Metabolomics in Multiple Sclerosis: Advances, Challenges, and Clinical Perspectives—A Systematic Review
Abstract
1. Introduction
Added Value Compared with Prior Reviews
2. Methods
2.1. Search Strategy
2.2. Eligibility Criteria
2.3. Information Sources
2.4. Quality and Risk of Bias Assessment
2.5. Technical Validation and Quality Control
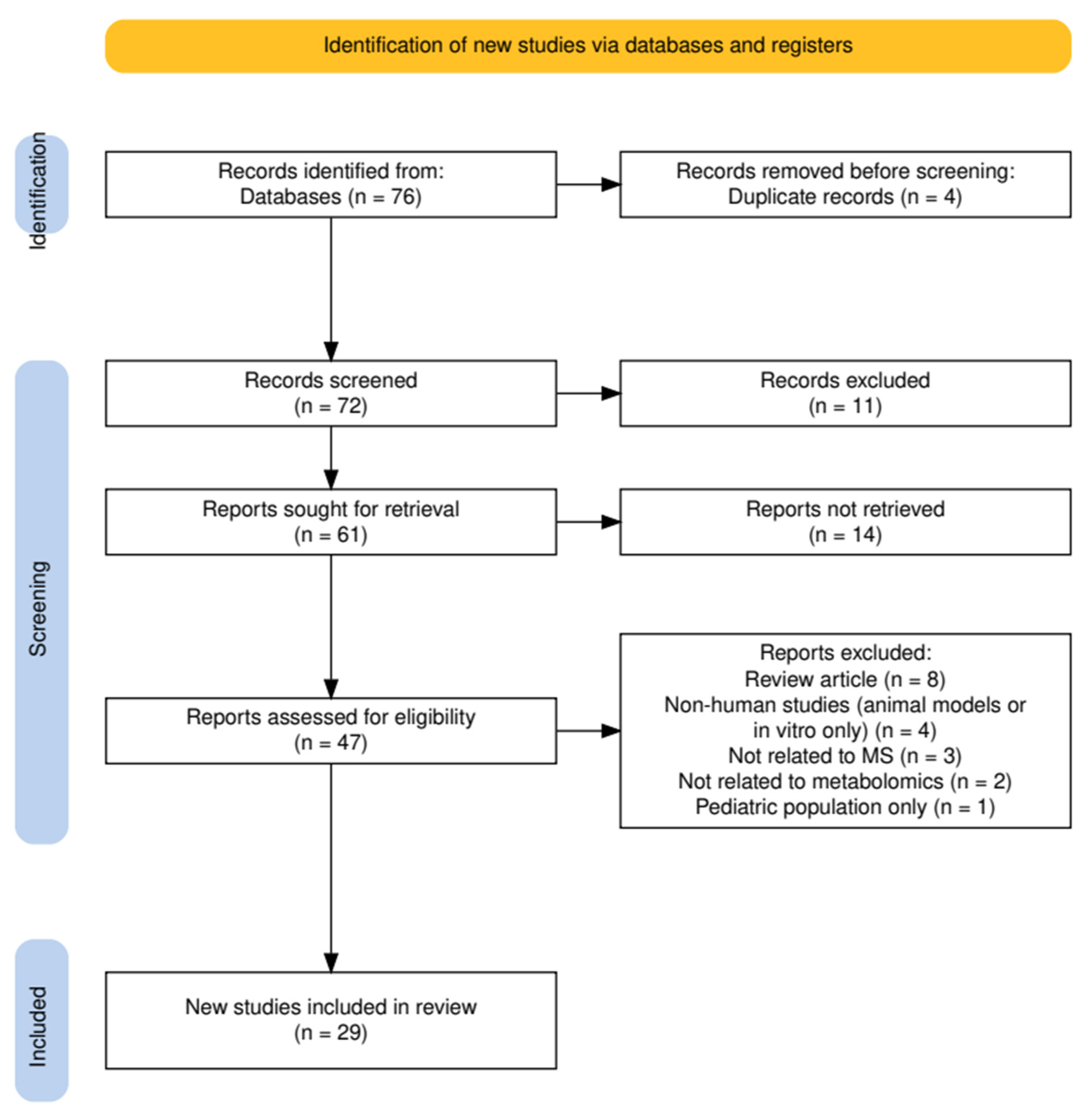
2.6. Study Selection
2.7. Data Synthesis
3. Results
3.1. The Kynurenine Pathway
3.2. Energy Metabolism
3.2.1. Ketone Bodies
3.2.2. Central Carbon Metabolism: Glycolysis, Pyruvate, and TCA Cycle
3.2.3. Metabolic Reprogramming vs. Transcriptional Regulation
3.2.4. Inflammation-Linked Energy Metabolites and Mitochondrial Cofactors
3.3. Lipid Metabolism
3.3.1. Lipid Mediators in RRMS and Progressive MS
3.3.2. Effect of DMTs and Exercise on Lipid Metabolism
3.3.3. Endocannabinoid System and Neurosteroids
3.3.4. Phospholipids, Sphingolipids, and Membrane Structure
3.3.5. Short-Chain Fatty Acids and Gut–Brain Crosstalk
3.3.6. Early and Postmortem Lipid Alterations
3.4. Nitrogen Metabolism
3.4.1. Amino Acid Disturbances and Disease Phenotype
3.4.2. Amino Acids as Relapse and Disability Markers
3.4.3. Polyamine and Biogenic Amine Metabolism
3.4.4. Peptide Metabolism
3.4.5. Nucleotide Metabolism
3.5. Other Metabolites
3.6. Metabolomics as a Diagnostic Tool in Multiple Sclerosis
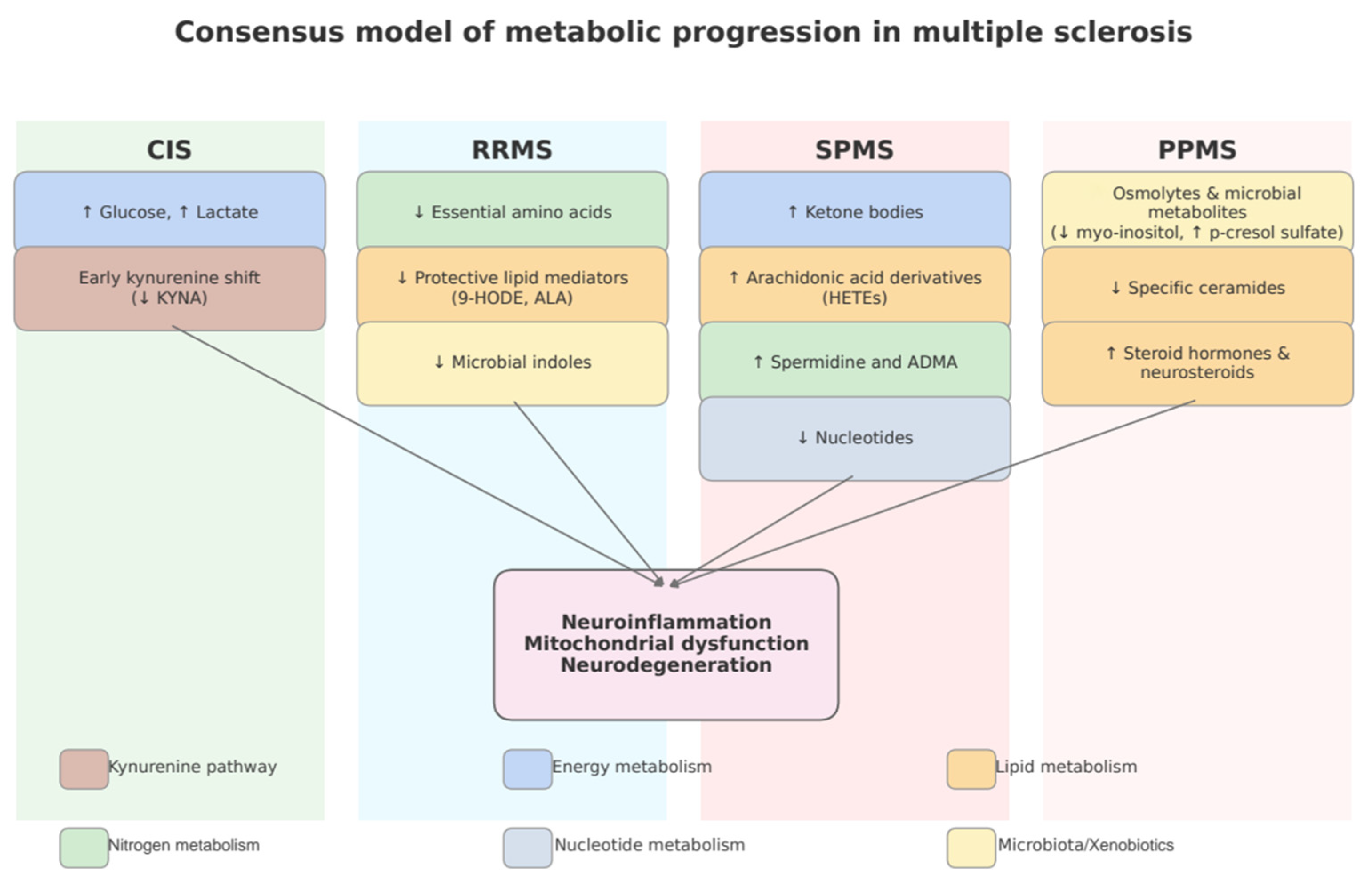
3.7. Subgroup Analysis by MS Phenotype
4. Discussion
Limitations
5. Conclusions
- Longitudinal, phenotype-stratified metabolomic studies that capture dynamic metabolic trajectories across disease stages, including conversion from CIS to MS and progression from RRMS to SPMS/PPMS;
- Validation of candidate biomarkers (e.g., myo-inositol, lysine, acetate) in multicenter cohorts using harmonized protocols;
- Integration of multi-omics platforms, including transcriptomics, microbiomics, and neuroimaging, to build mechanistic models of MS pathophysiology;
- While metabolomics-based machine-learning classifiers demonstrate promising discriminatory performance (typically 70–80% accuracy), these approaches are not yet sufficiently robust for clinical decision-making. Their current role should be regarded as exploratory, requiring validation in larger, multicenter cohorts before they can be translated into practice;
- Exploration of therapeutic modulation, assessing how metabolic pathways respond to DMTs (e.g., fingolimod, ocrelizumab, IFNβ) or interventions like resistance training and probiotic supplementation;
- To improve reproducibility and comparability of metabolomic findings, future guidelines should mandate transparent reporting of pre-analytical procedures and support the development of SOPs and pre-analytical validation studies;
- Future studies should adopt harmonized standard operating procedures and transparent reporting of calibration, QC design, and internal standards, ideally aligned with the Metabolomics Standards Initiative, to improve reproducibility and facilitate clinical translation in multiple sclerosis;
- Future studies should harmonize inclusion and exclusion criteria and systematically document comorbidities, medications, and lifestyle factors to minimize confounding and enhance reproducibility;
- With continued advances in analytical technologies and systems-level approaches, metabolomics shows considerable promise for advancing personalized medicine in MS, particularly through biomarker discovery that may eventually support diagnosis, monitoring of disease progression, and development of targeted therapeutic strategies. However, clinical validation in large, standardized cohorts is still required before translation into practice.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Ghasemi, N.; Razavi, S.; Nikzad, E. Multiple Sclerosis: Pathogenesis, Symptoms, Diagnoses and Cell-Based Therapy. Cell J. 2017, 19, 1–10. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dighriri, I.M.; Aldalbahi, A.A.; Albeladi, F.; Tahiri, A.A.; Kinani, E.M.; Almohsen, R.A.; Alamoudi, N.H.; Alanazi, A.A.; Alkhamshi, S.J.; Althomali, N.A.; et al. An Overview of the History, Pathophysiology, and Pharmacological Interventions of Multiple Sclerosis. Cureus 2023, 15, e33242. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lassmann, H. Multiple Sclerosis Pathology. Cold Spring Harb. Perspect. Med. 2018, 8, a028936. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lublin, F.D.; Reingold, S.C.; Cohen, J.A.; Cutter, G.R.; Sørensen, P.S.; Thompson, A.J.; Wolinsky, J.S.; Balcer, L.J.; Banwell, B.; Barkhof, F.; et al. Defining the clinical course of multiple sclerosis: The 2013 revisions. Neurology 2014, 83, 278–286. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tafti, D.; Ehsan, M.; Xixis, K.L. Multiple Sclerosis. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar] [PubMed]
- Kapica-Topczewska, K.; Kulakowska, A.; Kochanowicz, J.; Brola, W. Epidemiology of multiple sclerosis: Global trends, regional differences, and clinical implications. Neurol. I Neurochir. Pol. 2025. ahead of print. [Google Scholar] [CrossRef]
- Shi, M.; Liu, Y.; Gong, Q.; Xu, X. Multiple Sclerosis: An Overview of Epidemiology, Risk Factors, and Serological Biomarkers. Acta Neurol. Scand. 2024, 2024, 7372789. [Google Scholar] [CrossRef]
- Mao, P.; Reddy, P.H. Is multiple sclerosis a mitochondrial disease? Biochim. Biophys. Acta 2010, 1802, 66–79. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- GBD 2016 Multiple Sclerosis Collaborators. Global, regional, and national burden of multiple sclerosis 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019, 18, 269–285. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Safiri, S.; Ghaffari Jolfayi, A.; Mousavi, S.E.; Nejadghaderi, S.A.; Sullman, M.J.M.; Kolahi, A.A. Global burden of multiple sclerosis and its attributable risk factors, 1990–2019. Front. Neurol. 2024, 15, 1448377. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Thompson, A.J.; Banwell, B.L.; Barkhof, F.; Carroll, W.M.; Coetzee, T.; Comi, G.; Correale, J.; Fazekas, F.; Filippi, M.; Freedman, M.S.; et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018, 17, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Waters, J.; Rui, B. Metabolomics as a promising tool for improving understanding of multiple sclerosis: A review of recent advances. Biomed. J. 2022, 45, 594–606. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Broos, J.Y.; van der Burgt, R.T.M.; Konings, J.; Rijnsburger, M.; Werz, O.; de Vries, H.E.; Giera, M.; Kooij, G. Arachidonic acid-derived lipid mediators in multiple sclerosis pathogenesis: Fueling or dampening disease progression? J. Neuroinflammation 2024, 21, 21. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Otto, C.; Kalantzis, R.; Kübler-Weller, D.; Kühn, A.A.; Böld, T.; Regler, A.; Strathmeyer, S.; Wittmann, J.; Ruprecht, K.; Heelemann, S. Comprehensive analysis of the cerebrospinal fluid and serum metabolome in neurological diseases. J. Neuroinflammation 2024, 21, 234. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pousinis, P.; Begou, O.; Boziki, M.K.; Grigoriadis, N.; Theodoridis, G.; Gika, H. Recent Advances in Metabolomics and Lipidomics Studies in Human and Animal Models of Multiple Sclerosis. Metabolites 2024, 14, 545. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Alwahsh, M.; Nimer, R.M.; Dahabiyeh, L.A.; Hamadneh, L.; Hasan, A.; Alejel, R.; Hergenröder, R. NMR-based metabolomics identification of potential serum biomarkers of disease progression in patients with multiple sclerosis. Sci. Rep. 2024, 26, 14806. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wicks, T.R.; Shalaurova, I.; Wolska, A.; Browne, R.W.; Weinstock-Guttman, B.; Zivadinov, R.; Remaley, A.T.; Otvos, J.D.; Ramanathan, M. Endogenous Ketone Bodies Are Associated with Metabolic Vulnerability and Disability in Multiple Sclerosis. Nutrients 2025, 17, 640. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Siavoshi, F.; Ladakis, D.C.; Muller, A.; Nourbakhsh, B.; Bhargava, P. Ocrelizumab alters the circulating metabolome in people with relapsing-remitting multiple sclerosis. Ann. Clin. Transl. Neurol. 2024, 11, 2485–2498. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Probert, F.; Yeo, T.; Zhou, Y.; Sealey, M.; Arora, S.; Palace, J.; Claridge, T.D.W.; Hillenbrand, R.; Oechtering, J.; Leppert, D.; et al. Integrative biochemical, proteomics and metabolomics cerebrospinal fluid biomarkers predict clinical conversion to multiple sclerosis. Brain Commun. 2021, 3, fcab084. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Radford-Smith, D.E.; Yates, A.G.; Kacerova, T.; Nylund, M.; Sucksdorff, M.; Matilainen, M.; Willemse, E.; Oechtering, J.; Maleska Maceski, A.; Leppert, D.; et al. Integrating TSPO-PET imaging with metabolomics for enhanced prognostic accuracy in multiple sclerosis. BMJ Neurol. Open 2025, 7, e001026. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Keller, J.; Zackowski, K.; Kim, S.; Chidobem, I.; Smith, M.; Farhadi, F.; Bhargava, P. Exercise leads to metabolic changes associated with improved strength and fatigue in people with MS. Ann. Clin. Transl. Neurol. 2021, 8, 1308–1317. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Broos, J.Y.; Loonstra, F.C.; de Ruiter, L.R.J.; Gouda, M.; Fung, W.H.; Schoonheim, M.M.; Heijink, M.; Strijbis, E.M.M.; Teunissen, C.; Killestein, J.; et al. Association of Arachidonic Acid-Derived Lipid Mediators with Disease Severity in Patients with Relapsing and Progressive Multiple Sclerosis. Neurology 2023, 101, e533–e545. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Meier, P.; Glasmacher, S.; Salmen, A.; Chan, A.; Gertsch, J. Comparative targeted lipidomics between serum and cerebrospinal fluid of multiple sclerosis patients shows sex and age-specific differences of endocannabinoids and glucocorticoids. Acta Neuropathol. Commun. 2024, 12, 160. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Duscha, A.; Gisevius, B.; Hirschberg, S.; Yissachar, N.; Stangl, G.I.; Dawin, E.; Bader, V.; Haase, S.; Kaisler, J.; David, C.; et al. Propionic Acid Shapes the Multiple Sclerosis Disease Course by an Immunomodulatory Mechanism. Cell 2020, 180, 1067–1080.e16. [Google Scholar] [CrossRef] [PubMed]
- Schwerdtfeger, L.A.; Montini, F.; Lanser, T.B.; Ekwudo, M.N.; Zurawski, J.; Tauhid, S.; Glanz, B.I.; Chu, R.; Bakshi, R.; Chitnis, T.; et al. Gut microbiota and metabolites are linked to disease progression in multiple sclerosis. Cell Rep. Med. 2025, 6, 102055. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Datta, I.; Zahoor, I.; Ata, N.; Rashid, F.; Cerghet, M.; Rattan, R.; Poisson, L.M.; Giri, S. Utility of an untargeted metabolomics approach using a 2D GC-GC-MS platform to distinguish relapsing and progressive multiple sclerosis. Metabolites 2024, 14, 493. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yeo, T.; Sealey, M.; Zhou, Y.; Saldana, L.; Loveless, S.; Claridge, T.D.W.; Robertson, N.; DeLuca, G.; Palace, J.; Anthony, D.C.; et al. A blood-based metabolomics test to distinguish relapsing-remitting and secondary progressive multiple sclerosis: Addressing practical considerations for clinical application. Sci. Rep. 2020, 10, 12381. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Carlsson, H.; Abujrais, S.; Herman, S.; Khoonsari, P.E.; Åkerfeldt, T.; Svenningsson, A.; Burman, J.; Kultima, K. Targeted metabolomics of CSF in healthy individuals and patients with secondary progressive multiple sclerosis using high-resolution mass spectrometry. Metabolomics 2020, 16, 26. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ladakis, D.C.; Pedrini, E.; Reyes-Mantilla, M.I.; Sanjayan, M.; Smith, M.D.; Fitzgerald, K.C.; Pardo, C.A.; Reich, D.S.; Absinta, M.; Bhargava, P. Metabolomics of Multiple Sclerosis Lesions Demonstrates Lipid Changes Linked to Alterations in Transcriptomics-Based Cellular Profiles. Neurol. Neuroimmunol. Neuroinflammation 2024, 11, e200219. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Amatruda, M.; Petracca, M.; Wentling, M.; Inbar, B.; Castro, K.; Chen, E.Y.; Kiebish, M.A.; Edwards, K.; Inglese, M.; Casaccia, P. Retrospective unbiased plasma lipidomic of progressive multiple sclerosis patients-identifies lipids discriminating those with faster clinical deterioration. Sci. Rep. 2020, 10, 15644. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Židó, M.; Kačer, D.; Valeš, K.; Zimová, D.; Štětkářová, I. Metabolomics of Cerebrospinal Fluid Amino and Fatty Acids in Early Stages of Multiple Sclerosis. Int. J. Mol. Sci. 2023, 24, 16271. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yang, F.; Wu, S.C.; Ling, Z.X.; Chao, S.; Zhang, L.J.; Yan, X.M.; He, L.; Yu, L.M.; Zhao, L.Y. Altered Plasma Metabolic Profiles in Chinese Patients with Multiple Sclerosis. Front. Immunol. 2021, 12, 792711. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gaetani, L.; Boscaro, F.; Pieraccini, G.; Calabresi, P.; Romani, L.; Di Filippo, M.; Zelante, T. Host and Microbial Tryptophan Metabolic Profiling in Multiple Sclerosis. Front. Immunol. 2020, 11, 157. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zahoor, I.; Suhail, H.; Datta, I.; Ahmed, M.E.; Poisson, L.M.; Waters, J.; Rashid, F.; Bin, R.; Singh, J.; Cerghet, M.; et al. Blood-based untargeted metabolomics in relapsing-remitting multiple sclerosis revealed the testable therapeutic target. Proc. Natl. Acad. Sci. USA 2022, 119, e2123265119. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Levi, I.; Gurevich, M.; Perlman, G.; Magalashvili, D.; Menascu, S.; Bar, N.; Godneva, A.; Zahavi, L.; Chermon, D.; Kosower, N.; et al. Potential role of indolelactate and butyrate in multiple sclerosis revealed by integrated microbiome-metabolome analysis. Cell Rep. Med. 2021, 2, 100246. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yeo, T.; Probert, F.; Sealey, M.; Saldana, L.; Geraldes, R.; Höeckner, S.; Schiffer, E.; Claridge, T.D.W.; Leppert, D.; DeLuca, G.; et al. Objective biomarkers for clinical relapse in multiple sclerosis: A metabolomics approach. Brain Commun. 2021, 3, fcab240, Erratum in Brain Commun. 2021, 3, fcab283. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Staats Pires, A.; Krishnamurthy, S.; Sharma, S.; Chow, S.; Klistorner, S.; Guillemin, G.J.; Klistorner, A.; You, Y.; Heng, B. Dysregulation of the Kynurenine Pathway in Relapsing Remitting Multiple Sclerosis and Its Correlations with Progressive Neurodegeneration. Neurol. Neuroimmunol. Neuroinflammation 2025, 12, e200372. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fung, W.H.; van Lingen, M.R.; Broos, J.Y.; Lam, K.H.; van Dam, M.; Fung, W.K.; Noteboom, S.; Koubiyr, I.; de Vries, H.E.; Jasperse, B.; et al. 9-HODE associates with thalamic atrophy and predicts white matter damage in multiple sclerosis. Mult. Scler. Relat. Disord. 2024, 92, 105946. [Google Scholar] [CrossRef] [PubMed]
- Murgia, F.; Lorefice, L.; Noto, A.; Spada, M.; Frau, J.; Fenu, G.; Coghe, G.; Gagliano, A.; Atzori, L.; Cocco, E. Metabolomic changes in patients affected by multiple sclerosis and treated with fingolimod. Metabolites 2023, 13, 428. [Google Scholar] [CrossRef]
- Židó, M.; Kačer, D.; Valeš, K.; Svobodová, Z.; Zimová, D.; Štětkárová, I. Metabolomics of Cerebrospinal Fluid in Multiple Sclerosis Compared with Healthy Controls: A Pilot Study. Front. Neurol. 2022, 13, 874121. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Olsson, A.; Gustavsen, S.; Nguyen, T.D.; Nyman, M.; Langkilde, A.R.; Hansen, T.H.; Sellebjerg, F.; Oturai, A.B.; Bach Søndergaard, H. Serum Short-Chain Fatty Acids and Associations with Inflammation in Newly Diagnosed Patients with Multiple Sclerosis and Healthy Controls. Front. Immunol. 2021, 12, 661493. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fitzgerald, K.C.; Smith, M.D.; Kim, S.; Sotirchos, E.S.; Kornberg, M.D.; Douglas, M.; Nourbakhsh, B.; Graves, J.; Rattan, R.; Poisson, L.; et al. Multi-omic evaluation of metabolic alterations in multiple sclerosis identifies shifts in aromatic amino acid metabolism. Cell. Rep. Med. 2021, 2, 100424. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Penkert, H.; Lauber, C.; Gerl, M.J.; Klose, C.; Damm, M.; Fitzner, D.; Flierl-Hecht, A.; Kümpfel, T.; Kerschensteiner, M.; Hohlfeld, R.; et al. Plasma lipidomics of monozygotic twins discordant for multiple sclerosis. Ann. Clin. Transl. Neurol. 2020, 7, 2461–2466. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Waddington, K.E.; Papadaki, A.; Coelewij, L.; Adriani, M.; Nytrova, P.; Kubala Havrdova, E.; Fogdell-Hahn, A.; Farrell, R.; Dönnes, P.; Pineda-Torra, I.; et al. Using Serum Metabolomics to Predict Development of Anti-drug Antibodies in Multiple Sclerosis Patients Treated with IFNβ. Front. Immunol. 2020, 11, 1527. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rispoli, M.G.; Valentinuzzi, S.; De Luca, G.; Del Boccio, P.; Federici, L.; Di Ioia, M.; Digiovanni, A.; Grasso, E.A.; Pozzilli, V.; Villani, A.; et al. Contribution of Metabolomics to Multiple Sclerosis Diagnosis, Prognosis and Treatment. Int. J. Mol. Sci. 2021, 22, 11112. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Byun, J.; Pennathur, S. Analytical approaches to metabolomics and applications to systems biology. Semin. Nephrol. 2010, 30, 500–511. [Google Scholar] [CrossRef] [PubMed]
- Munjal, Y.; Tonk, R.K.; Sharma, R. Analytical Techniques Used in Metabolomics: A Review. Syst. Rev. Pharm. 2022, 13, 515–521. [Google Scholar]
- Segers, K.; Declerck, S.; Mangelings, D.; Heyden, Y.V.; Eeckhaut, A.V. Analytical Techniques for Metabolomic Studies: A Review. Bioanalysis 2019, 11, 2297–2318. [Google Scholar] [CrossRef]
- Chen, Y.; Guillemin, G.J. Kynurenine pathway metabolites in humans: Disease and healthy States. Int. J. Tryptophan Res. 2009, 2, IJTR-S2097. [Google Scholar] [CrossRef]
- Lovelace, M.D.; Varney, B.; Sundaram, G.; Franco, N.F.; Ng, M.L.; Pai, S.; Lim, C.K.; Guillemin, G.J.; Brew, B.J. Current Evidence for a Role of the Kynurenine Pathway of Tryptophan Metabolism in Multiple Sclerosis. Front. Immunol. 2016, 7, 246. [Google Scholar] [CrossRef]
- Braidy, N.; Grant, R. Kynurenine pathway metabolism and neuroinflammatory disease. Neural Regen. Res. 2017, 12, 39–42. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Robertson, D.; Moreo, N. Disease-Modifying Therapies in Multiple Sclerosis: Overview and Treatment Considerations. Fed. Pract. 2016, 33, 28–34. [Google Scholar] [PubMed] [PubMed Central]
- Gębka-Kępińska, B.; Adamczyk, B.; Gębka, D.; Czuba, Z.; Szczygieł, J.; Adamczyk-Sowa, M. Cytokine Profiling in Cerebrospinal Fluid of Patients with Newly Diagnosed Relapsing-Remitting Multiple Sclerosis (RRMS): Associations between Inflammatory Biomarkers and Disease Activity. Int. J. Mol. Sci. 2024, 25, 7399. [Google Scholar] [CrossRef]
- Disanto, G.; Barro, C.; Benkert, P.; Naegelin, Y.; Schädelin, S.; Giardiello, A.; Zecca, C.; Blennow, K.; Zetterberg, H.; Leppert, D.; et al. Serum Neurofilament light: A biomarker of neuronal damage in multiple sclerosis. Ann. Neurol. 2017, 81, 857–870. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Niedziela, N.; Nowak-Kiczmer, M.; Malciene, L.; Stasiołek, M.; Zalejska-Fiolka, J.; Czuba, Z.P.; Niedziela, J.T.; Szczygieł, J.; Lubczyński, M.; Adamczyk-Sowa, M. Can Selected Parameters of Brain Injury Reflect Neuronal Damage in Smoldering Multiple Sclerosis? Diagnostics 2024, 14, 1993. [Google Scholar] [CrossRef]
- Ayrignac, X.; Le Bars, E.; Duflos, C.; Hirtz, C.; Maleska Maceski, A.; Carra-Dallière, C.; Charif, M.; Pinna, F.; Prin, P.; Menjot de Champfleur, N.; et al. Serum GFAP in multiple sclerosis: Correlation with disease type and MRI markers of disease severity. Sci. Rep. 2020, 10, 10923. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jafari, A.; Babajani, A.; Rezaei-Tavirani, M. Multiple Sclerosis Biomarker Discoveries by Proteomics and Metabolomics Approaches. Biomark Insights 2021, 16, 11772719211013352. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Oppong, A.E.; Coelewij, L.; Robertson, G.; Martin-Gutierrez, L.; Waddington, K.E.; Dönnes, P.; Nytrova, P.; Farrell, R.; Pineda-Torra, I.; Jury, E.C. Blood metabolomic and transcriptomic signatures stratify patient subgroups in multiple sclerosis according to disease severity. iScience 2024, 27, 109225. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- De Keersmaecker, A.V.; Van Doninck, E.; Popescu, V.; Willem, L.; Cambron, M.; Laureys, G.; D’ Haeseleer, M.; Bjerke, M.; Roelant, E.; Lemmerling, M.; et al. A metformin add-on clinical study in multiple sclerosis to evaluate brain remyelination and neurodegeneration (MACSiMiSE-BRAIN): Study protocol for a multi-center randomized placebo controlled clinical trial. Front. Immunol. 2024, 15, 1362629. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hammer, A.; Waschbisch, A.; Kuhbandner, K.; Bayas, A.; Lee, D.H.; Duscha, A.; Haghikia, A.; Gold, R.; Linker, R.A. The NRF2 pathway as potential biomarker for dimethyl fumarate treatment in multiple sclerosis. Ann. Clin. Transl. Neurol. 2018, 5, 668–676. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nijland, P.G.; Witte, M.E.; van het Hof, B.; van der Pol, S.; Bauer, J.; Lassmann, H.; van der Valk, P.; de Vries, H.E.; van Horssen, J. Astroglial PGC-1alpha increases mitochondrial antioxidant capacity and suppresses inflammation: Implications for multiple sclerosis. Acta Neuropathol. Commun. 2014, 2, 170. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
| Author (Year) | MS Phenotype | Number of Participants | Mean Age ± SD | Sex (F/M) | Biological Sample | Analysis Type | Analytical Technique |
|---|---|---|---|---|---|---|---|
| Staats Pires (2025) [38] | RRMS | RRMS—98 HC—39 | RRMS: 42.3 (10.8) HC: 36 (6.3) | RRMS 72F/26M HC 28F/11M | serum | targeted | HPLC/GCLC |
| Wicks (2025) [17] | RRMS, PMS | RRMS—184 PMS—91 HC—152 | RRMS: 44.2 (9.7) PMS: 54.1 (8.82) HC: 45.7 (13.9) | RRMS 139F/45M PMS 66F/25M HC 85F/67M | serum | targeted | NMR |
| Radford-Smith (2025) [20] | RRMS (progressors vs. non-progressors) | Non-progressors—52 Progressors—23 | Non-progressors: 46.4 Progressors: 48.9 | Non-progressors 43F/9M Progressors 16F/7M | serum | untargeted | NMR |
| Schwerdtfeger (2025) [25] | RRMS (progressors vs. non-progressors), | RRMS, stable—124 RRMS progression—68 | RRMS, stable: 51.5 (9.8) RRMS progression: 54.1 (8.5) | RRMS stable 94F/30M RRMS progression 52F/16M | serum + feces | untargeted | HPLC |
| Alwahsh (2024) [16] | RRMS, PPMS, SPMS | RRMS—30 PPMS—30 SPMS—30 HC—30 | RRMS: 37.0 (8.6) PPMS: 33.4 (7.0) SPMS: 37.9 (10.5) HC: 35.6 (2.5) | RRMS 19F/11M PPMS 15F/15M SPMS 15F/15M HC 15F/15M | serum | untargeted | NMR |
| Siavoshi (2024) [18] | RRMS (pre/post-ocrelizumab) | RRMS—31 | 40.84 (10.34) | 21F/10M | serum | untargeted | LC-MS/CG-MS |
| Ladakis (2024) [30] | MS lesions | SPMS—5 Controls—6 | Not reported | Not reported | brain tissue | untargeted | LC-MS + WGCNA |
| Fung (2024) [39] | RRMS | RRMS—28 HC—31 | RRMS: 41.4 (9.1) HC: 41.9 (9.5) | RRMS 21F/7M HC 23F/8M | serum | targeted | HPLC-MS/MS |
| Datta (2024) [26] | RRMS, PPMS | RRMS—41 PMS—31 RRMS-HC—44 PPMS-HC—47 | RRMS median: 39 (range 34–48) PPMS median: 49 (range 46–58) RRMS-HC median: 39.5 (range 33.45–39) PPMS-HC median: 53 (range 46.5–60.5) | RRMS 29F/12M PMS 22F/9M RRMS-HC 31F/13M PPMS-HC 35F/12M | serum | untargeted | 2D GC × GC-MS |
| Meier (2024) [23] | RRMS, PPMS, SPMS | RRMS—58 SPMS—9 PPMS—4 Undefined—3 HC—80 | MS: 43.6 (14.1) HC: 44.7 (16.1) | RRMS 42F/16M SPMS 6F/3M PPMS 2F/2M Undefined: 2F/1M HC 48F/32M | serum and CSF | targeted | LC-ESI-MS/MS |
| Murgia (2023) [40] | RRMS (fingolimod treatment) | RRMS—42 HC—22 | MS: 39.0 (8.7) HC: 40.8 (13.8) | MS 23F/19M HC 17F/5M | plasma | untargeted | NMR |
| Broos (2023) [22] | RRMS, PMS | RRMS—170 PMS—115 HC—125 | RRMS: 52.9 (0.9) PMS: 53.0 (0.9) HC: 52.9 (1.2) | RRMS 139F/31M PMS 65F/50M HC 92F/33M | serum | targeted | HPLC-MS/MS |
| Židó (2023) [32] | Early MS | MS—40 HC—33 | MS median: 34 (range 18–54) HC: not reported | MS 31F/9M HC 26F/7M | CSF | untargeted/targeted | HPLC-MS/MS |
| Židó (2022) [41] | Early SM | MS—19 HC—19 | MS: 36 HC: 35 | MS 16F/3M HC 16F/3M | CSF | untargeted/targeted | HPLC-MS/MS |
| Zahoor (2022) [35] | RRMS | RRMS—35 HC—14 | RRMS: 45 HC: 40 | RRMS 22F/13M HC 9F/5M | serum | untargeted | UPLC-MS/GCLC/MS |
| Yang (2021) [33] | RRMS, PPMS | RRMS—20 PPMS—2 HC—21 | MS: 34.8 (7.5) HC: 33.3 (8.5) | MS 14F/8M HC 13F/8M | plasma | untargeted | LC-MS/MS |
| Olsson (2021) [42] | RRMS/CIS | RRMS/CIS—58 HC—50 | RRMS/CIS median: 34 (range 27–40) HC median: 33 (range 28–39) | RRMS/CIS 44F/14M HC 34F/16M | serum | targeted | UPLC-MS/MS, |
| Fitzgerald (2021) [43] | MS | MS—514 HC—241 | MS: 42.54 (14.91) HC: 35.85 (15.71) | MS 468F/47M HC 224F/17M | plasma | untargeted | GC/MS, LC/MS/MS |
| Levi (2021) [36] | MS | MS—129 HC—58 | MS: 38.3 (11.8) HC: 45.8 (12.5) | MS 93F/36M HC 29F/29M | serum | untargeted | LC/MS |
| Probert (2021) [19] | CIS converters vs. non-converters | Converters—22 Non-converters—32 | Converters: 31.3 (9.9) Non-converters: 36.4 (11.2) | Converters 17F/5M Non-converters 21F/11M | CSF | untargeted | NMR, |
| Keller (2021) [21] | MS (PRT treatment) | MS—14 HC—13 | MS: 42 (13) HC: 39 (14) | MS 12F/2M HC 11F/2M | serum | untargeted | UPLC-MS/MS |
| Yeo (2021) [37] | RRMS | RRMS relapse—28 RRMS last relapse in 1–6 months—28 RRMS last relapse in 6–24 months—34 RRMS last relapse in over 24 months—101 | RRMS relapse: 38.3 (9.5) RRMS last relapse in 1–6 months: 38.7 (7.0) RRMS last relapse in 6–24 months: 43.5 (9.7) RRMS last relapse in over 24 months: 44.2 (9.9) | RRMS relapse 27F/11M RRMS last relapse in 1–6 months 23F/5M RRMS last relapse in 6–24 months 22F/12M RRMS last relapse in over 24 months 73F/28M | serum | untargeted/targeted | NRM |
| Yeo (2020) [27] | RRMS, SPSM | RRMS—31 SPMS—28 | RRMS: 43.5 (9.7) SPMS: 58.1 (9.6) | RRMS 23F/8M SPMS 20F/8M | serum | untargeted | NMR |
| Gaetani (2020) [34] | RRMS | RRMS—47 HC—43 | RRMS: 31.8 (9.7) HC: 32.7 (10.6) | RRMS 40F/7M HC 27F/16M | urine | targeted | HPLC-MS/MS |
| Carlsson (2020) [29] | SPMS | SPMS—12 HC—12 | SPMS: 58.7 (7.5) HC: 54.2 (6.1) | SPMS 7F/5M HC: 7F/5M | CSF | targeted | LC-HRMS, FIA-HRMS |
| Duscha (2020) [24] | MS | RRMS—161 SPMS—103 PPMS—39 HC—68 NMO—1 | RRMS: 45.8 (12.7) SPMS: 57.8 (9.4) PPMS: 60 (11.4) HC: 48.5 (14.5) NMO: 55 | RRMS 102F/59M SPMS 62F/39M PPMS 16F/23M HC 32F/36M NMO 1M | serum, feces | targeted | LC-MS/MS |
| Penkert (2020) [44] | HC vs. MS/CIS (Monozygotic twins) | MS—73 HC—73 | 40.8 (12.1) | 110F/36M | plasma | untargeted | shotgun lipidomics (LC-MS/MS) |
| Amatruda (2020) [31] | PPMS | PPMS—19 HC—8 Validation cohort: SPMS—11 RRMS—24 | PPMS: 49.8 (11.1) HC: 42.1 (8.4) validation cohort: not reported | PPMS 11F/8M HC 4F/4M validation cohort-not reported | plasma | untargeted | LC-MS/MS, |
| Waddington (2020) [45] | RRMS (IFNβ-treated), CIS | ADA (−)—52 ADA (+)—30 | ADA (−): 34.6 (9.3) ADA (+): 37.9 (9.8) | ADA (−) 37F/15M ADA (+) 19F/11M | serum, PBMCs | untargeted | NMR |
| Author (Year) | Clinical Groups | Metabolite | Direction | Fold-Change | Comparison | p-Value | FDR |
|---|---|---|---|---|---|---|---|
| Staats Pires (2025) [38] | RRMS, HC | KYNA | ↓ | 1.2-fold (19%) | RRMS vs. HC | p < 0.05 | FDR = 0.0394 |
| 3HK | ↓ | 1.5-fold (32%) | RRMS vs. HC | p < 0.05 | FDR = 0.0008 | ||
| AA | ↑ | 3.1-fold (212%) | RRMS vs. HC | p < 0.0001 | FDR < 0.0001 | ||
| KYN/TRP | ↑ | 1.15-fold (15%) | RRMS vs. HC | p < 0.01 | FDR = 0.0187 | ||
| 3HK/KYN | ↓ | 1.5-fold (33%) | RRMS vs. HC | p < 0.001 | FDR < 0.0001 | ||
| QUIN/KYNA | ↑ | 1.27-fold (27%) | RRMS vs. HC | p < 0.05 | NR | ||
| 3HAA/AA | ↓ | 1.8-fold (44%) | RRMS vs. HC | p < 0.0001 | FDR < 0.0001 | ||
| Wicks (2025) [17] | RRMS, PMS and HC | β-hydroxybutyrate | ↑ | 1.24-fold (24%) | RRMS vs. HC | NR | NR |
| β-hydroxybutyrate | ↓ | 0.87-fold (13%) | RRMS vs. PMS | NR | NR | ||
| β-hydroxybutyrate | ↑ | 1.43-fold (43%) | PMS vs. HC | p = 0.005 | NR | ||
| β-hydroxybutyrate | ↑ | 1.15-fold (15%) | RRMS vs. RRMS | NR | NR | ||
| Acetoacetate | ↑ | 1.15-fold (15%) | RRMS vs. HC | NR | NR | ||
| Acetoacetate | ↓ | 0.88-fold (12%) | RRMS vs. PMS | p = NR | NR | ||
| Acetoacetate | ↑ | 1.31-fold (31%) | PMS vs. HC | p = 0.013 | NR | ||
| Acetoacetate | ↑ | 1.13-fold (13%) | PMS vs. RRMS | p = NR | NR | ||
| Radford-Smith (2025) [20] | RRMS (progressors vs. non-progressors) | Glucose | ↑ | - | progressors vs. non-progressors | p = 0.0021 (discovery cohort); p = 0.0027 (validation cohort) | NR |
| Glutamate | ↑ | - | progressors vs. non-progressors | p = 0.0006 (discovery cohort); p = 0.0409 (validation cohort) | NR | ||
| Glutamine | ↑ | - | progressors vs. non-progressors | p = 0.0052 | NR | ||
| Schwerdtfeger (2025) [25] | RRMS (progressors vs. non-progressors) | Spermidine (serum) | ↑ | - | progressors vs. non-progressors | p < 0.05 | NR |
| p-Cresol sulfate (serum) | ↑ | - | progressors vs. non-progressors | p < 0.05 | NR | ||
| Nicotinate (feces) | ↓ | - | progressors vs. non-progressors | p < 0.05 | NR | ||
| Phenyllactate (feces) | ↓ | - | progressors vs. non-progressors | p < 0.05 | NR | ||
| Protoporphyrin IX (feces) | ↓ | - | progressors vs. non-progressors | p < 0.05 | NR | ||
| N-Oleoyltaurine (feces) | ↓ | - | progressors vs. non-progressors | p < 0.05 | NR | ||
| Alwahsh (2024) [16] | RRMS, PPMS, SPMS and HC | Tryptophan | ↑ | 3.34-fold | RRMS vs. HC | p = 4.39 × 10−7 | FDR < 0.05 |
| Succinate | ↑ | 1.58-fold | RRMS vs. HC | p = 2.50 × 10−6 | FDR < 0.05 | ||
| ATP | ↑ | 1.98-fold | RRMS vs. HC | p = 1.11 × 10−4 | FDR < 0.05 | ||
| Formate | ↑ | 2.47-fold | RRMS vs. HC | p = 5.65 × 10−4 | FDR < 0.05 | ||
| Inosine | ↑ | 1.61-fold | RRMS vs. HC | p = 1.24 × 10−3 | FDR < 0.05 | ||
| Histidine | ↑ | 1.90-fold | RRMS vs. HC | p = 2.90 × 10−3 | FDR < 0.05 | ||
| Glutathione | ↑ | 1.47-fold | RRMS vs. HC | p = 3.29 × 10−3 | FDR < 0.05 | ||
| Pantothenate | ↑ | 1.37-fold | RRMS vs. HC | p = 5.54 × 10−3 | FDR < 0.05 | ||
| Lysine | ↓ | 0.48-fold | RRMS vs. HC | p = 3.11 × 10−19 | FDR < 0.001 | ||
| Myo-inositol | ↓ | 0.36-fold | RRMS vs. HC | p = 6.13 × 10−18 | FDR < 0.001 | ||
| Glutamate | ↓ | 0.57-fold | RRMS vs. HC | p = 1.56 × 10−16 | FDR < 0.001 | ||
| Threonine | ↓ | 0.38-fold | RRMS vs. HC | p = 7.64 × 10−16 | FDR < 0.001 | ||
| Glycine | ↓ | 0.45-fold | RRMS vs. HC | p = 9.51 × 10−15 | FDR < 0.001 | ||
| Tyrosine | ↓ | 0.61-fold | RRMS vs. HC | p = 3.33 × 10−12 | FDR < 0.001 | ||
| Choline | ↓ | 0.50-fold | RRMS vs. HC | p = 5.18 × 10−12 | FDR < 0.001 | ||
| O-phosphocholine | ↓ | 0.54-fold | RRMS vs. HC | p = 1.79 × 10−9 | FDR < 0.001 | ||
| Serine | ↓ | 0.71-fold | RRMS vs. HC | p = 7.47 × 10−8 | FDR < 0.001 | ||
| Cysteine | ↓ | 0.65-fold | RRMS vs. HC | p = 1.67 × 10−3 | FDR < 0.001 | ||
| sn-Glycero-3-phosphocholine | ↓ | 0.77-fold | RRMS vs. HC | p = 1.10 × 10−6 | FDR < 0.05 | ||
| Creatine | ↓ | 0.46-fold | RRMS vs. HC | p = 1.64 × 10−5 | FDR < 0.001 | ||
| Tryptophan | ↑ | - | RRMS vs. PPMS | p = 1.42 × 10−5 | FDR < 0.05 | ||
| Ascorbate | ↑ | - | RRMS vs. PPMS | p < 0.05 | FDR < 0.01 | ||
| Histidine | ↓ | - | RRMS vs. PPMS | p = 5.48 × 10−5 | FDR < 0.05 | ||
| Proline | ↓ | - | RRMS vs. PPMS | p = 8.03 × 10−7 | FDR < 0.001 | ||
| Phenylalanine | ↓ | - | RRMS vs. PPMS | p = 1.24 × 10−6 | FDR < 0.001 | ||
| Cysteine | ↓ | - | RRMS vs. PPMS | p = 3.93 × 10−5 | FDR < 0.001 | ||
| Formate | ↓ | - | RRMS vs. PPMS | p = 1.49 × 10−3 | FDR < 0.05 | ||
| Fumarate | ↓ | - | RRMS vs. PPMS | p = 2.33 × 10−5 | FDR < 0.001 | ||
| Inosine | ↑ | - | RRMS vs. SPMS | p = 5.23 × 10−3 | FDR < 0.05 | ||
| NAD+ | ↑ | - | RRMS vs. SPMS | p = 9.88 × 10−3 | FDR < 0.05 | ||
| Siavoshi (2024) [18] | RRMS (pre/post—ocrelizumab) | Pregnenediol disulfate | ↓ | - | pre- vs. post-ocrelizumab | p = 1.78 × 10−7 | FDR = 1.90 × 10−4 |
| Pregnenetriol disulfate | ↓ | - | pre- vs. post-ocrelizumab | p = 5.29 × 10−5 | FDR = 1.13 × 10−2 | ||
| Androstene-diol disulfate | ↓ | - | pre- vs. post-ocrelizumab | p = 1.15 × 10−4 | FDR = 1.76 × 10−2 | ||
| Taurocholenate sulfate | ↓ | - | pre- vs. post-ocrelizumab | p = 2.43 × 10−4 | FDR = 2.16 × 10−2 | ||
| 1-Linoleoyl-GPA (18:2) | ↓ | - | pre- vs. post-ocrelizumab | p = 2.39 × 10−4 | FDR = 2.16 × 10−2 | ||
| 2′-Deoxyuridine | ↓ | - | pre- vs. post-ocrelizumab | p = 1.64 × 10−5 | FDR = 4.38 × 10−3 | ||
| PFOS | ↓ | - | pre- vs. post-ocrelizumab | p = 1.17 × 10−6 | FDR = 6.23 × 10−4 | ||
| PFOA | ↓ | - | pre- vs. post-ocrelizumab | p = 2.20 × 10−4 | FDR = 2.16 × 10−2 | ||
| Serine | ↓ | - | pre- vs. post-ocrelizumab | p = 1.43 × 10−4 | FDR = 1.92 | ||
| Lactate | ↓ | - | pre- vs. post-ocrelizumab | p = 1.87 × 10−4 | FDR = 2.16 × 10−2 | ||
| N-acetyl-β-alanine | ↑ | - | pre- vs. post-ocrelizumab | p = 6.85 × 10−5 | FDR = 1.22 × 10−2 | ||
| Propyl | ↑ | - | pre- vs. post-ocrelizumab | p = 1.02 × 10−5 | FDR = 3.64 × 10−3 | ||
| Propyl 4-hydroxybenzoate sulfate | ↑ | - | pre- vs. post-ocrelizumab | p = 1.02 × 10−5 | FDR = 3.64 × 10−3 | ||
| Ladakis (2024) [30] | MS lesions (SPMS) and HC | Sphingomyelins | ↑ | - | MS lesions vs. control | p = 1.26 × 10−8 | FDR = 2.15 × 10−7 |
| Ceramides | ↑ | - | MS lesions vs. control | p = 1.26 × 10−8 | FDR = 2.15 × 10−7 | ||
| Sphingosine | ↑ | - | MS lesions vs. control | p = 2.54 × 10−6 | FDR = 2.88 × 10−5 | ||
| Dipeptides | ↑ | - | MS lesions vs. control | p = 0.04 | FDR = 0.10 | ||
| Lysophospholipids | ↓ | - | MS lesions vs. control | p = 5.48 × 10−9 | FDR = 1.86 × 10−7 | ||
| Monoacylglycerols | ↓ | - | MS lesions vs. control | p = 0.008 | FDR = 0.04 | ||
| Hexosylceramides | ↓ | - | MS lesions vs. control | p = 0.002 | FDR = 0.06 | ||
| Guanosine | ↓ | - | MS lesions vs. control | p = 0.00014 | FDR = 0.001 | ||
| Pyridoxamine phosphate | ↓ | - | MS lesions vs. control | p = 0.00014 | FDR = 0.001 | ||
| Glutamate-γ-methyl ester | ↓ | - | MS lesions vs. control | p = 0.00014 | FDR = 0.001 | ||
| Nucleotides | ↓ | - | MS lesions vs. control | p = 0.01 | FDR = 0.05 | ||
| UFAs | ↓ | - | MS lesions vs. control | p = 0.02 | FDR = 0.06 | ||
| Endocannabinoids | ↓ | - | MS lesions vs. control | p < 0.001 | FDR = 5.2 × 10−9 | ||
| Fung (2024) [39] | RRMS and HC | 9-HODE | ↑ | 2.6-fold | preserved white matter integrity vs. white matter injury | p = 0.008 | FDR ≈ 0.05 |
| ALA/GLA | ↑ | 1.17-fold | preserved white matter integrity vs. white matter injury | p = 0.04 | FDR ≈ 0.05 | ||
| 9-HOTrE | ↑ | 4.2-fold | preserved white matter integrity vs. white matter injury | p = 0.003 | FDR ≈ 0.05 | ||
| 9,10-EpOME | ↑ | - | white matter injury vs. preserved white matter | p = 0.01 | FDR ≈ 0.05 | ||
| 9-HOTrE | ↓ | - | white matter injury vs. preserved white matter | p = 0.003 | FDR ≈ 0.05 | ||
| Datta (2024) [26] | RRMS, PPMS and HS | 2-Ethylhexanoic acid | ↑ | - | RRMS vs. HC | p < 0.05 | FDR ≈ 0.3 (ns) |
| Ribose | ↑ | - | RRMS vs. HC | p < 0.05 | FDR ≈ 0.3 (ns) | ||
| Erythrose | ↑ | - | RRMS vs. HC | p < 0.05 | FDR ≈ 0.3 (ns) | ||
| 3-Indolepropionic acid | ↓ | - | RRMS vs. HC | p < 0.05 | FDR ≈ 0.3 (ns) | ||
| α-D-Glucopyranose | ↑ | - | RRMS vs. HC | p < 0.05 | FDR ≈ 0.3 (ns) | ||
| D-Glucuronic acid lactone | ↑ | - | RRMS vs. HC | p < 0.05 | FDR ≈ 0.3 (ns) | ||
| Heptanoic acid | ↑ | - | RRMS vs. HC | p < 0.05 | FDR ≈ 0.3 (ns) | ||
| L-Threose | ↑ | - | RRMS vs. HC | p < 0.05 | FDR ≈ 0.3 (ns) | ||
| Lanthionine | ↑ | - | RRMS vs. HC | p < 0.05 | FDR ≈ 0.3 (ns) | ||
| Linoleic acid | ↑ | - | RRMS vs. HC | p < 0.05 | FDR ≈ 0.3 (ns) | ||
| 11,14-Eicosadienoic acid | ↑ | - | RRMS vs. HC | p < 0.05 | FDR ≈ 0.3 (ns) | ||
| Succinic acid | ↓ | - | RRMS vs. HC | p < 0.05 | FDR ≈ 0.3 (ns) | ||
| Meier (2024) [23] | RRMS, PPMS, SPMS and HC | 2-AG (CSF) | ↑ | - | Male RRMS vs. female RRMS and female HC | p < 0.05 | FDR ≈ 0.05 |
| AEA (CSF) | ↑ | - | RRMS < 39 yrs vs. age/sex-matched HC | p < 0.05 | FDR ≈ 0.05 | ||
| 2-AG (CSF) | ↑ | - | RRMS < 39 yrs vs. age/sex-matched HC | p < 0.05 | FDR ≈ 0.05 | ||
| AA (CSF) | ↑ | - | male vs. female RRMS | p < 0.05 | FDR ≈ 0.05 | ||
| Cortisol (CSF) | ↑ | - | PMS (PPMS and SPMS) vs. RRMS and HC | p < 0.05 | FDR ≈ 0.05 | ||
| Corticosterone (CSF) | ↑ | - | PMS (PPMS and SPMS) vs. HC | p < 0.05 | FDR ≈ 0.05 | ||
| 2-OG (CSF) | ↑ | - | RRMS ≥39 yrs vs. HC | p < 0.05 | FDR ≈ 0.05 | ||
| PGE2 (serum) | ↑ | - | RRMS vs. HC | p < 0.05 | FDR ≈ 0.05 | ||
| SEA (serum) | ↑ | - | MS (all subtypes) vs. HC | p < 0.05 | FDR ≈ 0.05 | ||
| Murgia (2023) [40] | RRMS (after fingolimod treatment) and HC | Alanine | ↑ | - | RRMS after fingolimod treatment vs. baseline | p < 0.05 | FDR ≈ 0.05 |
| Phenylalanine | ↑ | - | RRMS after fingolimod treatment vs. baseline | p < 0.05 | FDR ≈ 0.05 | ||
| Glycine | ↑ | - | RRMS after fingolimod treatment vs. baseline | p < 0.05 | FDR ≈ 0.05 | ||
| Pyroglutamic acid | ↑ | - | RRMS after fingolimod treatment vs. baseline | p < 0.05 | FDR ≈ 0.05 | ||
| Tryptophan | ↑ | - | RRMS after fingolimod treatment vs. baseline | p < 0.05 | FDR ≈ 0.05 | ||
| Fructose | ↑ | - | RRMS after fingolimod treatment vs. baseline | p < 0.05 | FDR ≈ 0.05 | ||
| Glucose | ↑ | - | RRMS after fingolimod treatment vs. baseline | p < 0.05 | FDR ≈ 0.05 | ||
| 2-Hydroxyisovalerate | ↑ | - | RRMS after fingolimod treatment vs. baseline | p < 0.05 | FDR ≈ 0.05 | ||
| Creatinine | ↑ | - | RRMS after fingolimod treatment vs. baseline | p < 0.05 | FDR ≈ 0.05 | ||
| Lactate | ↓ | - | RRMS after fingolimod treatment vs. baseline | p < 0.05 | FDR ≈ 0.05 | ||
| Isoleucine | ↓ | - | RRMS after fingolimod treatment vs. baseline | p < 0.05 | FDR ≈ 0.05 | ||
| Glutamate | ↓ | - | RRMS after fingolimod treatment vs. baseline | p < 0.05 | FDR ≈ 0.05 | ||
| Lysine | ↑ | - | responders vs. non-responders | p < 0.05 | FDR ≈ 0.05 | ||
| Lactate | ↑ | - | responders vs. non-responders | p < 0.05 | FDR ≈ 0.05 | ||
| Glucose | ↑ | - | non-responders vs. responders | p < 0.05 | FDR ≈ 0.05 | ||
| Broos (2023) [22] | RRMS, PMS, and HC | 11,12-DiHET | ↑ | - | RRMS vs. HC | p < 0.05 | FDR < 0.05 |
| DPAn-3 | ↑ | - | RRMS vs. HC | p < 0.05 | FDR < 0.05 | ||
| 15-HETE | ↑ | - | PMS vs. HC | p < 0.05 | FDR (ns) | ||
| 8-HETE | ↑ | - | PMS vs. HC | p < 0.05 | FDR (ns) | ||
| 5-HETE | ↑ | - | PMS vs. HC | p < 0.05 | FDR (ns) | ||
| 11,12-DiHETE | ↑ | - | PMS vs. HC | p < 0.05 | FDR < 0.05 | ||
| 20-HETE | ↑ | - | PMS vs. HC | p < 0.05 | FDR < 0.05 | ||
| ARA | ↑ | - | PMS vs. HC | p < 0.05 | FDR < 0.05 | ||
| DGLA | ↑ | - | PMS vs. HC | p < 0.05 | FDR < 0.05 | ||
| AdA | ↑ | - | PMS vs. HC | p < 0.05 | FDR < 0.05 | ||
| 9-HODE | ↓ | - | PMS vs. HC | p < 0.05 | FDR < 0.05 | ||
| 13-HODE | ↓ | - | PMS vs. HC | p < 0.05 | FDR < 0.05 | ||
| 14,15-DiHETE | ↓ | - | PMS vs. HC | p < 0.05 | FDR < 0.05 | ||
| 19,20-DiHDPA | ↓ | - | PMS vs. HC | p < 0.05 | FDR < 0.05 | ||
| Židó (2023) [32] | Early MS and HC | Arginine | ↓ | - | Early MS vs. HC | p = 0.0037 | FDR < 0.05 |
| Histidine | ↓ | - | Early MS vs. HC | p = 0.0058 | FDR < 0.05 | ||
| Glutamate | ↓ | - | Early MS vs. HC | p = 0.0145 | FDR < 0.05 | ||
| Choline | ↓ | - | Early MS vs. HC | p = 0.0233 | FDR < 0.05 | ||
| Tyrosine | ↓ | - | Early MS vs. HC | p = 0.0313 | FDR < 0.05 | ||
| Serine | ↓ | - | Early MS vs. HC | p = 0.0473 | FDR < 0.05 | ||
| Methionine | ↑ | - | Early MS vs. HC | p < 0.001 | FDR < 0.05 | ||
| Linoleic acid | ↓ | - | Early MS vs. HC | p = 0.001 | FDR < 0.05 | ||
| Stearic acid | ↓ | - | Early MS vs. HC | p = 0.029 | FDR < 0.05 | ||
| Spermidine | ↑ | - | Early MS vs. HC | p = 0.0124 | FDR < 0.05 | ||
| Oleic acid | ↑ | - | Early MS vs. HC | p = 0.015 | FDR < 0.05 | ||
| Židó (2022) [41] | Early MS and HC | Arginine | ↓ | - | Early MS vs. HC | p = 0.007 | FDR < 0.05 |
| Histidine | ↓ | - | Early MS vs. HC | p = 0.012 | FDR < 0.05 | ||
| Palmitic acid | ↑ | - | Early MS vs. HC | p = 0.039 | FDR ≈ 0.05 | ||
| Zahoor (2022) [35] | RRMS and HC | Phosphoethanolamine | ↑ | - | RRMS vs. HC | p < 0.05 | FDR < 0.10 (ns) |
| Lactic acid | ↑ | - | RRMS vs. HC | p < 0.05 | FDR < 0.10 (ns) | ||
| Fumaric acid | ↑ | - | RRMS vs. HC | p < 0.05 | FDR < 0.10 (ns) | ||
| 3-Hydroxybutyrate | ↑ | - | RRMS vs. HC | p < 0.05 | FDR < 0.10 (ns) | ||
| Oleoylethanolamide (OEA) | ↑ | - | RRMS vs. HC | p < 0.05 | FDR < 0.10 (ns) | ||
| Sphingosine-1-phosphate (S1P) | ↑ | - | RRMS vs. HC | p < 0.05 | FDR < 0.10 (ns) | ||
| Yang (2021) [33] | RRMS, PPMS and HC | L-Tyrosine | ↓ | - | RRMS and PPMS vs. HC | p < 0.001 | FDR < 0.05 |
| L-Tryptophan | ↓ | - | RRMS and PPMS vs. HC | p = 0.015 | FDR < 0.05 | ||
| L-Phenylalanine | ↓ | - | RRMS and PPMS vs. HC | p = 0.033 | FDR < 0.05 | ||
| L-Leucine | ↓ | - | RRMS and PPMS vs. HC | p = 0.0049 | FDR < 0.05 | ||
| L-Isoleucine | ↓ | - | RRMS and PPMS vs. HC | p < 0.001 | FDR < 0.05 | ||
| Sphingosine-1-phosphate | ↓ | - | RRMS and PPMS vs. HC | p < 0.001 | FDR < 0.05 | ||
| Sphinganine-1-phosphate | ↓ | - | RRMS and PPMS vs. HC | p < 0.001 | FDR < 0.05 | ||
| Phytosphingosine | ↓ | - | RRMS and PPMS vs. HC | p < 0.01 | FDR < 0.05 | ||
| 17α-Estradiol | ↓ | - | RRMS and PPMS vs. HC | p < 0.001 | FDR < 0.05 | ||
| Methyl jasmonate | ↑ | - | RRMS and PPMS vs. HC | p < 0.001 | FDR < 0.05 | ||
| Myo-inositol | ↓ | - | RRMS and PPMS vs. HC | p < 0.001 | FDR < 0.05 | ||
| Oleic acid | ↑ | - | RRMS and PPMS vs. HC | p = 0.046 | FDR < 0.05 | ||
| Palmitic acid | ↑ | - | RRMS and PPMS vs. HC | p < 0.01 | FDR < 0.05 | ||
| Arachidonic acid | ↑ | - | RRMS and PPMS vs. HC | p = 0.015 | FDR < 0.05 | ||
| β-Alanyl-L-arginine | ↑ | - | RRMS and PPMS vs. HC | p < 0.01 | FDR < 0.05 | ||
| 4-Oxoglutaramate | ↑ | - | RRMS and PPMS vs. HC | p < 0.01 | FDR < 0.05 | ||
| Isocitric acid | ↑ | - | RRMS and PPMS vs. HC | p < 0.05 | FDR < 0.05 | ||
| O-Phosphoethanolamine | ↑ | - | RRMS and PPMS vs. HC | p < 0.01 | FDR < 0.05 | ||
| Sorbitol | ↑ | - | RRMS and PPMS vs. HC | p < 0.01 | FDR < 0.05 | ||
| Spermidine | ↑ | - | RRMS and PPMS vs. HC | p < 0.05 | FDR < 0.05 | ||
| Homovanillic acid | ↓ | - | RRMS and PPMS vs. HC | p < 0.05 | FDR < 0.05 | ||
| Deoxyuridine | ↓ | - | RRMS and PPMS vs. HC | p < 0.01 | FDR < 0.0 | ||
| L-Arogenate | ↓ | - | RRMS and PPMS vs. HC | p < 0.05 | FDR < 0.05 | ||
| Pseudouridine | ↓ | - | RRMS and PPMS vs. HC | p < 0.01 | FDR < 0.05 | ||
| Uridine | ↓ | - | RRMS and PPMS vs. HC | p < 0.001 | FDR < 0.05 | ||
| Dodecanoic acid | ↓ | - | RRMS and PPMS vs. HC | p < 0.05 | FDR < 0.05 | ||
| Niacinamide | ↓ | - | RRMS and PPMS vs. HC | p < 0.05 | FDR < 0.05 | ||
| α-Dimorphecolic acid | ↓ | - | RRMS and PPMS vs. HC | p < 0.01 | FDR < 0.05 | ||
| cis-4-Hydroxy-D-proline | ↓ | - | RRMS and PPMS vs. HC | p = 0.004 | FDR < 0.05 | ||
| N-Acetyl-L-asparagine | ↓ | - | RRMS and PPMS vs. HC | p < 0.01 | FDR < 0.05 | ||
| L-Valine | ↑ | - | RRMS and PPMS vs. HC | p < 0.05 | FDR < 0.05 | ||
| Glutamic acid | ↑ | - | RRMS and PPMS vs. HC | p < 0.001 | FDR < 0.05 | ||
| Myristic acid | ↓ | - | RRMS and PPMS vs. HC | p < 0.05 | FDR < 0.05 | ||
| DHEA | ↓ | - | RRMS and PPMS vs. HC | p < 0.05 | FDR < 0.05 | ||
| Creatinine | ↓ | - | RRMS and PPMS vs. HC | p < 0.01 | FDR < 0.05 | ||
| Nicotinuric acid | ↓ | - | RRMS and PPMS vs. HC | p < 0.05 | FDR < 0.05 | ||
| Trans-cinnamic acid | ↓ | - | RRMS and PPMS vs. HC | p < 0.01 | FDR < 0.05 | ||
| 2-Dehydropantoate | ↓ | - | RRMS and PPMS vs. HC | p < 0.05 | FDR < 0.05 | ||
| Olsson (2021) [42] | RRMS, CIS and HC | Acetate | ↓ | - | RRMS and CIS vs. HC | p = 0.021 | FDR = 0.067 (ns) |
| Valine | ↓ | - | RRMS and CIS vs. HC | p = 0.005 | FDR > 0.05 (ns) | ||
| L-Methionine | ↓ | - | RRMS and CIS vs. HC | p = 0.025 | FDR > 0.05 (ns) | ||
| Kynurenic acid (KYNA) | ↓ | - | RRMS and CIS vs. HC | p = 0.019 | FDR > 0.05 (ns) | ||
| Creatine | ↑ | - | RRMS and CIS vs. HC | p = 0.022 | FDR > 0.05 (ns) | ||
| Pantothenic acid | ↑ | - | RRMS and CIS vs. HC | p = 0.021 | FDR > 0.05 (ns) | ||
| D-Glucuronic acid | ↑ | - | RRMS and CIS vs. HC | p = 0.031 | FDR > 0.05 (ns) | ||
| 3-Hydroxyanthranilic acid (3HAA) | ↑ | - | RRMS and CIS vs. HC | p = 0.040 | FDR > 0.05 (ns) | ||
| Acetate/Butyrate ratio | ↓ | - | RRMS and CIS vs. HC | p = 0.005 | FDR = 0.06 (ns) | ||
| Acetate/(Propionate + Butyrate) ratio | ↓ | - | RRMS and CIS vs. HC | p = 0.010 | FDR = 0.06 (ns) | ||
| Fitzgerald (2021) [43] | MS and HC | Phenyllactate (PLA) | ↓ | - | MS vs. HC | p < 1.0 × 10−19 | FDR < 0.001 |
| 3-(4-Hydroxyphenyl)lactate | ↓ | - | MS vs. HC | p < 1.0 × 10−17 | FDR < 0.001 | ||
| Indolelactate | ↓ | - | MS vs. HC | p < 1.0 × 10−15 | FDR < 0.001 | ||
| Imidazole lactate | ↓ | - | MS vs. HC | p < 1.0 × 10−10 | FDR < 0.001 | ||
| Tyrosine | ↓ | - | MS vs. HC | p < 1.0 × 10−6 | FDR < 0.001 | ||
| Tryptophan | ↓ | - | MS vs. HC | p < 1.0 × 10−10 | FDR < 0.001 | ||
| Phenylpyruvate | ↓ | - | MS vs. HC | p < 1.0 × 10−6 | FDR < 0.001 | ||
| Kynurenine | ↓ | - | MS vs. HC | p < 0.05 | FDR ≈ 0.05 | ||
| Phenylacetylglutamine | ↑ | - | MS vs. HC | p < 1.0 × 10−4 | FDR < 0.05 | ||
| p-Cresol glucuronide | ↑ | - | MS vs. HC | p < 1.0 × 10−4 | FDR < 0.05 | ||
| p-Cresol sulfate | ↑ | - | MS vs. HC | p < 0.001 | FDR < 0.05 | ||
| 4-Hydroxyphenylpyruvate | ↓ | - | MS vs. HC | p < 1.0 × 10−9 | FDR < 0.001 | ||
| Xanthurenate | ↓ | - | MS vs. HC | p < 1.0 × 10−4 | FDR < 0.05 | ||
| Levi (2021) [36] | MS and HC | β-Hydroxyasparagine | ↑ | - | MS vs. HC | p < 0.004 | FDR < 0.05 |
| Sphingosine-1-phosphate (S1P) | ↓ | - | MS vs. HC | p < 0.007 | FDR < 0.05 | ||
| Carnitine | ↓ | - | MS vs. HC | p < 0.007 | FDR < 0.05 | ||
| Indolepropionate | ↓ | - | MS vs. HC | p < 0.03 | FDR < 0.05 | ||
| Indolelactate | ↓ | - | MS vs. HC | p < 0.03 | FDR < 0.05 | ||
| p-Cresol sulfate | ↑ | - | MS vs. HC | p = 0.12 | FDR (ns) | ||
| Stachydrine | ↑ | - | MS vs. HC | NR | FDR < 0.05 | ||
| 3-Hydroxyhippurate | ↑ | - | MS vs. HC | NR | FDR < 0.05 | ||
| Probert (2021) [19] | CIS converters and non-converters | Glucose | ↑ | - | CIS converters vs. non-converters | p < 0.05 | FDR (ns) |
| Lactate | ↑ | - | CIS converters vs. non-converters | p < 0.05 | FDR (ns) | ||
| Myo-inositol | ↓ | - | CIS converters vs. non-converters | p < 0.05 | FDR (ns) | ||
| Creatine | ↓ | - | CIS converters vs. non-converters | p < 0.05 | FDR (ns) | ||
| Keller (2021) [21] | MS and HC (after PRT treatment) | Myristate | ↑ | - | MS vs. HC after PRT | p < 0.001 | FDR (ns) |
| Palmitate | ↑ | - | MS vs. HC after PRT | p < 0.001 | FDR (ns) | ||
| Arachidonate | ↑ | - | MS vs. HC after PRT | p = 0.0012 | FDR (ns) | ||
| Oleate/Vaccinate | ↑ | - | MS vs. HC after PRT | p = 0.0012 | FDR (ns) | ||
| Linoleate | ↑ | - | MS vs. HC after PRT | p = 0.0017 | FDR (ns) | ||
| 5-Dodecenoate | ↑ | - | MS vs. HC after PRT | p = 0.003 | FDR (ns) | ||
| 1-Palmitoleoyl-GPC | ↑ | - | MS vs. HC after PRT | p = 0.00012 | FDR (ns) | ||
| 1-Stearoyl-GPC | ↑ | - | MS vs. HC after PRT | p = 0.00061 | FDR (ns) | ||
| 1-1-Enyl-palmitoyl-GPC | ↑ | - | MS vs. HC after PRT | p = 0.0012 | FDR (ns) | ||
| Margarate | ↑ | - | MS vs. HC after PRT | p = 0.00061 | FDR (ns) | ||
| Dihomolinoleate | ↑ | - | MS vs. HC after PRT | p = 0.0017 | FDR (ns) | ||
| Stearidonate | ↑ | - | MS vs. HC after PRT | p = 0.0017 | FDR (ns) | ||
| 3-Hydroxyisobutyrate | ↑ | - | MS vs. HC after PRT | p = 0.00037 | FDR (ns) | ||
| Glycerol | ↑ | - | MS vs. HC after PRT | p = 0.0031 | FDR (ns) | ||
| 3-Aminoisobutyrate | ↑ | - | MS vs. HC after PRT | p = 0.0052 | FDR (ns) | ||
| Hydantoin-5-propionate | ↑ | - | MS vs. HC after PRT | p = 0.00061 | FDR (ns) | ||
| DHEAS | ↑ | - | MS vs. HC after PRT | p = 0.03 | FDR (ns) | ||
| Acylcarnitines | ↑ | - | MS vs. HC after PRT | p = 0.018 | FDR (ns) | ||
| Stearate | ↑ | - | MS vs. HC after PRT | p = 0.0040 | FDR (ns) | ||
| Yeo (2021) [37] | RRMS | Lysine | ↑ | - | RRMS, relapse vs. long remission (LR ≥ 24 M) | p < 0.05 | FDR (ns) |
| Asparagine | ↑ | - | RRMS, relapse vs. long remission (LR ≥ 24 M) | p < 0.05 | FDR (ns) | ||
| Isoleucine | ↓ | - | RRMS, relapse vs. long remission (LR ≥ 24 M) | p < 0.01 | FDR (ns) | ||
| Leucine | ↓ | - | RRMS, relapse vs. long remission (LR ≥ 24 M) | p < 0.01 | FDR (ns) | ||
| Yeo (2020) [27] | RRMS and SPMS | Lipoproteins | ↓ | - | SPMS vs. RRMS | NR | FDR < 0.05 |
| Choline | ↓ | - | SPMS vs. RRMS | NR | FDR < 0.05 | ||
| 3-Hydroxybutyrate | ↓ | - | SPMS vs. RRMS | NR | FDR < 0.05 | ||
| Glucose | ↑ | - | SPMS vs. RRMS | NR | FDR < 0.05 | ||
| N-acetylated glycoproteins | ↑ | - | SPMS vs. RRMS | NR | FDR < 0.05 | ||
| Gaetani (2020) [34] | RRMS and HC | Kynurenine | ↓ | - | RRMS vs. HC | p = 0.01 | NR |
| K/T ratio | ↓ | - | RRMS vs. HC | p = 0.04 | NR | ||
| Tryptophan | ↑ | - | RRMS vs. HC | p = 0.001 | NR | ||
| Indole-3-propionic acid | ↑ | - | RRMS vs. HC | p < 0.001 | NR | ||
| Indole-3-propionic acid | ↑ | - | RRMS with recent relapse (<30 days) vs. stable RRMS | p = 0.04 | NR | ||
| K/A ratio | ↓ | - | RRMS with recent relapse (<30 days) vs. stable RRMS | p = 0.03 | NR | ||
| Anthranilate | ↑ | - | RRMS with recent relapse (<30 days) vs. stable RRMS | p = 0.02 | NR | ||
| Carlsson (2020) [29] | SPMS and HC | Glycine | ↑ | - | SPMS vs. HC | p = 0.016 | FDR < 0.05 |
| ADMA | ↑ | - | SPMS vs. HC | p = 0.009 | FDR < 0.05 | ||
| PC-O (34:0) | ↑ | - | SPMS vs. HC | p = 0.046 | FDR < 0.05 | ||
| Hexoses | ↑ | - | SPMS vs. HC | p = 0.010 | FDR < 0.05 | ||
| Duscha (2020) [24] | MS | Propionic acid | ↓ | - | MS vs. HC | p = 0.0016 | FDR < 0.05 |
| Penkert (2020) [44] | HC and MS/CIS (Monozygotic twins) | Ether phosphatidylcholines (PC O-) | ↓ | - | MS vs. HC co-twins | p = 0.00081 | FDR = 0.0095 |
| Ether phosphatidylethanolamines (PE O-) | ↓ | - | MS vs. HC co-twins | p = 0.0015 | FDR = 0.0095 | ||
| Phosphatidylcholines (PC) | ↓ | - | MS vs. HC co-twins | p = 0.017 | FDR = 0.074 | ||
| PC O- species with DPA (C22:5) | ↓ | - | MS vs. HC co-twins | p = 0.00047 | FDR = 0.011 | ||
| PC O- species with other PUFA acyl chains (C22:4, C20:3, C20:4) | ↓ | - | MS vs. HC co-twins | p < 0.05 | FDR (ns) | ||
| PC O- species with ether-bound alkyl chains (O-16:0;0, O-16:1;0, O-18:1;0) | ↓ | - | MS vs. HC co-twins | p < 0.05 | FDR (ns) | ||
| PC O-16:1;0/20:3;0 | ↓ | - | MS vs. HC co-twins | p = 0.00006 | FDR = 0.015 | ||
| Multiple PC O- species with PUFA side chains | ↓ | - | MS vs. HC co-twins | p < 0.05 | FDR (ns) | ||
| Amatruda (2020) [31] | PPMS, HC, validation cohort: SPMS and RRMS | DiHexCer(d18:1/18:2) | ↓ | - | PPMS vs. HC | p < 0.05 | FDR (ns) |
| DiHexCer(d18:1/18:3) | ↓ | - | PPMS vs. HC | p < 0.05 | FDR (ns) | ||
| SM(d18:1/14:0) | ↓ | - | PPMS vs. HC | p < 0.01 | FDR (ns) | ||
| MonoHexCer(d18:1/20:0) | ↑ | - | PPMS vs. HC | p < 0.01 | FDR (ns) | ||
| LPA-18:2 | ↓ | - | PPMS progressors vs. PPMS non-progressors and HC | p < 0.05 | FDR (ns) | ||
| LPA-18:2 | ↓ | - | SPMS progressors vs. SPMS non-progressors | p < 0.05 | FDR (ns) | ||
| Waddington (2020) [45] | RRMS (IFNβtreated), CIS | VLDL-PA | ↑ | - | ADA+ vs. ADA− | p < 0.05 | FDR (ns) |
| Cholesteryl esters in VLDL | ↑ | - | ADA+ vs. ADA− | p < 0.05 | FDR (ns) | ||
| TG/PG ratio | ↑ | - | ADA+ vs. ADA− | p < 0.05 | FDR (ns) | ||
| Free cholesterol | ↑ | - | ADA+ vs. ADA− | p < 0.05 | FDR (ns) |
| Author (Year) | MS Phenotype | Algorithm | Validation Strategy | Performance Metrics |
|---|---|---|---|---|
| Datta (2024) [26] | RRMS vs. Progressive | PLS-DA, SVM, Random Forest (biosigner feature selection/classification) | Train–test split; classifiers evaluated on independent test set; feature tiers assigned by biosigner. | Prediction accuracy reported; AUROC NR. |
| Yeo (2020) [27] | RRMS vs. SPMS | OPLS-DA | External 10-fold cross-validation with repetition and permutation testing; ensemble of 1000 models; independent test sets | Accuracy, sensitivity, specificity reported |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smusz, J.; Mojsak, P.; Matys, P.; Mirończuk, A.; Tarasiuk, J.; Grubczak, K.; Starosz, A.; Kochanowicz, J.; Kułakowska, A.; Ruszczyńska, K.; et al. Metabolomics in Multiple Sclerosis: Advances, Challenges, and Clinical Perspectives—A Systematic Review. Int. J. Mol. Sci. 2025, 26, 9207. https://doi.org/10.3390/ijms26189207
Smusz J, Mojsak P, Matys P, Mirończuk A, Tarasiuk J, Grubczak K, Starosz A, Kochanowicz J, Kułakowska A, Ruszczyńska K, et al. Metabolomics in Multiple Sclerosis: Advances, Challenges, and Clinical Perspectives—A Systematic Review. International Journal of Molecular Sciences. 2025; 26(18):9207. https://doi.org/10.3390/ijms26189207
Chicago/Turabian StyleSmusz, Jan, Patrycja Mojsak, Paulina Matys, Anna Mirończuk, Joanna Tarasiuk, Kamil Grubczak, Aleksandra Starosz, Jan Kochanowicz, Alina Kułakowska, Katarzyna Ruszczyńska, and et al. 2025. "Metabolomics in Multiple Sclerosis: Advances, Challenges, and Clinical Perspectives—A Systematic Review" International Journal of Molecular Sciences 26, no. 18: 9207. https://doi.org/10.3390/ijms26189207
APA StyleSmusz, J., Mojsak, P., Matys, P., Mirończuk, A., Tarasiuk, J., Grubczak, K., Starosz, A., Kochanowicz, J., Kułakowska, A., Ruszczyńska, K., & Kapica-Topczewska, K. (2025). Metabolomics in Multiple Sclerosis: Advances, Challenges, and Clinical Perspectives—A Systematic Review. International Journal of Molecular Sciences, 26(18), 9207. https://doi.org/10.3390/ijms26189207







