Stratifying ALS Patients by Mode of Inheritance Reveals Transcriptomic Signatures Specific to sALS and fALS
Abstract
1. Introduction
2. Results
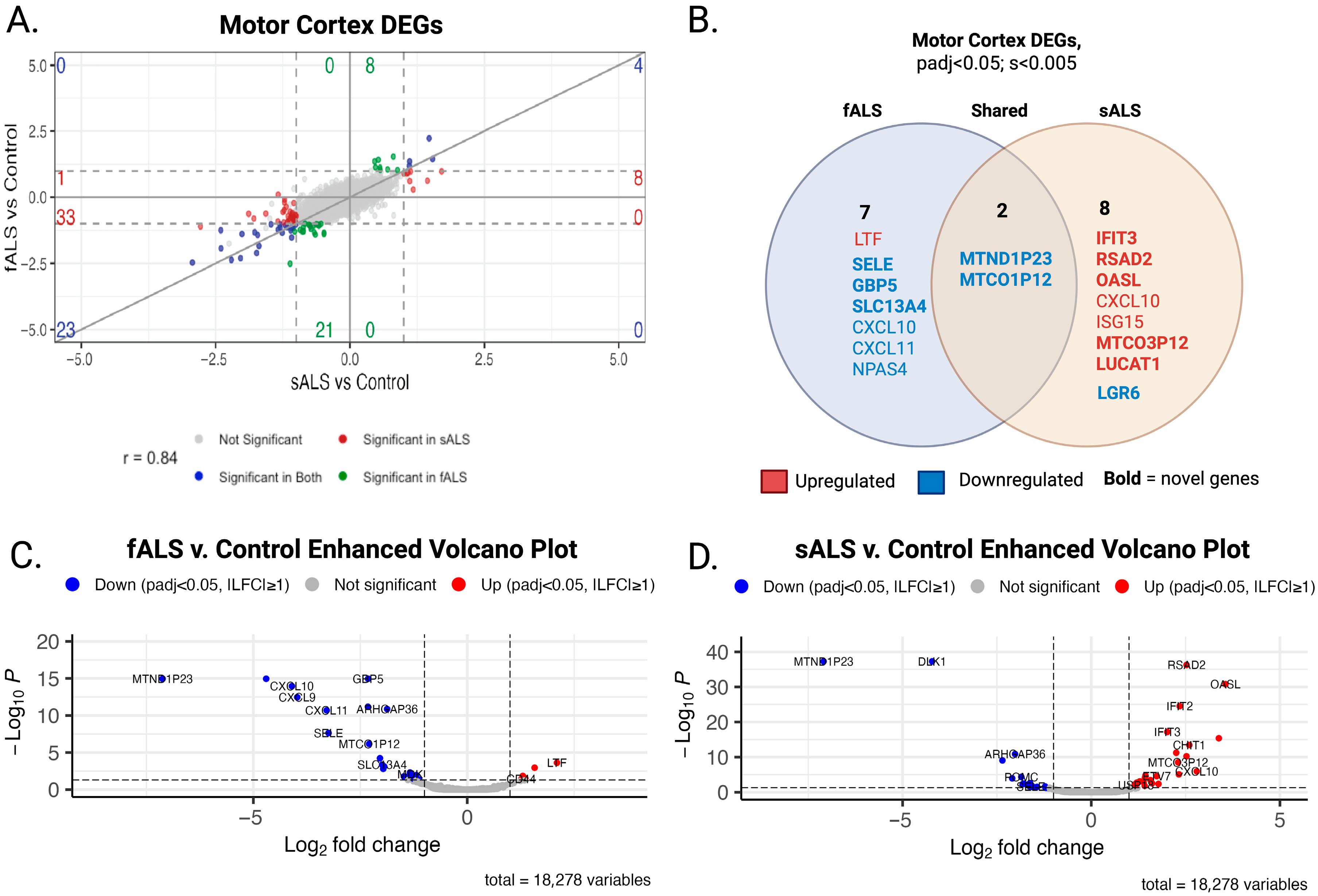
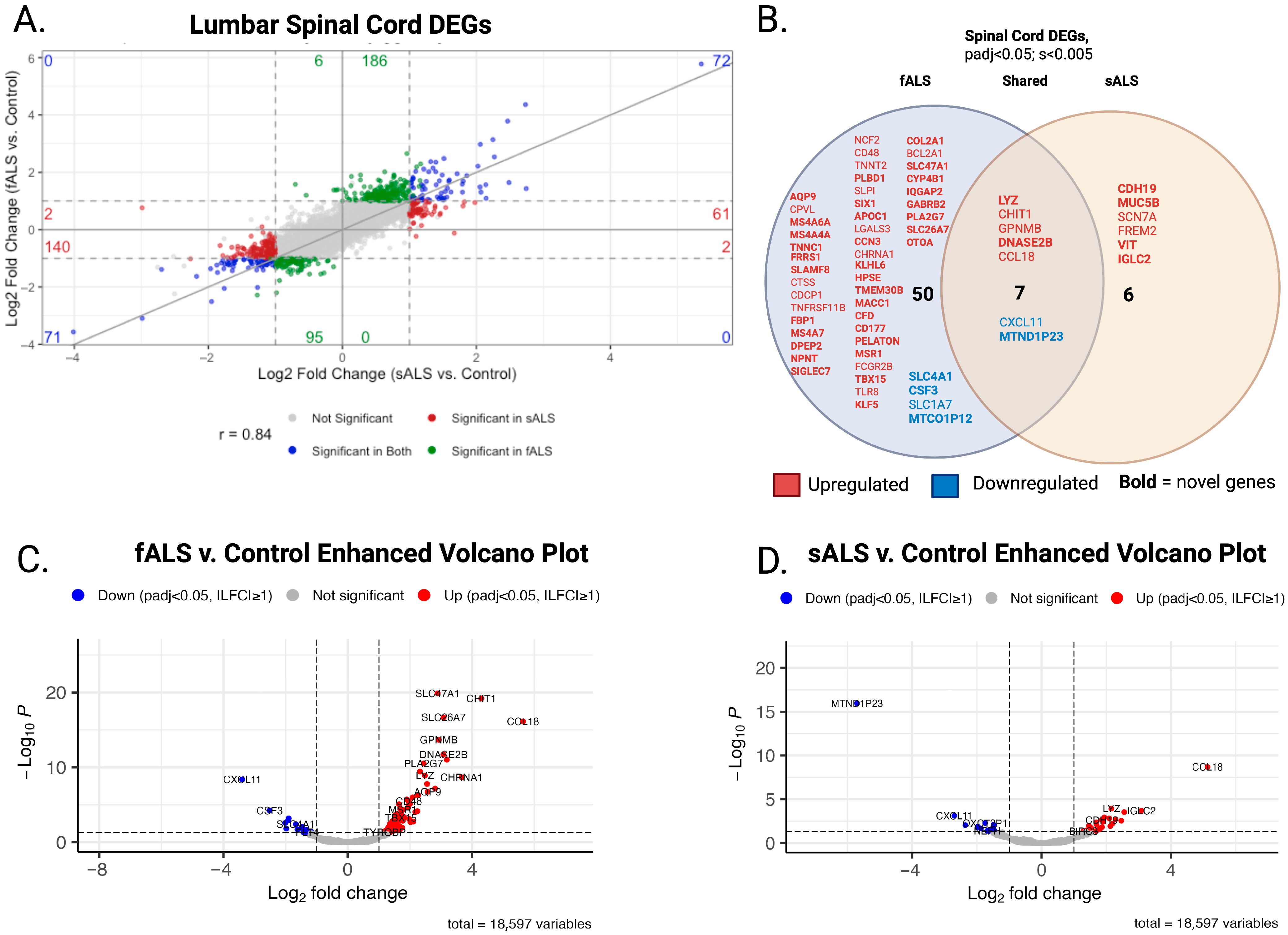
2.1. DESeq2
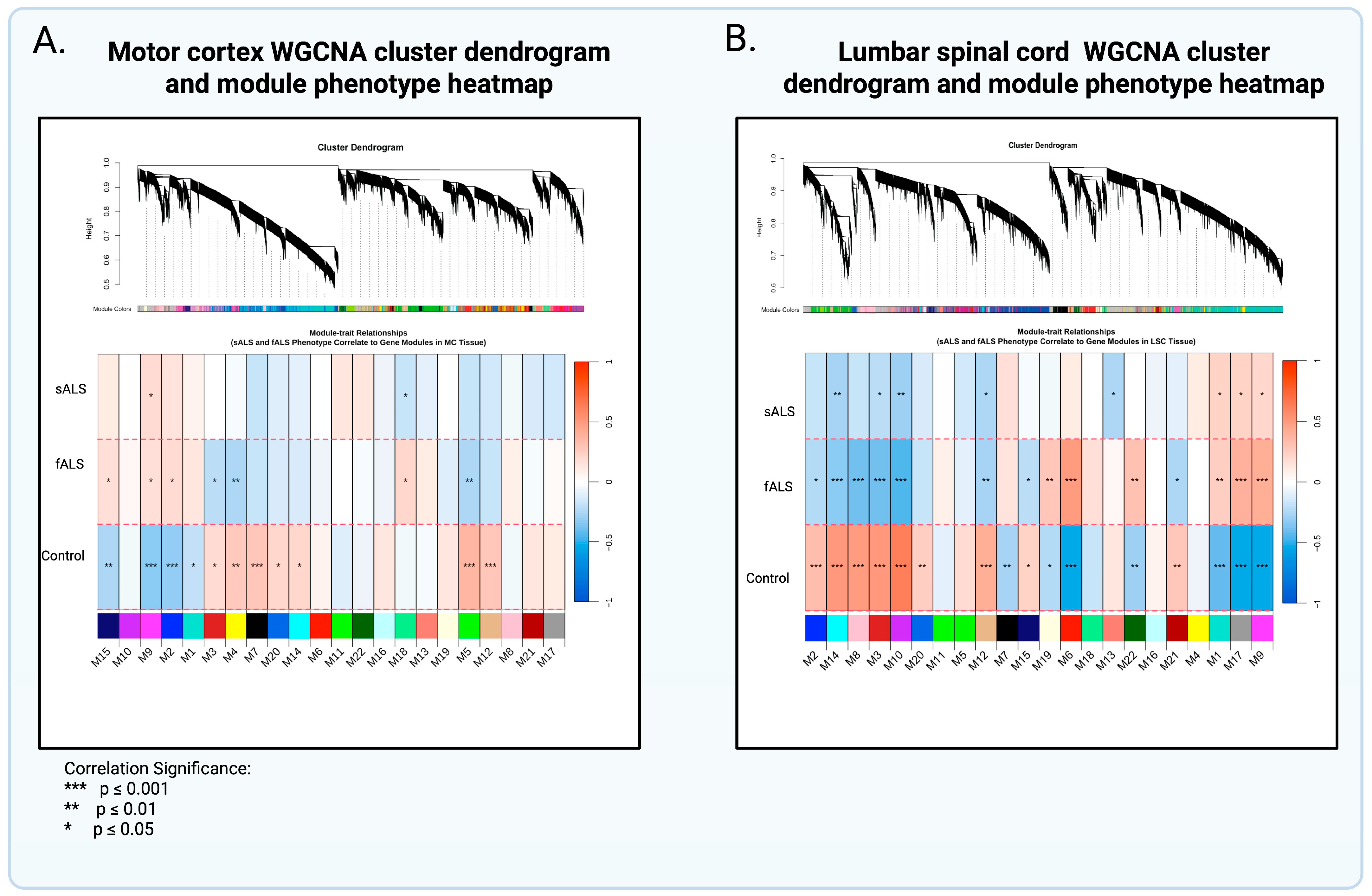
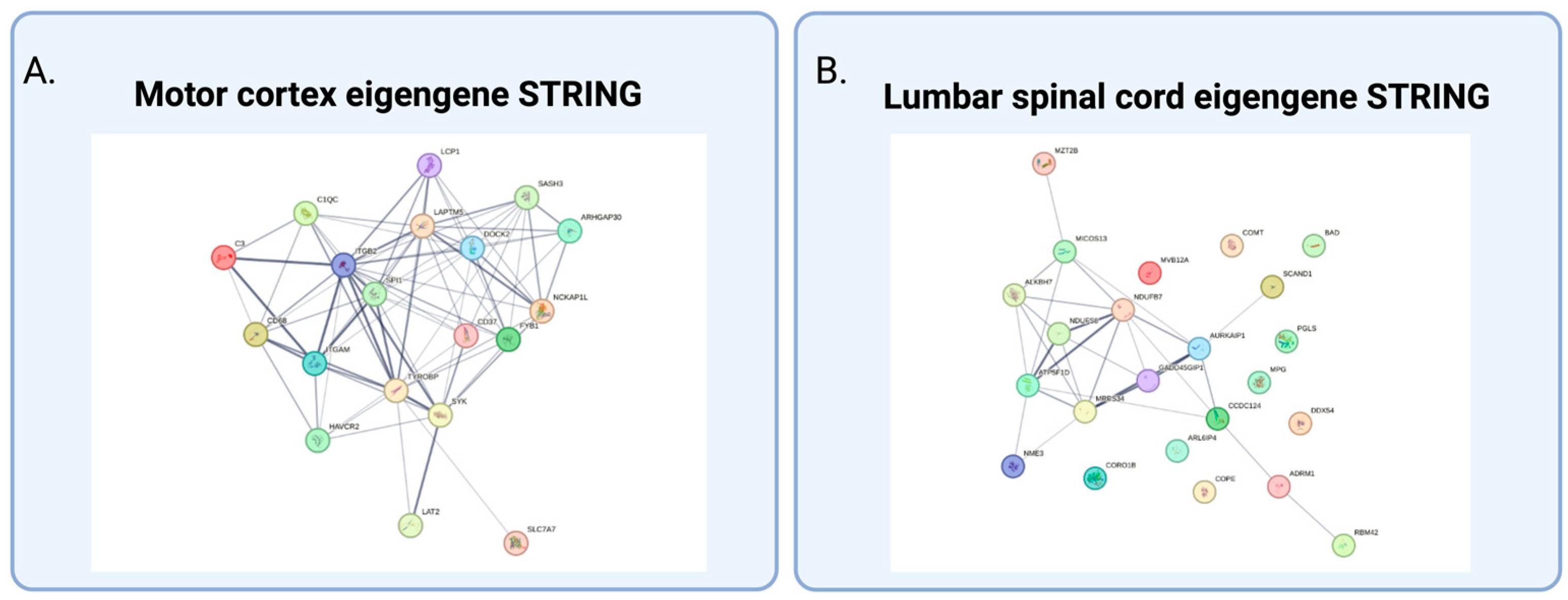
2.2. Weighted Gene Coexpression Analysis and Gene Ontology
3. Discussion
3.1. Gene Ontology and Interpretation of Modules Specific to sALS
3.2. Differential Expression Analysis Revealed Novel Genes Enriched in sALS and fALS
4. Methods and Materials
4.1. Data Acquisition and ALS Consortium
4.2. Data Management, Processing, Quality Control, and Verification
4.3. Cohort Filtering
4.4. Differential Gene Expression Analysis
Differential Expression Design
countData = ctsMatrix,
colData = metaTissue,
design = ~FamHist)
dds.Tissue <– DESeq (object = dds.Tissue).
4.5. Binary Construction
4.6. Correlation Analysis Using WGCNA
4.7. Gene Ontology (GO)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALS | Amyotrophic Lateral Sclerosis |
| fALS | Familial Amyotrophic Lateral Sclerosis |
| sALS | Sporadic Amyotrophic Lateral Sclerosis |
| pALS | Persons with ALS |
| WGCNA | Weighted gene co-expression network analysis |
| DESeq2 | Differential expression sequencing |
| DEGS | Differentially expressed genes |
| GO | Gene Ontology |
| MC | Motor cortex |
| LSP | Lumbar spinal cord |
| mcMod | Motor cortex module |
| scMod | Lumber spinal cord module |
References
- Hardiman, O.; Al-Chalabi, A.; Chio, A.; Corr, E.M.; Logroscino, G.; Robberecht, W.; Shaw, P.J.; Simmons, Z.; van den Berg, L.H. Amyotrophic lateral sclerosis. Nat. Rev. Dis. Primers 2017, 3, 17085. [Google Scholar] [CrossRef]
- Mehta, P.; Raymond, J.; Nair, T.; Han, M.; Berry, J.; Punjani, R.; Larson, T.; Mohidul, S.; Horton, D.K. Amyotrophic lateral sclerosis estimated prevalence cases from 2022 to 2030, data from the national ALS Registry. Amyotroph. Lateral Scler. Front. Degener. 2025, 26, 290–295. [Google Scholar] [CrossRef]
- Mehta, P.; Raymond, J.; Punjani, R.; Larson, T.; Bove, F.; Kaye, W.; Nelson, L.M.; Topol, B.; Han, M.; Muravov, O.; et al. Prevalence of amyotrophic lateral sclerosis (ALS), United States, 2016. Amyotroph. Lateral Scler. Front. Degener. 2022, 23, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Mehta, P.; Raymond, J.; Zhang, Y.; Punjani, R.; Han, M.; Larson, T.; Muravov, O.; Lyles, R.H.; Horton, D.K. Prevalence of amyotrophic lateral sclerosis in the United States, 2018. Amyotroph. Lateral Scler. Front. Degener. 2023, 24, 702–708. [Google Scholar] [CrossRef]
- Berry, J.D.; Blanchard, M.; Bonar, K.; Drane, E.; Murton, M.; Ploug, U.; Ricchetti-Masterson, K.; Savic, N.; Worthington, E.; Heiman-Patterson, T. Epidemiology and economic burden of amyotrophic lateral sclerosis in the United States: A literature review. Amyotroph. Lateral. Scler. Front. Degener. 2023, 24, 436–448. [Google Scholar] [CrossRef] [PubMed]
- Wong, W. Managed care considerations to improve health care utilization for patients with ALS. Am. J. Manag. Care 2023, 29 (Suppl. 7), S120–S126. [Google Scholar]
- Brown, R.H.; Al-Chalabi, A. Amyotrophic Lateral Sclerosis. N. Engl. J. Med. 2017, 377, 162–172. [Google Scholar] [CrossRef]
- Ruf, W.P.; Boros, M.; Freischmidt, A.; Brenner, D.; Grozdanov, V.; de Meirelles, J.; Meyer, T.; Grehl, T.; Petri, S.; Grosskreutz, J.; et al. Spectrum and frequency of genetic variants in sporadic amyotrophic lateral sclerosis. Brain Commun. 2023, 5, fcad152. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Haider, S.; Kumar, R.; Malik, Z.; Singh, M.; Rachana, R.; Mani, S. Amyotrophic Lateral Sclerosis Risk Genes and Suppressor. Curr. Gene Ther. 2023, 23, 148–162. [Google Scholar] [CrossRef]
- Renton, A.E.; Chio, A.; Traynor, B.J. State of play in amyotrophic lateral sclerosis genetics. Nat. Neurosci. 2014, 17, 17–23. [Google Scholar] [CrossRef]
- Goutman, S.A.; Savelieff, M.G.; Jang, D.-G.; Hur, J.; Feldman, E.L. The amyotrophic lateral sclerosis exposome: Recent advances and future directions. Nat. Rev. Neurol. 2023, 19, 617–634. [Google Scholar] [CrossRef]
- McCann, E.P.; Henden, L.; Fifita, J.A.; Zhang, K.Y.; Grima, N.; Bauer, D.C.; Fat, S.C.M.; Twine, N.A.; Pamphlett, R.; Kiernan, M.C.; et al. Evidence for polygenic and oligogenic basis of Australian sporadic amyotrophic lateral sclerosis. J. Med. Genet. 2020, 58, 87–95. [Google Scholar] [CrossRef]
- Benatar, M.; Heiman-Patterson, T.D.; Cooper-Knock, J.; Brickman, D.; Casaletto, K.B.; Goutman, S.A.; Vinceti, M.; Dratch, L.; Arias, J.J.; Swidler, J.; et al. Guidance for clinical management of pathogenic variant carriers at elevated genetic risk for ALS/FTD. J. Neurol. Neurosurg. Psychiatry 2025, 96, 209–218. [Google Scholar] [CrossRef]
- Duranti, E.; Villa, C. Molecular Investigations of Protein Aggregation in the Pathogenesis of Amyotrophic Lateral Sclerosis. Int. J. Mol. Sci. 2022, 24, 704. [Google Scholar] [CrossRef] [PubMed]
- Grad, L.I.; Rouleau, G.A.; Ravits, J.; Cashman, N.R. Clinical Spectrum of Amyotrophic Lateral Sclerosis (ALS). Cold Spring Harb Perspect Med. 2017, 7, a024117. [Google Scholar] [CrossRef] [PubMed]
- Konopka, A.; Atkin, J.D. The Emerging Role of DNA Damage in the Pathogenesis of the C9orf72 Repeat Expansion in Amyotrophic Lateral Sclerosis. Int. J. Mol. Sci. 2018, 19, 3137. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.J.; McKeown, S.R.; Rashid, S. Mutant SOD1 mediated pathogenesis of Amyotrophic Lateral Sclerosis. Gene 2016, 577, 109–118. [Google Scholar] [CrossRef]
- Tafuri, F.; Ronchi, D.; Magri, F.; Comi, G.P.; Corti, S. SOD1 misplacing and mitochondrial dysfunction in amyotrophic lateral sclerosis pathogenesis. Front. Cell. Neurosci. 2015, 9, 336. [Google Scholar] [CrossRef]
- Beland, L.C.; Markovinovic, A.; Jakovac, H.; De Marchi, F.; Bilic, E.; Mazzini, L.; Kriz, J.; Munitic, I. Immunity in amyotrophic lateral sclerosis: Blurred lines between excessive inflammation and inefficient immune responses. Brain Commun. 2020, 2, fcaa124. [Google Scholar] [CrossRef]
- Puentes, F.; Malaspina, A.; van Noort, J.M.; Amor, S. Non-neuronal Cells in ALS: Role of Glial, Immune cells and Blood-CNS Barriers. Brain Pathol. 2016, 26, 248–257. [Google Scholar] [CrossRef]
- Ashhurst, J.F.; Tu, S.; Timmins, H.C.; Kiernan, M.C. Progress, development, and challenges in amyotrophic lateral sclerosis clinical trials. Expert Rev. Neurother. 2022, 22, 905–913. [Google Scholar] [CrossRef]
- Plowman, E.K.; Tabor, L.C.; Wymer, J.; Pattee, G. The evaluation of bulbar dysfunction in amyotrophic lateral sclerosis: Survey of clinical practice patterns in the United States. Amyotroph. Lateral Scler. Front. Degener. 2017, 18, 351–357. [Google Scholar] [CrossRef]
- Bradley, W.G.; Andrew, A.S.; Traynor, B.J.; Chiò, A.; Butt, T.H.; Stommel, E.W. Gene-Environment-Time Interactions in Neurodegenerative Diseases: Hypotheses and Research Approaches. Ann. Neurosci. 2018, 25, 261–267. [Google Scholar] [CrossRef]
- Vasta, R.; Chia, R.; Traynor, B.J.; Chiò, A. Unraveling the complex interplay between genes, environment, and climate in ALS. EBioMedicine 2022, 75, 103795. [Google Scholar] [CrossRef]
- Granit, V.; Grignon, A.; Wuu, J.; Katz, J.; Walk, D.; Hussain, S.; Hernandez, J.; Jackson, C.; Caress, J.; Yosick, T.; et al. Harnessing the power of the electronic health record for ALS research and quality improvement: CReATe CAPTURE-ALS and the ALS Toolkit. Muscle Nerve 2022, 65, 154–161. [Google Scholar] [CrossRef]
- Berger, A.; Locatelli, M.; Arcila-Londono, X.; Hayat, G.; Olney, N.; Wymer, J.; Gwathmey, K.; Lunetta, C.; Heiman-Patterson, T.; Ajroud-Driss, S.; et al. The natural history of ALS: Baseline characteristics from a multicenter clinical cohort. Amyotroph. Lateral Scler. Front. Degener. 2023, 24, 625–6339. [Google Scholar] [CrossRef]
- O’Neill, K.; Shaw, R.; Bolger, I.; Tam, O.H.; Phatnani, H.; Hammell, M.G. ALS molecular subtypes are a combination of cellular and pathological features learned by deep multiomics classifiers. Cell Rep. 2025, 44, 115402. [Google Scholar] [CrossRef] [PubMed]
- Tam, O.H.; Rozhkov, N.V.; Shaw, R.; Kim, D.; Hubbard, I.; Fennessey, S.; Propp, N.; Fagegaltier, D.; Harris, B.T.; Ostrow, L.W.; et al. Postmortem Cortex Samples Identify Distinct Molecular Subtypes of ALS: Retrotransposon Activation, Oxidative Stress, and Activated Glia. Cell Rep. 2019, 29, 1164–1177.e5. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.; Ibrahim, J.G.; Love, M.I. Heavy-tailed prior distributions for sequence count data: Removing the noise and preserving large differences. Bioinformatics 2019, 35, 2084–2092. [Google Scholar] [CrossRef]
- Oldham, M.C.; Konopka, G.; Iwamoto, K.; Langfelder, P.; Kato, T.; Horvath, S.; Geschwind, D.H. Functional organization of the transcriptome in human brain. Nat. Neurosci. 2008, 11, 1271–1282. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. Fast R Functions for Robust Correlations and Hierarchical Clustering. J. Stat. Softw. 2012, 46, 1–17. [Google Scholar] [CrossRef]
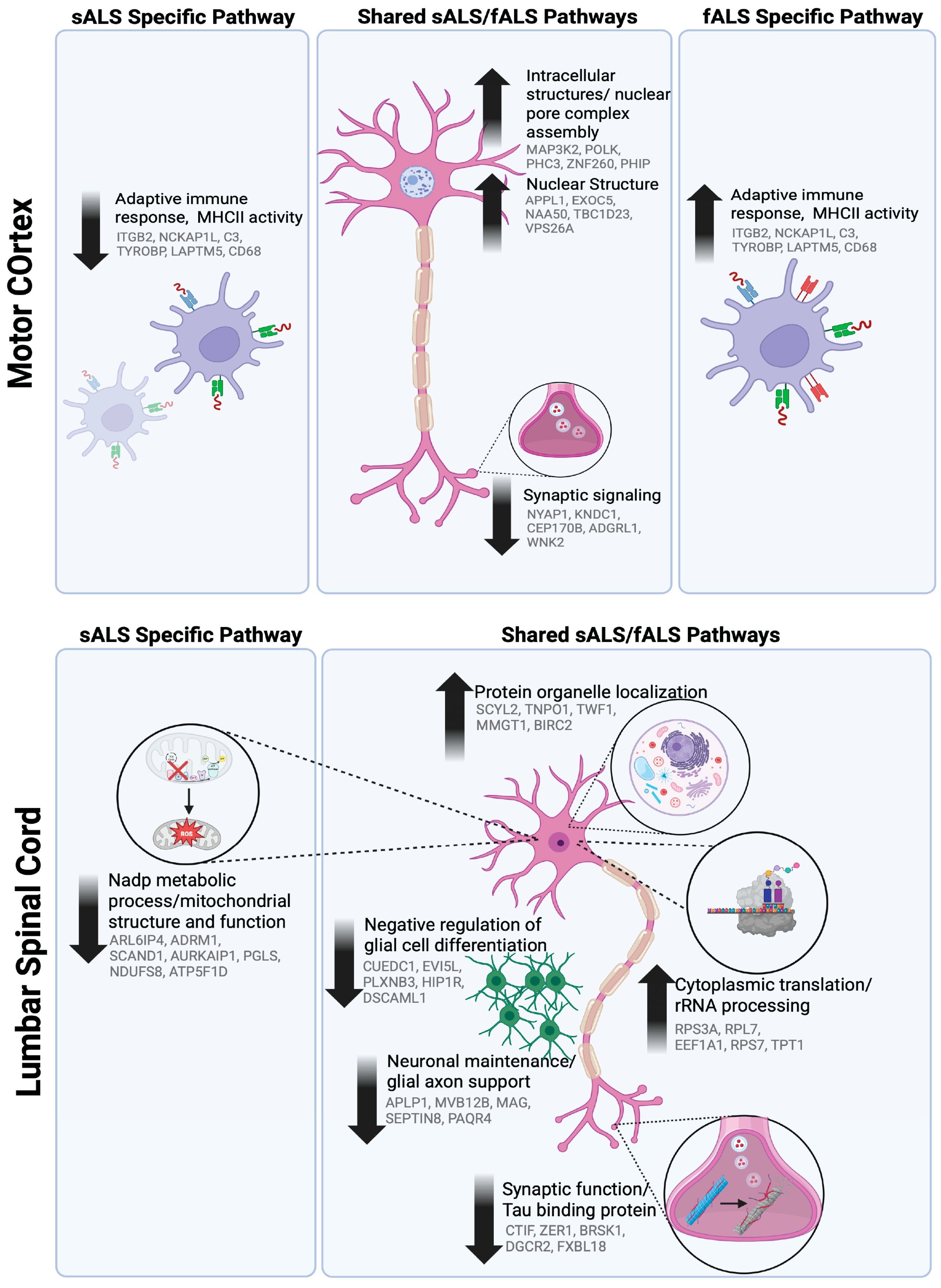
| Motor Cortex Module | Gene Ontology | Hub Genes | MEs ANOVA p-Value: | |
|---|---|---|---|---|
| Shared Gene Networks | ||||
| Upregulated | ||||
| mcM9 | Intracellular structures/nuclear pore complex assembly | MAP3K2; POLK; PHC3; ZNF260; PHIP | 0.02 | |
| mcM2 | Nuclear structure | APPL1; EXOC5; NAA50; TBC1D23; VPS26A | 0.05 | |
| Downregulated | mcM5 | Synaptic signaling | NYAP1; KNDC1; CEP170B; ADGRL1; WNK2 | 0.05 |
| Upregulated in fALS Downregulated in sALS | mcM18 | Immune and metabolic processes; MHCII and compliment system | ITGB2, NCKAP1L, C3, TYROBP, LAPTM5, CD68 | 0.09 |
| Spinal Cord Module | Gene Ontology | Hub Genes |
MEs ANOVA p-Value: | |
|---|---|---|---|---|
| Shared Gene Networks | ||||
| Upregulated | ||||
| scM1 | Protein organelle localization | SCYL2; TNPO1; TWF1; MMGT1; BIRC2 | 0.02 | |
| scM17 | Cytoplasmic translation/rRNA processing | RPS3A; RPL7; EEF1A1; RPS7; TPT1 | 0.04 | |
| Downregulated | scM14 | Synaptic function/Tau binding protein | CTIF; ZER1; BRSK1; DGCR2; FXBL18 | 0.01 |
| scM3 | Negative regulation of glial cell differentiation | CUEDC1; EVI5L; PLXNB3; HIP1R; DSCAML1 | 0.05 | |
| scM10 | Neuronal maintenance/ glial-axon support | APLP1; MVB12B; MAG; SEPTIN8; PAQR4 | 0.01 | |
| scM12 | Regulation of neuronal synaptic plasticity | NEURL1; BSN; JPH3; JPH4; IQSEC3 | 0.01 | |
| sALS Network | ||||
| Downregulated | scM13 | Nadp metabolic process/mitochondrial structure and function | ARL6IP4; ADRM1; SCAND1; AURKAIP1; PGLS; NDUFS8; ATP5F1D | 0.02 |
| Course Region | fALS | sALS |
Non-Neurological Control | Total n |
|---|---|---|---|---|
| Motor Cortex | 48 | 31 | 69 | 148 |
| Lumbar Spinal Cord | 32 | 14 | 53 | 99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Awai, A.; Johnson, E.L.; Leng, T.; Patrickson, J.; Zody, M.C.; Lillard, J.W., Jr.; on behalf of the NYGC ALS Consortium. Stratifying ALS Patients by Mode of Inheritance Reveals Transcriptomic Signatures Specific to sALS and fALS. Int. J. Mol. Sci. 2025, 26, 9234. https://doi.org/10.3390/ijms26189234
Awai A, Johnson EL, Leng T, Patrickson J, Zody MC, Lillard JW Jr., on behalf of the NYGC ALS Consortium. Stratifying ALS Patients by Mode of Inheritance Reveals Transcriptomic Signatures Specific to sALS and fALS. International Journal of Molecular Sciences. 2025; 26(18):9234. https://doi.org/10.3390/ijms26189234
Chicago/Turabian StyleAwai, Alexandria, Erica L. Johnson, Tiandong Leng, John Patrickson, Michael C. Zody, James W. Lillard, Jr., and on behalf of the NYGC ALS Consortium. 2025. "Stratifying ALS Patients by Mode of Inheritance Reveals Transcriptomic Signatures Specific to sALS and fALS" International Journal of Molecular Sciences 26, no. 18: 9234. https://doi.org/10.3390/ijms26189234
APA StyleAwai, A., Johnson, E. L., Leng, T., Patrickson, J., Zody, M. C., Lillard, J. W., Jr., & on behalf of the NYGC ALS Consortium. (2025). Stratifying ALS Patients by Mode of Inheritance Reveals Transcriptomic Signatures Specific to sALS and fALS. International Journal of Molecular Sciences, 26(18), 9234. https://doi.org/10.3390/ijms26189234





