcia-Romero, N.; Carrion-Navarro, J.; Esteban-Rubio, S.; Lazaro-Ibanez, E.; Peris-Celda, M.; Alonso, M.M.; Guzman-De-Villoria, J.; Fernandez-Carballal, C.; de Mendivil, A.O.; Garcia-Duque, S.; et al. DNA sequences within glioma-derived extracellular vesicles can cross the intact blood-brain barrier and be detected in peripheral blood of patients. Oncotarget 2017, 8, 1416–1428. [Google Scholar] [CrossRef]
- Salviano-Silva, A.; Wollmann, K.; Brenna, S.; Reimer, R.; Neumann, J.E.; Dottermusch, M.; Woythe, L.; Maire, C.L.; Puig, B.; Schuller, U.; et al. Extracellular Vesicles Carrying Tenascin-C are Clinical Biomarkers and Improve Tumor-Derived DNA Analysis in Glioblastoma Patients. ACS Nano 2025, 19, 9844–9859. [Google Scholar] [CrossRef]
- Santos, V.; Freitas, C.; Fernandes, M.G.; Sousa, C.; Reboredo, C.; Cruz-Martins, N.; Mosquera, J.; Hespanhol, V.; Campelo, R. Liquid biopsy: The value of different bodily fluids. Biomark. Med. 2022, 16, 127–145. [Google Scholar] [CrossRef] [PubMed]
- Tivey, A.; Church, M.; Rothwell, D.; Dive, C.; Cook, N. Circulating tumour DNA—Looking beyond the blood. Nat. Rev. Clin. Oncol. 2022, 19, 600–612. [Google Scholar] [CrossRef] [PubMed]
- Cornelli, L.; Van Paemel, R.; Ferro Dos Santos, M.R.; Roelandt, S.; Willems, L.; Vandersteene, J.; Baert, E.; Mus, L.M.; Van Roy, N.; De Wilde, B.; et al. Diagnosis of pediatric central nervous system tumors using methylation profiling of cfDNA from cerebrospinal fluid. Clin. Epigenetics 2024, 16, 87. [Google Scholar] [CrossRef]
- Miller, A.M.; Szalontay, L.; Bouvier, N.; Hill, K.; Ahmad, H.; Rafailov, J.; Lee, A.J.; Rodriguez-Sanchez, M.I.; Yildirim, O.; Patel, A.; et al. Next-generation sequencing of cerebrospinal fluid for clinical molecular diagnostics in pediatric, adolescent and young adult brain tumor patients. Neuro Oncol. 2022, 24, 1763–1772. [Google Scholar] [CrossRef] [PubMed]
- Benayas, B.; Morales, J.; Egea, C.; Armisen, P.; Yanez-Mo, M. Optimization of extracellular vesicle isolation and their separation from lipoproteins by size exclusion chromatography. J. Extracell. Biol. 2023, 2, e100. [Google Scholar] [CrossRef]
- De Sousa, K.P.; Rossi, I.; Abdullahi, M.; Ramirez, M.I.; Stratton, D.; Inal, J.M. Isolation and characterization of extracellular vesicles and future directions in diagnosis and therapy. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol 2023, 15, e1835. [Google Scholar] [CrossRef]
- O’Halloran, K.; Crotty, E.E.; Christodoulou, E.; Leary, S.E.; Miller, A.; Paulson, V.A.; Lockwood, C.M.; Margol, A.S.; Biegel, J.A. Targeted detection of sequence variants in cell-free DNA from cerebrospinal fluid in pediatric central nervous system tumors. Front. Oncol. 2024, 14, 1513073. [Google Scholar] [CrossRef]
- Benedikter, B.J.; Bouwman, F.G.; Vajen, T.; Heinzmann, A.C.A.; Grauls, G.; Mariman, E.C.; Wouters, E.F.M.; Savelkoul, P.H.; Lopez-Iglesias, C.; Koenen, R.R.; et al. Ultrafiltration combined with size exclusion chromatography efficiently isolates extracellular vesicles from cell culture media for compositional and functional studies. Sci. Rep. 2017, 7, 15297. [Google Scholar] [CrossRef] [PubMed]
- Droste, M.; Tertel, T.; Jeruschke, S.; Dittrich, R.; Kontopoulou, E.; Walkenfort, B.; Borger, V.; Hoyer, P.F.; Buscher, A.K.; Thakur, B.K.; et al. Single Extracellular Vesicle Analysis Performed by Imaging Flow Cytometry and Nanoparticle Tracking Analysis Evaluate the Accuracy of Urinary Extracellular Vesicle Preparation Techniques Differently. Int. J. Mol. Sci. 2021, 22, 12436. [Google Scholar] [CrossRef] [PubMed]
- Gerashchenko, T.; Skitchenko, R.; Korobeynikova, A.; Kuanysheva, K.; Khozyainova, A.; Vorobiev, R.; Rodionov, E.; Miller, S.; Topolnitsky, E.; Shefer, N.; et al. Whole-exome sequencing reveals an association of rs112065068 in TGOLN2 gene with distant metastasis of non-small cell lung cancer. Gene 2024, 920, 148507. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Nikanjam, M.; Kato, S.; Kurzrock, R. Liquid biopsy: Current technology and clinical applications. J. Hematol. Oncol. 2022, 15, 131. [Google Scholar] [CrossRef]
- O’Brien, K.; Ughetto, S.; Mahjoum, S.; Nair, A.V.; Breakefield, X.O. Uptake, functionality, and re-release of extracellular vesicle-encapsulated cargo. Cell Rep. 2022, 39, 110651. [Google Scholar] [CrossRef]
- Ter-Ovanesyan, D.; Norman, M.; Lazarovits, R.; Trieu, W.; Lee, J.H.; Church, G.M.; Walt, D.R. Framework for rapid comparison of extracellular vesicle isolation methods. Elife 2021, 10, 70725. [Google Scholar] [CrossRef]
- Ghadami, S.; Dellinger, K. The lipid composition of extracellular vesicles: Applications in diagnostics and therapeutic delivery. Front. Mol. Biosci. 2023, 10, 1198044. [Google Scholar] [CrossRef]
- Lischnig, A.; Bergqvist, M.; Ochiya, T.; Lässer, C. Quantitative proteomics identifies proteins enriched in large and small extracellular vesicles. Mol. Cell. Proteom. 2022, 21, 100273. [Google Scholar] [CrossRef]
- Herzog, M.; Verdenik, I.; Kobal, B.; Černe, K. Size distribution of extracellular vesicles in pretreatment ascites and plasma is correlated with primary treatment outcome in advanced high-grade serous carcinoma. Sci. Rep. 2025, 15, 4500. [Google Scholar] [CrossRef]
- Garcia-Martin, R.; Brandao, B.B.; Thomou, T.; Altindis, E.; Kahn, C.R. Tissue differences in the exosomal/small extracellular vesicle proteome and their potential as indicators of altered tissue metabolism. Cell Rep. 2022, 38, 110277. [Google Scholar] [CrossRef]
- Sochocka, M.; Diniz, B.S.; Leszek, J. Inflammatory response in the CNS: Friend or foe? Mol. Neurobiol. 2017, 54, 8071–8089. [Google Scholar] [CrossRef]
- Biller, S.J.; Coe, A.; Arellano, A.A.; Dooley, K.; Silvestri, S.M.; Gong, J.S.; Yeager, E.A.; Becker, J.W.; Chisholm, S.W. Environmental and taxonomic drivers of bacterial extracellular vesicle production in marine ecosystems. Appl. Environ. Microbiol. 2023, 89, e0059423. [Google Scholar] [CrossRef]
- Tóth, E.Á.; Turiák, L.; Visnovitz, T.; Cserép, C.; Mázló, A.; Sódar, B.W.; Försönits, A.I.; Petővári, G.; Sebestyén, A.; Komlósi, Z.; et al. Formation of a protein corona on the surface of extracellular vesicles in blood plasma. J. Extracell. Vesicles 2021, 10, e12140. [Google Scholar] [CrossRef]
- Försönits, A.I.; Tóth, E.Á.; Jezsoviczky, S.; Bárkai, T.; Khamari, D.; Galinsoga, A.; Királyhidi, P.; Kittel, Á.; Fazakas, J.; Lenzinger, D.; et al. Improved accessibility of extracellular vesicle surface molecules upon partial removal of the protein corona by high ionic strength. J. Extracell. Vesicles 2025, 14, e70124. [Google Scholar] [CrossRef] [PubMed]
- Suresh, P.S.; Zhang, Q. Comprehensive comparison of methods for isolation of extracellular vesicles from human plasma. J. Proteome Res. 2025, 24, 2956–2967. [Google Scholar] [CrossRef]
- Escudero-Cernuda, S.; Eiro, N.; Fraile, M.; Vizoso, F.J.; Fernández-Colomer, B.; Fernández-Sánchez, M.L. Limitations and challenges in the characterization of extracellular vesicles from stem cells and serum. Microchim. Acta 2025, 192, 311. [Google Scholar] [CrossRef]
- Hanelova, K.; Raudenska, M.; Masarik, M.; Balvan, J. Protein cargo in extracellular vesicles as the key mediator in the progression of cancer. Cell Commun. Signal 2024, 22, 25. [Google Scholar] [CrossRef]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Li, Y.; Zhang, S.; Wang, X.; Dou, H.; Yu, X.; Zhang, Z.; Yang, S.; Xiao, M. Extracellular matrix remodeling in tumor progression and immune escape: From mechanisms to treatments. Mol. Cancer 2023, 22, 48. [Google Scholar] [CrossRef] [PubMed]
- Trombetta-Lima, M.; Rosa-Fernandes, L.; Angeli, C.B.; Moretti, I.F.; Franco, Y.M.; Mousessian, A.S.; Wakamatsu, A.; Lerario, A.M.; Oba-Shinjo, S.M.; Pasqualucci, C.A.; et al. Extracellular Matrix Proteome Remodeling in Human Glioblastoma and Medulloblastoma. J. Proteome Res. 2021, 20, 4693–4707. [Google Scholar] [CrossRef] [PubMed]
- Linke, F.; Aldighieri, M.; Lourdusamy, A.; Grabowska, A.M.; Stolnik, S.; Kerr, I.D.; Merry, C.L.; Coyle, B. 3D hydrogels reveal medulloblastoma subgroup differences and identify extracellular matrix subtypes that predict patient outcome. J. Pathol. 2021, 253, 326–338. [Google Scholar] [CrossRef]
- Karamanos, N.K.; Theocharis, A.D.; Piperigkou, Z.; Manou, D.; Passi, A.; Skandalis, S.S.; Vynios, D.H.; Orian-Rousseau, V.; Ricard-Blum, S.; Schmelzer, C.E.H.; et al. A guide to the composition and functions of the extracellular matrix. FEBS J. 2021, 288, 6850–6912. [Google Scholar] [CrossRef] [PubMed]
- Barros, C.S.; Franco, S.J.; Muller, U. Extracellular matrix: Functions in the nervous system. Cold Spring Harb. Perspect. Biol. 2011, 3, a005108. [Google Scholar] [CrossRef]
- Xu, S.; Xu, H.; Wang, W.; Li, S.; Li, H.; Li, T.; Zhang, W.; Yu, X.; Liu, L. The role of collagen in cancer: From bench to bedside. J. Transl. Med. 2019, 17, 309. [Google Scholar] [CrossRef]
- Szvicsek, Z.; Oszvald, A.; Szabo, L.; Sandor, G.O.; Kelemen, A.; Soos, A.A.; Paloczi, K.; Harsanyi, L.; Tolgyes, T.; Dede, K.; et al. Extracellular vesicle release from intestinal organoids is modulated by Apc mutation and other colorectal cancer progression factors. Cell Mol. Life Sci. 2019, 76, 2463–2476. [Google Scholar] [CrossRef]
- Chen, X.; Ma, C.; Li, Y.; Liang, Y.; Chen, T.; Han, D.; Luo, D.; Zhang, N.; Zhao, W.; Wang, L.; et al. COL5A1 promotes triple-negative breast cancer progression by activating tumor cell-macrophage crosstalk. Oncogene 2024, 43, 1742–1756. [Google Scholar] [CrossRef]
- Zhu, H.; Hu, X.; Feng, S.; Jian, Z.; Xu, X.; Gu, L.; Xiong, X. The Hypoxia-Related Gene COL5A1 Is a Prognostic and Immunological Biomarker for Multiple Human Tumors. Oxid. Med. Cell Longev. 2022, 2022, 6419695. [Google Scholar] [CrossRef]
- Gu, S.; Peng, Z.; Wu, Y.; Wang, Y.; Lei, D.; Jiang, X.; Zhao, H.; Fu, P. COL5A1 Serves as a Biomarker of Tumor Progression and Poor Prognosis and May Be a Potential Therapeutic Target in Gliomas. Front. Oncol. 2021, 11, 752694. [Google Scholar] [CrossRef]
- Wang, H.; Liu, Z.; Li, A.; Wang, J.; Liu, J.; Liu, B.; Lian, X.; Zhang, B.; Pang, B.; Liu, L.; et al. COL4A1 as a novel oncogene associated with the clinical characteristics of malignancy predicts poor prognosis in glioma. Exp. Ther. Med. 2021, 22, 1224. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Sampson, C.; Liu, C.; Piao, H.L.; Liu, H.X. Integrin signaling in cancer: Bidirectional mechanisms and therapeutic opportunities. Cell Commun. Signal 2023, 21, 266. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef]
- Guo, D.; Zhang, D.; Ren, M.; Lu, G.; Zhang, X.; He, S.; Li, Y. THBS4 promotes HCC progression by regulating ITGB1 via FAK/PI3K/AKT pathway. FASEB J. 2020, 34, 10668–10681. [Google Scholar] [CrossRef]
- Xie, J.; Guo, T.; Zhong, Z.; Wang, N.; Liang, Y.; Zeng, W.; Liu, S.; Chen, Q.; Tang, X.; Wu, H.; et al. ITGB1 Drives Hepatocellular Carcinoma Progression by Modulating Cell Cycle Process Through PXN/YWHAZ/AKT Pathways. Front. Cell Dev. Biol. 2021, 9, 711149. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, W.; Lin, K.; Chen, X.; Lin, G.; Li, Y.; Cui, C. Cancer-associated fibroblasts promote the proliferation and metastasis of colon cancer by mediating the RLIM/PML axis through paracrine COMP. J. Gastroenterol. Hepatol. 2024, 39, 2677–2689. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Chen, P.; Wei, B.; Tan, H.L.; Zhao, Y.X.; Ai, L.; Li, N.; Jiang, Y.K.; Lin, J.; Li, S.J.; et al. FN1 shapes the behavior of papillary thyroid carcinoma through alternative splicing of EDB region. Sci. Rep. 2025, 15, 327. [Google Scholar] [CrossRef]
- Tang, Y.; Nan, N.; Gui, C.; Zhou, X.; Jiang, W.; Zhou, X. Blockage of PD-L1 by FERMT3-mediated Wnt/beta-catenin signalling regulates chemoresistance and immune evasion of colorectal cancer cells. Clin. Exp. Pharmacol. Physiol. 2022, 49, 988–997. [Google Scholar] [CrossRef]
- Corona, A.; Blobe, G.C. The role of the extracellular matrix protein TGFBI in cancer. Cell. Signal. 2021, 84, 110028. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Jiang, J.; Zhou, P.; Deng, K.; Liu, Z.; Yang, M.; Yang, X.; Li, J.; Li, R.; Xia, J. H3K18 lactylation-mediated VCAM1 expression promotes gastric cancer progression and metastasis via AKT-mTOR-CXCL1 axis. Biochem. Pharmacol. 2024, 222, 116120. [Google Scholar] [CrossRef]
- He, Z.C.; Liu, Q.; Yang, K.D.; Chen, C.; Zhang, X.N.; Wang, W.Y.; Zeng, H.; Wang, B.; Liu, Y.Q.; Luo, M.; et al. HOXA5 is amplified in glioblastoma stem cells and promotes tumor progression by transcriptionally activating PTPRZ1. Cancer Lett. 2022, 533, 215605. [Google Scholar] [CrossRef] [PubMed]
- Tentler, D.; Lomert, E.; Novitskaya, K.; Barlev, N.A. Role of ACTN4 in Tumorigenesis, Metastasis, and EMT. Cells 2019, 8, 1427. [Google Scholar] [CrossRef]
- Zheng, P.; Wang, Q.; Teng, J.; Chen, J. Calumenin and fibulin-1 on tumor metastasis: Implications for pharmacology. Pharmacol. Res. 2015, 99, 11–15. [Google Scholar] [CrossRef]
- Phoenix, T.N.; Patmore, D.M.; Boop, S.; Boulos, N.; Jacus, M.O.; Patel, Y.T.; Roussel, M.F.; Finkelstein, D.; Goumnerova, L.; Perreault, S.; et al. Medulloblastoma Genotype Dictates Blood Brain Barrier Phenotype. Cancer Cell 2016, 29, 508–522. [Google Scholar] [CrossRef]
- Sinha, A.; Singh, V.; Tandon, R.; Mohan Srivastava, L. Dichotomy of complement system: Tumorigenesis or destruction. Immunol. Lett. 2020, 223, 89–96. [Google Scholar] [CrossRef]
- Huang, T.Y.; Piunti, A.; Lulla, R.R.; Qi, J.; Horbinski, C.M.; Tomita, T.; James, C.D.; Shilatifard, A.; Saratsis, A.M. Detection of Histone H3 mutations in cerebrospinal fluid-derived tumor DNA from children with diffuse midline glioma. Acta Neuropathol. Commun. 2017, 5, 28. [Google Scholar] [CrossRef]
- Zuccato, J.A.; Patil, V.; Mansouri, S.; Voisin, M.; Chakravarthy, A.; Shen, S.Y.; Nassiri, F.; Mikolajewicz, N.; Trifoi, M.; Skakodub, A.; et al. Cerebrospinal fluid methylome-based liquid biopsies for accurate malignant brain neoplasm classification. Neuro Oncol. 2023, 25, 1452–1460. [Google Scholar] [CrossRef]
- Shao, G.; Chen, R.; Li, M.; Liu, Y.; Zhang, K.; Zhan, Q. Direct SERS profiling of small extracellular vesicles in cerebrospinal fluid for pediatric medulloblastoma detection and treatment monitoring. Anal. Bioanal. Chem. 2025. Available online: https://link.springer.com/article/10.1007/s00216-025-05970-5 (accessed on 19 September 2025). [CrossRef]
- Hughes, C.S.; Foehr, S.; Garfield, D.A.; Furlong, E.E.; Steinmetz, L.M.; Krijgsveld, J. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 2014, 10, 757. [Google Scholar] [CrossRef]
- Hughes, C.S.; Moggridge, S.; Muller, T.; Sorensen, P.H.; Morin, G.B.; Krijgsveld, J. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 2019, 14, 68–85. [Google Scholar] [CrossRef]
- Thompson, A.; Wolmer, N.; Koncarevic, S.; Selzer, S.; Bohm, G.; Legner, H.; Schmid, P.; Kienle, S.; Penning, P.; Hohle, C.; et al. TMTpro: Design, Synthesis, and Initial Evaluation of a Proline-Based Isobaric 16-Plex Tandem Mass Tag Reagent Set. Anal. Chem. 2019, 91, 15941–15950. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Franken, H.; Mathieson, T.; Childs, D.; Sweetman, G.M.; Werner, T.; Togel, I.; Doce, C.; Gade, S.; Bantscheff, M.; Drewes, G.; et al. Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat. Protoc. 2015, 10, 1567–1593. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Huber, W.; von Heydebreck, A.; Sultmann, H.; Poustka, A.; Vingron, M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 2002, 18, S96–S104. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bandla, C.; Kundu, D.J.; Kamatchinathan, S.; Bai, J.; Hewapathirana, S.; John, N.S.; Prakash, A.; Walzer, M.; Wang, S.; et al. The PRIDE database at 20 years: 2025 update. Nucleic Acids Res. 2025, 53, D543–D553. [Google Scholar] [CrossRef]
- Anagnostopoulos, A.K.; Papathanassiou, C.; Karamolegou, K.; Anastasiadou, E.; Dimas, K.S.; Kontos, H.; Koutsopoulos, A.; Prodromou, N.; Tzortzatou-Stathopoulou, F.; Tsangaris, G.T. Proteomic studies of pediatric medulloblastoma tumors with 17p deletion. J. Proteome Res. 2015, 14, 1076–1088. [Google Scholar] [CrossRef]
- Rivero-Hinojosa, S.; Lau, L.S.; Stampar, M.; Staal, J.; Zhang, H.; Gordish-Dressman, H.; Northcott, P.A.; Pfister, S.M.; Taylor, M.D.; Brown, K.J.; et al. Proteomic analysis of Medulloblastoma reveals functional biology with translational potential. Acta Neuropathol. Commun. 2018, 6, 48. [Google Scholar] [CrossRef]
- Lilley, L.M.; Sanche, S.; Moore, S.C.; Salemi, M.R.; Vu, D.; Iyer, S.; Hengartner, N.W.; Mukundan, H. Methods to capture proteomic and metabolomic signatures from cerebrospinal fluid and serum of healthy individuals. Sci. Rep. 2022, 12, 13339. [Google Scholar] [CrossRef]
- Bruschi, M.; Petretto, A.; Cama, A.; Pavanello, M.; Bartolucci, M.; Morana, G.; Ramenghi, L.A.; Garre, M.L.; Ghiggeri, G.M.; Panfoli, I.; et al. Potential biomarkers of childhood brain tumor identified by proteomics of cerebrospinal fluid from extraventricular drainage (EVD). Sci. Rep. 2021, 11, 1818. [Google Scholar] [CrossRef]
- Bruschi, M.; Kajana, X.; Petretto, A.; Bartolucci, M.; Pavanello, M.; Ghiggeri, G.M.; Panfoli, I.; Candiano, G. Weighted Gene Co-Expression Network Analysis and Support Vector Machine Learning in the Proteomic Profiling of Cerebrospinal Fluid from Extraventricular Drainage in Child Medulloblastoma. Metabolites 2022, 12, 724. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reetz, L.; Ghanam, J.; Chetty, V.K.; Barthel, L.; Tippelt, S.; Fleischhack, G.; Böckmann, M.; Reinhardt, K.; Thakur, B.K. Cerebrospinal Fluid-Derived Small Extracellular Vesicles May Better Reflect Medulloblastoma Proteomes than Those from Blood Plasma. Int. J. Mol. Sci. 2025, 26, 9279. https://doi.org/10.3390/ijms26199279
Reetz L, Ghanam J, Chetty VK, Barthel L, Tippelt S, Fleischhack G, Böckmann M, Reinhardt K, Thakur BK. Cerebrospinal Fluid-Derived Small Extracellular Vesicles May Better Reflect Medulloblastoma Proteomes than Those from Blood Plasma. International Journal of Molecular Sciences. 2025; 26(19):9279. https://doi.org/10.3390/ijms26199279
Chicago/Turabian StyleReetz, Laura, Jamal Ghanam, Venkatesh K. Chetty, Lennart Barthel, Stephan Tippelt, Gudrun Fleischhack, Marie Böckmann, Katarina Reinhardt, and Basant K. Thakur. 2025. "Cerebrospinal Fluid-Derived Small Extracellular Vesicles May Better Reflect Medulloblastoma Proteomes than Those from Blood Plasma" International Journal of Molecular Sciences 26, no. 19: 9279. https://doi.org/10.3390/ijms26199279
APA StyleReetz, L., Ghanam, J., Chetty, V. K., Barthel, L., Tippelt, S., Fleischhack, G., Böckmann, M., Reinhardt, K., & Thakur, B. K. (2025). Cerebrospinal Fluid-Derived Small Extracellular Vesicles May Better Reflect Medulloblastoma Proteomes than Those from Blood Plasma. International Journal of Molecular Sciences, 26(19), 9279. https://doi.org/10.3390/ijms26199279





