Integration of Repeatome and Cytogenetic Data on Tandem DNAs in a Medicinal Plant Polemonium caeruleum L.
Abstract
1. Introduction
2. Results
2.1. Identification of DNA Repeats by RepeatExplorer/TAREAN Pipelines
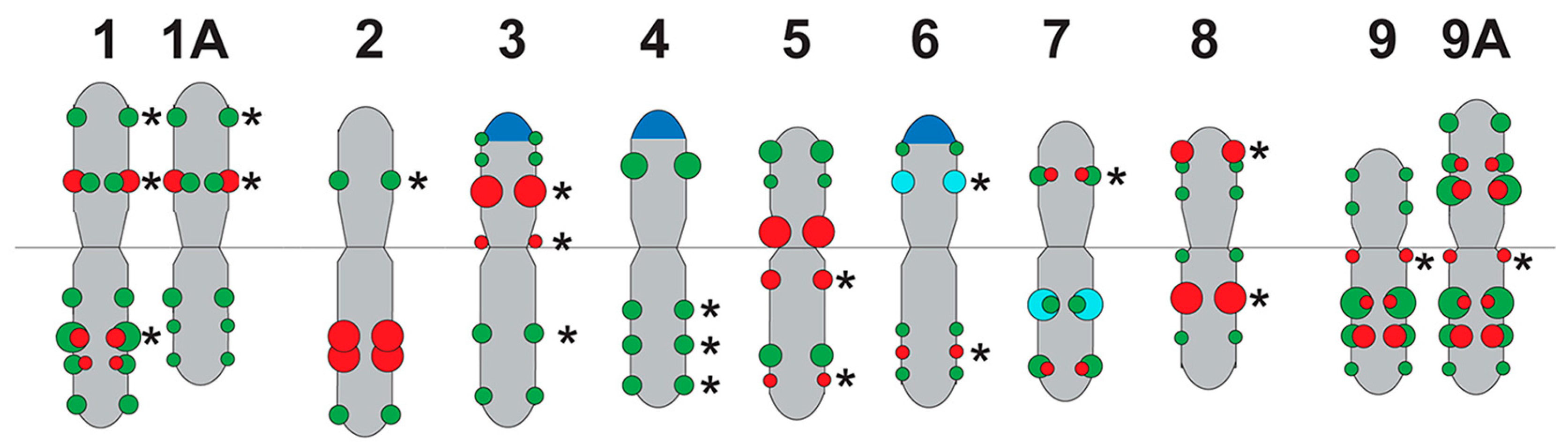
2.2. Chromosomal Localization of Tandem DNAs
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Sequence Analysis and Identification of DNA Repeats
4.3. Chromosome Spread Preparation
4.4. FISH Procedure
4.5. Analysis of Chromosome Preparations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Malyshev, L.I. (Ed.) Flora of Siberia. Vol. 11. Pyrolaceae—Lamiaceae; CRC Press: Boca-Raton, FL, USA, 2006; 310p. [Google Scholar] [CrossRef]
- Davidson, J.F. The genus Polemonium [Tournefort] L. Univ. Calif. Publ. Bot. 1950, 23, 209–282. [Google Scholar]
- Grant, V. Taxonomy of the tufted alpine and subalpine polemoniums (Polemoniaceae). Bot. Gaz. 1989, 150, 158–169. [Google Scholar] [CrossRef]
- Bożek, M. Characteristics of blooming, pollen production, and insect visitors of Polemonium caeruleum L.—A species with a potential to enrich pollinator-friendly urban habitats. Acta Agrobotanica 2019, 72, 1795. [Google Scholar] [CrossRef]
- Ryniewicz, J.; Skłodowski, M.; Chmur, M.; Bajguz, A.; Roguz, K.; Roguz, A.; Zych, M. Intraspecific Variation in Nectar Chemistry and Its Implications for Insect Visitors: The Case of the Medicinal Plant, Polemonium caeruleum, L. Plants 2020, 9, 1297. [Google Scholar] [CrossRef]
- Golyak, Y.A.; Khishova, O.M.; Dubashinskaya, N.V.; Kukhareva, L.V. Quantitative determination of total triterpenoid saponins in Polemonium caeruleum rhizomes and roots. Pharm. Chem. J. 2008, 42, 456–459. [Google Scholar] [CrossRef]
- Marzio, L.D.; Ventura, C.A.; Cosco, D.; Paolino, D.; Stefano, A.D.S.; Stancanelli, R.; Tommasini, S.; Cannavà, C.; Celia, C.; Fresta, M. Nanotherapeutics for anti-inflammatory delivery. J. Drug Deliv. Sci. Technol. 2016, 32, 174–191. [Google Scholar] [CrossRef]
- Łaska, G.; Sieniawska, E.; Świątek, Ł.; Zjawiony, J.; Khan, S.; Boguszewska, A.; Stocki, M.; Angielczyk, M.; Polz-Dacewicz, M. Phytochemistry and biological activities of Polemonium caeruleum L. Phytochem. Lett. 2019, 30, 314–323. [Google Scholar] [CrossRef]
- Frolova, L.; Kovaleva, E.; Shelestova, V.; Kuteynikov, V.; Flisyuk, E.; Pozharitskaya, O.; Shikov, A. Comparison of Analytical Methods Used for Standardization of Triterpenoid Saponins in Herbal Monographs Included in the Russian and Other Pharmacopeias. Phytochem. Anal. 2025; ahead of print. [Google Scholar] [CrossRef]
- Hiller, K.; Paulick, A.; Friedrich, E. Zur Hemmwirkung von Polemonium saponin gegenüber Pilzen [On the inhibitory activity of Polemonium saponin against fungi (author’s transl)]. Die Pharm. 1981, 36, 133–134. [Google Scholar]
- Hazieva, F.; Korotkikh, I.; Kovalev, N.; Romashkina, S.; Samatadze, T. Growth regulators and microfertilizer on Polemonium caeruleum L. application: Effectiveness for medicinal raw material and seeds yield increasing. Sib. J. Life Sci. Agric. 2022, 14, 90–104. [Google Scholar] [CrossRef]
- Samatadze, T.E.; Yurkevich, O.Y.; Khazieva, F.M.; Basalaeva, I.V.; Konyaeva, E.A.; Burova, A.E.; Zoshchuk, S.A.; Morozov, A.I.; Amosova, A.V.; Muravenko, O.V. Agro-Morphological and Cytogenetic Characterization of Colchicine-Induced Tetraploid Plants of Polemonium caeruleum L. (Polemoniaceae). Plants 2022, 11, 2585. [Google Scholar] [CrossRef]
- Vassiljev, V.; Polemonium, L. Flora of the U.S.S.R., Volume XIX Tubiflorae; Shishkin, B.K., Ed.; Israel Program for Scientific Translations: Jerusalem, Israel, 1974; pp. 58–69. [Google Scholar]
- Feulner, M.; Möseler, B.M.; Nezadal, W. Introgression und morphologische variabilitt bei der Blauen Himmelsleiter, Polemonium caeruleum L. in Nordbayern, Deutschland. Feddes Repert. 2001, 112, 231–246. [Google Scholar] [CrossRef]
- Rose, J.P.; Toledo, C.A.P.; Lemmon, E.M.; Lemmon, A.R.; Sytsma, K.J. Out of Sight, Out of Mind: Widespread Nuclear and Plastid-Nuclear Discordance in the Flowering Plant Genus Polemonium (Polemoniaceae) Suggests Widespread Historical Gene Flow Despite Limited Nuclear Signal. Syst. Biol. 2021, 70, 162–180. [Google Scholar] [CrossRef]
- Matoba, H.; Inaba, K.; Nagano, K.; Uchiyama, H. Use of RAPD analysis to assess the threat of interspecific hybridization to the critically endangered Polemonium kiushianum in Japan. J. Plant Res. 2011, 124, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Ito, K. Polemonium in Hokkaido, the Kuriles and Sakhalin. Environ. Sci. Hokkaido 1983, 6, 247–280. [Google Scholar]
- Worley, A.C.; Ghazvini, H.; Schemske, D.W. A Phylogeny of the Genus Polemonium Based on Amplified Fragment Length Polymorphism (AFLP) Markers. Syst. Bot. 2009, 34, 149–161. [Google Scholar] [CrossRef]
- Index to Plant Chromosome Numbers (IPCN). Missouri Botanical Garden: St. Louis, MO, USA. Available online: http://www.tropicos.org/Project/IPCN (accessed on 18 September 2025).
- Rice, A.; Mayrose, I. The Chromosome Counts Database (CCDB); Methods in Molecular Biology; Humana: New York, NY, USA, 2023; Volume 2703, pp. 123–129. [Google Scholar] [CrossRef]
- Louzada, S.; Lopes, M.; Ferreira, D.; Adega, F.; Escudeiro, A.; Gama-Carvalho, M.; Chaves, R. Decoding the Role of Satellite DNA in Genome Architecture and Plasticity-An Evolutionary and Clinical Affair. Genes 2020, 11, 72. [Google Scholar] [CrossRef]
- Garrido-Ramos, M.A. Satellite DNA: An evolving topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef]
- Plohl, M.; Meštrovic, N.; Mravinac, B. Satellite DNA evolution. In Repetitive DNA; Garrido-Ramos, M.A., Ed.; Karger: Granada, Spain, 2012; pp. 126–152. [Google Scholar]
- Schmidt, T.; Heslop-Harrison, J.S. Genomes, genes and junk: The large-scale organization of plant chromosomes. Trends Plant Sci. 1998, 3, 195–199. [Google Scholar] [CrossRef]
- Mehrotra, S.; Goyal, V. Repetitive sequences in plant nuclear DNA: Types, distribution, evolution and function. Genom. Proteom. Bioinform. 2014, 12, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Campomayor, N.B.; Waminal, N.E.; Kang, B.Y.; Nguyen, H.; Lee, S.S.; Huh, J.H.; Kim, H.H. Subgenome discrimination in Brassica and Raphanus allopolyploids using microsatellites. Cells 2021, 10, 2358. [Google Scholar] [CrossRef]
- Amosova, A.V.; Yurkevich, O.Y.; Bolsheva, N.L.; Samatadze, T.E.; Zoshchuk, S.A.; Muravenko, O.V. Repeatome Analyses and Satellite DNA Chromosome Patterns in Deschampsia sukatschewii, D. cespitosa, and D. antarctica (Poaceae). Genes 2022, 13, 762. [Google Scholar] [CrossRef]
- Yurkevich, O.Y.; Samatadze, T.E.; Zoshchuk, S.A.; Semenov, A.R.; Morozov, A.I.; Selyutina, I.Y.; Amosova, A.V.; Muravenko, O.V. Repeatome Analysis and Satellite DNA Chromosome Patterns in Hedysarum Species. Int. J. Mol. Sci. 2024, 25, 12340. [Google Scholar] [CrossRef]
- Kalnyuk, J.V.; Yurkevich, O.Y.; Badaeva, E.D.; Semenov, A.R.; Zoshchuk, S.A.; Amosova, A.V.; Muravenko, O.V. Taxonomy, Phylogeny, Genomes, and Repeatomes in the Subgenera Salvia, Sclarea, and Glutinaria (Salvia, Lamiaceae). Int. J. Mol. Sci. 2025, 26, 6436. [Google Scholar] [CrossRef]
- Neumann, P.; Novák, P.; Hoštáková, N.; Macas, J. Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mobile DNA 2019, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Samoluk, S.S.; Vaio, M.; Ortíz, A.M.; Chalup, L.M.I.; Robledo, G.; Bertioli, D.J.; Seijo, G. Comparative repeatome analysis reveals new evidence on genome evolution in wild diploid Arachis (Fabaceae) species. Planta 2022, 256, 50. [Google Scholar] [CrossRef]
- He, B.; Liu, W.; Li, J.; Xiong, S.; Jia, J.; Lin, Q.; Liu, H.; Cui, P. Evolution of Plant Genome Size and Composition. Genom. Proteom. Bioinform. 2024, 22, qzae078. [Google Scholar] [CrossRef]
- Lee, S.I.; Kim, N.S. Transposable elements and genome size variations in plants. Genom. Inform. 2014, 12, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Ma, J. Co-evolution of plant LTR-retrotransposons and their host genomes. Protein Cell 2013, 4, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.F.; Zhu, X.G.; Hutang, G.R.; Li, J.Y.; Tian, J.Q.; Jiang, X.H.; Zhang, D.; Gao, L.Z. Genome Size Variation and Evolution Driven by Transposable Elements in the Genus Oryza. Front. Plant Sci. 2022, 13, 921937. [Google Scholar] [CrossRef]
- Vitte, C.; Panaud, O. LTR retrotransposons and flowering plant genome size: Emergence of the increase/decrease model. Cytogenet. Genome Res. 2005, 110, 91–107. [Google Scholar] [CrossRef]
- Zhou, S.S.; Yan, X.M.; Zhang, K.F.; Liu, H.; Xu, J.; Nie, S.; Jia, K.H.; Jiao, S.Q.; Zhao, W.; Porth, I. A comprehensive annotation dataset of intact LTR retrotransposons of 300 plant genomes. Sci. Data 2021, 8, 174. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, P.; Wang, S.; Yang, J.; Dai, L.; Zheng, J.; Wang, Y. The Dynamics of Long Terminal Repeat Retrotransposon Proliferation and Decay Drive the Evolution of Genome Size Variation in Capsicum. Plants 2025, 14, 2136. [Google Scholar] [CrossRef]
- Ishiguro, S.; Taniguchi, S.; Schmidt, N.; Jost, M.; Wanke, S.; Heitkam, T.; Ohmido, N. Repeatome landscapes and cytogenetics of hortensias provide a framework to trace Hydrangea evolution and domestication. Ann. Bot. 2025, 135, 549–564. [Google Scholar] [CrossRef]
- Han, D.; Li, W.; Hou, Z.; Lin, C.; Xie, Y.; Zhou, X.; Gao, Y.; Huang, J.; Lai, J.; Wang, L.; et al. The Chromosome-Scale Assembly 518 of the Salvia Rosmarinus Genome Provides Insight into Carnosic Acid Biosynthesis. Plant J. 2023, 113, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Stitzer, M.C.; Anderson, S.N.; Springer, N.M.; Ross-Ibarra, J. The genomic ecosystem of transposable elements in maize. PLoS Genet. 2019, 17, e1009768. [Google Scholar] [CrossRef]
- Morales-Díaz, N.; Sushko, S.; Campos-Dominguez, L.; Kopalli, V.; Golicz, A.A.; Castanera, R.; Casacuberta, J.M. Tandem LTR-retrotransposon structures are common and highly polymorphic in plant genomes. Mob. DNA 2025, 16, 10. [Google Scholar] [CrossRef] [PubMed]
- Nicolau, M.; Picault, N.; Moissiard, G. The Evolutionary Volte-Face of Transposable Elements: From Harmful Jumping Genes to Major Drivers of Genetic Innovation. Cells 2021, 10, 2952. [Google Scholar] [CrossRef]
- Neumann, P.; Navrátilová, A.; Koblížková, A.; Kejnovský, E.; Hřibová, E.; Hobza, R.; Widmer, A.; Doležel, J.; Macas, J. Plant centromeric retrotransposons: A structural and cytogenetic perspective. Mob. DNA 2011, 2, 4. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.H.; Mokhtar, M.M.; El Allali, A. Transposable elements: Multifunctional players in the plant genome. Front. Plant Sci. 2024, 14, 1330127. [Google Scholar] [CrossRef]
- Yu, Z.; Huang, Y.; Wu, J.; Zhang, M.; Deng, Z. Diversity Chromosome Evolution of Ty1-copia Retrotransposons in Pennisetum purpureum Revealed by FISH. Agronomy 2022, 12, 1312. [Google Scholar] [CrossRef]
- Sattler, M.C.; Silva, J.C.; Oliveira, S.C.; Clarindo, W.R. Chromosome distribution of four LTR retrotransposons and 18 S rDNA in Coffea eugenioides. Sci. Rep. 2025, 15, 3768. [Google Scholar] [CrossRef] [PubMed]
- Ugarkovic, D. Functional elements residing within satellite DNAs. EMBO Rep. 2005, 6, 1035–1039. [Google Scholar] [CrossRef]
- Feliciello, I.; Akrap, I.; Brajković, J.; Zlatar, I.; Ugarković, Đ. Satellite DNA as a driver of population divergence in the red flour beetle Tribolium castaneum. Genome Biol. Evol. 2014, 7, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ruano, F.; López-León, M.; Cabrero, J.; Camacho, J.P.M. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci. Rep. 2016, 6, 28333. [Google Scholar] [CrossRef]
- Šatović-Vukšić, E.; Plohl, M. Satellite DNAs-From Localized to Highly Dispersed Genome Components. Genes 2023, 14, 742. [Google Scholar] [CrossRef]
- Thakur, J.; Packiaraj, J.; Henikoff, S. Sequence, Chromatin and Evolution of Satellite DNA. Int. J. Mol. Sci. 2021, 22, 4309. [Google Scholar] [CrossRef]
- Kroupin, P.Y.; Badaeva, E.D.; Sokolova, V.M.; Chikida, N.N.; Belousova, M.K.; Surzhikov, S.A.; Nikitina, E.A.; Kocheshkova, A.A.; Ulyanov, D.S.; Ermolaev, A.S.; et al. Aegilops crassa Boiss. repeatome characterized using low-coverage NGS as a source of new FISH markers: Application in phylogenetic studies of the Triticeae. Front. Plant Sci. 2022, 13, 980764. [Google Scholar] [CrossRef]
- Schmidt, N.; Sielemann, K.; Breitenbach, S.; Fuchs, J.; Pucker, B.; Weisshaar, B.; Holtgräwe, D.; Heitkam, T. Repeat turnover meets stable chromosomes: Repetitive DNA sequences mark speciation and gene pool boundaries in sugar beet and wild beets. Plant J. Cell Mol. Biol. 2024, 118, 171–190. [Google Scholar] [CrossRef]
- Huang, K.; Rieseberg, L.H. Frequency, Origins, and Evolutionary Role of Chromosomal Inversions in Plants. Front. Plant Sci. 2020, 11, 296. [Google Scholar] [CrossRef] [PubMed]
- Deon, G.A.; Dos Santos, R.Z.; Sassi, F.M.C.; Moreira-Filho, O.; Vicari, M.R.; Porto-Foresti, F.; Utsunomia, R.; Cioffi, M.B. The role of satellite DNAs in the chromosomal rearrangements and the evolution of the rare XY1Y2 sex system in Harttia (Siluriformes: Loricariidae). J. Hered. 2024, 115, 541–551. [Google Scholar] [CrossRef]
- Lv, R.; Gou, X.; Li, N.; Zhang, Z.; Wang, C.; Wang, R.; Wang, B.; Yang, C.; Gong, L.; Zhang, H.; et al. Chromosome translocation affects multiple phenotypes, causes genome-wide dysregulation of gene expression, and remodels metabolome in hexaploid wheat. Plant J. Cell Mol. Biol. 2023, 115, 1564–1582. [Google Scholar] [CrossRef]
- Schubert, I.; Lysak, M.A. Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet. 2011, 27, 207–216. [Google Scholar] [CrossRef]
- Rodionov, A. Tandem duplications, eupolyploidy and secondary diploidization—Genetic mechanisms of plant speciation and progressive evolution. Turczaninowia 2022, 25, 87–121. [Google Scholar] [CrossRef]
- Porter, J.M.; Johnson, L.A. A Phylogenetic Classification of Polemoniaceae. Aliso J. Syst. Florist. Bot. 2000, 19, 55–91. [Google Scholar] [CrossRef]
- Pustahija, F.; Bašić, N.; Siljak-Yakovlev, S. Karyotype Variability in Wild Narcissus poeticus L. Populations from Different Environmental Conditions in the Dinaric Alps. Plants 2024, 13, 208. [Google Scholar] [CrossRef] [PubMed]
- Amosova, A.V.; Bolsheva, N.L.; Samatadze, T.E.; Twardovska, M.O.; Zoshchuk, S.A.; Andreev, I.O.; Badaeva, E.; Kunakh, V.A.; Muravenko, O.V. Molecular Cytogenetic Analysis of Deschampsia antarctica Desv. (Poaceae), Maritime Antarctic. PLoS ONE 2015, 10, e0138878. [Google Scholar] [CrossRef]
- Rogers, S.O.; Bendich, A.J. Ribosomal RNA genes in plants: Variability in copy number and in intergenic spacer. Plant Mol. Biol. 1987, 9, 509–520. [Google Scholar] [CrossRef]
- Bai, C.; Alverson, W.S.; Follansbee, A.; Waller, D.M. New reports of nuclear DNA content for 407 U.S. plant species. Ann. Bot. 2012, 110, 1623–1629. [Google Scholar] [CrossRef]
- Leitch, I.J.; Johnston, E.; Pellicer, J.; Hidalgo, O.; Bennett, M.D. Plant DNA C-Values Database (Release 7.1, April 2019). Royal Botanic Gardens, Kew. Available online: https://cvalues.science.kew.org/ (accessed on 17 April 2025).
- Clausen, J. Genetic studies in Polemonium. III. Preliminary account on the cytology of species and specific hybrids. Hereditas 1931, 15, 62–66. [Google Scholar] [CrossRef]
- Novak, P.; Robledillo, L.A.; Koblizkova, A.; Vrbova, I.; Neumann, P.; Macas, J. TAREAN: A computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acid Res. 2017, 45, e111. [Google Scholar] [CrossRef]
- Novák, P.; Neumann, P.; Macas, J. Global analysis of repetitive DNA from unassembled sequence reads using RepeatExplorer2. Nat. Protoc. 2020, 15, 3745–3776. [Google Scholar] [CrossRef]
- Novák, P.; Hoštáková, N.; Neumann, P.; Macas, J. DANTE and DANTE_LTR: Lineage-centric annotation pipelines for long terminal repeat retrotransposons in plant genomes. NAR Genom. Bioinform. 2024, 6, lqae113. [Google Scholar] [CrossRef]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, 71–74. [Google Scholar] [CrossRef]
- Gerlach, W.L.; Bedbrook, J.R. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 1979, 7, 1869–1885. [Google Scholar] [CrossRef] [PubMed]
- Gerlach, W.L.; Dyer, T.A. Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Res. 1980, 8, 4851–4855. [Google Scholar] [CrossRef] [PubMed]
- Amosova, A.V.; Yurkevich, O.Y.; Semenov, A.R.; Samatadze, T.E.; Sokolova, D.V.; Artemyeva, A.M.; Zoshchuk, S.A.; Muravenko, O.V. Genome Studies in Amaranthus cruentus L. and A. hypochondriacus L. Based on Repeatomic and Cytogenetic Data. Int. J. Mol. Sci. 2024, 25, 13575. [Google Scholar] [CrossRef] [PubMed]

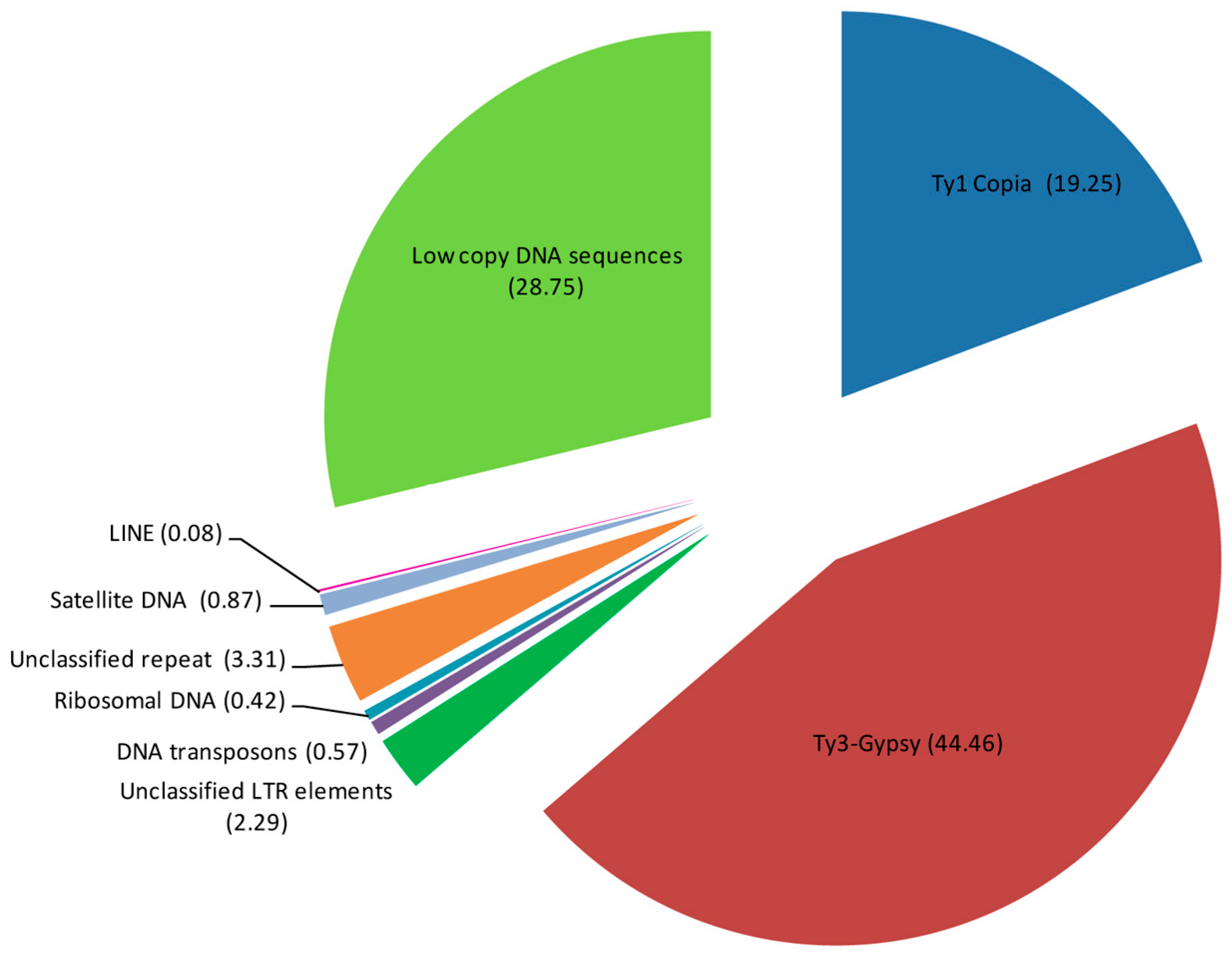
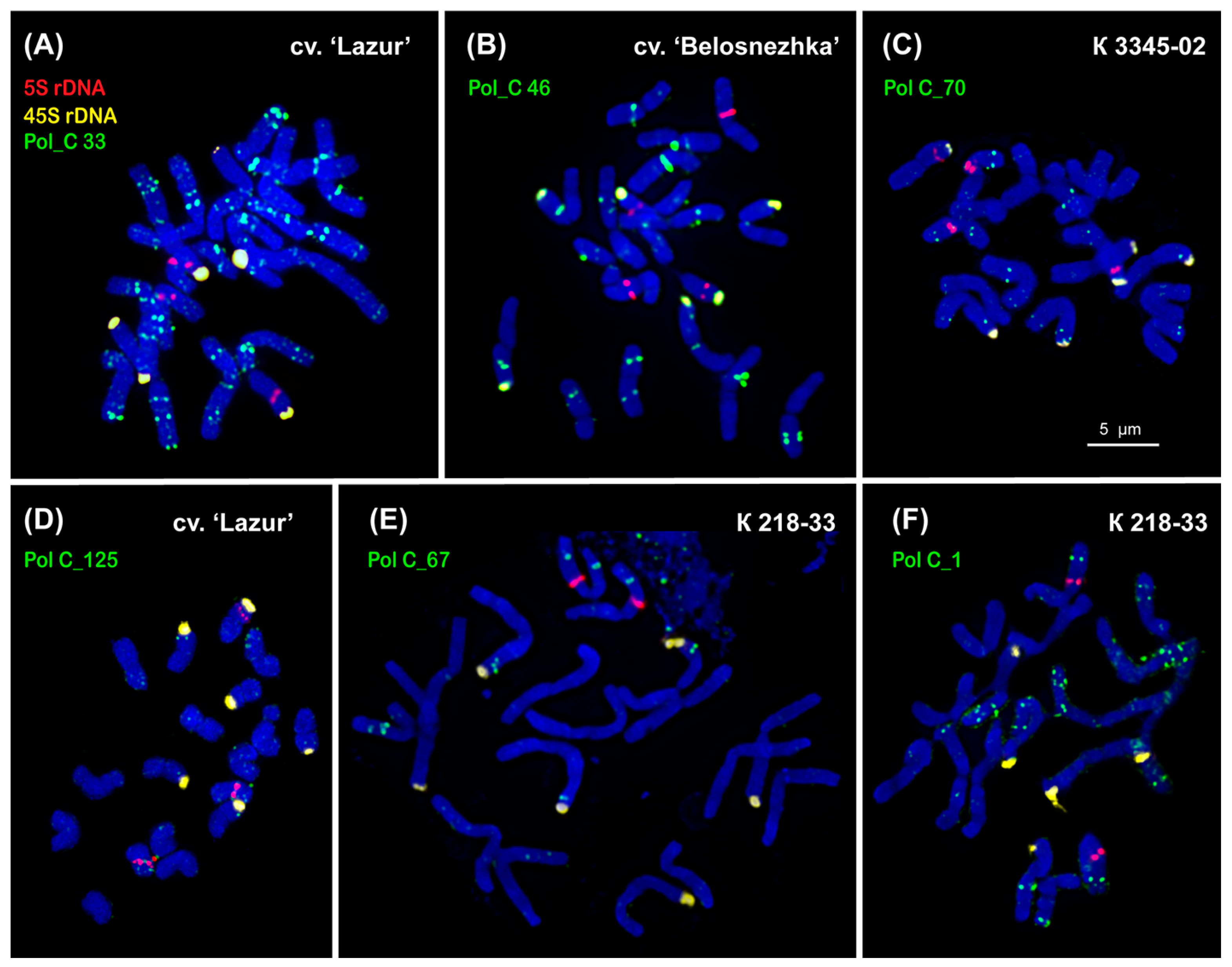
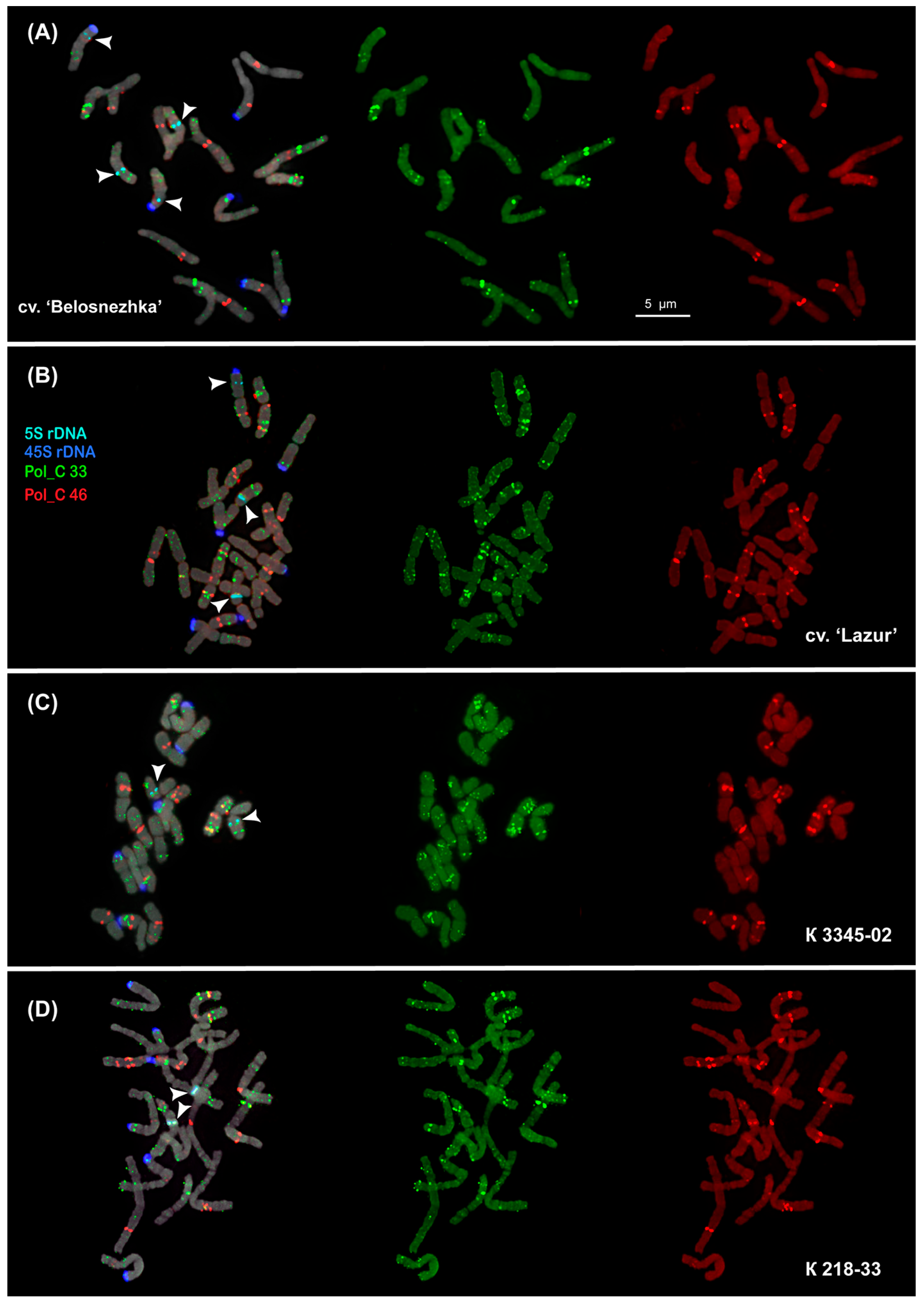
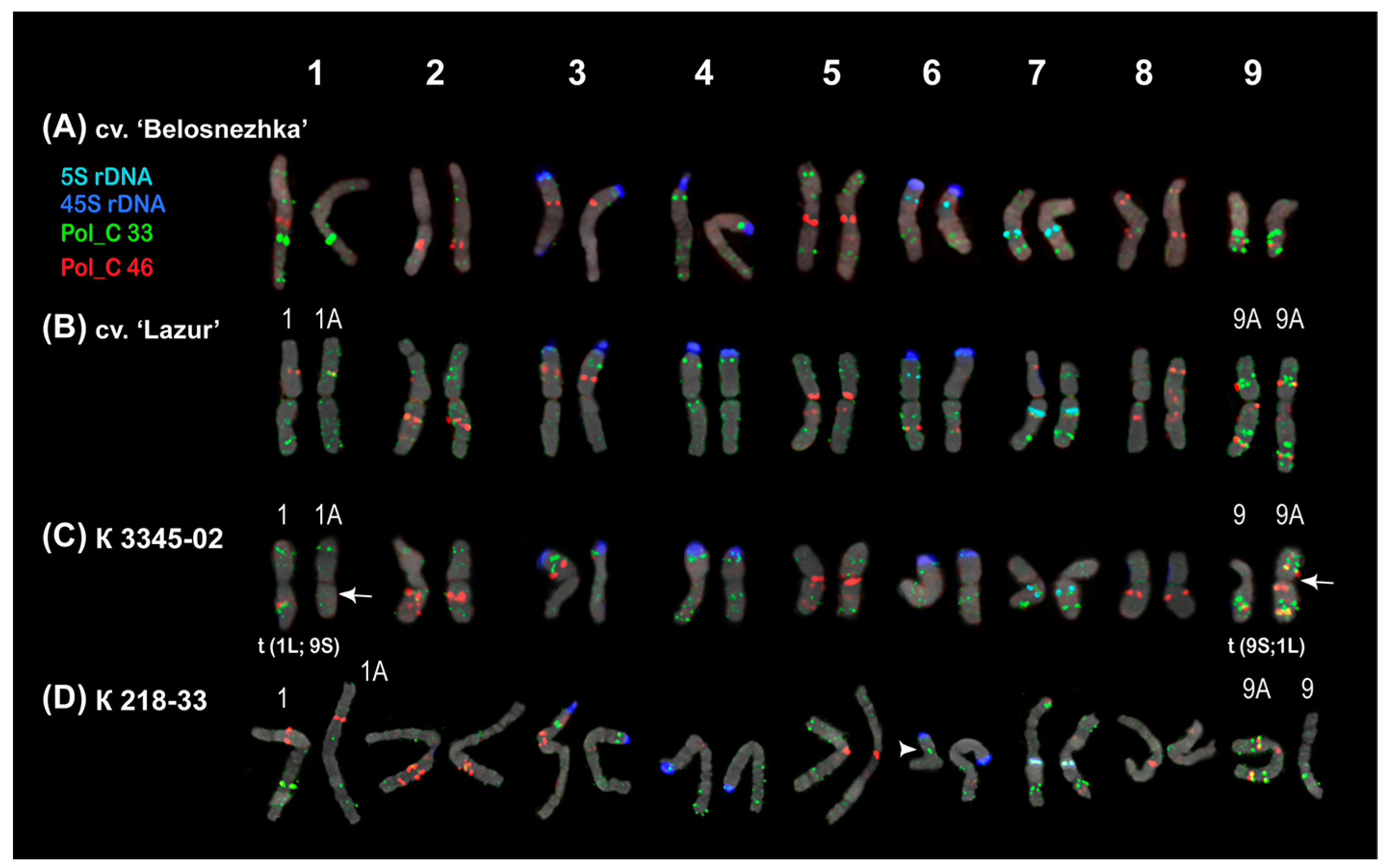
| Repeat Name | Genome Proportion (%) |
|---|---|
| Retrotransposons (Class I) | 66.08 |
| Ty1 Copia | 19.25 |
| Ale | 0.24 |
| Ikeros | 0.06 |
| Angela | 7.66 |
| SIRE | 10.70 |
| TAR | 0.31 |
| Tork | 0.28 |
| Ty3-Gypsy | 44.46 |
| non-chromovirus Athila | 10.90 |
| non-chromovirus Tat- Ogre | 0.86 |
| chromovirus CRM | 0.40 |
| chromovirus Galadriel | 0.06 |
| chromovirus Tekay | 32.19 |
| chromovirus Reina | 0.05 |
| LINE | 0.08 |
| Unclassified LTR elements | 2.29 |
| Transposons (Class II) | 0.57 |
| Cacta | 0.17 |
| hAT | 0.04 |
| MuDR_Mutator | 0.36 |
| rDNA | 0.42 |
| Unclassified repeat | 3.31 |
| DNA satellite | 0.87 |
| Repetitive DNA | 71.25 |
| Putative satellites | 6 high confident 3 low confident |
| SatDNA/Genome Proportion, %/Repeat Length, bp | Blast Homology | ||
|---|---|---|---|
| ‘Lazur’ | ERR5555406 | ERR5555143 | |
| Pol_C 33/0.44/508 | CL67/0.42/509 | CL59/0.54/411 | 93%/92% of coverage/identity with CL67 in ERR5555406 sample 99%/92% of coverage/identity with CL59 in ERR5555143 sample |
| Pol_C 46/0.24/191 | CL90/0.18/192 | CL86/0.22/191 | 99%/100% of coverage/identity with CL90 in ERR5555406 sample 95%/100% of coverage/identity with CL86 in ERR5555143 sample |
| Pol_C 67/0.12/89 | CL84/0.22/89 | CL89/0.2/90 | 100%/100% of coverage/identity with CL84 in ERR5555406 sample 79%/100% of coverage/identity with CL89 in ERR5555143 sample |
| Pol_C 70/0.092/393 | CL124/0.05/393 | CL117/0.062/393 | 100%/98% of coverage/identity with CL124 in ERR5555406 sample 100%/98% of coverage/identity with CL117 in ERR5555143 sample |
| Pol_C 125/0.016/83 | no | no | no |
| Pol_C 134/0.014/364 | CL159/0.022/364 | CL152/0.021/364 | 100%/100% of coverage/identity with CL159 in ERR5555406 sample 100%/99% of coverage/identity with CL152 in ERR5555143 sample |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muravenko, O.V.; Amosova, A.V.; Semenov, A.R.; Kalnyuk, J.V.; Khazieva, F.M.; Korotkikh, I.N.; Basalaeva, I.V.; Badaeva, E.D.; Zoshchuk, S.A.; Yurkevich, O.Y. Integration of Repeatome and Cytogenetic Data on Tandem DNAs in a Medicinal Plant Polemonium caeruleum L. Int. J. Mol. Sci. 2025, 26, 9240. https://doi.org/10.3390/ijms26189240
Muravenko OV, Amosova AV, Semenov AR, Kalnyuk JV, Khazieva FM, Korotkikh IN, Basalaeva IV, Badaeva ED, Zoshchuk SA, Yurkevich OY. Integration of Repeatome and Cytogenetic Data on Tandem DNAs in a Medicinal Plant Polemonium caeruleum L. International Journal of Molecular Sciences. 2025; 26(18):9240. https://doi.org/10.3390/ijms26189240
Chicago/Turabian StyleMuravenko, Olga V., Alexandra V. Amosova, Alexey R. Semenov, Julia V. Kalnyuk, Firdaus M. Khazieva, Irina N. Korotkikh, Irina V. Basalaeva, Ekaterina D. Badaeva, Svyatoslav A. Zoshchuk, and Olga Yu. Yurkevich. 2025. "Integration of Repeatome and Cytogenetic Data on Tandem DNAs in a Medicinal Plant Polemonium caeruleum L." International Journal of Molecular Sciences 26, no. 18: 9240. https://doi.org/10.3390/ijms26189240
APA StyleMuravenko, O. V., Amosova, A. V., Semenov, A. R., Kalnyuk, J. V., Khazieva, F. M., Korotkikh, I. N., Basalaeva, I. V., Badaeva, E. D., Zoshchuk, S. A., & Yurkevich, O. Y. (2025). Integration of Repeatome and Cytogenetic Data on Tandem DNAs in a Medicinal Plant Polemonium caeruleum L. International Journal of Molecular Sciences, 26(18), 9240. https://doi.org/10.3390/ijms26189240








